Abstract
Background
Neuroblastoma is a very heterogeneous pediatric tumor of the sympathetic nervous system showing clinically significant patterns of genetic alterations. Favorable tumors usually have near-triploid karyotypes with few structural rearrangements. Aggressive stage 4 tumors often have near-diploid or near-tetraploid karyotypes and structural rearrangements. Whole genome approaches for analysis of genome-wide copy number have been used to analyze chromosomal abnormalities in tumor samples. We have used array-based copy number analysis using oligonucleotide single nucleotide polymorphisms (SNP) arrays to analyze the chromosomal structure of a large number of neuroblastoma tumors of different clinical and biological subsets.
Results
Ninety-two neuroblastoma tumors were analyzed with 50 K and/or 250 K SNP arrays from Affymetrix, using CNAG3.0 software. Thirty percent of the tumors harbored 1p deletion, 22% deletion of 11q, 26% had MYCN amplification and 45% 17q gain. Most of the tumors with 1p deletion were found among those with MYCN amplification. Loss of 11q was most commonly seen in tumors without MYCN amplification. In the case of MYCN amplification, two types were identified. One type displayed simple continuous amplicons; the other type harbored more complex rearrangements. MYCN was the only common gene in all cases with amplification. Complex amplification on chromosome 12 was detected in two tumors and three different overlapping regions of amplification were identified. Two regions with homozygous deletions, four cases with CDKN2A deletions in 9p and one case with deletion on 3p (the gene RBMS3) were also detected in the tumors.
Conclusion
SNP arrays provide useful tools for high-resolution characterization of significant chromosomal rearrangements in neuroblastoma tumors. The mapping arrays from Affymetrix provide both copy number and allele-specific information at a resolution of 10–12 kb. Chromosome 9p, especially the gene CDKN2A, is subject to homozygous (four cases) and heterozygous deletions (five cases) in neuroblastoma tumors.
Background
Neuroblastoma (NB) is the most common pediatric solid tumor. It arises from primitive sympathetic nervous cells and is characterized by clinical heterogeneity, including spontaneously regressing tumors, as well as aggressive malignant tumors. Common chromosomal abnormalities include partial deletion of the short arm of chromosome 1 (1p deletion) in 30–35% of NB tumors, additional genetic material from the long arm of chromosome 17 (17q gain) in more than 50%, amplification of the proto-oncogene MYCN (25–30%) and deletion of chromosome 11q and 14q [1-7]. Whole-genome array-based approaches to analyse genomic rearrangements and chromosomal abnormalities have been employed for a variety of tumors, including NB tumors. Initially, array comparative genomic hybridization (aCGH) was used. Different studies of NB have been conducted using bacterial artificial chromosome (BAC) arrays and custom-made cDNA arrays [8-15]. Based on these previous investigations, NB has been categorized into three major subtypes; types 1, 2A and 2B. Subtype 1 comprises favorable NB with near triploidy and a predominance of numerical gains and losses, mostly representing non-metastatic NB stages 1, 2 and 4S. Subtypes 2A and 2B are found in unfavorable widespread NB, stages 3 and 4, with 11q loss and 17q gain without MYCN amplification (subtype 2A) or with MYCN amplification often together with 1p deletions and 17q gain (subtype 2B) [16]. More recently commercially available high-density oligonucleotide based SNP arrays have been employed in whole-genome copy number analyses of human tumors. They have provided accurate and rapid identification of genome abnormalities at high resolution. A few groups have used commercial oligonucleotide arrays to analyze NB tumors [11,17]. We present a comprehensive genome-wide analysis of DNA copy number in 92 NB tumors using 50 K and/or 250 K gene chip arrays from Affymetrix.
Results
Ninety-two NB tumors and four NB cell lines was analyzed with SNP arrays from Affymetrix. For a representative tumor, see Figure 1. Figure 1A–C shows chromosomal rearrangements analyzed with the CNAG3.0 software.
Figure 1.
Representative views of the technologies used. (A) Chromosome view from the CNAG3.0 software showing a representative NB tumor. (B) 1p deletion, MYCN amplification and 17q gain are indicated by arrows. (C) Heterozygous deletion in chromosome 9p, in the CDKN2A and CDKN2B region.
Regions with common hemizygous deletions
Chromosome 1p deletion
Loss of parts of the short arm of chromosome 1 (1p) was found in 28/92 (30%) of the tumors; 52/92 (57%) presented with intact chromosome 1. The other 12 tumors harbored other rearrangements, such as 1q gain. Seventeen of the 28 tumors with deletions also had MYCN amplification, whereas 11 did not (p < 2E-06). The tumors with MYCN amplification generally had larger 1p deletions than tumors without MYCN amplification (the median size of deletion for MYCN-amplified tumors was 84 Mb and for non-amplified 46 Mb). The five smallest deletions including the terminal of the short arm were found in tumors without amplification of the MYCN gene (p < 0.005), see Figure 2A. The consensus loss in the tumors with MYCN amplification was from position 17.2 to 37.0 Mb and, in tumors without MYCN amplification, from the terminal of 1p to 10.4 Mb.
Figure 2.
Deletions of chromosome 1p and 11q. Bars illustrate the deleted region; red bars for tumors with MYCN amplification and blue for tumors without. The positions of the breakpoints are indicated in megabases. (A) Deletions of chromosome 1p. (B) Deletions of chromosome 11q.
Loss of chromosome 11q
Loss of the whole of chromosome 11 was detected in 15% (14/92) of the NB tumors. Partial loss of 11q was found in 20/92 (22%), see Figure 2B. Loss of 11q was most commonly seen in tumors without MYCN amplification; of the 20 tumors with 11q loss, 18 were not MYCN amplified. The consensus loss in the two tumors with MYCN amplification was 24.4 Mb (from 110.1 Mb to 134.5 Mb/qter), while it was 50 Mb (from 84.5 Mb to 134.5/qter) in tumors without MYCN amplification.
