Abstract
Background
Lipocalins are a large and diverse family of small, mostly extracellular proteins implicated in many important functions. This family has been studied in bacteria, invertebrate and vertebrate animals but little is known about these proteins in plants. We recently reported the identification and molecular characterization of the first true lipocalins from plants, including the Apolipoprotein D ortholog AtTIL identified in the plant model Arabidopsis thaliana. This study aimed to determine its physiological role in planta.
Results
Our results demonstrate that the AtTIL lipocalin is involved in modulating tolerance to oxidative stress. AtTIL knock-out plants are very sensitive to sudden drops in temperature and paraquat treatment, and dark-grown plants die shortly after transfer to light. These plants accumulate a high level of hydrogen peroxide and other ROS, which causes an oxidative stress that is associated with a reduction in hypocotyl growth and sensitivity to light. Complementation of the knock-out plants with the AtTIL cDNA restores the normal phenotype. On the other hand, overexpression enhances tolerance to stress caused by freezing, paraquat and light. Moreover, this overexpression delays flowering and maintains leaf greenness. Microarray analyses identified several differentially-regulated genes encoding components of oxidative stress and energy balance.
Conclusion
This study provides the first functional evidence that a plant lipocalin is involved in modulating tolerance to oxidative stress. These findings are in agreement with recently published data showing that overexpression of ApoD enhances tolerance to oxidative stress and increases life span in mice and Drosophila. Together, the three papers strongly support a similar function of lipocalins in these evolutionary-distant species.
Background
Lipocalins are small ligand-binding proteins found in bacteria and in invertebrate and vertebrate animals. Over 40 lipocalin members have been identified from all kingdoms [1]. They show a simple tertiary structure which gives them the ability to bind small, generally hydrophobic, molecules. Animal lipocalins play important roles in the regulation of immunological and developmental processes, and they are involved in the responses of organisms to various stress factors and in signal transduction pathways. It was recently shown that the insect glial Lazarillo lipocalin (GLaz) possesses a protective role against oxidative stress conditions and that its absence increases lipid peroxydation, reduces life span and accelerates neurodegeneration in Drosophila [2]. On the other hand, its overexpression protects against the effects of starvation, hypoxia and hyperoxia, and extends the fly's life span [3].
Data mining of genomic databases and bioinformatic predictions allowed us to determine that plants also possess lipocalins, which were classified as temperature-induced lipocalins (TILs) and chloroplastic lipocalins (CHLs) [4]. The TIL genes are induced by high and low temperature (LT) and their level of expression is associated with the plant's capacity to develop freezing tolerance (FT). Furthermore, the accumulation of TIL transcripts is limited to photosynthetic tissues and oscillates during the diurnal cycle. TILs do not show any targeting peptide, but the proteins are found at the plasma membrane. Sequence, structure and phylogenetic analyses revealed that TIL proteins share homology with three evolutionary related lipocalins: the bacterial lipocalin (Blc), the mammalian apolipoprotein D (ApoD), and the GLaz proteins.
In spite of the accumulating knowledge regarding the molecular features of plant lipocalins, very little is known about their function at the cellular and biochemical levels. However, the TIL properties, their association with the plasma membrane in photosynthetic tissues, and their accumulation in response to temperature stress support the hypothesis that these proteins may act as scavengers of potentially harmful molecules known to be induced by temperature stress and excess light [5]. In this report, we characterized knock-out, complementation and overexpression lines to determine the cellular and biochemical functions of the AtTIL lipocalin.
Results and discussion
AtTIL delays flowering and maintains leaf greenness
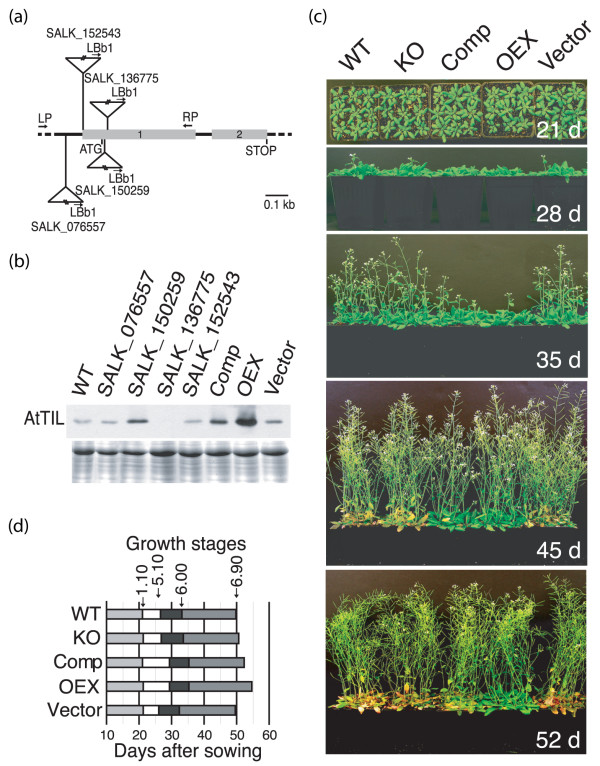
For functional analyses, four T-DNA lines carrying insertions in the AtTIL gene were analyzed to identify knock-out (KO) lines (Fig. 1a) [see Additional file 1] [6]. The SALK_136775 line carries a single insertion in the first exon of the AtTIL gene and shows no detectable AtTIL expression (Fig. 1b). Complementation of this KO line restored AtTIL protein accumulation to a level about two-fold higher than the wild-type (WT) plants (Fig. 1b, Comp). Overexpression of the AtTIL cDNA resulted in a 4-fold accumulation of the protein compared to the WT (Fig. 1b, OEX). Under normal growth conditions, the downregulation of AtTIL expression (KO) has no visible effect on plant growth and development compared to the WT (Fig. 1c). In contrast, the Comp and OEX lines show a delay in flowering and a stay-green phenotype clearly visible at 45 and 52 days, respectively (Fig. 1c and 1d, Table 1). These data suggest that an increased AtTIL level extends the vegetative phase and maintains leaf greenness. To determine if AtTIL protein accumulation is modulated during development, a western blot analysis was performed on samples prepared from tissues collected throughout the WT plant's life cycle. The results show that apart from the dry seeds stage, AtTIL is expressed at approximately the same level at all stages of development (Fig. 2a). Since there is no association between the protein accumulation and normal flowering, and because there is already a high level of protein before and during bolt elongation, we hypothesize that an excess of AtTIL may disrupt certain molecular events that ultimately delay the transition from the vegetative to reproductive phase.
Figure 1.
Modulation of AtTIL protein level affects development. (a) Genomic organization of Arabidopsis SALK lines carrying T-DNA insertions in the AtTIL gene [GenBank:BAB10998; Locus:At5g58070]. Boxes 1 and 2 are exons. The primers used for PCR analysis of the genomic DNA are indicated (see also Methods). (b) AtTIL protein levels in leaf extracts. Top panel: A rabbit anti-AtTIL antibody was used for immunoblot analysis. Bottom panel: Coomassie Brilliant Blue (CBB)-stained gel. (c) Plants were grown under normal conditions of temperature and photoperiod. Overhead (21 d) or lateral (28, 35, 45 d, 52 d) views are shown. (d) Developmental growth stages expressed according to Boyes et al. [31]. 1.10: 10 rosette leaves; 5.10: appearance of the first flower buds; 6.00: opening of the first flower; 6.90: completion of flowering. At least 30 plants per line per assay were monitored, and the experiment was repeated 3 times. WT, wild type Col-0 plants; SALK_XXXXXX, AtTIL T-DNA insertion lines from the SALK collection; Comp, SALK_136775 KO plant complemented by overexpression of AtTIL; OEX, an AtTIL overexpressing line; Vector, Col-0 transformed with a binary vector that does not carry the AtTIL cDNA (negative control).
