Abstract
Bacteriophage T4 UvsY is a recombination mediator protein that promotes assembly of the UvsX-ssDNA presynaptic filament. UvsY helps UvsX to displace T4 gene 32 protein (gp32) from ssDNA, a reaction necessary for proper formation of the presynaptic filament. Here we use DNA stretching to examine UvsY interactions with single DNA molecules in the presence and absence of gp32 and a gp32 C-terminal truncation (*I), and show that in both cases UvsY is able to destabilize gp32-ssDNA interactions. In these experiments UvsY binds more strongly to dsDNA than ssDNA due to its inability to wrap ssDNA at high forces. To support this hypothesis, we show that ssDNA created by exposure of stretched DNA to glyoxal is strongly wrapped by UvsY, but wrapping occurs only at low forces. Our results demonstrate that UvsY interacts strongly with stretched DNA in the absence of other proteins. In the presence of gp32 and *I, UvsY is capable of strongly destabilizing gp32-DNA complexes in order to facilitate ssDNA wrapping, which in turn prepares the ssDNA for presynaptic filament assembly in the presence of UvsX. Thus, UvsY mediates UvsX binding to ssDNA by converting rigid gp32-DNA filaments into a structure that can be strongly bound by UvsX.
Introduction
Bacteriophage T4 provides an excellent model system for studying the mechanism of DNA replication because it encodes its own replication proteins in a relatively small genome. There are two general modes of DNA replication during T4 infection in E.coli, origin-dependent and recombination-dependent 1; 2. Origin dependent replication occurs at early times in infection and is dependent upon particular DNA sequences. As the infection progresses into later times, recombination-dependent replication (RDR) predominates. T4 RDR requires all of the major phage–encoded DNA replication and recombination enzymes including: gp43 (DNA polymerase), gp45 (sliding clamp), gp44/62 (clamp loader), gp32 (single stranded DNA [ssDNA] binding protein or SSB), gp61 (primase), gp41 (DNA helicase), gp59 (helicase loader; replication mediator protein or RMP), UvsX (general recombinase), UvsY (recombination mediator protein or RMP), and gp46/47 (recombination exonuclease).
gp32 binds cooperatively to ssDNA and, on a thermodynamic basis, should destabilize secondary structure in the DNA 3. Because SSBs are found in viruses and all domains of life 4, and are essential for virtually all DNA functions 5, it is important to understand how SSBs destabilize double-stranded DNA. In our previous studies, we examined the mechanism of helix-destabilization by gp32 and its truncated forms in different salt concentrations 6–9.These experiments allowed us to obtain the DNA helix-coil transition free energy in the presence of gp32 and two of its truncated forms, *I, which lacks the C-terminal domain (residues 254–301), and *III (core domain, residues 22–253), which lacks both the C-terminal domain and the N-terminal domain (residues 1–21) 6, as shown schematically in Figure 1. Note that *III contains the ssDNA binding site, the N-terminal domain is involved in homotypic protein-protein interactions with the core domain of an adjacent DNA-bound protein, thus bringing about cooperativity, and the C-terminal domain has been implicated in heterotypic protein-protein interactions and a conformational change, discussed below.
Fig. 1.
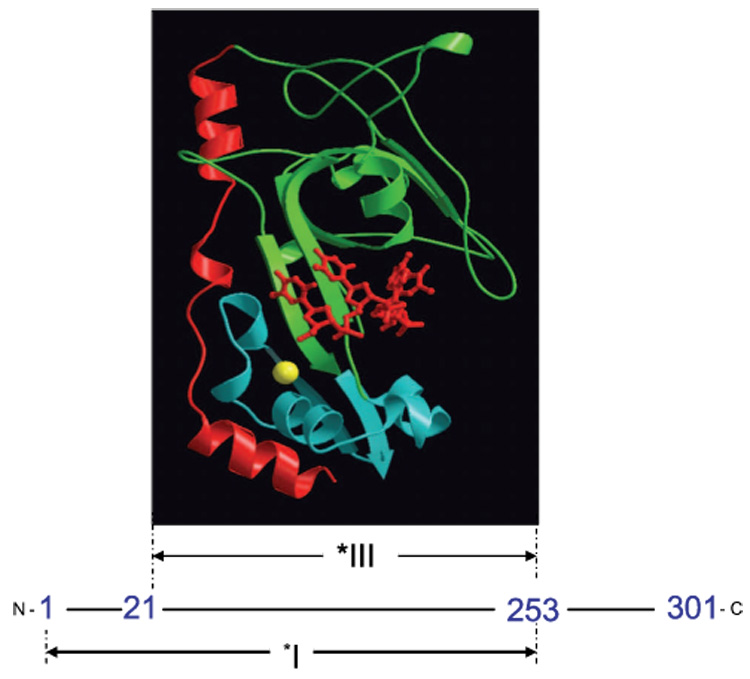
Proteolytic fragments of gene 32 protein. *I is obtained by trypsin cleavage of full length gp32 at residue 253, while *III results from cleavage at residues 21 and 253. A MOLSCRIPT 63 representation of a *III-oligonucleotide complex is shown at its location within the protein sequence. The protein is pictured in ribbon mode, with the major lobe green, the minor (Zn-containing) lobe blue, and the residue 198–239 flap red. The bound oligonucleotide, in sticks mode, is red, and the coordinated Zn2+, in space-filling mode, is yellow. The position of the oligodeoxynucleotide, pTTAT, is approximate; it was modeled by Shamoo et al. to maximally overlap excess electron density in the trough 64. The Protein Data Bank entry for core domain (without the oligonucleotide) is 1gpc.pdb.
To examine gp32 binding to DNA, we have recently developed a new method for measuring the thermodynamics and kinetics of DNA-protein binding 6–8; 10. To do this, we capture single DNA molecules in an optical tweezers instrument and stretch the molecules a fixed distance while measuring the force exerted by the optical trap. A schematic of the optical tweezers experiment is shown in Fig. 2A. The resulting force vs. extension curve for the DNA molecule is strongly dependent on solution conditions and is altered by the presence of proteins or other DNA binding ligands that may stabilize DNA by binding preferentially to dsDNA or destabilize the DNA helix by binding preferentially to ssDNA. Specifically, as the DNA is stretched, the measured force-extension data follows the wormlike chain model, represented by the leftmost solid line. However, at about 65 pN, the measured data differ substantially from the expected polymer model, representing a structural transition in DNA. The transition can be explained as a force-induced melting transition, in which dsDNA is converted into ssDNA as the DNA is extended 11; 12. In this process in the absence of binding ligand, melting likely occurs in the middle of the DNA molecule, at AT-rich regions, with only minor fraying from the ends 13. This hypothesis was initially tested in a series of experiments in which the DNA overstretching force, now referred to as the DNA melting force, was measured as a function of solution conditions such as ionic strength 14, pH 15, and temperature 16. In each case, the dependence of the melting force on these factors followed that expected based on thermal melting experiments. These studies suggested a new method for measuring DNA-ligand binding. Ligands that bind to dsDNA are expected to stabilize the DNA helix and therefore the DNA melting free energy should increase in the presence of such ligands. In contrast, ligands that bind preferentially to ssDNA should lower the melting force. Several examples of these effects have been demonstrated. For example, studies of DNA binding drugs have shown behavior that parallels DNA melting studies 17–20. Similarly, studies of single-stranded DNA binding proteins have demonstrated strong DNA helix-destabilization, as expected for such proteins. In particular, our recent studies of T4 gene 32 protein have allowed us to use DNA force-induced melting to quantify both ssDNA and dsDNA binding by gp32. Our quantitative binding results agree with the available bulk measurements, and we also were able to extend these measurements to solution conditions not achievable in previous bulk studies 6–8. In addition to the thermodynamic evidence in support of force-induced melting, in a recent study it was also shown that the DNA base pairs are exposed during DNA overstretching21. Finally, several recent theoretical studies are also consistent with this model 22–24. Taken together, all of these studies strongly support the DNA force-induced melting model, which we use here to examine additional DNA-protein interactions.
Fig. 2.
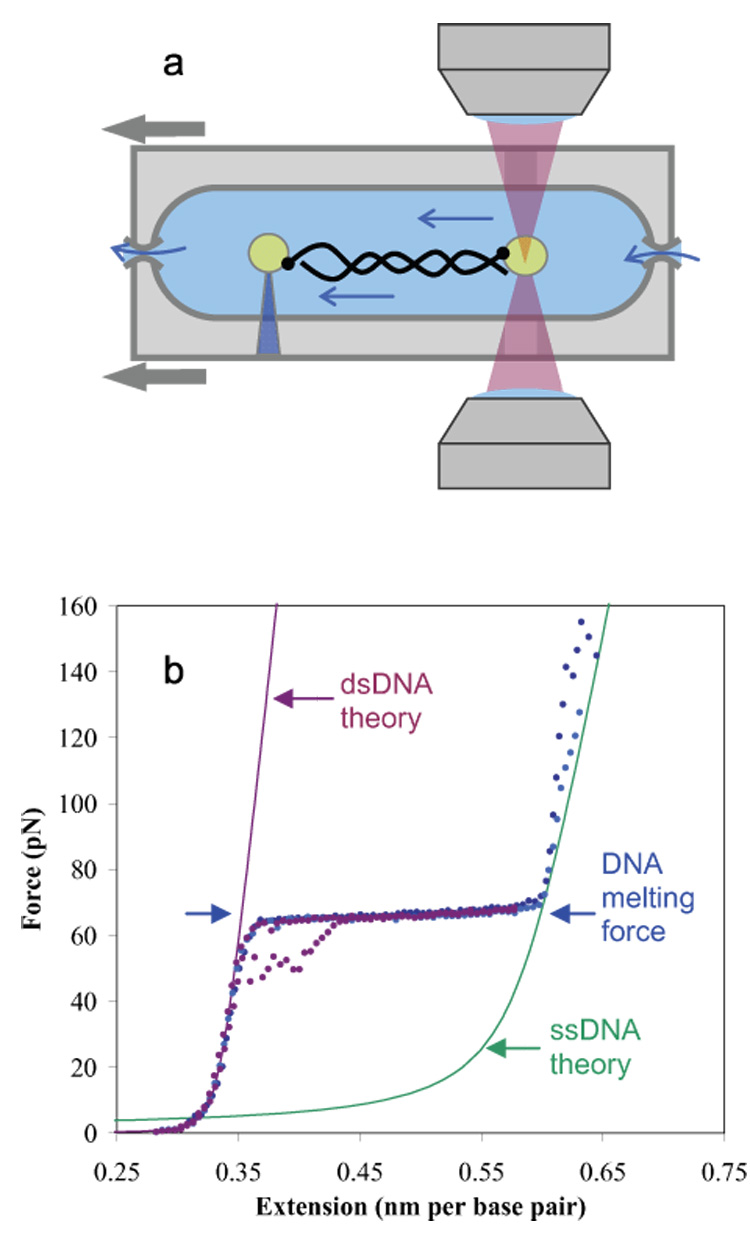
a) In a dual beam optical tweezers instrument, two laser beams are focused to a small spot, creating an optical trap that attracts polystyrene beads. Single DNA molecules are attached at one end to a bead in the trap, while the other end is attached to another bead held by a glass micropipette. As the DNA molecule is stretched by moving the micropipette, the resulting force on the bead in the trap is measured. b) Typical force extension curves for double stranded DNA are shown as dotted lines. As the stretching force is increased, dsDNA reveals an entropic elastic response, followed by the overstretching region. The data in purple shows typical data for a full cycle of extension and relaxation, including some hysteresis upon reannealing. The data in blue and cyan show the response of the resulting single strands to yet higher forces, as the strands finally separate near 150 pN (thus there are no relaxation curves). The solid lines are DNA models for ssDNA and dsDNA, as described previously 65.