Regions with homozygous and heterozygous deletions
Deletions in chromosome 3p
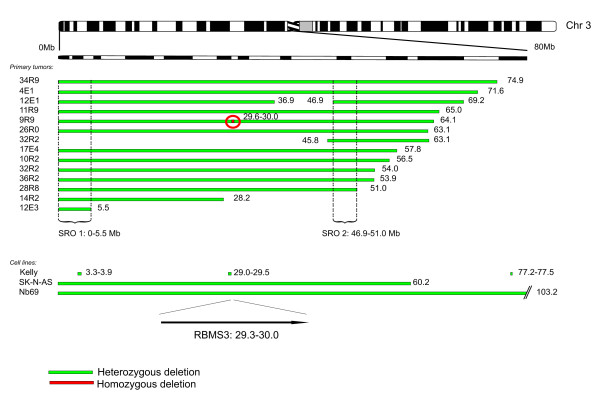
In 9R9, a stage 3 tumor, one homozygous deletion on the short arm of chromosome 3, 29.6–30.0 Mb, was detected (Figure 3). This region contains exons 4–11 of the RBMS3 gene; in a patient with unfavorable outcome. In addition, 14 of 92 tumors (15%) with heterozygous deletions were detected; 12 stage 4 tumors, one stage 2 dead of disease (DOD) and the tumor mention above, see Additional file 1. Two regions with overlap of deletions were identified among these 14 tumors. The shortest region of overlap, SRO 1, located at 0–5.5 Mb, was identified in 13 of the tumors and SRO 2, from 46.9 to 51.0 Mb, in 12 of the tumors. Moreover, three cell lines had deletions of regions covering the RBMS3 gene. SK-N-AS and NB69 had large deletions, whereas Kelly harbored four small deletions, one of which resided in RBMS3. Kelly also had a homozygous deletion at region 116.7–118.5, covering the gene LSAMP.
Figure 3.
Deletions of chromosome 3p. Green bars illustrate heterozygous deletions and the red mark indicates a homozygous deletion in tumor 9R9 covering the gene RBMS3. Two regions of overlap of deletions were identified in the primary tumors.
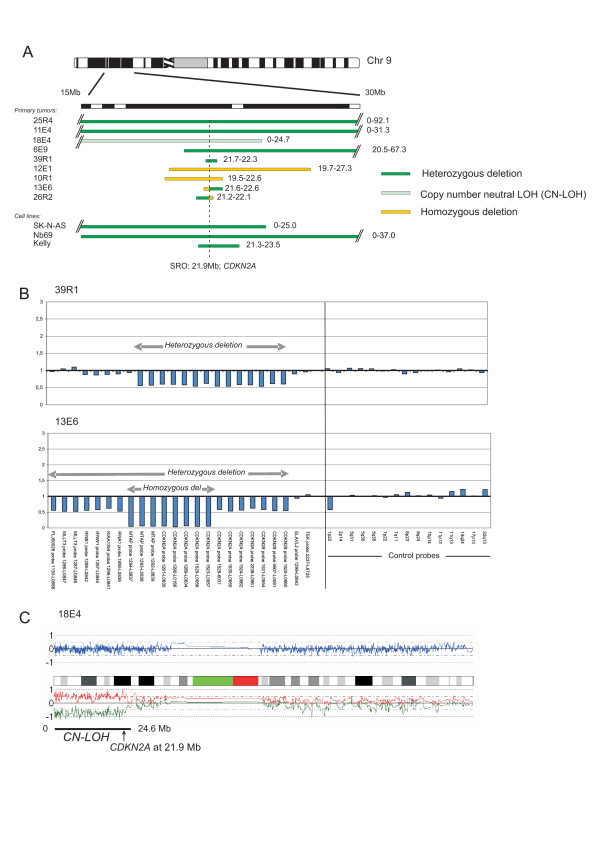
Deletions in chromosome 9p
Homozygous deletions were also detected in chromosome region 9p in four NB tumors. The shortest region of overlapping deletions, at 21.9 Mb, resided in the gene CDKN2A. Four tumors with heterozygous deletions and one with a copy neutral loss of heterozygosity (CN-LOH) were also detected, see Figure 4A and 4C. The tumors with deletions were either high-stage NB or from patients with unfavorable outcome, see Additional file 1. The cell lines SK-N-AS, NB69 and Kelly also had heterozygous deletions in this region. In all cases, the homozygous or heterozygous losses in the CDKN2A/B region could be verified by multiplex ligation-dependent probe amplification (MLPA), see Figure 4B.
Figure 4.
Deletions of chromosome 9p. (A) Array-copy number analyses of chromosome 9 deletions. Green bars illustrate heterozygous deletions, yellow bars homozygous deletions and light green CN-LOH. (B) Example of the MLPA analysis of the CDKN2A/B region. The SRO of deletions resides in the gene CDKN2A. (C) CN-LOH of 9p in tumor 18E4.
Presence of copy-neutral LOH
In the total set of genome profiles from the 92 tumors, only three cases of CN-LOH could be detected; (i) the earlier mentioned case 18E4 concerning chromosome 9p (Figure 4; see above); (ii) a case of partial 5q loss in 9R9 and (iii) CN-LOH of the entire chromosome 11 in 6R9.
Regions with amplification or gain
Amplification on chromosome 2p
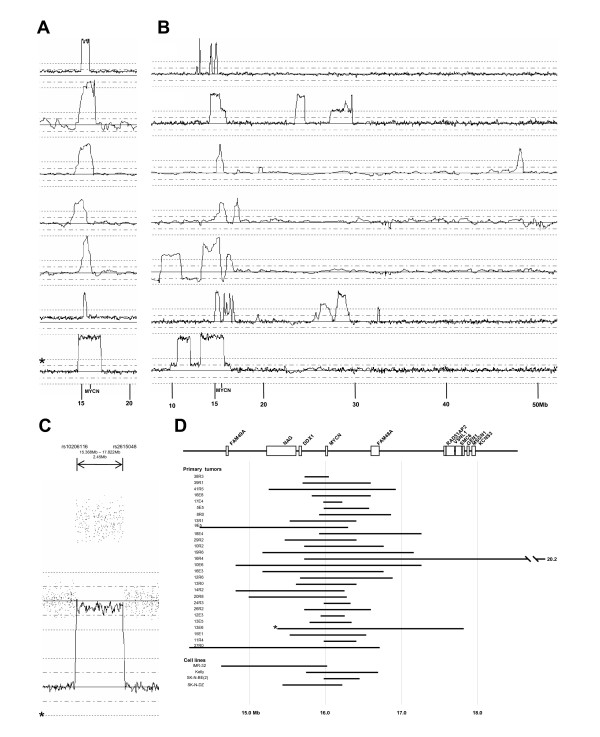
Twenty-four of the 92 tumors (26%) exhibited 2p amplification without gain and 15 tumors (16%) had gains of parts of 2p without amplification, with 8/92 (9%) having both amplification and gain. Two types of MYCN-amplified tumor were identified. One type displayed simple amplicons, where a continuous region in and around MYCN was amplified. The other type harbored more complex rearrangements, where several discontinuous amplification regions were included in the amplified fragment (see Figure 5A and 5B). Apart from MYCN, no other genes were found to be amplified in all cases with amplification (Figure 5D).