Table 1.
Rosette leaf number at time of bolting in wild-type and AtTIL mutant plants
| WT | KO | Comp | OEX | Vector |
| 9.9 ± 0.7 | 9.8 ± 0.8 | 11.0 ± 0.7 * | 12.2 ± 0.8 ** | 10.0 ± 0.9 |
Values shown are the mean number ± SD of rosette leaves at the time of flowering. At least 12 plants of each line were used, and the experiments were repeated at least twice. Statistical analysis was performed by one-way ANOVA. The asterisks (**) and (*) indicate differences that are significant at the P < 0.001 and P < 0.01 levels respectively. Acronyms are as described in Fig. 1.
Figure 2.
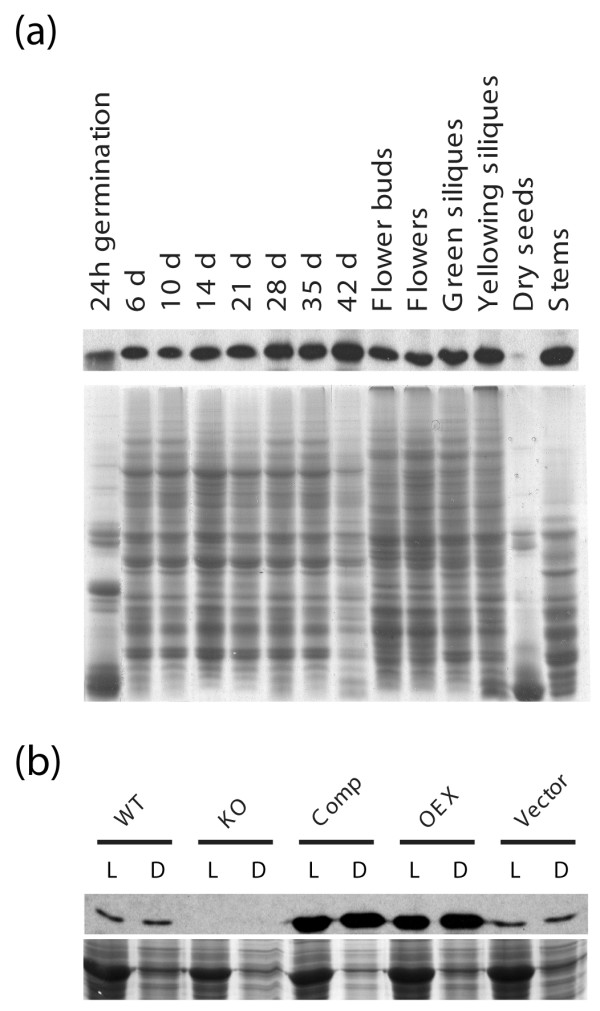
Accumulation of the AtTIL protein immunodetected with the anti-AtTIL antibody. (a) Proteins were purified from wild type plants grown under normal conditions, at various stages of their life cycle. (b) AtTIL protein levels in 7 day-old seedlings grown under light (L) or dark (D) conditions. Bottom panels: CBB-stained gel. WT, wild type Col-0 plants; KO, AtTIL T-DNA insertion lines from the SALK collection; Comp, KO plant complemented by overexpression of AtTIL; OEX, an AtTIL overexpressing line; Vector, Col-0 transformed with a binary vector that does not carry the AtTIL cDNA (negative control).
AtTIL enhances tolerance to freezing and oxidative stress
There are no noticeable morphological differences between the different Arabidopsis lines when they are compared before (non-acclimated; NA) or after cold acclimation (CA) (Fig. 3a). After subjecting NA plants to a freezing test at -6°C, a survival rate of 75% was obtained for the wild-type (WT) and control (vector) plants (Fig. 3b). In contrast, only 50% of the KO plants survived while the survival rate of the Comp and OEX plants was more than 90%. Freezing treatment caused typical damage and necrosis to the leaves of surviving WT and KO plants. On the other hand, leaves of OEX plants showed no damage after freezing and resumed vigourous growth when transferred to normal growth conditions at 20°C (Fig. 3a). After 7 days of cold acclimation (CA7), the level of AtTIL protein increases in all lines except in KO plants (Fig. 3c). This indicates that AtTIL accumulation is associated with an increased freezing tolerance that provides protection against damages caused by a sudden drop in temperature. On the other hand, the difference in sensitivity to freezing between NA WT and KO plants is not observed when plants are cold-acclimated prior to exposure to freezing (CA7 -10°C, Fig. 3a). Additional freezing tests performed down to -13°C did not reveal any differences between WT and KO plants [see Additional file 2], suggesting that cold acclimation is sufficient to provide maximal FT, and that AtTIL is likely not essential when plants undergo the full acclimation process. Protoplasts isolated from non acclimated lipocalin transgenic plants showed a higher survival rate compared to WT after freezing to -10°C. This increase in survival rate was associated with a decrease in the incidence of dehydration-induced injury [7].
Figure 3.
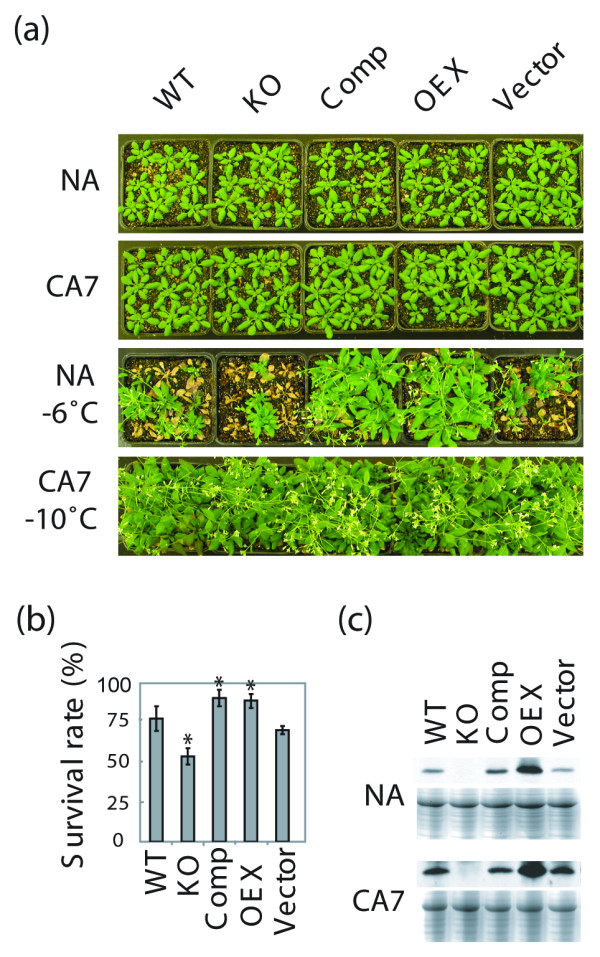
AtTIL enhances tolerance to freezing stress. (a) Plants were grown for 3 weeks at 22°C (NA) or grown at 22°C then transferred at 4°C for 7 days (CA7), and pictures were captured. The same plants were subjected to a freeze test performed at -6°C or -10°C for the NA and CA plants, respectively, and pictures were captured after a recovery period of 3 weeks. (b) Survival rate after freezing of NA plants to -6°C, expressed as a percent of surviving plants. Statistical analysis was performed by one-way ANOVA, and the asterisks (*) indicate differences that are significant at the P < 0.001 level. (c) AtTIL protein accumulation in leaves of NA and CA plants. Proteins were extracted and analyzed by immunoblotting. CBB-stained gels are shown as loading controls. Results are representative of at least three independent assays involving 18 plants per line per assay.