In previous single molecule studies of gp32-DNA interactions, we measured the DNA melting force as a function of gp32 concentration. By globally fitting this dependence and using our model for DNA force-induced melting, we were able to determine the equilibrium binding constant of gp32 to ssDNA Kss as a function of salt concentration 8; 9. gp32 and *I show a very steep dependence of binding affinity on salt above ~0.2 M NaCl. However, at lower [NaCl], there is virtually no salt dependence of gp32 binding to ssDNA, while *I shows a significantly greater dlogKss/dlog[Na+] of ~-3 8, though reduced from the slope observed at higher salt.
As proposed previously, there is a required conformational change in gp32 prior to its binding to ssDNA. 25–29 Our results are consistent with intramolecular association of the C-terminal domain to the core domain in pre-equilibrium to the binding of gp32 to both dsDNA and ssDNA 8. As the salt concentration is lowered, the expected increase in protein-DNA binding affinity is counteracted by the stronger binding of the acidic flap to the core domain, effectively regulating the protein’s capability to destabilize DNA 8; 9; 30.
In this study, we use the method of DNA force-induced melting to study the helix-destabilization capabilities of gp32 in the presence of UvsY, a recombination mediator protein (RMP) of T4 31–34. UvsY binds tightly but noncooperatively to ssDNA 35 and shows a weaker affinity for dsDNA, when measured in bulk binding studies (H. Xu, H. T.H. Beernink, and S. W. Morrical, submitted). Like other RMP proteins participating in recombination, UvsY facilitates the assembly of the recombinase onto ssDNA and overcomes the thermodynamic and kinetic blocks imposed by gp32 36. In addition to its ssDNA binding capability, UvsY interacts with other proteins of the T4 recombination machinery, including recombinase and gp32.
At physiological salt concentrations, UvsY exists predominately as a hexamer of identical 15.8 kDa subunits and binds to ssDNA in this form 37. Wrapping or other distortions of ssDNA structure are proposed to explain the observed destabilization of gp32-ssDNA interactions brought about by UvsY 38. The reduction of gp32-ssDNA affinity and /or cooperativity within a tripartite UvsY-gp32-ssDNA intermediate may facilitate the local displacement of gp32 from the lattice by incoming UvsX, resulting in a UvsY-mediated nucleation event for presynaptic filament formation 36; 38; 39. UvsY-ssDNA interactions are sufficient to destabilize gp32-ssDNA complexes 38. Other results suggest that UvsY-gp32, UvsY-UvsX, and UvsY-UvsY interactions are also required for efficient presynaptic filament formation and for the recombination functions of the presynaptic filament 31; 40.
We also examine two site-directed mutants of UvsY, UvsYK58A and UvsYK58A,R60A, which contain single and double missense mutations, respectively, within a conserved LKARLDY motif 41. These mutants show severely reduced affinities for ssDNA and dsDNA (H. Xu, H. T.H. Beernink, and S. W. Morrical, submitted). However, they retain self- and heteroprotein association activities similar to wild type UvsY. Both mutant proteins are partially, but not totally defective in stimulating UvsX-catalyzed reactions, including ssDNA-dependent ATP hydrolysis and DNA strand exchange 41.
In this work, we show that UvsY binds more strongly to dsDNA than ssDNA in high protein and low salt concentrations, in contrast to measurements obtained in bulk solution studies 33; 35; 42. We explain this discrepancy in the context of a previous model for UvsY activity, which suggests that UvsY significantly wraps ssDNA upon binding. Our study supports this model because wrapping of ssDNA is expected to be much weaker in an experiment in which ssDNA is stretched, and therefore binding to ssDNA in our experiment should be weaker if the model holds. Equilibrium binding to dsDNA is not expected to involve wrapping of the DNA, where the persistence length of the double helix precludes this type of interaction (see below). To test this hypothesis, we directly measure UvsY wrapping of ssDNA by DNA stretching, and find that wrapping only initiates at very low forces. Thus, measurements of the binding of UvsY to dsDNA in our stretching experiments agree well with bulk measurements.
We also determine the apparent equilibrium binding constants of gp32 and *I to ssDNA (KssωApp) in the presence of UvsY protein at different salt concentrations. Our measurements show that UvsY destabilizes the interactions between gp32-ssDNA and gp32-dsDNA. This effect is very strong, despite the fact that UvsY binding to ssDNA is weakened by stretching. This may be due to the ability of UvsY to wrap gp32-ssDNA filaments created at the ends of the DNA molecule. Our results demonstrate that UvsY interacts strongly with stretched DNA in the absence of gp32. In the presence of gp32 and *I, UvsY is capable of strongly destabilizing gp32-DNA complexes in order to facilitate ssDNA wrapping, which in turn prepares the ssDNA for presynaptic filament assembly in the presence of UvsX 43.
Results
Force spectroscopy of single DNA molecules in the presence of UvsY and its mutants
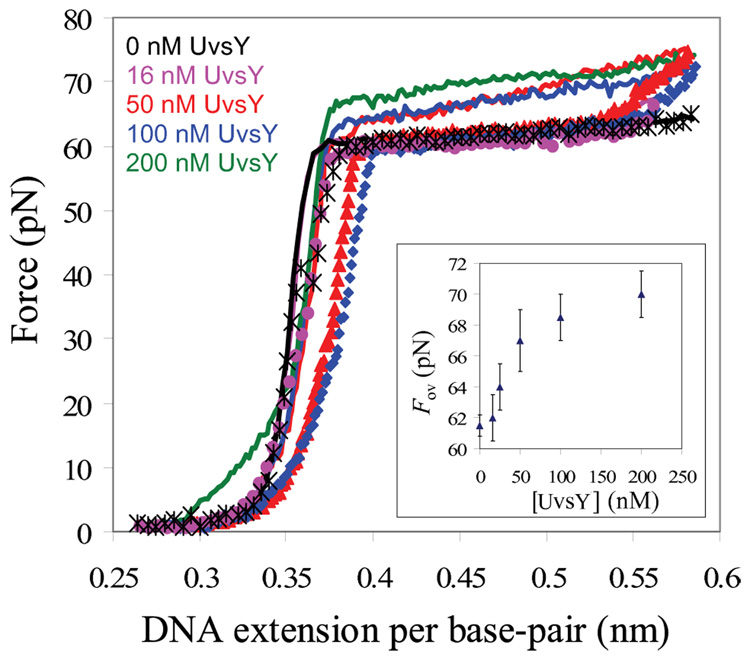
To determine the binding activity of UvsY and its mutants, we measured the force extension curve of double-stranded bacteriophage λ-DNA (used for all subsequent data) over a range of salt and protein concentrations. In 0.1M Na+, there is a slight increase in the DNA melting force, ~ 1pN, over a range of UvsY concentrations (data not shown). Fig. 3 shows the results obtained for DNA stretching in the presence of UvsY at the lower Na+ concentration of 0.05 M. The constant force region of the stretching curve, which represents DNA force-induced melting, as discussed in the Introduction, rises significantly with increasing UvsY concentration. The observed increase in melting force begins to saturate at about 50 nM UvsY, as shown in the inset of Fig. 3. In the presence of 50 nM UvsY, the elevation in DNA melting force is about 8 pN, as shown in the main panel of Figure 3. These results suggest that UvsY binds preferentially to double stranded DNA (dsDNA) at low salt and high protein concentration 18; 19. However, UvsY has been shown in bulk studies to bind both single-stranded DNA (ssDNA) and dsDNA, with higher affinity for the former 32; 34; 35; 42. Detailed solution studies showed that UvsY binds non-cooperatively to ssDNA, with a site size of four nucleotide residues per UvsY monomer, and an intrinsic binding constant comparable to that of gp32 at physiological ionic strength 35; 44. Fig. 3 also shows relaxation curves, in which DNA is stretched through the force-induced melting transition (solid lines) and then returned to lower extension at the same rate used for pulling (symbols). Here we see that the relaxation curves do not match the stretching curves, but parallel the stretching curves at a slightly lower force. This hysteresis represents the fact that the DNA is unable to reanneal immediately on the timescale of the relaxation experiment, but the DNA does eventually reanneal when the force reaches that observed for the melting plateau in the absence of protein. In the absence of protein, very little hysteresis is observed. The fact that we see additional hysteresis in the presence of UvsY likely indicates that some UvsY is removed as the DNA is melted by force, providing further support for preferential binding of UvsY to dsDNA in these experiments. However, the hysteresis is very small, and complete reannealing occurs when the B-form contour length of 0.34 nm/bp is reached, so any fraying of the single strands from the ends, which would be followed by wrapping of ssDNA by UvsY, is negligible in the presence of UvsY alone.
Fig. 3.