Figure 5.
Amplification of MYCN. (A) Representative tumors with simple continuous amplicons amplified. (B) Tumors with complex rearrangements. (C) The sample marked with an asterisk from the A panel in more detail. The figure shows how precisely the amplification borders can be defined using this technique. (D) The common region of amplification in tumors and cell lines. No genes other than MYCN were found in all cases with amplification.
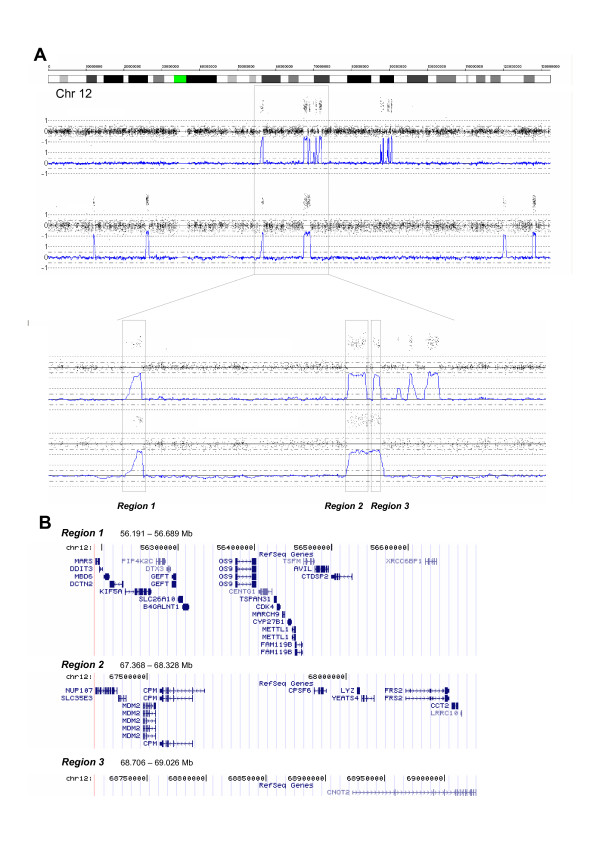
Amplification on chromosome 12
Complex amplification on chromosome 12 was detected in two tumors, one of which also had MYCN amplification. Three different regions on chromosome 12 were amplified in both cases. Region I contained the genes GLI1, OS9 and CDK4, among others, while MDM2 and YEATS4 were two of the genes located in region II; see Figure 6 for more details.
Figure 6.
Amplification on chromosome 12. (A) Complex amplifications in two tumors. The common region is enhanced in the lower panel. (B) Genes located in the amplified regions.
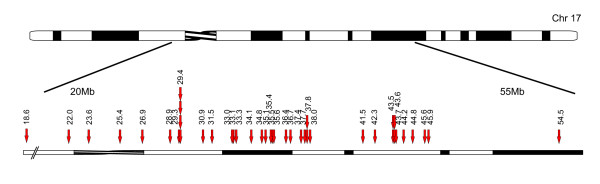
Gain of chromosome 17q
Gain of chromosome 17q was observed in 41 of 92 cases (45%). The region of gain always included the terminal of the q-arm, the smallest being 24.3 Mb (54.5–78.8/qter), see Figure 7.
Figure 7.
Gain of chromosome 17q. Arrows indicate the proximal border of the gained region which always includes the terminal of 17q. The shortest gain to be identified was located from 54.5 Mb to the terminal.
Eighteen of the 92 NB tumors (20%) showed only numerical rearrangements and 16/92 tumors (17%) had no structural or numerical rearrangements on any chromosome.
Discussion
The used technique enabled high resolution detection and mapping of all numerical and structural genomic changes in the tumor material. We could also pinpoint several previously undetected rearrangements and map them in detail. These include five new cases of homozygous deletion, which is only infrequently reported in primary neuroblastoma tumors (se discussion below). The technique proved to be fast, robust, reproducible and reliable and it is likely to be a valuable tool in future studies of neuroblastoma tumors, both in research and in the clinical setting.
Regions of deletions
Thirty percent of the tumors had 1p deletion and those were significantly more often MYCN amplified. Tumors with MYCN amplification had generally larger 1p deletions than tumors without MYCN amplification and the five smallest deletions including the p-terminal were found in tumors without MYCN amplification, which confirms an earlier study [18]. So, when identifying the SRO in 1p deletions in NB, this will be delineated by the tumors without MYCN amplification showing the most distal breakpoints. It is possible that different sets of 1p-deleted genes are important for the biological behavior of the MYCN amplified and the non-MYCN amplified cases, respectively. The SRO of deletions in tumors without MYCN amplification was located from 0 to 10.4 Mb. For the non-amplified tumors, an interstitial SRO located between 17.2 Mb to 37.0 Mb (covering 19.8 Mb) was defined. It has previously been reported that tumors with MYCN amplification have 1p deletions extending proximal to 1p36 whereas non-amplified tumors more often have small terminal deletions of 1p36 [19]. Many groups have previously tried to narrow down the shortest region of overlap of deletions on chromosome 1p [20-30]. In the largest study [31], the smallest region of consistent deletion (SRD) in all but one NB tumor was located between 5.3 Mb and 6.1 Mb which resides inside our SRO for tumors without MYCN amplification.
There was a significant inverse correlation between 11q loss and the amplification of MYCN. Only 2 of the 20 tumors with a loss of chromosome 11 had MYCN amplification. The smallest region on chromosome 11q that was lost was detected in the two tumors that also had MYCN amplification (Figure 2B), the smallest being 24.4 Mb, from 110.1 Mb to 134.5/qter. The SRO in the tumors without MYCN amplification was defined as being 50 Mb, from 84.5 Mb to 134.5/qter. The fact that 11q deletions occurs predominantly in tumors without MYCN amplification is in agreement with previous studies [15,32].