The freeze-induced lesions may be caused by harmful reactive oxygen species (ROS) generated when plants are exposed to freezing. To test the effect of ROS, plants were treated with the oxidant paraquat, a powerful herbicide that generates ROS [8-10]. After 7 days of treatment, KO plants showed more necrotic lesions than WT plants, whereas OEX plants were more resistant (Fig. 4a and 4b).
Figure 4.
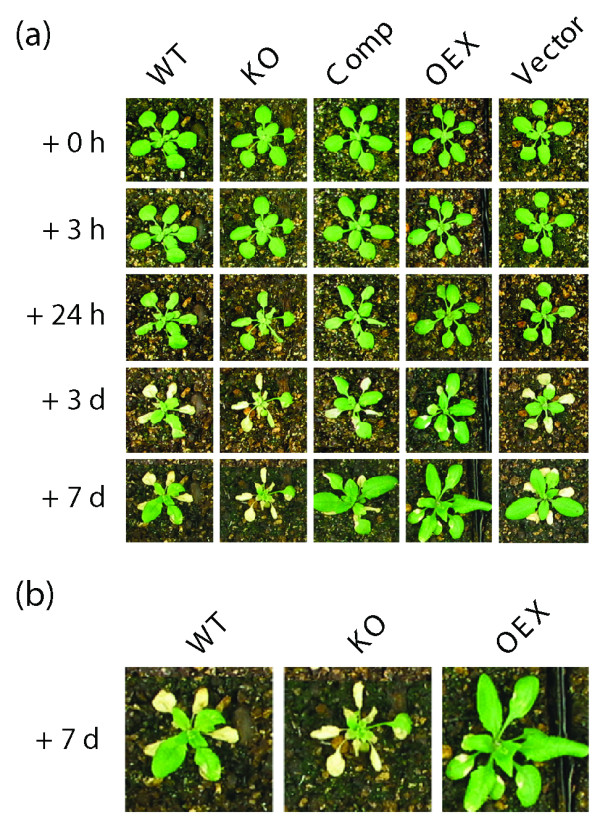
The level of AtTIL accumulation influences oxidative stress tolerance of Arabidopsis. (a) Plants were grown under normal conditions for 3 weeks and then sprayed until run off with either water or a 15 μM paraquat solution. Pictures were captured at the indicated time after treatment. Only the paraquat-treated plants are shown since no effect was observed for plants sprayed with water. (b) A close-up is shown to better show the necrotic lesions. Results are representative of at least three independent assays involving 32 plants per line per assay.
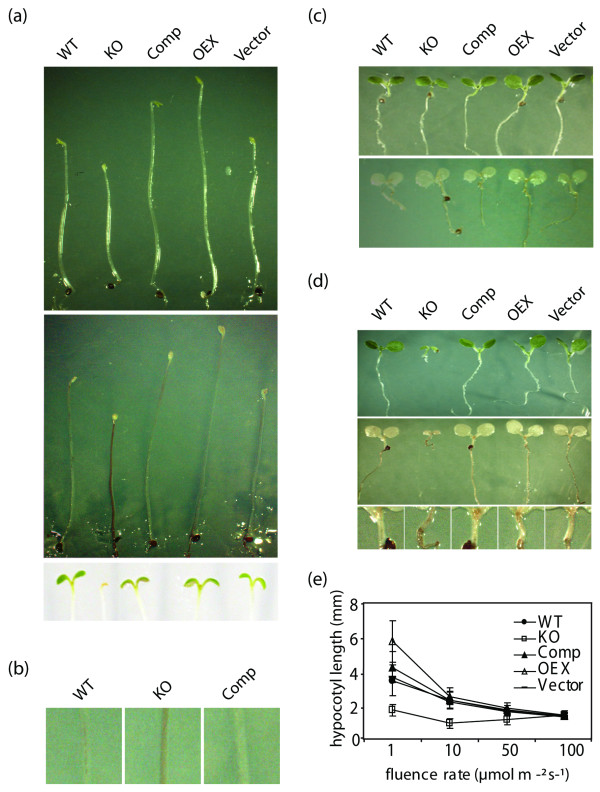
The effect of light stress on the different lines was monitored under different fluence rates (Fig. 5). Dark-grown KO plants show a reduction in hypocotyl elongation that is reversed in the Comp line, while OEX plants show longer hypocotyls (Fig. 5a, upper panel). There is no difference in AtTIL accumulation between plants grown under light or dark conditions (Fig. 2b). When dark-grown WT, Comp, OEX and control plants are transfered to normal photoperiod and light conditions, greening occurs and typical development resumes. In contrast, dark-grown KO plants transferred to light do not accumulate chlorophyll and die shortly after (Fig. 5a, lower panel). This suggests that the absence of lipocalin impairs the plants' ability to adapt to a sudden light exposure. There was no difference in growth and development when plants of the different lines were grown under a normal light-dark cycle (Fig. 5c, upper panel). Under a continuous moderate fluence of 100 μmol-1 sec-2, the different lines show similar hypocotyl elongation but KO plants have smaller cotyledons (Fig. 5d, upper panel). These data indicate that in the absence of AtTIL, plants cannot tolerate the stress generated by continuous light and that a dark period is needed for plants to recover. Staining with 3,3'-diaminobenzidine (DAB) revealed that KO plants grown under stress conditions (dark or continuous light conditions) accumulate hydrogen peroxide and other ROS (Fig. 5a middle panel, Fig. 5b, Fig. 5d middle and lower panels) while KO plants grown under normal growth conditions do not (Fig. 5c lower panel). A differential growth sensitivity was observed when plants were grown under continuous low light. However, at moderate fluence of 50 and 100 μmol-1 sec-2, the different lines show similar hypocotyl elongation (Fig. 5e), but the KO plants have small cotyledons as shown in Fig. 5D. It has been shown that hydrogen peroxide accumulation causes growth inhibition in maize and pine hypocotyls [11]. We speculate that the higher basal level of peroxide and other ROS in the KO plants is likely responsible for the higher sensitivity to oxidative stress, which leads to a reduction in hypocotyl growth and increased sensitivity to light. It was reported that the inability to eliminate excess peroxide increases sensitivity towards environmental stresses and induces the expression of several stress-related genes [12].