Stretching (solid line) and relaxation curves (symbols) for λ-DNA and UvsY in 10mM Hepes, 50 mM Na+ (45 mM NaCl and 5 mM NaOH) and pH 7.5. Main panel : Absence of protein (black), 16nM UvsY (pink), 50nM UvsY (red) 100 nM UvsY (blue) and 200nM (green, stretching curve only). Inset: Overstretching force of DNA (Fov) as a function of UvsY concentration in 50mM Na+. While each curve shown on the main panel represents one stretching experiment, the error bars on the insert show the standard deviation of measured forces for at least three stretches, demonstrating that the stretching results are reproducible within an error of approximately ± 1.5 pN.
To quantify these results, we utilize the theory of force-induced melting of single DNA molecules to measure the equilibrium binding constant of UvsY to dsDNA. The equilibrium melting force of DNA, Fm, changes from its value in the absence of the protein, , as the protein is added. In this model 11; 12, the melting free energy variations due to either force or temperature are considered to be equivalent, such that
| 1 |
where δT and δF are the temperature and the force variations, and ΔS and Δx are the entropy and elongation per base pair upon DNA melting, respectively. According to equation 1, the change in the DNA melting force in the presence of DNA binding proteins should be related to the corresponding change in the melting temperature as
| 2 |
Thus, measurements of the shift in the equilibrium DNA melting force are equivalent to measurements of the shift in DNA melting temperature, which can be used to determine binding constants to dsDNA and ssDNA. As shown by McGhee 45,
| 3 |
where Kss and Kds are equilibrium association constants to ssDNA and dsDNA, respectively, and Tm are the melting temperatures of DNA in the absence and presence of protein, nss and nds are the occluded binding site sizes of the protein on single and double strands, i.e. the number of nucleotides or base pairs covered by each protein monomer, C is the protein concentration, and ΔH and ΔS are the enthalpy and entropy of DNA melting. The approximation is valid for low protein concentration, under conditions Kds·C≪1 and KssC≪1. Unless Kds is equal to Kss to within an order of magnitude, the contribution of the lower binding constant will be small. Thus, at low protein concentration, preferential binding to dsDNA will cause the melting temperature to increase linearly with protein concentration, while preferential binding to ssDNA will cause the melting temperature to decrease linearly. According to equation 2, the melting force will change linearly at low protein concentration, such that
| 4 |
Based on the large increase in Fm with protein concentration in this experiment, and therefore assuming Kds ≫ Kss, we find Kds is about 107 M−1 in 0.05M Na+ and 106 M−1 in 0.1 M Na+. These order of magnitude estimates are in agreement with recent quantitative DNA-cellulose chromatography measurements of dsDNA binding by UvsY in 0.05 M and 0.1M Na+. (H. Xu, H. T.H. Beernink, and S. W. Morrical, submitted). Note that here we have made the approximation that the melting forces measured in the presence of UvsY are equilibrium, but there is some hysteresis. However, the amount of hysteresis is quite small and would likely only affect the measured forces by a few pN, which is within the measurement error.
Since UvsY has high affinity for dsDNA in 0.05 M Na+, we measured the overstretching force of DNA in the presence of different concentrations of two site-directed UvsY mutant proteins, UvsYK58A and UvsYK58A,R60A 41. These mutants of UvsY do not have any measurable effect on the overstretching force of DNA in 0.05M Na+, even at protein concentrations as high as 200nM. Thus, the association constant for these proteins to bind to dsDNA or ssDNA in 50 mM Na+ must be less than 106 M−1. This is in agreement with quantitative DNA chromatography data (H. Xu, H. T.H. Beernink, and S. W. Morrical, submitted).
The increase in the DNA melting force observed in these studies suggests that under these salt conditions UvsY binds more strongly to dsDNA than to ssDNA. This is in contrast to previous ensemble solution studies, which showed preferential binding to ssDNA 32; 34; 35; 42. In addition, quantitative DNA-cellulose chromatography measurements of ssDNA binding by UvsY (H. Xu, H. T.H. Beernink, and S. W. Morrical, submitted) show an affinity of UvsY for ssDNA that is at least three orders of magnitude greater than its binding to dsDNA at NaCl concentration ≤ 200 mM. These ensemble studies 38 suggest that the interaction involves ssDNA wrapping around hexameric UvsY. ssDNA, which has a persistence length of less than 1 nm 46; 47 should be capable of wrapping around the protein in order to maximize cationic binding to the highly positively-charged UvsY hexamer. However, this is not expected to occur on short time scales in our single molecule experiment, where the ssDNA segments which form upon stretching are formed at AT-rich regions in the middle of the DNA molecule. An alternative model would suggest that wrapping could occur from the ends of the DNA molecule (as will occur in the presence of cooperatively binding gp32). However, such wrapping would not allow rapid reannealing of ssDNA to form dsDNA, as is observed in Fig. 3. Thus, binding due to wrapping of ssDNA around the protein is not observed in this experiment. In contrast, wrapping of dsDNA around UvsY would not be likely to occur in either an ensemble or single-molecule experiment since dsDNA, with a persistence length of 50 nm 14, is rigid on the length scale of the size of a UvsY hexamer. The reduced binding to ssDNA in single molecule as compared to bulk studies, and the reversal of the preference for ss vs. dsDNA in the single molecule experiments supports a model in which binding of UvsY to ssDNA is strongly enhanced (a factor of 103) by the wrapping of ssDNA around the hexameric UvsY protein. The wrapping model is further supported by the observation that a truncated, monomeric form of UvsY binds to ssDNA with 104-fold lower affinity than wild-type 48.
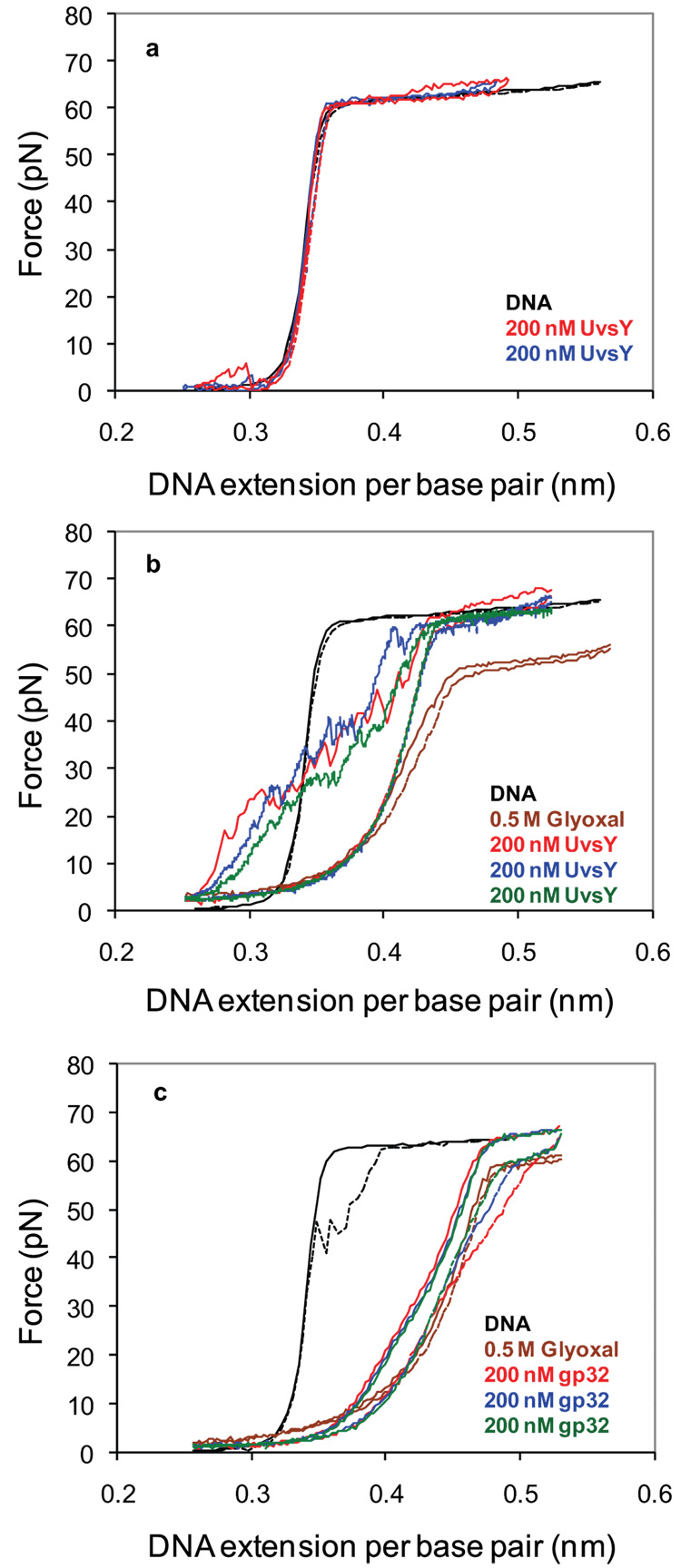
The expected signature of DNA wrapping is a strong decrease in the observed contour length on a DNA force-extension curve. Fig. 4a shows that even after waiting for 15 minutes at an extension of 0.25 nm/bp, UvsY does not alter the contour length of dsDNA measured in subsequent force-extension curves, and it is therefore unable to wrap dsDNA. To further test the hypothesis that UvsY preferentially wraps ssDNA, but is unable to wrap stretched ssDNA in the above experiments, we have performed DNA stretching experiments on ssDNA in the presence of UvsY. To obtain ssDNA, dsDNA is first stretched to an extension of 0.45 nm/bp, or about halfway through the DNA melting transition. To stabilize the partially melted DNA into the configuration that appears upon force-induced melting, we add the chemical glyoxal, a chemical that forms a stable DNA adduct with exposed guanine residues. It introduces an additional ring to the G base, sterically preventing GC base pair reannealing 49. This method has previously been used to demonstrate the creation of melted ssDNA upon DNA overstretching 21. We then wait 30 minutes at 0.45 nm/bp to allow all of the force-melted DNA to become essentially permanently melted. Subsequent stretching and relaxation curves illustrate that the DNA contour length has increased by the amount expected for a molecule that is about half ssDNA and half dsDNA, as shown in Fig. 4b (brown curve). The resulting ssDNA will not base pair but will have the same short persistence length required for wrapping by UvsY. To test the effect of UvsY on the newly created ssDNA, we then exchange the solution surrounding the molecule to replace the glyoxal solution with a solution containing 200 nM UvsY. After waiting at an extension of 0.25 nm/bp for 15 minutes, subsequent force-extension curves show an immediate decrease in the DNA contour length to less than the dsDNA contour length (Fig. 4b, red, green, and blue curves). Regardless of the pulling rate used, the force immediately rises, showing that the DNA has become shorter due to ssDNA wrapping. After the DNA extension reaches 0.45 nm/bp, the overstretching transition reappears. The relaxation curve in each case follows the relaxation curve of the partially ssDNA in the absence of UvsY. Therefore, at these high forces, UvsY is unable to initiate wrapping of ssDNA. However, subsequent stretching curves after waiting at 0.25 nm/bp always show wrapping. The jumps in the UvsY stretching curves of Fig. 4b likely represent the release of wrapped ssDNA-UvsY complexes. These curves very much resemble the type of force-extension curves obtained upon stretching chromatin fibers, which also contain DNA wrapped by protein 50; 51. DNA stretching studies of chromatin fibers also observed that nucleosome assembly by DNA wrapping was strongly inhibited by forces above 10 pN 52. As a further control, we performed the same experiment in the presence of gp32. To do this, we first create stabilized ssDNA using glyoxal, demonstrated by the brown stretching and relaxation curves in Fig. 4c. We then exchange the solution surrounding the DNA molecule for a solution containing 200 nM gp32. While the presence of gp32 slightly alters the shape of the ssDNA, very little reduction in contour length is observed and the jagged stretching curves characteristic of the release of wrapped ssDNA are not observed. Taken together, these results show that wrapping of ssDNA does not occur in the presence of gp32, but strong wrapping of ssDNA does occur in the presence of UvsY. One might argue that we have only shown that UvsY wraps ssDNA that has been altered by glyoxal. However, while it is possible for the presence of the small glyoxal adduct to hinder wrapping by UvsY, it is highly unlikely that the adduct could facilitate DNA wrapping by the protein. Therefore, our results clearly demonstrate that UvsY wraps ssDNA for the first time.