Homozygous deletions are rare events in primary NB tumors. Only a few have been reported in single cases; the deletion or homozygous gene inactivation of NF1 [26,33], the deletion of CDKN2A [34], PTEN and DMBDT1 [35]. In addition, homozygous deletions in chromosome regions 1p36 [36], 3p22.3 [8] and 2q33 (CASP8) [37] have been detected in NB cell lines. In our material, we detected homozygous deletions in the CDKN2A gene in chromosome 9p21 in four tumors. This region is frequently deleted in a wide range of malignancies [38]. CDKN2A encodes the transcripts p16INK4a and p14ARF in alternative reading frames. p16INK4a is an inhibitor to the cell cycle activators CDK4 and CDK6, which inactivate the tumor suppressor protein pRB, and p14ARF binds and inactivates MDM2, which is responsible for the degradation of TP53, thereby leading to the stabilization of TP53 (for a review, see Sharpless et al. [39]). A second region of homozygous deletion was discovered in one NB tumor, located in chromosome region 3p24.1, harboring the gene RBMS3. The protein encoded by this gene is a member of a protein family which binds single-stranded DNA/RNA. We also detected two homozygous deletions in the NB cell line Kelly, one in chromosome 3p, covering the gene LSAMP, and one in the gene PTPRD in chromosome 9p. LSAMP is a neuronal surface glycoprotein that has been identified as a putative tumor suppressor gene in ovarian and renal carcinomas [40,41], also reported to be diminished in Kelly and SK-N-AS by Stallings et al. [42]. PTPRD is a candidate tumor suppressor gene that encodes a receptor type protein tyrosine phosphatase. This confirms the finding by Stallings et al. who have previously reported that this gene is heterozygously deleted in some NB tumors and cell lines, as well as being homozygously deleted in Kelly [42]. We identified two regions of SRO on chromosome 3p in our material; SRO 1 from 0–5.5 Mb and SRO 2 from 46.9–51.0. Our SRO 2 region overlaps one of the three SROs previously defined by Hoebeeck et al [43]. This region contains among others RASSF1A and ZMYND10; two candidate tumor suppressor genes that are epigenetically silenced in a proportion of NB tumors [44,45].
Although differences in dosage between the different alleles is very common in NB tumors, given that several tumors are in the triploid range, it is noteworthy that in the present investigation, only three of the 92 analyzed NBs showed a CN-LOH.
Regions of gain and amplification
The most common chromosomal abnormality found in our material was the gain of chromosome 17q, found in 45% of the tumors. The SRO of gains was located from 54.5 Mb to the terminal of the long arm, including the gene PPM1D located at 56.0 Mb. PPM1D has been reported to be the most likely target of the 17q23 gain in NB tumors [46]; this gene was included in the gained region in all our tumors with 17q gain. Recently, Vandesompele and coworkers proposed that 17q gains may target two segments with one large from 44.3 Mb, in cases with a single region of gain, and one region of superimposed gains located more distally around 60 Mb [47].
The amplification of chromosome 12 was detected in two NB tumors. The gene MDM2 is located in the amplified region. The overexpression of MDM2 can result in the excessive inactivation of TP53, thereby diminishing its tumor suppressor function. Another gene is YEATS4 (GAS41; glioma amplified sequence), which has high expression in the human brain and is frequently amplified in gliomas [48]. Genes in this region have also previously been found to be amplified in single NB tumor samples or cell lines [15,49,50].
TP53 is inactivated by mutations in approximately half of all human tumors, and is believed to be abrogated in most tumors [51], although TP53 mutations are rare in neuroblastoma tumors [52-55]. However, mechanisms other than TP53 mutations could prevent TP53 activation. Previous studies have shown that silencing CDKN2A by methylation or the deletion or amplification of MDM2 are mechanisms that are responsible for inactivating TP53 in human tumors [56-59]. PPM1D has also been reported as a candidate proto-oncogene that may be involved in tumorigenesis through the inactivation of TP53 [51]. In our study, we detected homozygous and heterozygous deletions of CDKN2A and the amplification of MDM2 and copy number gain of PPM1D, which shows that these genes can be involved in the initiation/progression of neuroblastoma through the inactivation of TP53.
The fact that 17% of the NB tumors presented with no rearrangements was probably due to the tumors not having any rearrangements that could be visualized with the arrays. However, the possibility that some of these tumor samples contained regions of normal stoma cells, in spite of our efforts to obtain pure tumor material for the studies, cannot be ruled out.
Conclusion
We have used oligonuceotide SNP arrays from Affymetrix to perform copy number analysis on chromosomal rearrangements in 92 primary NB tumors and four cell lines. The arrays, in combination with the CNAG software for analyses and visualization, make the technique very useful for analyses of tumor tissue. The mapping arrays provide both copy number and allele-specific information and have the capacity to detect duplications, amplifications, homozygous and hemizygous deletions and copy neutral LOH (genomic regions that have a normal gene copy number, albeit both gene copies originate from the same parental chromosome, i.e. uniparental disomy).
The most common structural abnormality in the tumors was the gain of 17q, which was identified in 45% of tumors, while 30% of the tumors harbored 1p deletion. Two regions of 1p-SRO deletions were identified, one larger for the tumors with MYCN amplification (17.2–37.0 Mb) and one smaller for those without (0–10.4 Mb). Most of the tumors with 1p deletion did also show MYCN amplification. Twenty-three percent of tumors had a loss of 11q; a feature most commonly seen in tumors without MYCN amplification. The smallest 11q deletions were found in the few tumors with amplification of MYCN (SRO of deletions from 110.1 Mb to 134.5/qter). Twenty-six percent of the NB had MYCN amplification. Two types of amplification were identified; one type displayed simple continuous amplicons, while the other type harbored more complex rearrangements. MYCN was the only common gene in all cases with amplification. Complex amplification on chromosome 12 was detected in two tumors and three different overlapping regions of amplification were identified. Two regions with homozygous deletions were detected indicating genes with tumor suppressor features. Four NB tumors had deletions in the CDKN2A gene region in 9p and one tumor had a deletion on 3p involving the RBMS3 gene.
Methods
Tumor material and DNA isolation
A panel of 92 primary NB tumors, 14 stage 1, eight stage 2, 15 stage 3, 47 stage 4 and four stage 4S, was used in this study (together with 5 tumors of unknown stage), see Additional file 1. Four NB cell lines (IMR-32, Kelly, SK-N-AS and NB69) were also used. Tumor cell content of the samples was histologically assessed in adjacent tumor tissue to that used for DNA extraction. Genomic DNA was extracted with a DNeasy blood and tissue kit (Qiagen, Hilden, Germany) according to the protocol provided by the supplier.