Figure 5.
AtTIL knock-out plants show reduced hypocotyl elongation, hydrogen peroxide and other ROS accumulation. (a) Representative seedlings grown under dark conditions (upper panel), then stained with DAB (middle panel) to detect H2O2 and other ROS, or returned to light conditions for 24 h (lower panel). (b) Close-up of hypocotyls from dark-grown plants stained with DAB. (c) Representative seedlings grown under light-dark cycles (upper panel), then stained with DAB (lower panel). (d) Representative seedlings grown under continuous light (upper panel), then stained with DAB (middle panel). A close-up is shown to better show the DAB staining (lower panel). (e) Fluence response curves of seedlings grown under continuous light. Dark-adapted seedlings were exposed 7 days in the dark or under various intensities of continuous light. Images were captured and hypocotyl length was measured with the Image J software.
Transcriptome analysis of AtTIL knock-out plants
Expression profiles were determined for WT and KO plants grown under NA and CA conditions (7 days at 4°C). No differences in gene expression were observed between the CA WT and CA KO, except for the absence of AtTIL transcript in the KO line. However, 66 genes were differentially regulated by more than 2-fold in the NA KO compared to the NA WT (51 up and 15 down-regulated; Table 2). Among these 66 genes, 49 are regulated in a similar manner (up or down) by LT exposure in WT plants while 5 are regulated in an opposite manner. These genes encode known or putative transcription factors or signal transduction proteins, heat shock proteins, enzymes involved in carbohydrate metabolism or oxidative stress responses, and senescence and circadian clock-associated proteins. The remaining 12 genes are regulated only in the KO line (* in Table 2; KO-specific genes) and encode proteins that are associated with stress responses. This analysis indicates that without exposure to LT, the absence of AtTIL mimicks part of the LT response. The accumulation of hydrogen peroxide and other ROS in the KO plants (Fig. 5) may act as a signal that triggers the changes observed in gene expression. A similar finding was reported for catalase-deficient plants under non-stressed conditions [13]. It is known that hydrogen peroxide accumulation results in differential gene expression [14].
Table 2.
Genes showing at least two-fold differential expression (induction/repression) in Arabidopsis AtTIL knock-out plants.
| Differential expression (absolute values) |
||||||||
| KO | KO 4°C | WT 4°C | KO 4°C | |||||
| vs | vs | vs | vs | Regulation | ||||
| Putative Function and Reference | Locus ID | ProbesetIDs | WT | KO | WT | WT 4°C | ||
| Upregulated Genes | ||||||||
| Transcription Factors: | ||||||||
| pseudo-response regulator 5 (APRR5) | At5g24470 | 249741_at | 5,3 | 6,3 | 33,8 | -1,0 | † | |
| zinc-binding family protein (DUF597) | At1g76590 | 259977_at | 2,9 | 2,1 | 5,7 | 1,1 | # | |
| WRKY family transcription factor (WRKY54) | At2g40750 | 257382_at | 2,8 | 2,2 | 4,8 | 1,3 | # | |
| DRE-binding protein (DREB1A)/CRT/DRE-binding factor 3 (CBF3) | At4g25480 | 254066_at | 2,3 | 4,0 | 12,9 | -1,4 | ||
| ABA-responsive element-binding protein (ABRE) | At1g49720 | 261613_at | 2,2 | 2,4 | 4,9 | 1,1 | ||
| sigA-binding protein | At3g56710 | 246293_at | 2,2 | 1,8 | 3,6 | 1,1 | # | |
| heat shock factor protein, putative (HSF5) (HSTF5) | At1g67970 | 259992_at | 2,1 | 2,2 | 4,0 | 1,2 | # | |
| gigantea protein (GI) | At1g22770 | 264211_at | 2,0 | 3,6 | 6,9 | 1,1 | † | |
| zinc finger (MYND type) family protein | At5g50450 | 248502_at | 2,1 | 2,2 | 4,4 | 1,1 | ||
| zinc finger (C3HC4-type RING finger) family protein | At1g49230 | 260753_at | 2,1 | 1,4 | 2,9 | 1,0 | # | |
| CONSTANS-LIKE 13 zinc finger (B-box type) family protein | At2g47890 | 266514_at | 2,1 | 3,5 | 6,5 | 1,1 | ||
| ABI3-interacting protein 1 (AIP1) (TOC1) | At5g61380 | 247525_at | 2,1 | 3,1 | 5,4 | 1,2 | † | |
| Signal Transduction: | ||||||||
| invertase/pectin methylesterase inhibitor family protein | At5g62360 | 247478_at | 5,2 | 1,3 | 5,8 | 1,1 | # | |
| cold circadian rhythm-RNA binding 2; CCR2 | At2g21660 | 263548_at | 4,4 | 3,2 | 13,4 | 1,1 | ||
| acid phosphatase class B family protein | At2g39920 | 267361_at | 4,0 | -1,1 | 3,4 | 1,0 | # | |
| auxin-regulated protein kinase, putative | At2g33830 | 267461_at | 3,2 | -1,6 | 1,7 | 1,1 | * | |
| peptidyl-prolyl cis-trans isomerase/cyclophilin (CYP2)/rotamase | At2g21130 | 264019_at | 2,3 | 2,9 | 6,4 | 1,1 | ||
| ankyrin repeat family protein (ACD6) | At4g14400 | 245265_at | 2,2 | 2,0 | 4,5 | -1,0 | ||
| protein kinase, putative BIK kinase | At3g55450 | 251789_at | 2,2 | 2,1 | 4,2 | 1,1 | # | |
| phosphorylase family protein | At4g24340 | 254163_s_at | 2,1 | -3,3 | -1,3 | -1,2 | * | |
| Stress Reponse, Cell Rescue and Defense: | ||||||||
| DNAJ heat shock N-terminal domain-containing protein | At5g23240 | 249850_at | 6,5 | 2,1 | 11,6 | 1,2 | # | |
| DNAJ heat shock N-terminal domain-containing protein | At1g56300 | 256221_at | 4,4 | -1,2 | 3,1 | 1,2 | # | |
| hydrophobic protein, low temperature and salt responsive protein | At4g30650 | 253627_at | 2,3 | 7,8 | 17,5 | 1,0 | ||
| senescence-associated protein-related (SAG102) | At1g53885 | 262226_at | 2,3 | -4,7 | -2,1 | 1,0 | ||
| hydrophobic protein, low temperature and salt responsive protein | At4g30660 | 253581_at | 2,1 | -2,7 | -1,1 | -1,2 | * | |
| universal stress protein (USP) family protein | At3g62550 | 251221_at | 2,0 | -2,4 | -1,3 | 1,1 | * | |
| heavy-metal-associated domain-containing protein | At1g51090 | 245749_at | 2,0 | 5,7 | 11,1 | 1,0 | ||
| pathogenesis-related thaumatin family protein (calcium storage) | At1g20030 | 261248_at | 2,0 | -1,7 | 1,0 | 1,0 | * | |
| cold-regulated protein (cor15b) | At2g42530 | 263495_at | 2,0 | 24,0 | 47,7 | 1,0 | ||
| pathogen and circadian controlled 1 (PCC1) | At3g22231 | 256766_at | 2,0 | 1,6 | 2,9 | 1,0 | # | |
| Metabolism: | ||||||||
| riboflavin biosynthesis protein, putative | At2g22450 | 264045_at | 2,8 | 1,2 | 3,2 | -1,0 | # | |
| pyruvate decarboxylase, putative | At5g54960 | 248138_at | 2,6 | 1,6 | 3,7 | 1,1 | # | |
| preprotein and amino acid transporter | At4g26670 | 253981_at | 2,4 | 1,8 | 4,0 | 1,1 | # | |
| succinate-semialdehyde dehydrogenase (SSADH1) | At1g79440 | 262892_at | 2,2 | 2,6 | 5,6 | 1,0 | ||
| aldose 1-epimerase family protein | At3g47800 | 252387_at | 2,2 | 1,6 | 3,7 | -1,1 | # | |
| nucellin protein, putative | At4g33490 | 253331_at | 2,1 | -1,1 | 1,7 | 1,1 | * | |
| auxin-responsive family protein | At5g35735 | 249719_at | 2,1 | 1,2 | 2,4 | 1,0 | # | |
| acyl CoA:diacylglycerol acyltransferase (DGAT) | At2g19450 | 267280_at | 2,1 | 3,0 | 5,9 | 1,1 | ||
| isoamylase, putative/starch debranching enzyme, putative | At4g09020 | 255070_at | 2,1 | 3,0 | 6,0 | 1,0 | ||
| 2-oxoacid-dependent oxidase, putative (DIN11) | At3g49620 | 252265_at | 2,1 | -2,8 | -1,2 | -1,1 | * | |
| OEP16-like protein | At2g28900 | 266225_at | 2,1 | 2,7 | 5,8 | -1,0 | ||
| starch phosphorylase, putative | At3g46970 | 252468_at | 2,1 | 1,6 | 2,9 | 1,1 | # | |
| glycoside hydrolase starch-binding domain-containing protein | At5g26570 | 246829_at | 2,0 | 2,5 | 4,9 | 1,0 | ||
| amino acid permease family protein or GABA permease | At2g01170 | 265790_at | 2,0 | 1,1 | 2,1 | 1,0 | # | |
| Expressed Protein | ||||||||
| expressed protein | At4g16146 | 245319_at | 4,0 | 4,2 | 14,7 | 1,2 | ||
| expressed protein | At1g53035 | 261318_at | 2,6 | 5,1 | 12,0 | 1,1 | ||
| expressed protein | At4g04330 | 255331_at | 2,6 | -1,6 | 1,5 | 1,1 | * | |
| expressed protein | At2g14560 | 265837_at | 2,5 | 2,9 | 6,1 | 1,2 | ||
| expressed protein | At2g15890 | 265478_at | 2,4 | -3,0 | -1,4 | 1,1 | * | |
| expressed protein | At1g70420 | 264314_at | 2,1 | -1,8 | 1,6 | -1,3 | * | |
| expressed protein | At1g14870 | 262832_at | 2,1 | 4,0 | 5,9 | 1,4 | ||
| Downregulated Genes | ||||||||
| Transcription Factors: | ||||||||
| late elongated hypocotyl (LHY) | At1g01060 | 261569_at | -3,2 | 6,2 | 2,1 | -1,1 | † | |
| CONSTANS-LIKE 2 (COL2) | At3g02380 | 258497_at | -2,4 | -2,0 | -3,2 | -1,5 | # | † |
| Signal Transduction: | ||||||||
| cysteine proteinase, putative | At2g27420 | 265665_at | -2,3 | 2,3 | 1,0 | -1,0 | * | |
| calcium-binding RD20 protein (RD20) | At2g33380 | 255795_at | -2,1 | -2,1 | -3,7 | -1,2 | # | |
| Stress response, Cell Rescue and Defense: | ||||||||
| At TIL, lipocalin | At5g58070 | 247851_at | -3,6 | -1,7 | 1,5 | -8,8 | * | |
| putative membrane related protein CP5 | At1g55960 | 260603_at | -2,2 | -2,0 | -4,3 | -1,0 | # | |
| phosphate-responsive protein, putative (EXO) | At4g08950 | 255064_at | -2,0 | -2,0 | -5,3 | 1,3 | ||
| Metabolism: | ||||||||
| dehydrodolichyl diphosphate synthase, putative | At5g58770 | 247780_at | -3,1 | -2,8 | -7,6 | -1,1 | ||
| trehalose-6-phosphate phosphatase (TPPA) | At5g51460 | 248404_at | -2,8 | -2,6 | -6,7 | -1,1 | ||
| starch synthase, putative | At1g32900 | 261191_at | -2,5 | 6,4 | 2,7 | -1,1 | ||
| carbonic anhydrase family protein | At3g52720 | 252011_at | -2,2 | -3,3 | -6,9 | -1,1 | ||
| glutathione S-transferase, putative (ERD9) | At1g10370 | 264436_at | -2,2 | 4,5 | 2,1 | -1,0 | ||
| adenosylmethionine decarboxylase family protein | At5g15950 | 246490_at | -2,0 | 6,6 | 3,5 | -1,1 | ||
| Expressed Protein | ||||||||
| expressed protein | At3g28270 | 256603_at | -2,3 | -1,9 | -4,0 | -1,1 | # | |
| expressed protein | At1g68440 | 259856_at | -2,1 | -2,5 | -4,6 | -1,1 | ||
* 12 genes regulated (induced or repressed) only in the KO line (not regulated by exposure of the WT to LT)
# 22 genes for which the regulation in KO is at least 50% of the regulation seen in WT exposed at 4°C
† key regulators of the circadian clock
Several genes related to the disease resistance pathway are overexpressed in the AtTIL KO plant. The WRKY54 gene is known for its association with the defense response and involves an increase in salicylic acid (SA), which accumulates during oxidative stress and mediates the induction of defense response genes [15,16]. Similarly, the SigA binding protein is a homologue of MSK1 which was shown to cause an accumulation of SA [17]. The At3g55450 protein is most homologous to the BIK1 kinase which is induced by ROS generators such as paraquat and proposed to play a role in regulating optimal SA levels [18]. Induction of the BIK1 kinase-like gene in AtTIL KO plants without exposure to any stress suggests that the level of ROS is higher than normal in these plants. The greater susceptibility to paraquat is also indicative that the AtTIL KO plants are less capable of fighting additional oxidative stress. Furthermore, the up-regulation of riboflavin biosynthesis protein in KO plants suggests that riboflavin becomes limiting when AtTIL is absent. Since riboflavin deficiency is associated with oxidative stress and is an important component of FAD-requiring enzymes such as glutathione reductase, it is possible that KO plants may try to fight oxidative stress by increasing their level of riboflavin.
The microarray analysis suggests that in the KO plants, starch synthesis is reduced while starch catabolism is accelerated to provide additional soluble sugars needed to fight oxidative stress. In addition, the Krebs cycle is inhibited under stress conditions, preventing the efficient utilization of the glycolysis products and thus affecting the respiratory machinery [19]. To overcome this situation, plants rely on the γ-aminobutyrate (GABA) shunt. Succinic semialdehyde dehydrogenase, the last enzyme of the GABA shunt, is upregulated in AtTIL KO plants. The GABA shunt is a bypass of the Krebs cycle which provides succinate and NADH to the respiratory machinery [20]. In plants, the activity of this pathway is enhanced in response to biotic and abiotic stresses [21,22] suggesting a potential role in reducing the impact of oxidative stress in mitochondria [19].
Animal lipocalins have been suggested to have a role in restoring membrane integrity caused by oxidative stress, likely by their binding to fatty acids [23]. This study provides evidence that the plasma membrane-associated AtTIL lipocalin plays a role in protecting plants against the oxidative stress induced by freezing, paraquat treatment and light. We hypothesize that during oxidative stress, lipocalins may bind and scavenge peroxidated lipids, and thus help restore membrane integrity. Our findings are in agreement with recently published data showing that overexpression of ApoD enhances tolerance to oxidative stress and increases life span in mice and Drosophila [24,25]. The available evidence therefore point towards a universal cellular function of lipocalins among species.
Methods
AtTIL lines
Arabidopsis ecotype Columbia (Col-0) was the genetic background of all the lines used. T-DNA knock-out (KO) lines for the AtTIL gene were obtained from the Salk Institute Genome Analysis Laboratory http://signal.salk.edu/. Seeds were sown on agar plates containing 50 μg/ml kanamycin, stratified for 2 days at 4°C and grown at 22°C. Kanamycin-resistant plants were propagated as individual lines on potting medium consisting of two parts Arabidopsis growing medium PM-15-13 (LEHLE seeds), one part vermiculite and one part black earth. Based on the segregation analysis of plants grown on antibiotic-containing plates, lines putatively homozygous for the T-DNA insertion were subjected to PCR analysis using T-DNA-specific (LBb1) and AtTIL-specific (RP) primers, and the resulting PCR fragments were sequenced to determine the precise insertion sites.
To generate the complementation (Comp) and overexpressing (OEX) lines, the AtTIL ORF was first cloned into pRTL2, a vector that contains a double cauliflower mosaic virus (CaMV) 35S promoter and a 35S terminator [26]. The resulting PRO35S:AtTIL:TERM35S cassette was then cloned into the binary plant expression vector pPZP121 [27] and electroporated into Agrobacterium GV3101. KO and WT Arabidopsis plants were transformed with this vector using the floral dipping method [28] to generate the Comp and OEX lines, respectively. Two independent OEX lines were selected for complete characterization. Since similar results were obtained, only one line is presented. To generate a negative control, WT plants were transformed with the pPZP121 vector that does not contain the AtTIL cassette. Transformed plants of all lines were selected by growth on gentamycin-containing medium. Homozygous plants of the fourth generation after transformation were used for the experiments.
Plant growth conditions and treatments
Surface-sterilized seeds were sown on sterile half strength Murashige and Skoog medium (Sigma-Aldrich). Seeds were stratified for 2 days at 4°C then germinated and grown at 22°C/18°C (day/night) with a 16 hr photoperiod and a photon flux density of 90 μmol m-2 sec-1. For soil-based analyses, seeds were sown directly on potting medium, stratified for 3 days at 4°C and grown for 11 days at 20°C under long day conditions (16 hr light/8 hr dark) at a photon flux density of 90 μmol m-2 sec-1. Seedlings were then transferred to 3.5 inches or 1.5 inches pots containing potting medium and grown under the same conditions as described above.
For low temperature treament, 3 week-old soil-grown plants were transferred to 4°C for 7 days under the same photoperiod conditions. For paraquat treatment, paraquat (Sigma) was dissolved in water at a concentration of 15 μM and plants were sprayed once with this solution until run off. Control plants were sprayed with distilled water. After spraying, plants were grown for 7 days under normal conditions of temperature and photoperiod.
Determination of freezing tolerance
A Caltec Scientific Ltd. Model 8–792 Large Capacity Temperature Stress Chamber was used to perform the freezing tolerance tests. This instrument consists of four major component systems: a Sanyo Model MDF-792 24.75 ft3 capacity ultra-low temperature chest freezer, a custom designed stainless steel plenum box with its integral blower and heater (provides air circulation and heating) and an Omega Engineering Inc. Model CN3002 programmable profile controller (monitors the test-chamber air temperature and controls the heating element). The controlled action of the heater combines with the constant cooling of the freezer to achieve the desired temperature at any given time.
Non-acclimated (NA) and cold-acclimated (CA) soil-grown plants (3 weeks-old) were subjected to the following freezing regime. The plants were equilibrated at -1°C for 2 hr and were seeded with ice chips to initiate freezing. The temperature was then lowered gradually (2°C per hr) to -6°C and maintained at this temperature for 6 hr. The temperature was then gradually increased (2°C per hr) to 4°C. To determine temperature variability in the freezer, temperatures were monitored by 6 independent thermocoupled T probes distributed in the freezer and connected to an Agilent 3497-0A data acquisition/switch unit. Freezing regimes that showed more than 0.5°C discrepancies between the different probes were rejected. To minimize light stress effects after the freezing treatment, plants were thawed at 4°C for 24 hr in the dark and away from direct light in the growth chamber (20°C) for an additional 24 hr before returning to normal light conditions. Pictures were taken 3 weeks after the freezing treatment. Eighteen plants were frozen per line per assay, and the experiment was repeated 3 times with independent biological replicates.
Hypocotyl analyses
Seeds were sown on sterile half strength Murashige and Skoog medium (Sigma-Aldrich) and stratified in the dark at 4°C for 6 days. They were then exposed to white light (90 μmol m-2 s-1) at 20°C for 60 min and returned to darkness for 23 hr before being exposed to varying fluence rates at 20°C for 7 days. Hypocotyl length was measured using the Image J 1.36b software (mean ± SD; n = 40). H2O2 and other ROS were detected in situ by 3,3'-diaminobenzidine (DAB) staining as described [29].
Protein isolation and immmunoblot analysis
The antibody raised against the wheat TaTIL lipocalin protein (4) does not cross react with the Arabidopsis AtTIL lipocalin, therefore it was necessary to raise a specific antibody against the latter. The AtTIL cDNA was cloned in the pTrc-HIS vector (Invitrogen) and electroporated into the E. coli strain DH5α. The recombinant HIS::AtTIL protein was produced by induction with 1 mM IPTG for 3 hr. The cells were collected, broken by lysozyme treatment and sonication, and HIS-tagged proteins were purified by immobilized metal affinity chromatography on Ni-NTA agarose (Novagen). The purified proteins were more than 90% pure and used to immunize a rabbit to obtain polyclonal antibodies.
Aerial parts of the Arabidopsis plants were cut and immediately frozen in liquid nitrogen. One hundred milligrams of leaf material was processed as one sample. Proteins were isolated using Tri Reagent according to the manufacturer's instructions (Sigma). Samples were separated on 12% SDS-PAGE gels and the rabbit anti-AtTIL antibody (1:10,000) was used for the immunoblot analysis. Detection was performed with a peroxidase-coupled anti-rabbit IgG secondary antibody (1:25,000) and the Western Lightning Chemiluminescence Reagent Plus (Perkin-Elmer).
PCR and Southern blot analyses
Genomic DNA was extracted from flower buds of the different Arabidopsis lines. PCR analysis was performed according to the SIGnAL protocol using the recommended primers designed with the SIGnAL iSect Primer Design sofware: LBb1 (5' GCGTGGACCGCTTGCTGCAACT 3'), LP (5' CTGGATCCAGAGATGAAGTCG 3') and RP (5' AAGACGTGTATGGTACCGTCG 3'). For Southern analysis, the DNA (5 μg) was digested by PciI (New England Biolabs) and fractionated by electrophoresis on a 0.7% agarose gel. After electrophoresis, the gel was transferred to a positively charged nylon membrane and hybridized with 32P-labeled probes corresponding to the T-DNA of pROK2 (vector used to generate the SALK lines) or the T-DNA of the 35S:AtTIL vector. The following primer pairs were used for amplification of the fragments used as probes: 5'-CAGCAAAATCACCAGTAGCACCATTACCAT-3' and 5'-GCGATAGAAAACAAAATATAGCGCGCAAAC-3' for the pROK2 T-DNA, and 5'-ACGAAACGTGGAGCAACGGGAAGAG-3' and 5'-TGCACATACAAATGGACGAACGGATAAACC -3' for the AtTIL T-DNA. All filters were washed at high stringency (0.1× SSC, 0.1% SDS), exposed to K screens (Kodak) and analyzed on a Molecular Imager FX (Bio-Rad).
Transcriptome analysis
Non-acclimated and cold-acclimated WT and KO plants were used for microarray analysis. For each sample, three independent biological replicates of 25 plants were harvested for RNA isolation. RNA quality assessment and hybridization to the Affymetrix GeneChip Arabidopsis ATH1 Genome Arrays were performed at the McGill University and Génome Québec Innovation Centre (Montreal QC Canada). Data accumulation and analysis were performed using the Robust Multi-array Average analysis (RMA) [30], and data was deposited in the ArrayExpress database [ArrayExpress:E-TABM-430]. An analysis of variance was performed using GraphPad InStat 3 to select genes that are significantly differentially expressed by at least two-fold under the conditions specified for each analysis (4°C vs 22°C and KO vs WT). Functional classification of the genes was done according to their annotation in the TAIR database http://www.arabidopsis.org/tools/bulk/go/index.jsp, by BLAST search of the NCBI database http://www.ncbi.nlm.nih.gov/blast/Blast.cgi and by literature search performed using the gene name and annotation. RT-PCR analyses of AtTIL and randomly selected transcripts were performed to confirm the absence of AtTIL expression in the KO plants and to validate the microarray results [see Additional file 3].
Abbreviations
ApoD: Apolipoprotein D; CA: cold acclimation; Comp: complementation; DAB 3: 3'-diaminobenzidine; FT: freezing tolerance; GLaz: insect Lazarillo protein; KO: knock-out; LT: low temperature; NA: non-acclimated; OEX: overexpression; ROS: reactive oxygen species; TIL: temperature-induced lipocalin; WT: wild-type.
Authors' contributions
JB performed all experimental procedures; MH performed the microarray analysis; JB and FS conceived the study; JB, FO, MH and FS participated in the experimental design and wrote the manuscript. All authors have read and approved the final manuscript.
Supplementary Material
Genomic organization of Arabidopsis SALK lines carrying T-DNA insertions in the AtTIL gene (At5g58070). (a) PCR analysis of genomic DNA extracted from the different lines, performed with primers indicated in Fig. 1. (b) Southern blot analysis of genomic DNA extracted from the different lines. Left panel, probe detecting the pROK2 T-DNA insertion present in the SALK lines; right panel, probe detecting both a portion of the AtTIL transgene T-DNA (including the right border) and the AtTIL endogenous gene. WT, wild type Col-0 plants; SALK_XXXXXX, AtTIL T-DNA insertion lines from the SALK collection; Comp, SALK_136775 KO plant complemented by overexpression of AtTIL; OEX, an AtTIL overexpressing line; Vector, Col-0 transformed with a binary vector that does not carry the AtTIL cDNA (negative control).
AtTIL does not enhance tolerance to freezing stress in cold acclimated plants. Plants were grown for 3 weeks at 22°C then transferred at 4°C for 7 days (CA7). The plants were subjected to a freeze test performed at various temperatures and pictures were captured after a recovery period of 3 weeks. Results are representative of at least two independent assays involving 11 plants per line per assay.
Validation of microarray data using reverse transcriptase-PCR (RT-PCR). Transcript levels of randomly selected genes were quantitated in the control and cold-treated WT and AtTIL KO plants by RT-PCR. UBQ10 was used as a constitutively expressed control transcript. Reactions were performed using three independent biological replicates. Only one replicate is presented. At1g32900, starch synthase; At3g52720, carbonic anhydrase; At5g51460, trehalose-6-phosphate phosphatase; At2g01170, amino acid permease; At1g79440, succinate-semialdehyde dehydrogenase; At2g19450, diacylglycerol O-acyltransferase; At4g09020, isoamylase; At5g54960, pyruvate decarboxylase.
Acknowledgments
Acknowledgements
We thank Guillaume Brault, Greg Cormack, Nadia Toudji, Karine Tremblay and Gabriel Lévesque Tremblay for technical assistance. We also thank Isobel Parkin (Agriculture and Agri-Food Canada, Saskatoon Canada) for the generation of the AtTIL OEX lines. This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada, Genome Canada and Génome Québec.
Contributor Information
Jean-Benoit F Charron, Email: jeanbenoit.frenettecharron@yale.edu.
Francois Ouellet, Email: ouellet.francois@uqam.ca.
Mario Houde, Email: houde.mario@uqam.ca.
Fathey Sarhan, Email: sarhan.fathey@uqam.ca.
References
- Sanchez D, Ganfornina MD, Gutierrez G, Gauthier-Jauneau AC, Risler JL, Salier JP. Lipocalins genes and their evolutionary history. In: Åkerström B and Lögdberg L, editor. Lipocalins. Georgetown TX, Landes Bioscience; 2006. pp. 5–16. [Google Scholar]
- Sanchez D, Lopez-Arias B, Torroja L, Canal I, Wang X, Bastiani MJ, Ganfornina MD. Loss of glial lazarillo, a homolog of apolipoprotein D, reduces lifespan and stress resistance in Drosophila. Curr Biol. 2006;16:680–686. doi: 10.1016/j.cub.2006.03.024. [DOI] [PubMed] [Google Scholar]
- Walker DW, Muffat J, Rundel C, Benzer S. Overexpression of a Drosophila homolog of apolipoprotein D leads to increased stress resistance and extended lifespan. Curr Biol. 2006;16:674–679. doi: 10.1016/j.cub.2006.01.057. [DOI] [PubMed] [Google Scholar]
- Charron JB, Ouellet F, Pelletier M, Danyluk J, Chauve C, Sarhan F. Identification, expression, and evolutionary analyses of plant lipocalins. Plant Physiol. 2005;139:2017–2028. doi: 10.1104/pp.105.070466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron JBF, Sarhan F. Plant lipocalins. In: Åkerström B and Lögdberg L, editor. Lipocalins. Georgetown TX, Landes Bioscience; 2006. pp. 41–48. [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Tominaga Y, Nakagawara C, Kawamura Y, Uemura M. Effect of plasma membrane-associated proteins on acquisition of freezing tolerance in Arabidopsis thaliana. In: Chen THH, Uemura M and Fujikawa S, editor. Cold hardiness in plants Molecular genetics, cell biology and physiology. Cambridge MA USA, CABI Publishing; 2006. pp. 235–249. [Google Scholar]
- Broadbent P, Creissen GP, Kular B, Wellburn AR, Mullineaux PM. Oxidative stress responses in transgenic tobacco containing altered levels of glutathione reductase activity. Plant J. 1995;8:247–255. doi: 10.1046/j.1365-313X.1995.08020247.x. [DOI] [Google Scholar]
- Perl-Treves R, Galun E. The tomato Cu,Zn superoxide dismutase genes are developmentally regulated and respond to light and stress. Plant Mol Biol. 1991;17:745–760. doi: 10.1007/BF00037058. [DOI] [PubMed] [Google Scholar]
- Tsang EW, Bowler C, Herouart D, Van Camp W, Villarroel R, Genetello C, Van Montagu M, Inze D. Differential regulation of superoxide dismutases in plants exposed to environmental stress. Plant Cell. 1991;3:783–792. doi: 10.1105/tpc.3.8.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedreira J, Sanz N, Pena MJ, Sanchez M, Queijeiro E, Revilla G, Zarra I. Role of apoplastic ascorbate and hydrogen peroxide in the control of cell growth in pine hypocotyls. Plant Cell Physiol. 2004;45:530–534. doi: 10.1093/pcp/pch059. [DOI] [PubMed] [Google Scholar]
- Vanderauwera S, Zimmermann P, Rombauts S, Vandenabeele S, Langebartels C, Gruissem W, Inze D, Van Breusegem F. Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiol. 2005;139:806–821. doi: 10.1104/pp.105.065896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V, Apel K, Inze D, Mittler R, Van Breusegem F. Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 2006;141:436–445. doi: 10.1104/pp.106.078717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell. 2005;17:1866–1875. doi: 10.1105/tpc.105.033589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journot-Catalino N, Somssich IE, Roby D, Kroj T. The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell. 2006;18:3289–3302. doi: 10.1105/tpc.106.044149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Amornsiripanitch N, Dong X. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2006;2:e123. doi: 10.1371/journal.ppat.0020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson E, Jenkins T, Brodersen P, Thorgrimsen S, Petersen NH, Zhu S, Qiu JL, Micheelsen P, Rocher A, Petersen M, Newman MA, Bjorn NH, Hirt H, Somssich I, Mattsson O, Mundy J. The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 2005;24:2579–2589. doi: 10.1038/sj.emboj.7600737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veronese P, Nakagami H, Bluhm B, Abuqamar S, Chen X, Salmeron J, Dietrich RA, Hirt H, Mengiste T. The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell. 2006;18:257–273. doi: 10.1105/tpc.105.035576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché N, Fait A, Bouchez D, Moller SG, Fromm H. Mitochondrial succinic-semialdehyde dehydrogenase of the gamma-aminobutyrate shunt is required to restrict levels of reactive oxygen intermediates in plants. Proc Natl Acad Sci USA. 2003;100:6843–6848. doi: 10.1073/pnas.1037532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelp BJ, Bown AW, McLean MD. Metabolism and functions of gamma-aminobutyric acid. Trends Plant Sci. 1999;4:446–452. doi: 10.1016/S1360-1385(99)01486-7. [DOI] [PubMed] [Google Scholar]
- Bown AW, Shelp BJ. The metabolism and functions of [gamma]-aminobutyric acid. Plant Physiol. 1997;115:1–5. doi: 10.1104/pp.115.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snedden WA, Fromm H. Regulation of the ã-aminobutyrate-synthesizing enzyme, glutamate decarboxylase, by calcium-calmodulin: a mechanism for rapid activation in response to stress. In: Lerner HR, editor. Plant responses to environmental stresses: from phytohormones to genome reorganization. New-York NY, Marcel Dekker; 1999. pp. 549–574. [Google Scholar]
- Bishop RE. The bacterial lipocalins. Biochim Biophys Acta. 2000;1482:73–83. doi: 10.1016/s0167-4838(00)00138-2. [DOI] [PubMed] [Google Scholar]
- Muffat J, Walker DW, Benzer S. Human ApoD, an apolipoprotein up-regulated in neurodegenerative diseases, extends lifespan and increases stress resistance in Drosophila. Proc Natl Acad Sci USA. 2008;105:7088–7093. doi: 10.1073/pnas.0800896105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganfornina MD, Carmo SD, Lora JM, Torres-Schumann S, Vogel M, Allhorn M, Gonzalez C, Bastiani MJ, Rassart E, Sanchez D. Apolipoprotein D is involved in the mechanisms regulating protection from oxidative stress. Aging Cell. 2008 doi: 10.1111/j.1474-9726.2008.00395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo MA, Freed DD, Carrington JC. Nuclear transport of plant potyviral proteins. Plant Cell. 1990;2:987–998. doi: 10.1105/tpc.2.10.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB. Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 1997;11:1187–1194. doi: 10.1046/j.1365-313X.1997.11061187.x. [DOI] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Gorlach J. Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell. 2001;13:1499–1510. doi: 10.1105/tpc.13.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genomic organization of Arabidopsis SALK lines carrying T-DNA insertions in the AtTIL gene (At5g58070). (a) PCR analysis of genomic DNA extracted from the different lines, performed with primers indicated in Fig. 1. (b) Southern blot analysis of genomic DNA extracted from the different lines. Left panel, probe detecting the pROK2 T-DNA insertion present in the SALK lines; right panel, probe detecting both a portion of the AtTIL transgene T-DNA (including the right border) and the AtTIL endogenous gene. WT, wild type Col-0 plants; SALK_XXXXXX, AtTIL T-DNA insertion lines from the SALK collection; Comp, SALK_136775 KO plant complemented by overexpression of AtTIL; OEX, an AtTIL overexpressing line; Vector, Col-0 transformed with a binary vector that does not carry the AtTIL cDNA (negative control).
AtTIL does not enhance tolerance to freezing stress in cold acclimated plants. Plants were grown for 3 weeks at 22°C then transferred at 4°C for 7 days (CA7). The plants were subjected to a freeze test performed at various temperatures and pictures were captured after a recovery period of 3 weeks. Results are representative of at least two independent assays involving 11 plants per line per assay.
Validation of microarray data using reverse transcriptase-PCR (RT-PCR). Transcript levels of randomly selected genes were quantitated in the control and cold-treated WT and AtTIL KO plants by RT-PCR. UBQ10 was used as a constitutively expressed control transcript. Reactions were performed using three independent biological replicates. Only one replicate is presented. At1g32900, starch synthase; At3g52720, carbonic anhydrase; At5g51460, trehalose-6-phosphate phosphatase; At2g01170, amino acid permease; At1g79440, succinate-semialdehyde dehydrogenase; At2g19450, diacylglycerol O-acyltransferase; At4g09020, isoamylase; At5g54960, pyruvate decarboxylase.