Fig. 4.
a) Stretching (solid line) and relaxation (dash line) curves for λ-DNA in the absence of protein (black) at a pulling rate of 250nm/s, 200nM UvsY (red and blue) at a pulling rate of 100nm/s. b) Stretching (solid line) and relaxation (dash line) curves for λ-DNA in the absence of protein (black) at a pulling rate of 250nm/s, 0.5mM Glyoxal (brown) at a pulling rate of 100nm/s, obtained after exposing DNA held at 0.45 nm/bp to Glyoxal for 30 minutes, 200nM UvsY (red) at a pulling rate of 100nm/s, 200nM UvsY (blue, green) at a pulling rate of 25nm/s. c) Stretching (solid line) and relaxation (dash line) curves for λ-DNA in the absence of protein (black) at a pulling rate of 250nm/s, 0.5mM Glyoxal (brown), obtained after exposing DNA held at 0.45 nm/bp for 30 minutes, 200nM gp32 (red, blue, green) at a pulling rate of 100nm/s. All data in the figure was obtained in 10mM Hepes, 100 mM Na+ (95 mM NaCl and 5 mM NaOH) and pH 7.5 and is representative data from a single DNA molecule chosen from a set of similar curves obtained on at least 3 different DNA molecules.
While it is likely that in its final state UvsY binds ssDNA in the proposed wrapped state, this may not always be possible in vivo. In the T4-infected cell, much of the available ssDNA will be bound by gp32. Such gp32-ssDNA filaments are most likely unable to wrap ssDNA, since gp32 has a strong tendency to stretch ssDNA. This is a property previously demonstrated by both electron microscopic 53 and linear dichroism measurements 54, and is comparable in its effect on DNA to the mechanical stretching that occurs in the present experiments. If UvsY were unable to bind stretched ssDNA, gp32-ssDNA filaments would not be bound by UvsY. However, if UvsY is capable of initially binding stretched ssDNA and then later wrapping ssDNA, then UvsY can be viewed as a protein that mediates the conversion of ssDNA from a stretched filament to a compact state. In the next section, we will test the capability of UvsY to destabilize gp32-ssDNA complexes.
Helix-destabilization capabilities of gp32 in the presence of UvsY
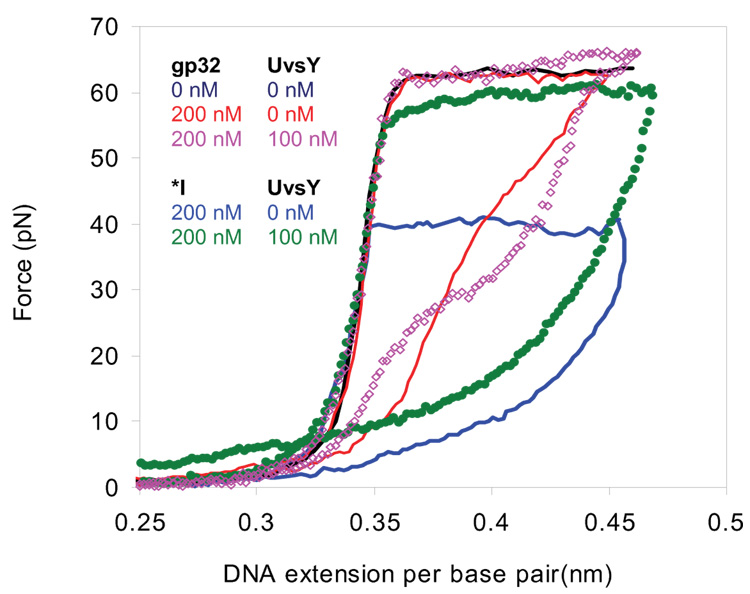
To determine the helix-destabilization capabilities of gp32 and its proteolytic fragment *I (which lacks the C-terminal domain) in the presence of UvsY, we measured the force-extension curve of λ-DNA over a range of salt and protein concentrations. In 0.1 M Na+, the presence of 200nM gp32 with or without 100nM UvsY has essentially no effect on the non-equilibrium DNA melting force (Fig. 4). However, in an analogous experiment with *I in place of gp32, the non-equilibrium DNA melting force is decreased by about 22 pN with *I alone, but this change is reduced to only about 3pN with the addition of UvsY (Fig. 5). When the salt is reduced to 0.05 M Na+, where 200 nM gp32 significantly reduced the non-equilibrium DNA melting force in the absence of UvsY, this gp32-induced reduction in the non-equilibrium DNA melting force is removed in the presence of UvsY, and the non-equilibrium melting force even increases at high UvsY concentrations (Fig. 6). The reduction in the helix-destabilizing capabilities of gp32 in the presence of UvsY agrees with previous bulk experiments 38. The analogous result with *I, which lacks the C-terminal domain but does not interact with UvsY 29; 40, indicates that the negative effect of UvsY on the helix-destabilizing activity of gp32 is not a function of specific protein–protein interactions between the two proteins. This result is in agreement with the experimental results of Sweezy and Morrical 38, but it contradicts Jiang et al. 40, who reported that protein-protein interactions between UvsY and the C-terminal domain of gp32 are necessary for loading UvsY onto gp32-covered DNA. The fact that UvsY reduces gp32’s helix-destabilizing capabilities in this experiment demonstrates that UvsY strongly alters gp32-DNA interactions.
Fig. 5.
Stretching and relaxation curves for λ-DNA and various combinations of UvsY, gp32 and *I in 10 mM Hepes,100 mM Na+ (95mM NaCl and 5mM NaOH) and pH 7.5. Data are shown in the absence of protein (dark blue), 200 nM gp32 (red), 200 nM gp32 and 100nM UvsY (pink open diamonds), 200nM *I (blue), and 200 nM of *I and 100 nM UvsY (green solid diamonds). The time for stretching and relaxation is approximately two minutes. The reproducibility of these measurements for individual stretching curves is similar to that obtained for the data in Fig. 3.
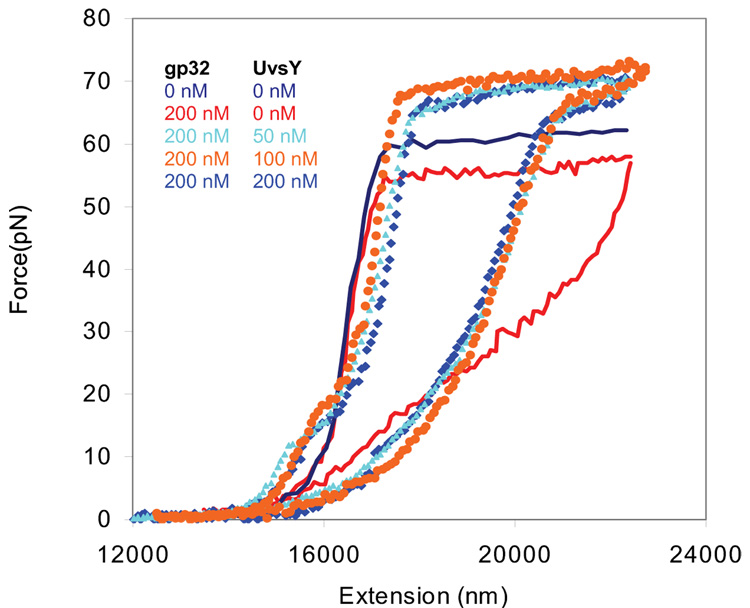
Fig. 6.
Stretching (solid line) and relaxation curves (symbols) for λ-DNA in 10 mM Hepes, pH 7.5, 50mM [Na+] (45 mM NaCl and 5 mM NaOH) in the absence of protein (dark blue) and in presence of 200 nM gp32 (red), 200 nM gp32 and 50nM UvsY (light blue), 200nM gp32 and 100nM UvsY( orange) and 200nM gp32 and 200nM UvsY (blue). The reproducibility of these measurements for individual stretching curves is similar to that obtained for the data in Fig. 3.
In the presence of gp32 and *I, there is always strong hysteresis observed. This hysteresis is due to the inability of the ssDNA created by force-induced melting to reanneal in the presence of these ssDNA binding proteins. The strong hysteresis observed in the stretching and relaxation curves of DNA in the presence of 200 nM gp32 in 0.05M Na+ decreases in the presence of 50 nM UvsY, as shown in Fig. 6. Increased concentration of UvsY has very little effect on the observed hysteresis and melting force of DNA (Fig. 6). The decrease in hysteresis and increase in the relaxation force in the presence of UvsY suggests that this protein promotes dissociation of gp32 from ssDNA and reannealing of the ssDNA. It is known that the UvsY protein itself possesses significant annealing activity 55, as does gp32 in high salt or in the presence of Mg2+ 25; 56. However, whether UvsY can stimulate the annealing of gp32-coated ssDNA is unclear since the rate of annealing of gp32-ssDNA complexes is more rapid than annealing by UvsY protein 55. The results described here show that under some conditions UvsY strongly facilitates annealing of ssDNA in the presence of gp32. In the absence of UvsY, we do not see any annealing of ssDNA in the presence of gp32 on the time scale of our single molecule experiments. This annealing capability of UvsY is likely a result of strong electrostatic interactions with DNA, which enhances removal of gp32 from ssDNA in favor of the formation of dsDNA.
Taken together, our results demonstrating destabilization of gp32-ssDNA complexes by UvsY as well as this protein’s ability to facilitate DNA annealing in the presence of gp32 suggest that it is capable of competing with gp32 for binding to ssDNA. The binding that results from these experiments may be binding to stretched ssDNA or ssDNA that is peeling from the ends and therefore not under tension. It is likely that both types of binding occur in these experiments. After the gp32-DNA interaction is destabilized, UvsY can then wrap ssDNA and prepare it for interaction with UvsX. This suggests a role for UvsY in mediating between the stretched and wrapped forms of ssDNA.
In 0.05 M Na+, 200nM of UvsYK58A and UvsYK58A,R60A, mutants of UvsY, do not affect the non-equilibrium melting force of DNA in the presence of 4 nM of *I (data not shown). At this concentration of wt UvsY and *I, the non-equilibrium DNA melting force is significantly increased relative to the force in the presence of *I alone. Our results suggest that these mutants of UvsY are unable to destabilize gp32-ssDNA interactions, which is consistent with the observation that they have a partial defect in mediated presynaptic filament assembly 41. The observed effect may be due simply to an overall reduction in binding of the mutants to DNA relative to the binding of wild type UvsY.
Equilibrium binding of gp32 to single stranded DNA in the presence of UvsY
To determine the apparent equilibrium binding constant to single stranded DNA (ssDNA) for gp32 and its proteolytic fragment *I in the presence of UvsY, we measured the force-extension curve of λ-DNA over a range of salt and protein concentrations. The melting transition force is decreased in the presence of gp32 and its fragment *I, while the transition appears as cooperative as it is in the absence of protein 6–8 .In contrast to the equilibrium nature of the transition force in the absence of protein, the transition force depends on the rate of pulling in the presence of *I and gp32. The non-equilibrium nature of the force is supported by the strong hysteresis observed in the released part of the DNA stretching cycle in the presence of gp32 and its fragment *I 6; 7 and in the presence of a mixture of gp32 proteins and UvsY (this work).The equilibrium DNA melting force (Fm) in the presence of protein was measured over a range of salt concentrations by monitoring the change in force as a function of time. In these experiments, single dsDNA molecules were each brought to a fixed position relative to the center of the optical trap (~0.42 nm/bp, where approximately one third of the DNA is melted during the initial stretch), and the force was measured in the presence of each protein as function of time 6.
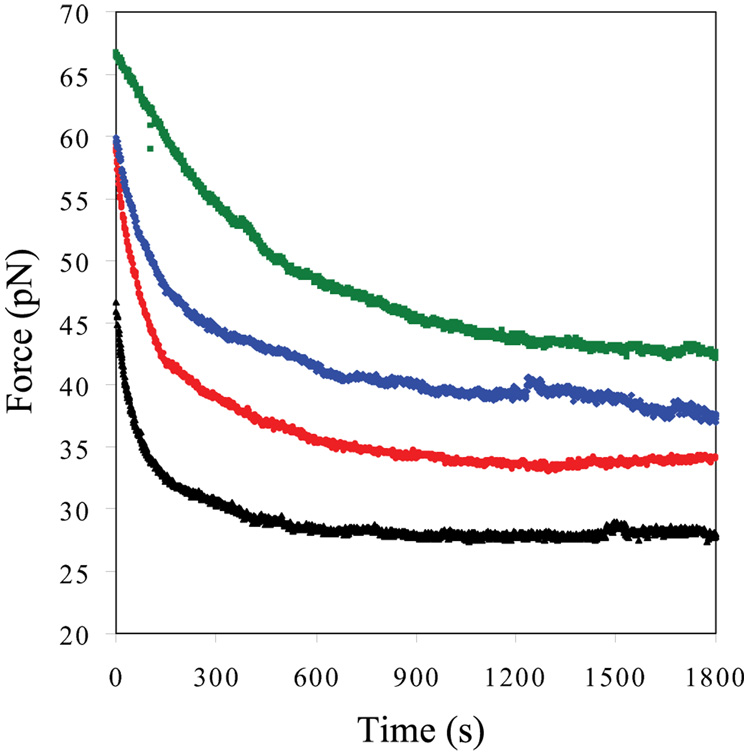
To determine the effect of UvsY on the DNA equilibrium melting force and the kinetics of secondary structure rearrangement by gp32, we measured the time dependence of the DNA melting force at constant position. Previously, we found that the DNA melting force decreases exponentially with time and reaches an equilibrium melting force in the presence of gp32 or *I in 0.1 M and 0.05 M Na+ 6; 8. We calculated the time constant of the exponential force decay for 200 nM of gp32 or *I in 0.1 M Na+ 6. The measured long time constants for both gp32 ( ~ 118 s) and *I ( ~ 38 s) reflect the time required to rearrange cooperatively bound sections of gp32 as DNA is melted by the protein. In 0.1 M Na+, the presence of 100 nM UvsY significantly increases the time constant of exponential decay for 200nM *I which is 80.0 ± 4.0 s. However, the presence of 100 nM UvsY has no significant effect on the time constant of DNA melting for 200nM of gp32 (data not shown). In 0.05 M Na+, the time constants for 100nM gp32 are 87±8 s and 230±37 s in the absence and presence of 50nM UvsY, respectively. This shows that 50nM UvsY is able to slow down the helix-destabilizing capabilities of 100 nM gp32 by a factor of 3. Increasing levels of UvsY (100nM, 200nM and 400nM) significantly increase the time constant of exponential decay for DNA melting in the presence of 50nM gp32. Similarly, in 0.05M Na+ and 4nM *I, the time constant (τ) for DNA melting by *I increases with increasing concentration of UvsY, as shown in Figure 7 and Table 1. These results suggest that UvsY slows down the rearrangement of cooperatively bound clusters of gp32 and *I.
Fig. 7.
Dependence of DNA melting force (F) as a function of time. Data was obtained in the presence of 4nM *I (black), 4 nM *I and 32 nM UvsY (red), 4 nM *I and 50 nM UvsY (blue), and 4 nM *I and 150nM UvsY (green) in 50 mM Na+, 10 mM Hepes, pH 7.5.
Table 1.
Equilibrium parameters of *I in the presence of UvsY in 0.05 M Na+
| Protein Concentration | Kssω (M−1) | τ(s) |
|---|---|---|
| [*I]=4 nM | 7.9 ± 1.2×109 | 116.1 ± 21.0 |
| [*I]=4 nM + [UvsY]=16 nM | 1.4 ± 0.2×108* | 128.7 ± 8.0 |
| [*I]=4 nM + [UvsY]=32 nM | 5.9 ± 0.9×108* | 165.0 ± 10.0 |
| [*I]=4 nM + [UvsY]=50 nM | 1.8 ± 0.3×108* | 210.2 ± 21.0 |
| [*I]=4 nM + [UvsY]=80 nM | 8.8 ± 0.1×107* | 278.8 ± 15.0 |
| [*I]=4 nM + [UvsY]=150 nM | 4.3 ± 0.7×107* | 363.0 ± 90.0 |
All measurements were performed in 10 mM Hepes pH 7.5, 50 mM [Na+]. Data are reported as mean ± standard error for n≥4.
Apparent binding constants
The equilibrium DNA melting force in the presence of protein can be expressed as 8
| 5 |
where ω is the cooperativity parameter. In previous work, we determined Kss for gp32 and *I in the range 0.05 M-0.2 M Na+ by fitting data to Eq. 5 8. The number of fitting parameters was minimized by taking into account all of the available information on gp32 and *I binding to ssDNA. We used the salt-independent parameters nss=7 and ω=1000 for both gp32 57 and *I 26, and the salt-dependent parameter from the work of Wenner et al. 14.
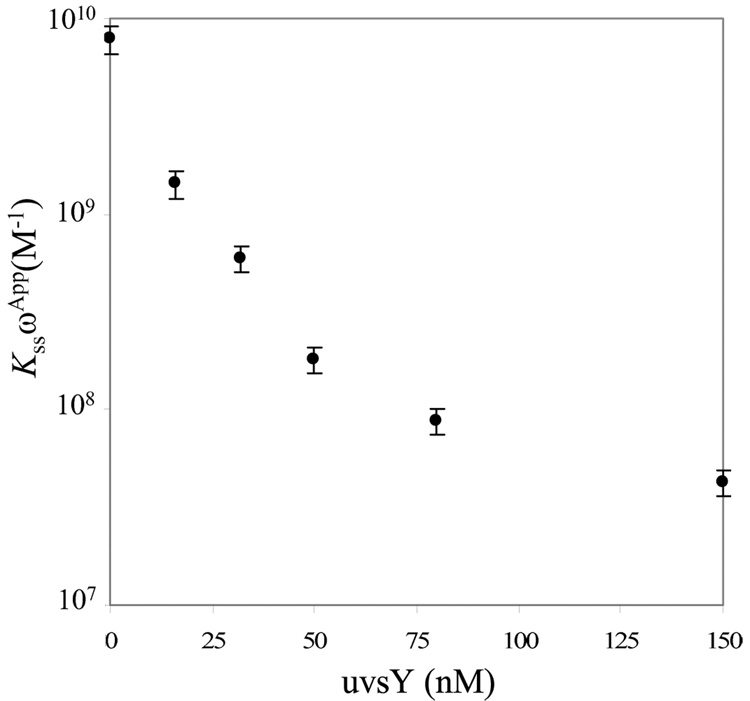
We now use this method to calculate Kss ωApp for gp32 and *I in the presence of UvsY. We again assume that the site size (nss) does not change in the presence of UvsY. The values of KssωApp for gp32 and *I are calculated using Eq.5 in the presence of UvsY. The concentrations of gp32 and *I used in this study are different because of their different helix-destablizing capabilities. In 0.1M Na+, it is found that 100 nM of UvsY has no significant effect on KssωApp for 200nM gp32 and *I (data not shown). In 0.05M Na+, it is found that KssωApp for 100nM of gp32 does not change in the presence of 50nM of UvsY (data not shown). However, KssωApp calculated for 4nM of *I decreases with increasing levels of UvsY (Table 1), and begins to saturate, as shown in Fig. 8. This result further supports the notion that protein-protein interactions between gp32 and UvsY are not necessary for destabilizing gp32-ssDNA interactions 38. Therefore, by quantifying equilibrium binding of *I in the presence of UvsY, we show that UvsY significantly reduces the affinity of *I for ssDNA under many conditions.
Fig. 8.
UvsY concentration dependence of the apparent cooperative equilibrium binding constant to ssDNA (KssωApp) of *I. The error bars are determined from the standard error of at least 3 measurements.
Discussion
This study significantly enhances our understanding of the interactions of UvsY with DNA in the presence of gene 32 protein. We have examined the effect of UvsY on the dsDNA helix-destabilizing activity of gp32. We have shown that UvsY binds strongly to dsDNA and reduces the capability of both gp32 and *I to destabilize dsDNA. We also show that UvsY can strongly facilitate DNA annealing, even in the presence of gp32. Finally, we demonstrate that UvsY slows down the rate at which gp32 binds to ssDNA and decreases the overall binding of gp32 to ssDNA.
In order to properly interpret the results of these experiments, we must consider the possible effect of force on the binding of each ligand to each form of DNA. For example, does DNA stretching affect the binding of gp32 and *I to dsDNA or ssDNA? This possibility has been examined in our previous work, in which we determined the salt-dependent binding of gp32 and *I to both dsDNA and ssDNA as a function of salt concentration. With the available bulk experimental data, we found excellent agreement with our single molecule binding studies of gp32 and *I to stretched dsDNA and ssDNA 8; 9. This is not surprising since gp32 neither wraps ssDNA nor kinks dsDNA upon binding. Thus, stretching forces would not be expected to alter binding. In contrast, UvsY is believed to wrap ssDNA but cannot do this to dsDNA, in concert with the observed weaker binding to ssDNA in our single molecule experiments relative to bulk studies and the results of Fig. 4. Finally, the application of force may alter the ability of UvsY to compete with the gp32 that is bound to ssDNA. However, we find that the equilibrium binding of *I to ssDNA is strongly reduced in the presence of UvsY (Fig. 8) and the rate at which clusters of *I cooperatively rearrange is also reduced, as shown in Fig. 7. Given that stretching can only reduce UvsY’s effectiveness at removing gp32, our results clearly show that UvsY can effectively compete with gp32 for ssDNA binding.
Similarly, and in contrast to previous bulk studies, we show in these DNA stretching experiments that UvsY binds more strongly to dsDNA than ssDNA. This apparent discrepancy is due to the absence of the UvsY-wrapped ssDNA binding mode under the conditions of the stretching experiment. We directly demonstrate that wrapping cannot occur with double-stranded DNA, where the persistence length is much higher than that of ssDNA 47 (see Fig. 4). Because only ssDNA can be wrapped by UvsY, as demonstrated in Fig. 4, affinities obtained from single molecule measurements of binding to ssDNA should be much lower than those observed in bulk experiments, while dsDNA binding for bulk and single molecule experiments should agree, as is observed here.
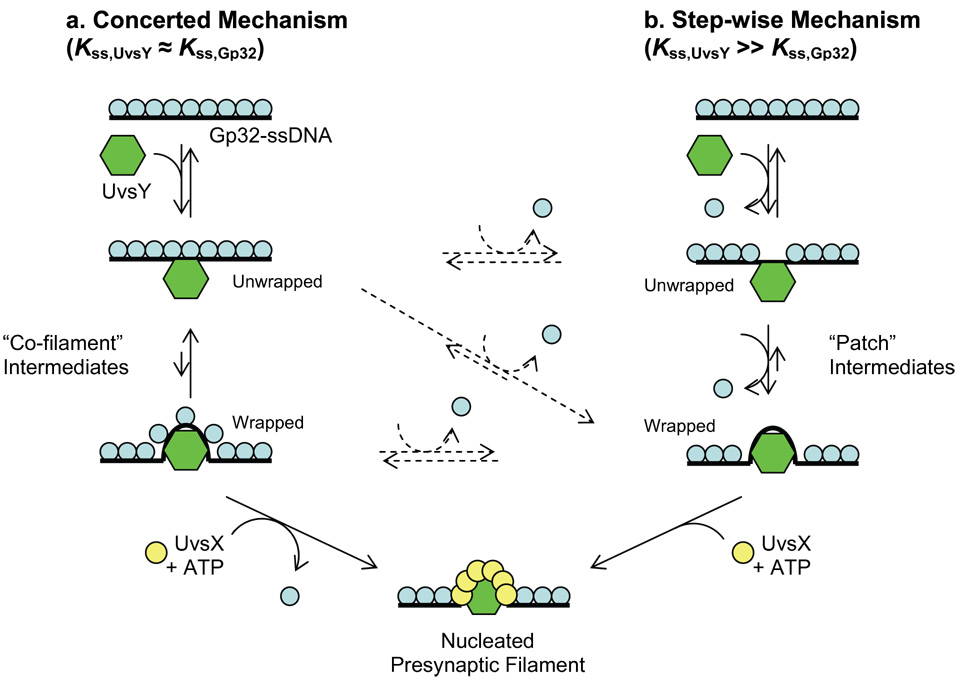
Bulk solution studies demonstrate that at moderate salt concentrations (e.g. 90–200 mM NaCl or KOAc), UvsY and gp32 form a “co-filament” structure in which both proteins co-occupy ssDNA without competing for binding sites 38; 40; 58. The co-filament appears to be a direct intermediate in UvsX recombinase assembly on ssDNA, and therefore in DNA strand exchange. Results of the current study indicate that at lower salt concentrations (e.g. 50 mM NaCl), UvsY is able to compete with and displace gp32 from DNA. The salt dependencies of UvsY-DNA and gp32-DNA interactions are markedly different. Below ~200 mM NaCl, KApp for gp32-DNA interactions do not change significantly with decreasing salt, while KApp for UvsY-DNA interactions increase by several orders of magnitude over the same range. Therefore, given that gp32 and UvsY are present in the cell in micromolar concentrations59; 60, it is possible that over the range of ionic conditions likely to be encountered in a T4-infected E. coli cell, salt-induced shifts in the relative DNA binding affinities of UvsY and gp32 could lead to different mechanisms of presynaptic filament assembly (Fig. 9). Conditions in which the ssDNA binding affinities of UvsY and gp32 are approximately balanced (Kss,UvsY ≈ Kss,Gp32) would favor a mechanism wherein UvsY and gp32 bind to ssDNA non-competitively, forming a co-filament (Fig. 9a). In this pathway, full wrapping of ssDNA by UvsY and displacement of gp32 may not occur until UvsX + ATP is recruited by UvsY, so this may be considered a concerted mechanism. In contrast, when Kss,UvsY ≫ Kss,gp32 a step-wise mechanism may prevail in which UvsY binding directly displaces gp32 from ssDNA, leading to wrapping and subsequently to UvsX + ATP recruitment (Fig. 9b). Conceivably, formation of the presynaptic filament could proceed via a combination of the two mechanisms. Initially a “co-filament” intermediate forms with gp32 and UvsY simultaneously occupying unwrapped DNA (Fig. 9a). This intermediate could then be converted to a wrapped intermediate, where the two proteins no longer simultaneously bind, and several formerly-bound gp32 molecules are released (Fig. 9b; the crossover between the two mechanisms is shown as a diagonal dotted arrow, and other potential crossover points are shown as horizontal dotted arrows). Note that even when gp32 remains bound to the wrapped DNA, this may necessarily occur in a non-cooperative mode. The geometry of the wrapped DNA may be inconsistent with what is required for the gp32-gp32 association that is responsible for cooperative binding.
Fig. 9.
Schematic diagram showing two mechanisms for the role of UvsY in synaptic filament formation, which are valid under different solution conditions. a) In the concerted mechanism, which is likely valid in high salt when Kss,UvsY ≈ Kss,gp32, UvsY initially binds to gp32-coated ssDNA and wraps the ssDNA, forming a cofilament structure to which gp32 remains bound. UvsX and ATP are subsequently required for removal of gp32 from the filament. b) In the step-wise mechanism, which is likely valid in low salt when Kss,UvsY ≫ Kss,gp32 , UvsY competes directly with gp32 for ssDNA binding, therefore removing gp32 from ssDNA. UvsY subsequently wraps the ssDNA tightly, forming patches of UvsY lacking gp32 completely. UvsX and ATP are then required to form the final synaptic filament. Pre-synaptic filament formation might occur via a combination of both mechanisms, denoted via dotted arrows (see Discussion for details).
While ssDNA wrapping clearly enhances UvsY binding to ssDNA, and therefore increases its ability to alter gp32-DNA binding, simple electrostatic binding by UvsY to both ssDNA and dsDNA has biological significance. In vivo, the wrapping of ssDNA by UvsY may be precluded by steric hindrance, kinetic factors, or even the presence of DNA stretching forces exerted by proteins or multimeric protein complexes 61; 62. These or other factors may influence the mechanistic path used by UvsY to remodel relatively rigid gp32-coated ssDNA molecules into the highly stable, wrapped UvsY-ssDNA complex that favors UvsX-ssDNA interactions 43.
Materials and Methods
The dual-beam optical tweezers instrument used in this study consists of two counter-propagating diode lasers focused to a small spot inside a liquid flow cell. One 5 µm diameter streptavidin-coated polystyrene bead (Bangs Labs, Fisher, IN) was held in the optical trap formed by the laser beams. Another streptavidin-coated bead was held on the end of a glass micropipette. To obtain force-extension measurements, a single double-stranded DNA molecule that had been labelled on the 3′ end of opposite strands with biotin was captured between the two beads 15. The DNA molecule was then stretched by moving the pipette and measuring the resulting calibrated force on the bead in the trap, as previously described 15; 16. Unless otherwise noted, the pulling rate for the DNA stretching curves was 100 nm/s. The buffer used in this study for capturing DNA was 10 mM HEPES (pH 7.5) with 45 mM NaCl and 5 mM NaOH (50 mM [Na+]) or 10 mM HEPES (pH 7.5) with 95 mM NaCl and 5mM NaOH (100 mM [Na+]).
The absolute extension of the DNA molecule was estimated by measuring the distance between the centers of the two beads using an image captured with a CCD camera. The change in position of the pipette was measured using a feedback-compensated piezoelectric translation stage that is accurate to 5 nm (Melles Griot, Irvine, CA). The position measurement was converted to a measurement of the molecular extension by correcting for the trap stiffness, which was 42 ± 3 pN/µm. To obtain the time dependence of the DNA overstretching force, the DNA molecule was stretched in 100 nm steps and then held at a constant position while the force as a function of time was measured.
T4 gene 32 protein and truncated forms *I and *III used in these experiments were prepared as described 25. UvsY and its two site directed mutants were prepared as described in 35 and 41 respectively. After capturing a single DNA molecule in the tethering buffer, the molecule was stretched to verify that the usual force-extension curve was obtained. To measure the effect of the protein on this transition, 4–5 cell volumes of a buffer solution containing a fixed amount of protein was added to the experimental cell until the buffer surrounding the captured DNA molecule was completely exchanged.
Acknowledgements
We wish to thank Elizabeth Flynn, Min Wu, Dr. Hang Xu, and Dr. Xiaoyan Chen for assistance with protein preparation. Micah McCauley is thanked for the design of Fig. 2. Funding for this project was provided by NIH (GM 52049, R.L.K., GM072462, M.C.W., and GM48847, S.W.M.) and NSF (MCB-0744456, M.C.W).
Abbreviations
- gp32
T4 Gene 32 protein
- UvsY
T4 recombination mediator protein
- ssDNA
single-stranded DNA
- dsDNA
double-stranded DNA
- SSB
single-stranded binding protein
- RMP
recombination mediator protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Luder A, Mosig G. Two alternative mechanisms for initiation of DNA replication forks in bacteriophage T4: priming by RNA polymerase and by recombination. Proc Natl Acad Sci U S A. 1982;79:1101–1105. doi: 10.1073/pnas.79.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kreuzer KN, Morrical SW. Intiation of T4 DNA replication. In: Karam JD, editor. Molecular Biology of Bacteriophage T4. Washigton, DC: ASM Press; 1994. pp. 28–42. [Google Scholar]
- 3.Williams KR, Spicer EK, LoPresti MB, Guggenheimer RA, Chase JW. Limited proteolysis studies on the Escherichia coli single-stranded DNA binding protein. Evidence for a functionally homologous domain in both the Escherichia coli and T4 DNA binding proteins. J. Biol. Chem. 1983;258:3346–3355. [PubMed] [Google Scholar]
- 4.Haseltine CA, Kowalczykoski SC. A distinctive single-strand DNA-binding protein from the Archaeon Sulfolobus solfataricus. Mol. Microbiol. 2002;43:1505–1515. doi: 10.1046/j.1365-2958.2002.02807.x. [DOI] [PubMed] [Google Scholar]
- 5.Acharya N, Varshney U. Biochemical properties of single-stranded DNA-binding protein from Mycobacterium smegmatis, a fast-growing mycobacterium and its physical and functional interaction with uracil DNA glycosylases. J. Mol. Biol. 2002;318:1251–1264. doi: 10.1016/s0022-2836(02)00053-0. [DOI] [PubMed] [Google Scholar]
- 6.Pant K, Karpel RL, Williams MC. Kinetic Regulation of Single DNA Molecule Denaturation by T4 Gene 32 Protein Structural Domains. J. Mol. Biol. 2003;327:571–578. doi: 10.1016/s0022-2836(03)00153-0. [DOI] [PubMed] [Google Scholar]
- 7.Pant K, Karpel RL, Rouzina I, Williams MC. Mechanical Measurement of Single -molecule Binding Rates: Kinetics of DNA Helix-destablization by T4 Gene 32 Protein. J. Mol. Biol. 2004;336:851–870. doi: 10.1016/j.jmb.2003.12.025. [DOI] [PubMed] [Google Scholar]
- 8.Pant K, Karpel RL, Rouzina I, Williams MC. Salt dependent binding of T4 gene 32 protein to single- and double-stranded DNA: Single molecule force spectroscopy measurements. J. Mol. Biol. 2005;349:317–330. doi: 10.1016/j.jmb.2005.03.065. [DOI] [PubMed] [Google Scholar]
- 9.Rouzina I, Pant K, Karpel RL, Williams MC. Theory of electrostatically regulated binding of T4 gene 32 protein to single- and double-stranded DNA. Biophys J. 2005;89:1941–1956. doi: 10.1529/biophysj.105.063776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shokri L, Marintcheva B, Richardson CC, Rouzina I, Williams MC. Single molecule force spectroscopy of salt-dependent bacteriophage T7 gene 2.5 protein binding to single-stranded DNA. J Biol Chem. 2006;281:38689–38696. doi: 10.1074/jbc.M608460200. [DOI] [PubMed] [Google Scholar]
- 11.Rouzina I, Bloomfield VA. Force-induced melting of the DNA double helix. 1. Thermodynamic analysis. Biophys. J. 2001;80:882–893. doi: 10.1016/S0006-3495(01)76067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rouzina I, Bloomfield VA. Force-induced melting of the DNA double helix. 2. Effect of solution conditions. Biophys. J. 2001;80:894–900. doi: 10.1016/S0006-3495(01)76068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams MC, Rouzina I, Bloomfield VA. Thermodynamics of DNA Interactions from Single Molecule Stretching Experiments. Acc. Chem. Res. 2002;35:159–166. doi: 10.1021/ar010045k. [DOI] [PubMed] [Google Scholar]
- 14.Wenner JR, Williams MC, Rouzina I, Bloomfield VA. Salt Dependence of the Elasticity and Overstretching Transition of Single DNA Molecules. Biophys. J. 2002;82:3160–3169. doi: 10.1016/S0006-3495(02)75658-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams MC, Wenner JR, Rouzina I, Bloomfield VA. The effect of pH on the overstretching transition of dsDNA: Evidence of force-induced DNA melting. Biophys. J. 2001;80:874–881. doi: 10.1016/S0006-3495(01)76066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams MC, Wenner JR, Rouzina I, Bloomfield VA. Entropy and Heat Capacity of DNA Melting from Temperature Dependence of Single Molecule Stretching. Biophys. J. 2001;80:1932–1939. doi: 10.1016/S0006-3495(01)76163-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krautbauer R, Fischerlander S, Allen S, Gaub HE. Mechanical fingerprints of DNA drug complexes. Single Mol. 2002;3:97–103. [Google Scholar]
- 18.Mihailović A, Vladescu I, McCauley M, Ly E, Williams MC, Spain EM, Nuñez ME. Exploring the Interaction of Ruthenium(II) Polypyridyl Complexes with DNA Using Single-Molecule Techniques. Langmuir. 2006;22:4699–4709. doi: 10.1021/la053242r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vladescu ID, McCauley MJ, Rouzina I, Williams MC. Mapping the phase diagram of single DNA molecule force-induced melting in the presence of ethidium. Phys Rev Lett. 2005;95:158102. doi: 10.1103/PhysRevLett.95.158102. [DOI] [PubMed] [Google Scholar]
- 20.Vladescu ID, McCauley MJ, Nunez ME, Rouzina I, Williams MC. Quantifying force-dependent and zero-force DNA intercalation by single-molecule stretching. Nat Methods. 2007;4:517–522. doi: 10.1038/nmeth1044. [DOI] [PubMed] [Google Scholar]
- 21.Shokri L, McCauley MJ, Rouzina I, Williams MC. DNA overstretching in the presence of glyoxal: Structural evidence of force–induced DNA melting. Biophys. J. 2008 doi: 10.1529/biophysj.108.132688. in press, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris SA, Sands ZA, Laughton CA. Molecular dynamics Simulations of duplex stretching reveal the importance of entropy in determining the biomechanical properties of DNA. Biophysical Journal. 2005;88:1684–1691. doi: 10.1529/biophysj.104.046912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heng JB, Aksimentiev A, Ho C, Marks P, Grinkova YV, Sligar S, Schulten K, Timp G. The electromechanics of DNA in a synthetic nanopore. Biophysical Journal. 2006;90:1098–1106. doi: 10.1529/biophysj.105.070672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piana S. Structure and energy of a DNA dodecamer under tensile load. Nucleic Acids Research. 2005;33:7029–7038. doi: 10.1093/nar/gki1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waidner L, Flynn E, Wu M, Li X, Karpel RL. Domain effects on the DNA-interactive properties of bacteriophage T4 gene 32 protein. J. Biol. Chem. 2001;276:2509–2516. doi: 10.1074/jbc.M007778200. [DOI] [PubMed] [Google Scholar]
- 26.Lonberg N, Kowalczykowski SC, Paul LS, von Hippel PH. Interactions of Bacteriophage T4-coded Gene 32 Protein with Nucleic Acids. III. Binding Properties of Two Specific Proteolytic Digestion Products of the Protein (G32P*I and G32P*III) J. Mol. Biol. 1981;145:123–138. doi: 10.1016/0022-2836(81)90337-5. [DOI] [PubMed] [Google Scholar]
- 27.Karpel RL. LAST motifs and SMART domains in gene 32 protein: an unfolding story of autoregulation? IUBMB Life. 2002;53:161–166. doi: 10.1080/15216540212332. [DOI] [PubMed] [Google Scholar]
- 28.Giedroc DP, Khan R, Barnhart K. Over-expression,purification and characterization of recombinant T4 Gene 32 protein22-301(g32P-B) J. Biol. Chem. 1990;265:11444–11455. [PubMed] [Google Scholar]
- 29.Hurley JM, Chervitz SA, Jarvis TC, Singer BS, Gold L. Assembly of the bacteriophage T4 replication machine requires the acidic carboxy terminus of gene 32 protein. J. Mol. Biol. 1993;229:398–418. doi: 10.1006/jmbi.1993.1042. [DOI] [PubMed] [Google Scholar]
- 30.Williams MC, Rouzina I, Karpel RL. Thermodynamics and kinetics of DNA-protein interactions from single molecule force spectroscopy measurements. Curr. Org. Chem. 2006;10:419–432. [Google Scholar]
- 31.Yassa DS, Chou KM, Morrical SW. Characterization of an amino-terminal fragment of the bacteriophage T4 uvsY recombination protein. Biochimie. 1997;79:275–285. doi: 10.1016/s0300-9084(97)83515-8. [DOI] [PubMed] [Google Scholar]
- 32.Yonesaki T, Minagawa T. T4 phage gene uvsX product catalyzes homologous DNA pairing. EMBO J. 1985;4:3321–3327. doi: 10.1002/j.1460-2075.1985.tb04083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morrical SW, Alberts BM. The UvsY protein of bacteriophage T4 modulates recombination-dependent DNA synthesis in vitro. J. Biol. Chem. 1990;265:15096–15103. [PubMed] [Google Scholar]
- 34.Kodadek T, Gan D, Stemke-Hale K. The phage T4 uvs Y recombination protein stabilizes presynaptic filaments. J. Biol. Chem. 1989;264:16451–16457. [PubMed] [Google Scholar]
- 35.Sweezy MA, Morrical SW. Single-stranded DNA binding properties of the uvsY recombination protein of bacteriophage T4. J. Mol. Biol. 1997;266:927–938. doi: 10.1006/jmbi.1996.0829. [DOI] [PubMed] [Google Scholar]
- 36.Beernink HT, Morrical SW. RMPs: recombination/replication mediator proteins. Trends Biochem. Sci. 1999;24:385–389. doi: 10.1016/s0968-0004(99)01451-6. [DOI] [PubMed] [Google Scholar]
- 37.Beernink HT, Morrical SW. The uvsY recombination protein of bacteriophage T4 forms hexamers in the presence and absence of single-stranded DNA. Biochemistry. 1998;37:5673–5681. doi: 10.1021/bi9800956. [DOI] [PubMed] [Google Scholar]
- 38.Sweezy MA, Morrical SW. Biochemical interactions within a ternary complex of the bacteriophage T4 recombination proteins uvsY and gp32 bound to single-stranded DNA. Biochemistry. 1999;38:936–944. doi: 10.1021/bi9817055. [DOI] [PubMed] [Google Scholar]
- 39.Bleuit JS, Xu H, Ma Y, Wang T, Liu J, Morrical SW. Mediator proteins orchestrate enzyme-ssDNA assembly during T4 recombination-dependent DNA replication and repair. Proc. Natl. Acad Sci. USA. 2001;98:8298–8305. doi: 10.1073/pnas.131007498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang H, Giedroc D, Kodadek T. The role of protein-protein interactions in the assembly of the presynaptic filament for T4 homologous recombination. J. Biol. Chem. 1993;268:7904–7911. [PubMed] [Google Scholar]
- 41.Bleuit JS, Ma Y, Munro J, Morrical SW. Mutations in a Conserved Motif Inhibit Single-stranded DNA Binding and Recombination Mediator Activities of Bacteriophage T4 UvsY Protein. J. Biol. Chem. 2004;279:6077–6086. doi: 10.1074/jbc.M311557200. [DOI] [PubMed] [Google Scholar]
- 42.Yonesaki T, Minagawa T. Synergistic action of three recombination gene products of bacteriophage T4, uvsX, uvsY, and gene 32 proteins. J. Biol. Chem. 1989;264:7814–7820. [PubMed] [Google Scholar]
- 43.Liu J, Bond JP, Morrical SW. Mechanism of presynaptic filament stabilization by the bacteriophage T4 UvsY recombination mediator protein. Biochemistry. 2006;45:5493–5502. doi: 10.1021/bi0525167. [DOI] [PubMed] [Google Scholar]
- 44.Ando RA, Morrical SW. Single-stranded DNA binding properties of the UvsX recombinase of bacteriophage T4: binding parameters and effects of nucleotides. J. Mol. Biol. 1998;283:785–796. doi: 10.1006/jmbi.1998.2124. [DOI] [PubMed] [Google Scholar]
- 45.McGhee JD. Theoretical calculations of the helix-coil transition of DNA in the presence of large, cooperatively binding ligands. Biopolymers. 1976;15:1345–1375. doi: 10.1002/bip.1976.360150710. [DOI] [PubMed] [Google Scholar]
- 46.Smith SB, Cui YJ, Bustamante C. Overstretching B-DNA: The elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 47.Williams MC, Rouzina I. Force Spectroscopy of Single DNA and RNA Molecules. Curr. Opin. Struct. Biol. 2002;12:330–336. doi: 10.1016/s0959-440x(02)00340-8. [DOI] [PubMed] [Google Scholar]
- 48.Ando RA, Morrical SW. Relationship between hexamerization and ssDNA binding affinity in the uvsY recombination protein of bacteriophage T4. Biochemistry. 1999;38:16589–16598. doi: 10.1021/bi991917h. [DOI] [PubMed] [Google Scholar]
- 49.Hutton JR, Wetmur JG. Effect of chemical modification on the rate of renaturation of deoxyribonucleic acid. Deaminated and glyoxalated deoxyribonucleic acid. Biochemistry. 1973;12:558–563. doi: 10.1021/bi00727a032. [DOI] [PubMed] [Google Scholar]
- 50.Bennink ML, Leuba SH, Leno GH, Zlatanova J, de Grooth BG, Greve J. Unfolding individual nucleosomes by stretching single chromatin fibers with optical tweezers. Nat Struct Biol. 2001;8:606–610. doi: 10.1038/89646. [DOI] [PubMed] [Google Scholar]
- 51.Brower-Toland BD, Smith CL, Yeh RC, Lis JT, Peterson CL, Wang MD. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc Natl Acad Sci U S A. 2002;99:1960–1965. doi: 10.1073/pnas.022638399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leuba SH, Karymov MA, Tomschik M, Ramjit R, Smith P, Zlatanova J. Assembly of single chromatin fibers depends on the tension in the DNA molecule: magnetic tweezers study. Proc Natl Acad Sci U S A. 2003;100:495–500. doi: 10.1073/pnas.0136890100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Delius H, Mantell NJ, Alberts B. Characterization by electron microscopy of the complex formed between T4 bacteriophage gene 32-protein and DNA. J Mol Biol. 1972;67:341–350. doi: 10.1016/0022-2836(72)90454-8. [DOI] [PubMed] [Google Scholar]
- 54.van Amerongen H, Kuil ME, Scheerhagen MA, van Grondelle R. Structure calculations for single-stranded DNA complexed with the single-stranded DNA binding protein GP32 of bacteriophage T4: a remarkable DNA structure. Biochemistry. 1990;29:5619–5625. doi: 10.1021/bi00475a029. [DOI] [PubMed] [Google Scholar]
- 55.Kantake N, Madiraju MV, Sugiyama T, Kowalczykowski SC. Escherichia coli RecO protein anneals ssDNA complexed with its cognate ssDNA-binding protein: A common step in genetic recombination. Proc. Natl. Acad. Sci. USA. 2002;99:15327–15332. doi: 10.1073/pnas.252633399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alberts BM, Frey L. T4 bacteriophage gene 32: a structural protein in the replication and recombination of DNA. Nature. 1970;227:1313–1318. doi: 10.1038/2271313a0. [DOI] [PubMed] [Google Scholar]
- 57.Kowalczykowski SC, Lonberg N, Newport JW, von Hippel PH. Interactions of bacteriophage T4-coded gene 32 protein with nucleic acids. I. Characterization of the binding interactions. J. Mol. Biol. 1981;145:75–104. doi: 10.1016/0022-2836(81)90335-1. [DOI] [PubMed] [Google Scholar]
- 58.Liu J, Qian N, Morrical SW. Dynamics of Bacteriophage T4 Presynaptic Filament Assembly from Extrinsic Fluorescence Measurements of Gp32-Single-stranded DNA Interactions. J Biol Chem. 2006;281:26308–26319. doi: 10.1074/jbc.M604349200. [DOI] [PubMed] [Google Scholar]
- 59.Cowan J, D’Acci K, Guttman B, Kutter E. Gel analysis of T4 prereplicative proteins. In: Karam JD, editor. Molecular Biology of Bacteriophage T4. Washington, DC: American Society for Microbiology Press; 1994. pp. 520–527. [Google Scholar]
- 60.von Hippel PH, Kowalczykowski SC, Lonberg N, Newport JW, Paul LS, Stormo GD, Gold L. Autoregulation of gene expression. Quantitative evaluation of the expression and function of the bacteriophage T4 gene 32 (single-stranded DNA binding) protein system. J. Mol. Biol. 1982;162:795–818. doi: 10.1016/0022-2836(82)90548-4. [DOI] [PubMed] [Google Scholar]
- 61.Cozzarelli NR, Cost GJ, Nöllmann M, Viard T, Stray JE. Giant proteins that move DNA: bullies of the genomic playground. Nat. Rev. Mol. Cell Biol. 2006;7:580. doi: 10.1038/nrm1982. [DOI] [PubMed] [Google Scholar]
- 62.Bustamante C, Bryant Z, Smith SB. Ten years of tension: single-molecule DNA mechanics. Nature. 2003;421:423–427. doi: 10.1038/nature01405. [DOI] [PubMed] [Google Scholar]
- 63.Kraulis PJ. MOLSCRIPT: A Program to Produce Both Detailed and Schematic Plots of Protein Structures. J. Appl. Crystall. 1991;24:946–950. [Google Scholar]
- 64.Shamoo Y, Friedman AM, Parsons MR, Konigsberg WH, Steitz TA. Crystal structure of a replication fork single-stranded DNA binding protein (T4 gp32) complexed to DNA. Nature. 1995;376:362–366. doi: 10.1038/376362a0. [DOI] [PubMed] [Google Scholar]
- 65.McCauley M, Hardwidge PR, Maher LJ, 3rd, Williams MC. Dual binding modes for an HMG domain from human HMGB2 on DNA. Biophys J. 2005;89:353–364. doi: 10.1529/biophysj.104.052068. [DOI] [PMC free article] [PubMed] [Google Scholar]