Microarray experiments
GeneChip® Human Mapping 50 K and 250 K assay
The Affymetrix 50 K Array used detects ~59,000 SNPs, while the 250 K detects ~262,000. These arrays were used to perform aCGH, where the samples were compared after the run to constitutional DNA from healthy individuals in silico. The array experiments were performed at our lab or at AROS Applied Biotechnology AS (Aros AB, Aarhus, Denmark) according to the protocol provided by the supplier (Affymetrix, Inc., Santa Clara, CA). Briefly, total genomic DNA (250 ng) was digested with the XbaI restriction enzyme for the 50 K array and NspI for the 250 K array and ligated to adaptors. After ligation, the template was subjected to PCR amplification using a generic primer that recognizes the adaptor sequence. The amplified DNA was fragmented with DNase I, labeled with biotin and hybridized to a GeneChip Human Mapping 50 K or 250 K array. The hybridized probes were washed using the Affymetrix Fluidics Station 450 and marked with streptavidin-phycoerythrin. The arrays were scanned using a confocal laser scanner, GeneChip Scanner 3000 (Affymetrix, Inc., Santa Clara, CA). Thirty-one NB tumors were analyzed with the 50 K array and 62 with the 250 K array (one tumor was analyzed with both the 50 K and the 250 K array).
Data analysis
Primary data analysis was performed using GDAS software (Affymetrix, Inc., Santa Clara, CA), while further statistical studies were performed using CNAG (Copy Number Analyzer for Affymetrix GeneChip Mapping arrays) software, version 3.0 (GenomeLaboratory, Tokyo University, http://www.genome.umin.jp) [60,61]. The UCSC genome browser, assembly March 2006 http://genome.ucsc.edu/, was used to visualize gene regions. Fisher's exact test, 2-sided, was used for statistical analysis.
Multiplex ligation-dependent probe amplification (MLPA)
MLPA analysis was performed using a probe mixture with 39 different probes and 5 control fragments. Twenty-one probes detect copy number changes in the CDKN2A/2B region at 9p21 (Salsa MLPA Kit P024B, MRC-Holland b.v., Amsterdam, the Netherlands). The analysis was performed according to the protocol provided by the supplier with some minor changes; the denaturation of the DNA was prolonged to 10 min and the polymerase mix was added while the samples were kept on ice. Briefly, 250 ng DNA in 5 μl TE was denaturated at 98°C and subsequently hybridized overnight (16 hours) with a mix of probes, each consisting of two parts that recognize adjacent target sequences. On day two, the hybridized probe parts were ligated with a thermostable ligase. After denaturation, PCR was performed with two universal PCR primers, amplifying all probe pairs in one reaction. The amplification products were separated by electrophoresis using an ABI 3730 Genetic Analyzer (Applied Biosystems, Foster City, CA).
Authors' contributions
The project was initiated by, and tumor collection was organized by TM, PK and JA. HC performed experimental analyses, data analyses and drafted the manuscript. JE and LO conducted experimental analyses. R–MS handled the tumor samples. JA and PK provided clinical data. TM conducted data analyses, helped to draft the manuscript and coordinated the study. All the authors reviewed and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Supplementary Material
Supplemental table
Acknowledgments
Acknowledgements
This work has been supported by grants from the Swedish Cancer Society, the Children's Cancer Foundation, the King Gustav V Jubilee Clinic Cancer Research Foundation, the Assar Gabrielsson Foundation, the Wilhelm and Martina Lundgren Research Foundation and the Sahlgrenska University Hospital Foundation. HC is a recipient of a fellowship from the Swedish Knowledge Foundation through the Industrial PhD program in Medical Bioinformatics at the Strategy and Development Office (SDO) at Karolinska Institutet and TM is a recipient of a senior cancer researcher position from the Swedish Cancer Society. We would like to thank the Swegene Gothenburg Genomics resource unit for providing access to the ABI 3730 Sequencer.
Contributor Information
Helena Carén, Email: helena.caren@clingen.gu.se.
Jennie Erichsen, Email: d02jener@student.his.se.
Linda Olsson, Email: linda.olsson@clingen.gu.se.
Charlotta Enerbäck, Email: charlotta.enerback@clingen.gu.se.
Rose-Marie Sjöberg, Email: rose-marie.sjoberg@clingen.gu.se.
Jonas Abrahamsson, Email: jonas.abrahamsson@vgregion.se.
Per Kogner, Email: per.kogner@ki.se.
Tommy Martinsson, Email: tommy.martinsson@clingen.gu.se.
References
- Gilbert F, Feder M, Balaban G, Brangman D, Lurie DK, Podolsky R, Rinaldt V, Vinikoor N, Weisband J. Human neuroblastomas and abnormalities of chromosomes 1 and 17. Cancer Res. 1984;44:5444–5449. [PubMed] [Google Scholar]
- Schwab M, Alitalo K, Klempnauer KH, Varmus HE, Bishop JM, Gilbert F, Brodeur G, Goldstein M, Trent J. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature. 1983;305:245–248. doi: 10.1038/305245a0. [DOI] [PubMed] [Google Scholar]
- Srivatsan ES, Ying KL, Seeger RC. Deletion of chromosome 11 and of 14q sequences in neuroblastoma. Genes Chromosomes Cancer. 1993;7:32–37. doi: 10.1002/gcc.2870070106. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Yokota J, Mugishima H, Okabe I, Ookuni M, Sugimura T, Terada M. Frequent loss of heterozygosity on chromosome 14q in neuroblastoma. Cancer Res. 1989;49:1095–1098. [PubMed] [Google Scholar]
- Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nature reviews. 2003;3:203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, Speleman F, Van Roy N, Laureys G, Brinskchmidt C, Christiansen H, Lampert F, Lastowska M, Bown N, Pearson A, Nicholson JC, Ross F, Combaret V, Delattre O, Feuerstein BG, Plantaz D. Multicentre analysis of patterns of DNA gains and losses in 204 neuroblastoma tumors: how many genetic subgroups are there? Med Pediatr Oncol. 2001;36:5–10. doi: 10.1002/1096-911X(20010101)36:1<5::AID-MPO1003>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Brodeur GM, Sekhon G, Goldstein MN. Chromosomal aberrations in human neuroblastomas. Cancer. 1977;40:2256–2263. doi: 10.1002/1097-0142(197711)40:5<2256::AID-CNCR2820400536>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Mosse YP, Greshock J, Weber BL, Maris JM. Measurement and relevance of neuroblastoma DNA copy number changes in the post-genome era. Cancer Lett. 2005;228:83–90. doi: 10.1016/j.canlet.2005.02.052. [DOI] [PubMed] [Google Scholar]
- Mosse YP, Diskin SJ, Wasserman N, Rinaldi K, Attiyeh EF, Cole K, Jagannathan J, Bhambhani K, Winter C, Maris JM. Neuroblastomas have distinct genomic DNA profiles that predict clinical phenotype and regional gene expression. Genes Chromosomes Cancer. 2007;46:936–949. doi: 10.1002/gcc.20477. [DOI] [PubMed] [Google Scholar]
- Scaruffi P, Valent A, Schramm A, Astrahantseff K, Eggert A, Tonini GP. Application of microarray-based technology to neuroblastoma. Cancer Lett. 2005;228:13–20. doi: 10.1016/j.canlet.2005.01.052. [DOI] [PubMed] [Google Scholar]
- Scaruffi P, Coco S, Cifuentes F, Albino D, Nair M, Defferrari R, Mazzocco K, Tonini GP. Identification and characterization of DNA imbalances in neuroblastoma by high-resolution oligonucleotide array comparative genomic hybridization. Cancer Genet Cytogenet. 2007;177:20–29. doi: 10.1016/j.cancergencyto.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Selzer RR, Richmond TA, Pofahl NJ, Green RD, Eis PS, Nair P, Brothman AR, Stallings RL. Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer. 2005;44:305–319. doi: 10.1002/gcc.20243. [DOI] [PubMed] [Google Scholar]
- De Preter K, Pattyn F, Berx G, Strumane K, Menten B, Van Roy F, De Paepe A, Speleman F, Vandesompele J. Combined subtractive cDNA cloning and array CGH: an efficient approach for identification of overexpressed genes in DNA amplicons. BMC Genomics. 2004;5:11. doi: 10.1186/1471-2164-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen QR, Bilke S, Khan J. High-resolution cDNA microarray-based comparative genomic hybridization analysis in neuroblastoma. Cancer Lett. 2005;228:71–81. doi: 10.1016/j.canlet.2004.12.056. [DOI] [PubMed] [Google Scholar]
- Chen QR, Bilke S, Wei JS, Whiteford CC, Cenacchi N, Krasnoselsky AL, Greer BT, Son CG, Westermann F, Berthold F, Schwab M, Catchpoole D, Khan J. cDNA array-CGH profiling identifies genomic alterations specific to stage and MYCN-amplification in neuroblastoma. BMC Genomics. 2004;5:70. doi: 10.1186/1471-2164-5-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels E, Vandesompele J, Hoebeeck J, Menten B, De Preter K, Laureys G, Van Roy N, Speleman F. Genome wide measurement of DNA copy number changes in neuroblastoma: dissecting amplicons and mapping losses, gains and breakpoints. Cytogenet Genome Res. 2006;115:273–282. doi: 10.1159/000095924. [DOI] [PubMed] [Google Scholar]
- Spitz R, Oberthuer A, Zapatka M, Brors B, Hero B, Ernestus K, Oestreich J, Fischer M, Simon T, Berthold F. Oligonucleotide array-based comparative genomic hybridization (aCGH) of 90 neuroblastomas reveals aberration patterns closely associated with relapse pattern and outcome. Genes Chromosomes Cancer. 2006;45:1130–1142. doi: 10.1002/gcc.20376. [DOI] [PubMed] [Google Scholar]
- Takeda O, Homma C, Maseki N, Sakurai M, Kanda N, Schwab M, Nakamura Y, Kaneko Y. There may be two tumor suppressor genes on chromosome arm 1p closely associated with biologically distinct subtypes of neuroblastoma. Genes Chromosomes Cancer. 1994;10:30–39. doi: 10.1002/gcc.2870100106. [DOI] [PubMed] [Google Scholar]
- Caron H, van Sluis P, de Kraker J, Bokkerink J, Egeler M, Laureys G, Slater R, Westerveld A, Voute PA, Versteeg R. Allelic loss of chromosome 1p as a predictor of unfavorable outcome in patients with neuroblastoma. N Engl J Med. 1996;334:225–230. doi: 10.1056/NEJM199601253340404. [DOI] [PubMed] [Google Scholar]
- Caron H, Peter M, van Sluis P, Speleman F, de Kraker J, Laureys G, Michon J, Brugieres L, Voute PA, Westerveld A. Evidence for two tumour suppressor loci on chromosomal bands 1p35-36 involved in neuroblastoma: one probably imprinted, another associated with N-myc amplification. Hum Mol Genet. 1995;4:535–539. doi: 10.1093/hmg/4.4.535. [DOI] [PubMed] [Google Scholar]
- Caron H, Spieker N, Godfried M, Veenstra M, van Sluis P, de Kraker J, Voute P, Versteeg R. Chromosome bands 1p35-36 contain two distinct neuroblastoma tumor suppressor loci, one of which is imprinted. Genes Chromosomes Cancer. 2001;30:168–174. doi: 10.1002/1098-2264(200102)30:2<168::AID-GCC1072>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Ejeskär K, Sjöberg R-M, Abel F, Kogner P, Ambros PF, Martinsson T. Fine mapping of a tumour suppressor candidate gene region in 1p36.2-3, commonly deleted in neuroblastomas and germ cell tumours. Med Pediatr Oncol. 2001;36:61–66. doi: 10.1002/1096-911X(20010101)36:1<61::AID-MPO1016>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Maris JM, Guo C, Blake D, White PS, Hogarty MD, Thompson PM, Rajalingam V, Gerbing R, Stram DO, Matthay KK, Seeger RC, Brodeur GM. Comprehensive analysis of chromosome 1p deletions in neuroblastoma. Med Pediatr Oncol. 2001;36:32–36. doi: 10.1002/1096-911X(20010101)36:1<32::AID-MPO1009>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Maris JM, White PS, Beltinger CP, Sulman EP, Castleberry RP, Shuster JJ, Look AT, Brodeur GM. Significance of chromosome 1p loss of heterozygosity in neuroblastoma. Cancer Res. 1995;55:4664–4669. [PubMed] [Google Scholar]
- Bauer A, Savelyeva L, Claas A, Praml C, Berthold F, Schwab M. Smallest region of overlapping deletion in 1p36 in human neuroblastoma: a 1 Mbp cosmid and PAC contig. Genes Chromosomes Cancer. 2001;31:228–239. doi: 10.1002/gcc.1139. [DOI] [PubMed] [Google Scholar]
- Martinsson T, Sjoberg RM, Hedborg F, Kogner P. Homozygous deletion of the neurofibromatosis-1 gene in the tumor of a patient with neuroblastoma. Cancer Genet Cytogenet. 1997;95:183–189. doi: 10.1016/S0165-4608(96)00259-2. [DOI] [PubMed] [Google Scholar]
- Martinsson T, Sjöberg R-M, Hedborg F, Kogner P. Deletion of chromosome 1p loci and microsatellite instability in neuroblastomas analyzed with short-tandem repeat polymorphisms. Cancer Res. 1995;55:5681–5686. [PubMed] [Google Scholar]
- Spieker N, Beitsma M, Van Sluis P, Chan A, Caron H, Versteeg R. Three chromosomal rearrangements in neuroblastoma cluster within a 300-kb region on 1p36.1. Genes Chromosomes Cancer. 2001;31:172–181. doi: 10.1002/gcc.1130. [DOI] [PubMed] [Google Scholar]
- White PS, Maris JM, Beltinger C, Sulman E, Marshall HN, Fujimori M, Kaufman BA, Biegel JA, Allen C, Hilliard C, et al. A region of consistent deletion in neuroblastoma maps within human chromosome 1p36.2-36.3. Proc Natl Acad Sci USA. 1995;92:5520–5524. doi: 10.1073/pnas.92.12.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PS, Thompson PM, Seifried BA, Sulman EP, Jensen SJ, Guo C, Maris JM, Hogarty MD, Allen C, Biegel JA, Matise TC, Gregory SG, Reynolds CP, Brodeur GM. Detailed molecular analysis of 1p36 in neuroblastoma. Med Pediatr Oncol. 2001;36:37–41. doi: 10.1002/1096-911X(20010101)36:1<37::AID-MPO1010>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- White PS, Thompson PM, Gotoh T, Okawa ER, Igarashi J, Kok M, Winter C, Gregory SG, Hogarty MD, Maris JM, Brodeur GM. Definition and characterization of a region of 1p36.3 consistently deleted in neuroblastoma. Oncogene. 2005;24:2684–2694. doi: 10.1038/sj.onc.1208306. [DOI] [PubMed] [Google Scholar]
- Guo C, White PS, Weiss MJ, Hogarty MD, Thompson PM, Stram DO, Gerbing R, Matthay KK, Seeger RC, Brodeur GM, Maris JM. Allelic deletion at 11q23 is common in MYCN single copy neuroblastomas. Oncogene. 1999;18:4948–4957. doi: 10.1038/sj.onc.1202887. [DOI] [PubMed] [Google Scholar]
- Origone P, Defferrari R, Mazzocco K, Lo Cunsolo C, De Bernardi B, Tonini GP. Homozygous inactivation of NF1 gene in a patient with familial NF1 and disseminated neuroblastoma. Am J Med Genet A. 2003;118:309–313. doi: 10.1002/ajmg.a.10167. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Maris JM, Hogarty MD, Seeger RC, Reynolds CP, Brodeur GM, White PS. Homozygous deletion of CDKN2A (p16INK4a/p14ARF) but not within 1p36 or at other tumor suppressor loci in neuroblastoma. Cancer Res. 2001;61:679–686. [PubMed] [Google Scholar]
- Munoz J, Lazcoz P, Inda MM, Nistal M, Pestana A, Encio IJ, Castresana JS. Homozygous deletion and expression of PTEN and DMBT1 in human primary neuroblastoma and cell lines. Int J Cancer. 2004;109:673–679. doi: 10.1002/ijc.20055. [DOI] [PubMed] [Google Scholar]
- Ohira M, Kageyama H, Mihara M, Furuta S, Machida T, Shishikura T, Takayasu H, Islam A, Nakamura Y, Takahashi M, Tomioka N, Sakiyama S, Kaneko Y, Toyoda A, Hattori M, Sakaki Y, Ohki M, Horii A, Soeda E, Inazawa J, Seki N, Kuma H, Nozawa I, Nakagawara A. Identification and characterization of a 500-kb homozygously deleted region at 1p36.2-p36.3 in a neuroblastoma cell line. Oncogene. 2000;19:4302–4307. doi: 10.1038/sj.onc.1203786. [DOI] [PubMed] [Google Scholar]
- Teitz T, Wei T, Valentine MB, Vanin EF, Grenet J, Valentine VA, Behm FG, Look AT, Lahti JM, Kidd VJ. Caspase 8 is deleted or silenced preferentially in childhood neuroblastomas with amplification of MYCN. Nat Med. 2000;6:529–535. doi: 10.1038/75007. [DOI] [PubMed] [Google Scholar]
- Rocco JW, Sidransky D. p16(MTS-1/CDKN2/INK4a) in cancer progression. Exp Cell Res. 2001;264:42–55. doi: 10.1006/excr.2000.5149. [DOI] [PubMed] [Google Scholar]
- Sharpless NE. INK4a/ARF: a multifunctional tumor suppressor locus. Mutat Res. 2005;576:22–38. doi: 10.1016/j.mrfmmm.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Chen J, Lui WO, Vos MD, Clark GJ, Takahashi M, Schoumans J, Khoo SK, Petillo D, Lavery T, Sugimura J, Astuti D, Zhang C, Kagawa S, Maher ER, Larsson C, Alberts AS, Kanayama HO, Teh BT. The t(1;3) breakpoint-spanning genes LSAMP and NORE1 are involved in clear cell renal cell carcinomas. Cancer Cell. 2003;4:405–413. doi: 10.1016/S1535-6108(03)00269-1. [DOI] [PubMed] [Google Scholar]
- Ntougkos E, Rush R, Scott D, Frankenberg T, Gabra H, Smyth JF, Sellar GC. The IgLON family in epithelial ovarian cancer: expression profiles and clinicopathologic correlates. Clin Cancer Res. 2005;11:5764–5768. doi: 10.1158/1078-0432.CCR-04-2388. [DOI] [PubMed] [Google Scholar]
- Stallings RL, Nair P, Maris JM, Catchpoole D, McDermott M, O'Meara A, Breatnach F. High-resolution analysis of chromosomal breakpoints and genomic instability identifies PTPRD as a candidate tumor suppressor gene in neuroblastoma. Cancer Res. 2006;66:3673–3680. doi: 10.1158/0008-5472.CAN-05-4154. [DOI] [PubMed] [Google Scholar]
- Hoebeeck J, Michels E, Menten B, Van Roy N, Eggert A, Schramm A, De Preter K, Yigit N, De Smet E, De Paepe A, Laureys G, Vandesompele J, Speleman F. High resolution tiling-path BAC array deletion mapping suggests commonly involved 3p21-p22 tumor suppressor genes in neuroblastoma and more frequent tumors. Int J Cancer. 2007;120:533–538. doi: 10.1002/ijc.22326. [DOI] [PubMed] [Google Scholar]
- Agathanggelou A, Dallol A, Zochbauer-Muller S, Morrissey C, Honorio S, Hesson L, Martinsson T, Fong KM, Kuo MJ, Yuen PW, Maher ER, Minna JD, Latif F. Epigenetic inactivation of the candidate 3p21.3 suppressor gene BLU in human cancers. Oncogene. 2003;22:1580–1588. doi: 10.1038/sj.onc.1206243. [DOI] [PubMed] [Google Scholar]
- Astuti D, Agathanggelou A, Honorio S, Dallol A, Martinsson T, Kogner P, Cummins C, Neumann HP, Voutilainen R, Dahia P, Eng C, Maher ER, Latif F. RASSF1A promoter region CpG island hypermethylation in phaeochromocytomas and neuroblastoma tumours. Oncogene. 2001;20:7573–7577. doi: 10.1038/sj.onc.1204968. [DOI] [PubMed] [Google Scholar]
- Saito-Ohara F, Imoto I, Inoue J, Hosoi H, Nakagawara A, Sugimoto T, Inazawa J. PPM1D is a potential target for 17q gain in neuroblastoma. Cancer Res. 2003;63:1876–1883. [PubMed] [Google Scholar]
- Vandesompele J, Michels E, De Preter K, Menten B, Schramm A, Eggert A, Ambros PF, Combaret V, Francotte N, Antonacci F, De Paepe A, Laureys G, Speleman F, Van Roy N. Identification of 2 putative critical segments of 17q gain in neuroblastoma through integrative genomics. Int J Cancer. 2008;122:1177–1182. doi: 10.1002/ijc.23156. [DOI] [PubMed] [Google Scholar]
- Munnia A, Schutz N, Romeike BF, Maldener E, Glass B, Maas R, Nastainczyk W, Feiden W, Fischer U, Meese E. Expression, cellular distribution and protein binding of the glioma amplified sequence (GAS41), a highly conserved putative transcription factor. Oncogene. 2001;20:4853–4863. doi: 10.1038/sj.onc.1204650. [DOI] [PubMed] [Google Scholar]
- Van Roy N, Forus A, Myklebost O, Cheng NC, Versteeg R, Speleman F. Identification of two distinct chromosome 12-derived amplification units in neuroblastoma cell line NGP. Cancer Genet Cytogenet. 1995;82:151–154. doi: 10.1016/0165-4608(95)00034-M. [DOI] [PubMed] [Google Scholar]
- Corvi R, Savelyeva L, Breit S, Wenzel A, Handgretinger R, Barak J, Oren M, Amler L, Schwab M. Non-syntenic amplification of MDM2 and MYCN in human neuroblastoma. Oncogene. 1995;10:1081–1086. [PubMed] [Google Scholar]
- Woods DB, Vousden KH. Regulation of p53 function. Exp Cell Res. 2001;264:56–66. doi: 10.1006/excr.2000.5141. [DOI] [PubMed] [Google Scholar]
- Hosoi G, Hara J, Okamura T, Osugi Y, Ishihara S, Fukuzawa M, Okada A, Okada S, Tawa A. Low frequency of the p53 gene mutations in neuroblastoma. Cancer. 1994;73:3087–3093. doi: 10.1002/1097-0142(19940615)73:12<3087::AID-CNCR2820731230>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Imamura J, Bartram CR, Berthold F, Harms D, Nakamura H, Koeffler HP. Mutation of the p53 gene in neuroblastoma and its relationship with N-myc amplification. Cancer Res. 1993;53:4053–4058. [PubMed] [Google Scholar]
- Komuro H, Hayashi Y, Kawamura M, Hayashi K, Kaneko Y, Kamoshita S, Hanada R, Yamamoto K, Hongo T, Yamada M, et al. Mutations of the p53 gene are involved in Ewing's sarcomas but not in neuroblastomas. Cancer Res. 1993;53:5284–5288. [PubMed] [Google Scholar]
- Vogan K, Bernstein M, Leclerc JM, Brisson L, Brossard J, Brodeur GM, Pelletier J, Gros P. Absence of p53 gene mutations in primary neuroblastomas. Cancer Res. 1993;53:5269–5273. [PubMed] [Google Scholar]
- Esteller M, Tortola S, Toyota M, Capella G, Peinado MA, Baylin SB, Herman JG. Hypermethylation-associated inactivation of p14(ARF) is independent of p16(INK4a) methylation and p53 mutational status. Cancer Res. 2000;60:129–133. [PubMed] [Google Scholar]
- Ho GH, Calvano JE, Bisogna M, Abouezzi Z, Borgen PI, Cordon-Cardo C, van Zee KJ. Genetic alterations of the p14ARF -hdm2-p53 regulatory pathway in breast carcinoma. Breast Cancer Res Treat. 2001;65:225–232. doi: 10.1023/A:1010686518990. [DOI] [PubMed] [Google Scholar]
- Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- Auerkari EI. Methylation of tumor suppressor genes p16(INK4a), p27(Kip1) and E-cadherin in carcinogenesis. Oral Oncol. 2006;42:5–13. doi: 10.1016/j.oraloncology.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Nannya Y, Sanada M, Nakazaki K, Hosoya N, Wang L, Hangaishi A, Kurokawa M, Chiba S, Bailey DK, Kennedy GC, Ogawa S. A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res. 2005;65:6071–6079. doi: 10.1158/0008-5472.CAN-05-0465. [DOI] [PubMed] [Google Scholar]
- Yamamoto G, Nannya Y, Kato M, Sanada M, Levine RL, Kawamata N, Hangaishi A, Kurokawa M, Chiba S, Gilliland DG, Koeffler HP, Ogawa S. Highly sensitive method for genomewide detection of allelic composition in nonpaired, primary tumor specimens by use of affymetrix single-nucleotide-polymorphism genotyping microarrays. Am J Hum Genet. 2007;81:114–126. doi: 10.1086/518809. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental table