Abstract
LpxC is a zinc metalloenzyme that catalyzes the first committed step in the biosynthesis of lipid A, a vital component of the outer membrane of Gram-negative bacteria. Accordingly, the inhibition of LpxC is an attractive strategy for the treatment of Gram-negative bacterial infections. Here, we report the 2.7 Å resolution X-ray crystal structure of LpxC from Aquifex aeolicus complexed with uridine 5′-diphosphate (UDP), and the 3.1 Å resolution structure of LpxC complexed with pyrophosphate. The X-ray crystal structure of the LpxC-UDP complex provides the first view of interactions likely to be exploited by the substrate UDP group in the “basic patch” of the active site. The diphosphate group of UDP makes hydrogen bond interactions with strictly conserved residue K239 as well as solvent molecules. The ribose moiety of UDP interacts with partially conserved residue E197. The UDP uracil group hydrogen bonds with both the backbone NH group and the backbone carbonyl group of E160, and with the backbone NH group of K162 through an intervening water molecule. Finally, the α-phosphate and uracil groups of UDP interact with R143 and R262 through intervening water molecules. The structure of LpxC complexed with pyrophosphate reveals generally similar intermolecular interactions in the basic patch. Unexpectedly, diphosphate binding in both complexes is accompanied by coordination to an additional zinc ion, resulting in the identification of a new metal-binding site termed the E-site. The structures of the LpxC-UDP and LpxC-pyrophosphate complexes provide new insights regarding substrate recognition in the basic patch and metal ion coordination in the active site of LpxC.
The zinc metalloenzyme UDP –{3-O-[(R)-3-hydroxymyristoyl]}-N-acetylglucosamine deacetylase (LpxC) catalyzes the first irreversible step in the biosynthesis of lipid A (Figure 1) (1-4), which is ultimately incorporated into the outer leaflet of the outer membrane of Gram-negative bacteria as the hydrophobic anchor of lipopolysaccharide (LPS) (5-9).1 The exterior sheath of LPS serves as a protective barrier that resists penetration by many common antibiotics such as erythromycin (8-14). Importantly, lipid A is the toxic component of LPS that triggers a severe immune response to Gram-negative bacterial infections, which can lead to septic shock accompanied by acute hypotension, multiple organ failure, and death (15-18). Accordingly, inhibitors of LpxC and lipid A biosynthesis represent a potential treatment option for Gram-negative sepsis.
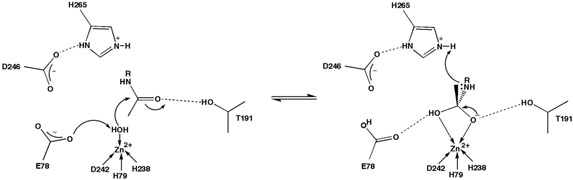
Figure 1.
Proposed catalytic mechanism of LpxC.
The structure of LpxC from Aquifex aeolicus was first determined by X-ray crystallography at 2.0 Å resolution and reveals a ~20 Å-deep active site located at the interface of two topologically similar α + β domains (19). Enzyme crystals were prepared in the presence of excess Zn2+, resulting in the formation of a binuclear zinc cluster composed of a catalytic zinc ion (Zn2+A, coordinated by H79, H238, D242, and a solvent molecule with tetrahedral geometry) and an inhibitory zinc ion (Zn2+B, coordinated by E78, H265, a solvent molecule, and a fatty acid with tetrahedral geometry) (19).22 The aliphatic tail of the fatty acid was bound in a ~15 Å-long hydrophobic tunnel leading out of the active site cleft and was hypothesized to represent the binding mode of the 3-O-(R-3-hydroxymyristoyl) group of the substrate (19). Consistent with this hypothesis, the subsequently determined NMR structure (20-21) and the X-ray crystal structure (22) of LpxC complexed with the substrate analogue inhibitor TU-514 (Figure 2) revealed the binding of the corresponding fatty acid substituent in the hydrophobic tunnel.
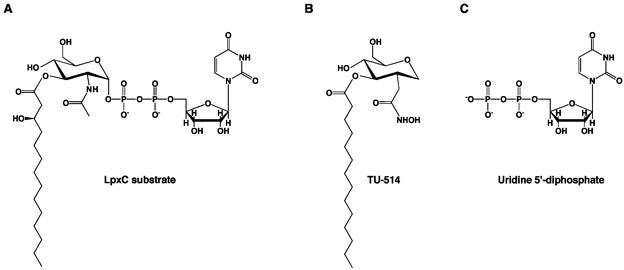
Figure 2.
(A) LpxC substrate, UDP –{3-O-[(R)-3-hydroxymyristoyl]}-N-acetylglucosamine. (B) Substrate analogue inhibitor, TU-514 (Ki ~ 650 nM (27)). (C) uridine 5′-diphosphate (UDP).
Experimental results acquired to date suggest that LpxC catalyzes substrate deacetylation through a mechanism involving a general acid-base catalyst pair (Figure 1) (23). In the first step of catalysis, E78 likely serves as a general base by abstracting a proton from a zinc-bound water molecule, thereby promoting nucleophilic attack at the substrate and formation of a tetrahedral oxyanion intermediate (19). Site directed mutagenesis studies (21, 23, 25) and X-ray crystal structures of LpxC-cacodylate and LpxC-palmitate complexes (23) are consistent with the general base function proposed for E78. The X-ray crystal structure of LpxC complexed with the tetrahedral cacodylate anion further implicates T191 and H265 in the stabilization of the tetrahedral intermediate and its flanking transition states (23). Protonation of the leaving amino group by general acid H265 (pKa ~ 8 (21)) facilitates the collapse of the tetrahedral intermediate to yield the final products (21, 23, 25).
The NMR (20-21) and X-ray crystal (22) structures of the LpxC–TU-514 complex yield generally similar inferences about substrate binding in the active site of LpxC despite significant differences in the zinc coordination polyhedron and intermolecular interactions of the hexose ring. The X-ray crystal structure reveals that the TU-514 hydroxamate moiety chelates Zn2+A to form a pentacoordinate zinc complex with square pyramidal coordination geometry (22). Hydrogen bond interactions between the hydroxamate OH group and D242 and H265, and the hydroxamate C=O group and T191, also stabilize the zinc complex. Hexose ring substituents hydrogen bond with E197 and H58, and K239 through a water-mediated hydrogen bond interaction. Although TU-514 is similar in structure to the actual LpxC substrate (Figure 2), it lacks the 3-hydroxy group of the 3-O myristoyl substituent and a UDP group (26, 27). Therefore, the structures of LpxC–TU-514 complexes (20-22) provide no information on the binding of the UDP portion of an intact substrate.
Based on analysis of the refined NMR structure of the LpxC–TU-514 complex (21), the negatively charged diphosphate group of UDP is hypothesized to interact with residues in the so-called “basic patch” defined by K239, R143, R262, and H265. The binding of a sulfate ion in the basic patch of LpxC complexed with cacodylate (PDB entry 1YHC) (23) supports this hypothesis (21). In order to explicitly define the role of the basic patch in substrate binding, we now report the crystal structure of the LpxC-UDP complex at 2.7 Å resolution. We also report the structure of LpxC complexed with pyrophosphate at 3.1 Å resolution. Surprisingly, both structures reveal that diphosphate binding in the basic patch is accommodated by a new metal ion in addition to an array of hydrogen bond interactions. Together, these structures provide key inferences on substrate recognition in the basic patch and changes in metal ion coordination that may accompany catalysis.
MATERIALS AND METHODS
Crystal structure of the LpxC-UDP complex
The C193A/ΔD284-L294 variant of LpxC from A. aeolicus (henceforth “LpxC”) was overexpressed in E. coli and purified as described (19, 28) and crystallized using previously reported conditions (23). To prepare the UDP complex, crystals were gradually transferred to 100 mM HEPES (pH 8.0), 180 mM NaCl, 14-16% PEG 3350, 0.5 mM ZnSO4, 2 mM MgCl2, and 30 mM UDP, and soaked for 16 hours. Diffraction data were measured to 2.7 Å resolution using a wavelength corresponding to the zinc K-edge (λ = 1.283 Å) at the National Synchrotron Light Source at Brookhaven National Laboratories (NSLS, beamline X12C, Upton, NY). Crystals were isomorphous with those of the zinc-inhibited enzyme (19) and belonged to space group P61 with unit cell parameters a = b = 101.2 Å, c = 122.2 Å (two monomers in the asymmetric unit). Data were indexed and merged using HKL2000 (29). The structure of zinc-inhibited LpxC (PDB entry 1P42) (19), excluding all zinc ions, solvent, and fatty acid molecules, was used as a search probe in molecular replacement calculations using AMoRe (30). Initial electron density maps showed that UDP was bound in the basic patch. Difference maps calculated using anomalous scattering data revealed a total of 9 zinc sites in the two LpxC monomers in the asymmetric unit. Seven of these sites corresponded to those observed in the zinc-inhibited enzyme (19) and two additional sites accommodated the binding of the UDP diphosphate groups (one per monomer). Iterative cycles of refinement and model building were performed with CNS (31) and O (32), respectively, to improve the structure as monitored by Rfree. Noncrystallographic symmetry (NCS) restraints were employed throughout refinement. Atomic coordinates of solvent molecules and UDP molecules were added during the last stages of refinement. Data collection and refinement statistics are reported in Table 1.
Table 1.
Data Collection and Refinement Statistics
| Complex | LpxC–UDP | LpxC pyrophosphate |
| Resolution range (Å) |
30.0-2.7 | 50-3.1 |
| Reflections (measured/ unique) |
123119/19510 | 33547/12560 |
| Completeness (%) (overall/ outer shell) |
99.9/100.0 | 99.3/100.0 |
| Rmergea (overall/ outer shell) |
0.128/0.333 | 0.134/0.393 |
| <I/σ> (overall/ outer shell) |
11.7/4.2 | 5.7/2.0 |
| Protein atoms (no.) b |
4298 | 4298 |
| Solvent atoms (no.) b |
83 | 24 |
| Metal ions (no.) b |
9 | 5 |
| Ligand atoms (no.) b |
92 | 54 |
| Reflections used in refinement (work/ free) |
17590/1920 | 11328/1232 |
| R/Rfree c | 0.213/0.241 | 0.230/0.277 |
| r.m.s. deviations | ||
| bonds (Å) | 0.007 | 0.008 |
| angles (deg.) | 1.3 | 1.3 |
| proper dihedral angles (deg.) |
23.2 | 23.3 |
| Improper dihedral angles (deg.) |
0.72 | 0.80 |
Rmerge = ∑|Ij - <Ij>|/ ∑Ij, where Ij is the observed intensity for reflection j and <Ij> is the average intensity calculated for reflection j from replicate data.
Per asymmetric unit.
R = ∑||Fo| - |Fc||/ ∑ |Fo|, where R and Rfree are calculated using the working and test reflection sets, respectively.
Crystal structure of the LpxC-pyrophosphate complex
Crystals of LpxC were prepared as described above and gradually transferred to 100 mM HEPES (pH 7.5), 180 mM NaCl, 12% PEG 3350, 0.5 mM ZnSO4, 5 mM MgCl2, and 10 mM tetrasodium pyrophosphate, and soaked for 16 hours. Diffraction data were measured to 3.1 Å resolution using an R-AXIS IV++ image plate area detector mounted on a Rigaku-200HB rotating anode X-ray generator operating at 50 kV and 100 mA. Crystals were isomorphous with those of the zinc-inhibited enzyme (19) and belonged to space group P61 with unit cell parameters a = b = 98.9 Å, c = 125.7 Å. Data were indexed and merged with d*Trek (33). The structure of zinc-inhibited LpxC (19), excluding all zinc ions, solvent, and fatty acid molecules was used as a search probe in molecular replacement calculations using AMoRe (30). Initial electron density maps showed that pyrophosphate was bound in the basic patch and coordinated to a metal ion interpreted as zinc. Interestingly, neither Zn2+B nor Zn2+C ions were observed (in the structure of the zinc-inhibited enzyme (19), Zn2+B is coordinated by E78 and H265, and Zn2+C is coordinated by H58 and H200). The catalytic zinc ion, Zn2+A, was evident, but the Zn2+D ion, which makes an interlattice contact in the structure of the zinc-inhibited enzyme (19), was only partially occupied. Iterative cycles of refinement and model building were performed with CNS (31) and O (32), respectively, to improve the structure as monitored by Rfree. Strict NCS constraints were employed initially and relaxed into appropriately weighted restraints as guided by Rfree. Atomic coordinates for pyrophosphate and solvent molecules were added during the last stages of refinement. Data collection and refinement statistics are reported in Table 1. All figures were prepared using PyMOL (34).
Results
LpxC-UDP complex
The transfer of LpxC crystals to a buffer solution containing 30 mM UDP results in the binding of UDP in the basic patch defined by residues K239, R143, R262, and H265. An electron density map of the LpxC-UDP complex is shown in Figure 3A and intermolecular interactions are illustrated in Figure 3B. The structure of LpxC in this complex is essentially identical to that of the zinc-inhibited enzyme (19) with an r.m.s. deviation of 0.235 Å for 267 Cα atoms; a superposition is shown in Figure 3C. UDP binds with full occupancy and makes extensive interactions within the basic patch. Although UDP binding is generally similar to each monomer in the asymmetric unit, some differences in hydrogen bond interactions are observed.
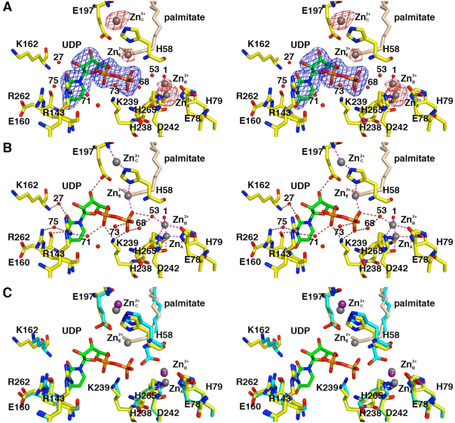
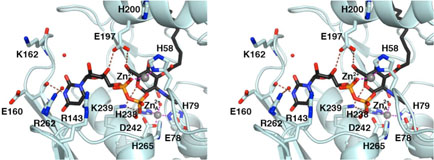
Figure 3.
LpxC-UDP complex (monomer A). (A) Omit electron density map contoured at 4σ (blue); Bijvoet difference Fourier map calculated with anomalous data collected at a wavelength corresponding to the zinc K-edge, contoured at 4σ (red). Atoms are color coded as follows: C = yellow, O = red, N = blue, P = orange; palmitate appears in light tan; zinc ions appear as dark gray spheres; water molecules appear as red spheres. Zinc occupancies are estimated as follows: Zn2+A′ 100%; Zn2+B′ 50%; Zn2+C′ 50%; Zn2+D′ 100% (not visible); Zn2+E′ 40%. (B) Intermolecular interactions in the LpxC-UDP complex. Dashed lines indicate zinc coordination (magenta) and hydrogen bond (brown) interactions. (C) Superposition with the zinc-inhibited structure of LpxC (19) (color coded as described above except that C = cyan and zinc ions appear as purple spheres) reveals minimal structural changes resulting from UDP binding.
In monomer A, both the α-and β-phosphate groups of UDP accept hydrogen bonds from strictly conserved residue K239 (Figure 3B). The α-phosphate also accepts a hydrogen bond from water molecule #71, which in turn accepts a hydrogen bond from R143; the β-phosphate also receives hydrogen bonds from water molecules #73, #68, and #53. The ribose 3′-OH group donates a hydrogen bond to E197. The uracil N3-H group is a bifurcated hydrogen bond donor to the backbone carbonyl of E160 and water molecule #75; in turn, water molecule #75 accepts a hydrogen bond from R262. The uracil 4-carbonyl group accepts a hydrogen bond from the backbone NH group of E160. The uracil 2-carbonyl group accepts a hydrogen bond from water molecule #27, which in turn accepts a hydrogen bond from the backbone NH group of K162.
In monomer B, both the α- and β-phosphate groups of UDP accept hydrogen bonds from strictly conserved residue K239; the β-phosphate also hydrogen bonds with water molecule #15 (data not shown). The ribose 3′-OH group donates a hydrogen bond to E197, which in turn, hydrogen bonds to water molecule #78. The uracil N3-H group donates a hydrogen bond to the backbone carbonyl of E160, and the uracil 4-carbonyl group accepts a hydrogen bond from the backbone NH group of E160. Thus, of the four residues hypothesized to interact with UDP (21), three are observed to make important interactions: K239 makes direct hydrogen bonds with both the α- and β-phosphate groups of UDP, and R143 and R262 interact with the α-phosphate group and uracil base, respectively, through bridging water molecules. No interactions are observed for H265.
As observed in the initial structure determination of LpxC prepared in the presence of excess zinc (19), several zinc ions are observed in the LpxC-UDP complex. The Bijvoet difference Fourier map in Figure 3A calculated using anomalous data collected at a wavelength corresponding to the zinc K-edge confirms the identities and locations of these zinc ions: some are functionally important, and others are artifacts of crystallization. The catalytic zinc ion, Zn2+A, is present with full occupancy in both monomers and is coordinated by H79, H238, D242, and a solvent molecule (#1 in monomer A, #13 in monomer B) with tetrahedral geometry. The inhibitory zinc ion, Zn2+B, is partially displaced by UDP binding and has an estimated occupancy of 50%. The Zn2+B ion blocks catalytic activity by complexation with the proposed general acid-base pair (23), H265 and E78. Solvent molecules #1 and #53 also coordinate to Zn2+B to complete a tetrahedral coordination polyhedron in monomer A; solvent molecule #13 is the only nonprotein ligand found in the Zn2+B coordination polyhedron in monomer B.
The Zn2+C ion is coordinated by the imidazole side chains of H58 and H200 and stabilizes the βαβ subdomain that frames the hydrophobic tunnel in the active site (19). The binding of the Zn2+C ion is likely an artifact of the ~5-fold molar excess of ZnSO4 used in the crystallization of LpxC, although it binds with an estimated occupancy of 50% in the LpxC-UDP complex. The Zn2+D ion binds with full occupancy, making an interlattice contact between E126 and the α-amino group of G2 in monomer A, and H29 and E95 of monomer B in an adjacent asymmetric unit. The Zn2+D ion is similarly an artifact of the crystallization conditions (19).
Unexpectedly, a newly-discovered zinc ion designated Zn2+E is coordinated by the UDP β-phosphate group and the Nδ atom of H58. Since H58 also coordinates to Zn2+C through its Nε atom, this interaction requires the imidazolate form of H58 if both metals are simultaneously coordinated by this residue. Alternatively, since Zn2+E binds with an estimated occupancy of 40% in monomer A and 50% in monomer B, and since Zn2+C binds with an estimated occupancy of 50% in both monomers, H58 could bind to either one or the other as the neutral imidazole if the binding of Zn2+E and Zn2+C is mutually exclusive. To summarize the zinc binding results, the active site of LpxC can accommodate three functional metal ions: the A-site metal ion activates a bound solvent molecule for catalysis, the B-site metal ion inhibits enzyme activity, and the E-site metal ion accommodates the binding of the substrate UDP diphosphate group. While it is conceivable that Mg2+ could fulfill the substrate binding function of Zn2+E, no evidence for Mg2+ binding to the E-site is apparent in the current study.
Interestingly, a palmitate molecule binds in the hydrophobic tunnel with its carboxylate head group clearly visible in electron density maps and oriented toward solvent at the outer end of the tunnel (data not shown). Consequently, the aliphatic tail of palmitate occupies the region near Zn2+E (Figure 3). Although at 2.7 Å resolution crystallographic occupancies are at best only rough estimates, palmitate appears to bind with reduced occupancy (~60% in monomer A, ~70% in monomer B) relative to the zinc-inhibited structure (19). Accordingly, palmitate binding and Zn2+E binding may be mutually exclusive. If the palmitate carboxylate was oriented toward the inner end of the hydrophobic tunnel, it could coordinate to Zn2+E; however, the electron density for palmitate is not consistent with this orientation.
LpxC-pyrophosphate complex
The transfer of crystals to a buffer solution containing 10 mM tetrasodium pyrophosphate results in the binding of pyrophosphate in the basic patch. An electron density map of the LpxC-pyrophosphate complex is shown in Figure 4A. The structure of the LpxC-pyrophosphate complex is essentially identical to that of the zinc-inhibited enzyme (19) with an r.m.s. deviation of 0.352 Å for 267 Cα atoms. Although this structure is determined at a relatively low resolution of 3.1 Å, general features of pyrophosphate interactions in the active site (Figure 4B) are consistent with those observed for diphosphate binding in the LpxC-UDP complex.
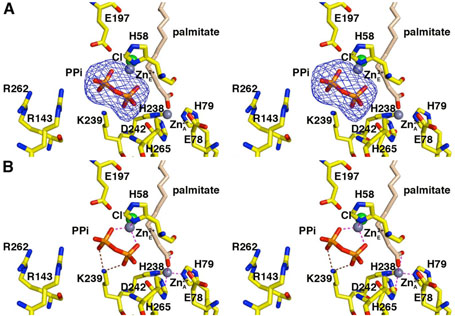
Figure 4.
LpxC–pyrophosphate complex (monomer A). (A) Omit electron density map contoured at 4σ; selected active site residues are indicated. Atoms are color coded as in Figure 3. Zinc occupancies are estimated as follows: Zn2+A′ 100%; Zn2+E′ 60%. (B) Intermolecular interactions in the LpxC-pyrophosphate complex. Dashed lines indicate zinc coordination (magenta) and hydrogen bond (brown) interactions.
In monomer A, both phosphates of the pyrophosphate group accept hydrogen bonds from K239. In monomer B, the phosphate group corresponding to the α-phosphate of the UDP diphosphate group interacts with K239 (3.4 Å) and accepts a hydrogen bond from water molecule #11. The phosphate group corresponding to the β-phosphate of the UDP diphosphate group also accepts a hydrogen bond from water molecule #11.
The pyrophosphate group also coordinates to a metal ion interpreted as Zn2+E by analogy with the LpxC-UDP complex. In both monomers A and B, Zn2+E occupancy is estimated to be 60%. In monomer A, the Zn2+E ion is tetrahedrally coordinated by H58, both oxygen atoms of the pyrophosphate group, and a chloride ion. In monomer B, Zn2+E is coordinated by the α-phosphate group, H58, and a chloride ion. In both monomers A and B, the Zn2+C ion, coordinated by H58 in the zinc-inhibited enzyme (19), is absent. The catalytic zinc ion (Zn2+A) is present and fully occupied, but the inhibitory zinc ion (Zn2+B) appears to be fully displaced by pyrophosphate binding and by a palmitate molecule that coordinates to Zn2+A. The Zn2+D ion, which makes an interlattice contact in the structure of the zinc-inhibited enzyme (19), is partially occupied (40%). The observation of partially occupied zinc ions is likely hindered by the relatively low resolution of this structure determination.
Discussion
Inferences on Substrate Binding
The structures of the LpxC-UDP and LpxC-pyrophosphate complexes provide the first view of intermolecular electrostatic interactions in the basic patch that may dominate the initial binding of enzyme and substrate in catalysis. Strictly conserved residue K239 accepts hydrogen bonds from both phosphate groups of UDP, and residues R143 and R262 (which usually appear as lysine residues in LpxC enzymes from other species) make water-mediated interactions with UDP (Figure 3B). Unexpectedly, the affinity of LpxC for UDP in the absence of substrate is quite low and the dissociation constant of this complex is estimated to be in the high millimolar range (35). Consistent with low apparent affinity, an NMR experiment with a 3:1 molar ratio of UDP to either A. aeolicus LpxC or the LpxC–TU-514 complex shows no chemical shift perturbation to indicate UDP binding (21). However, it is possible that the enzyme utilizes much of the favorable binding free energy to order the active site, since NMR studies of the native enzyme in solution indicate substantial active site mobility in the unliganded enzyme (20). It is also not clear how metal ion binding in the LpxC active site could influence these affinity measurements. Based on the low affinity of LpxC for UDP, it is likely that pyrophosphate binds with similar or even lower affinity.
Surprisingly, a new zinc-binding site is observed in both the UDP and pyrophosphate complexes: Zn2+E interacts with pyrophosphate and the diphosphate group of UDP, and Zn2+E may similarly interact with the UDP moiety of the actual LpxC substrate. Upon inspection of the initial difference electron density maps, this metal ion was originally hypothesized to be Mg2+, present in millimolar concentrations in the crystal stabilization buffers. The Mg2+ ion also accommodates the binding of diphosphate/pyrophosphate in a variety of enzyme systems, e.g., terpenoid cyclases (36). However, the Bijvoet difference Fourier map in Figure 3A conclusively demonstrates that zinc is bound at the E-site. Even so, Mg2+ could conceivably bind to the E-site in the absence of high Zn2+ concentrations. Although a histidine imidazolate would be unusual as a Mg2+ ligand, 325 examples of histidine-Mg2+ coordination interactions are found in the Metalloprotein Database (http://metallo.scripps.edu), and 9 examples of imidazole-Mg2+ coordination interactions are found in the Cambridge Structural Database (37), so there is ample structural precedent for such interactions. On the other hand, Zn2+ binds more frequently to nitrogen ligands than Mg2+ (38), and the results of the anomalous scattering experiment provide conclusive evidence for Zn2+E binding in the current study.
It is noteworthy that the newly identified Zn2+E site is accompanied by the partial displacement of the inhibitory zinc ion, Zn2+B, which is ~6 Å away. Possibly, Zn2+B moves to the E-site to accommodate UDP binding. Perhaps this could occur in catalysis, where at suitable zinc concentrations the enzyme would remain in a zinc-inhibited resting state until substrate binding, whereupon the inhibitory zinc ion would shift from the B-site to the E-site to stabilize the negative charge on the incoming UDP diphosphate group. A model of the substrate bound in the active site based on structures of the TU-514 complex (22) and the UDP complex with the novel zinc binding site is displayed in Figure 5.
Figure 5.
Model of the LpxC-substrate complex based on the X-ray crystal structures of the TU-514 complex (22) and the UDP complex (Figure 3). Atoms are color coded as follows: C= light cyan O = red, N = blue, P = orange; the substrate carbon atoms appear in black; zinc ions appears as grey spheres; solvent molecules appear as red spheres. Intermolecular interactions observed in the two complexes separately can be achieved by an intact molecule of the substrate, UDP –{3-O-[(R)-3-hydroxymyristoyl]}-N-acetylglucosamine (Figure 2A), bound in the active site.
Structural Aspects of Inhibitor Design
Intermolecular interactions of the UDP moiety and the pyrophosphate group in the active site point to specific interactions in the basic patch that could be exploited in the design of LpxC inhibitors. In order to develop broad-specificity inhibitors that target Gram-negative bacteria from a wide array of species, interactions with strictly conserved residues should be considered. For example, such residues include H265 and K239, which make key interactions with the tetrahedral intermediate and its flanking transition states (23) and UDP (Figure 3), respectively. Also in the basic patch, the positively charged guanidinium groups of R143 and R262 interact with UDP through bridging water molecules; since these residues usually appear as lysine in most other sequenced LpxC enzymes, they similarly could be targeted for enzyme-inhibitor charge-charge interactions. Finally, E197 makes key interactions with both the ribose ring of UDP (Figure 3) and the C-4 hydroxyl of the hexose ring of TU-514 (22). This residue also appears as aspartate or glutamine in most other sequenced LpxC enzymes, so an enzyme-inhibitor hydrogen bond interaction could target this residue.
The identification of a new zinc binding site could also be exploited inhibitor design. For instance, “two-prong” inhibitors could be designed to target the Zn2+A and Zn2+E binding sites simultaneously by incorporating a phosphate analogue or metal ligand designed to interact within the UDP diphosphate binding site. A similar approach has been adopted in the design of inhibitors of carbonic anhydrases I and II, in which a cupric iminodiacetate moiety is tethered to a sulfonamide group in order to simultaneously bind to the active site Zn2+ ion and a histidine residue ~8 Å away (39-41).
Previously, LpxC inhibitors have targeted two other features of the active site: the catalytic zinc ion (Zn2+A) and the hydrophobic tunnel (26-27, 42-45). It is possible that with modifications incorporated to target the UDP binding site, the binding affinity of inhibitors such as CHR-090 (Ki ~ 2nM (43)), could be enhanced even further. The X-ray crystal structure determinations of LpxC complexes with CHR-090 and other tight-binding inhibitors will facilitate the design of a new class of LpxC inhibitors targeting all three areas of molecular recognition in the active site: the catalytic zinc ion, the hydrophobic tunnel, and the UDP binding site.
In summary, this work provides the first view of UDP binding in the active site of LpxC. Data from previously determined structures of LpxC complexes with the substrate analogue inhibitor TU-514 (20-22), taken together with the structural information for UDP binding presented in this work, provide a complete view of intermolecular interactions important for enzyme-substrate association in catalysis, particularly with regard to the role of the newly discovered E-site metal ion in the molecular recognition of the substrate.
Acknowledgements
We thank the National Synchrotron Light Source (NSLS) for beamline access.
Footnotes
Abbreviations: LpxC, UDP –{3-O-[(R)-3-hydroxymyristoyl]}-N-acetylglucosamine deacetylase; LPS, lipopolysaccharide; TU-514, 1,5-anhydro-2-C-(carboxymethyl-N-hydroxamide)-2-deoxy-3-O-myristoyl-d-glucitol; UDP, uridine 5′-diphosphate; NCS, noncrystallographic symmetry; HEPES, 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid; PEG, polyethylene glycol.
Here, the numbering system for the well-studied LpxC enzyme from E. coli is adapted for the A. aeolicus enzyme. Based on an alignment performed with Clustal W, important residues conserved between E. coli/A. aeolicus correspond as follows: R58/H58, E78/E73, H79/H74, K143/R137, F161/F155, T191/T179, F192/F180, F194/F182, E197/D185, H238/H226, K239/K227, D242/D230, D246/D234, K262/R250, and H265/H253.
This work was supported by the National Institutes of Health grant GM49758.
The atomic coordinates of LpxC complexed with uridine 5′-diphosphate and pyrophosphate have been deposited in the Protein Data Bank, www.rcsb.org, with accession codes 2IER and 2IES, respectively.
References
- 1.Anderson MS, Bulawa CE, Raetz CR. The biosynthesis of Gram-negative endotoxin. Formation of lipid A precursors from UDP-GlcNAc in extracts of Escherichia coli. J. Biol. Chem. 1985;260:15536–15541. [PubMed] [Google Scholar]
- 2.Anderson MS, Robertson AD, Macher I, Raetz CRH. Biosynthesis of lipid A in Escherichia coli: identification of UDP-3-O-[(R)-3-hydroxymyristoyl]-alpha-D-glucosamine. Biochemistry. 1988;27:1908–1917. doi: 10.1021/bi00406a017. [DOI] [PubMed] [Google Scholar]
- 3.Anderson MS, Bull HG, Galloway SM, Kelly TM, Mohan S, Radika K, Raetz CRH. UDP-N-acetylglucosamine acyltransferase of Escherichia coli. The first step of endotoxin biosynthesis is thermodynamically unfavorable. J. Biol. Chem. 1993;268:19858–19865. [PubMed] [Google Scholar]
- 4.Young K, Silver LL, Bramhill D, Cameron P, Eveland SS, Raetz CRH, Hyland SA, Anderson MS. The envA permeability/cell division gene of Escherichia coli encodes the second enzyme of lipid A biosynthesis. UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase. J. Biol. Chem. 1995;270:30384–30391. doi: 10.1074/jbc.270.51.30384. [DOI] [PubMed] [Google Scholar]
- 5.Raetz CRH. Molecular genetics of membrane phospholipid synthesis. Ann. Rev. Genet. 1986;20:253–295. doi: 10.1146/annurev.ge.20.120186.001345. [DOI] [PubMed] [Google Scholar]
- 6.Raetz CRH. Biochemistry of endotoxins. Annu. Rev. Biochem. 1990;59:129–170. doi: 10.1146/annurev.bi.59.070190.001021. [DOI] [PubMed] [Google Scholar]
- 7.Raetz CRH. Bacterial endotoxins: extraordinary lipids that activate eukaryotic signal transduction. J. Bacteriol. 1993;175:5745–5753. doi: 10.1128/jb.175.18.5745-5753.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaara M. Endotoxin in health and disease. Marcel Dekker, Inc.; New York, NY: 1999. pp. 31–32. [Google Scholar]
- 9.Rick PD, Raetz CRH. Endotoxin in health and disease. Marcel Dekker, Inc.; New York, NY: 1999. pp. 283–304. [Google Scholar]
- 10.Nikaido H, Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaara M. Agents that increase the permeability of the outer membrane. Microbiol. Rev. 1992;56:395–411. doi: 10.1128/mr.56.3.395-411.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vuorio R, Vaara M. The lipid A biosynthesis mutation lpxA2 of Escherichia coli results in drastic antibiotic supersusceptibility. Antimicrob. Agents Chemother. 1992;36:826–829. doi: 10.1128/aac.36.4.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaara M. Outermembrane permeability barrier to azithromycin, clarithromycin, and roxithromycin in Gram-negative enteric bacteria. Antimicrob. Agents Chemother. 1993;37:354–356. doi: 10.1128/aac.37.2.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wyckoff TJ, Raetz CRH, Jackman JE. Antibacterial and antiinflammatory agents that target endotoxin. Trends Microbiol. 1998;6:154–159. doi: 10.1016/s0966-842x(98)01230-x. [DOI] [PubMed] [Google Scholar]
- 15.Parillo JE, Parker MM, Natanson C, Suffredini AF, Danner RL, Cunnion RE, Ognibene FP. Septic shock in humans. Advances in the understanding of pathogenesis, cardiovascular dysfunction, and therapy. Ann. Intern. Med. 1990;113:227–242. doi: 10.7326/0003-4819-113-3-227. [DOI] [PubMed] [Google Scholar]
- 16.Glauser MP, Zanetti G, Baumgartner J-D, Cohen J. Septic shock: pathogenesis. Lancet. 1991;338:732–736. doi: 10.1016/0140-6736(91)91452-z. [DOI] [PubMed] [Google Scholar]
- 17.Cohen J, Glauser MP. Septic shock treatment. Lancet. 1991;338:736–739. doi: 10.1016/0140-6736(91)91453-2. [DOI] [PubMed] [Google Scholar]
- 18.Parillo JE. Pathogenic mechanisms of septic shock. N. Eng. J. Med. 1993;328:1471–1477. doi: 10.1056/NEJM199305203282008. [DOI] [PubMed] [Google Scholar]
- 19.Whittington DA, Rusche KM, Shin H, Fierke CA, Christianson DW. Crystal structure of LpxC, a zinc-dependent deacetylase essential for endotoxin biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8146–8150. doi: 10.1073/pnas.1432990100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coggins BE, Li X, McClerrin AL, Hindsgaul O, Raetz CRH, Zhou P. Structure of the LpxC deacetylase with a bound substrate-analog inhibitor. Nat. Struct. Biol. 2003;10:645–651. doi: 10.1038/nsb948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coggins BE, McClerrin AL, Jiang L, Li X, Rudolph J, Hindsgaul O, Raetz CRH, Zhou P. Refined solution structure of the LpxC-TU-514 complex and pKa anlaysis of an active site histidine: insights into the mechanism and inhibitor design. Biochemistry. 2005;44:1114–1126. doi: 10.1021/bi047820z. [DOI] [PubMed] [Google Scholar]
- 22.Gennadios HA, Whittington DA, Li X, Fierke CA, Christianson DW. Mechanistic inferences from the binding of ligands to LpxC, a metaldependent deacetylase. Biochemistry. 2006;45:7940–7948. doi: 10.1021/bi060823m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hernick M, Gennadios HA, Whittington DA, Rusche KM, Christianson DW, Fierke CA. UDP-(3-O-((R)-3-hydroxymyristoyl))-N-acetylglucosamine deacetylase functions through a general acid-base catalyst pair mechanism. J. Biol. Chem. 2005;280:16969–16978. doi: 10.1074/jbc.M413560200. [DOI] [PubMed] [Google Scholar]
- 24.Hernick M, Fierke CA. Zinc hydrolases: the mechanisms of zinc-dependent deactylases. Arch. Biochem. Biophys. 2004;433:71–84. doi: 10.1016/j.abb.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 25.McClerren AL, Zhou P, Guan Z, Raetz CRH, Rudolph J. Kinetic analysis of the zinc-dependent deacetylase in the lipid A biosynthetic pathway. Biochemistry. 2005;44:1106–1113. doi: 10.1021/bi048001h. [DOI] [PubMed] [Google Scholar]
- 26.Jackman JE, Fierke CA, Tumey LN, Pirrung M, Uchiyama T, Tahir SH, Hindsgaul O, Raetz CRH. Antibacterial agents that target lipid A biosynthesis in Gram-negative bacteria. J. Biol. Chem. 2000;275:11002–11009. doi: 10.1074/jbc.275.15.11002. [DOI] [PubMed] [Google Scholar]
- 27.Li X, Uchiyama T, Raetz CRH, Hindsgaul O. Synthesis of a carbohydrate-derived hydroxamic acid inhibitor of the bacterial enzyme (LpxC) involved in lipid A biosythesis. Org. Lett. 2003;5:539–541. doi: 10.1021/ol027458l. [DOI] [PubMed] [Google Scholar]
- 28.Jackman JE, Raetz CRH, Fierke CA. UDP-(3-O-(R-3-hydroxymyristoyl))-N-acetylglucosamine deacetylase of Escherichia coli is a zinc metalloenzyme. Biochemistry. 1999;38:1902–1911. doi: 10.1021/bi982339s. [DOI] [PubMed] [Google Scholar]
- 29.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 30.Navaza J. AMoRe: an automated package for molecular replacement. Acta. Cryst. 1994;A50:157–163. [Google Scholar]
- 31.Brünger AT, Adams PD, Clore GM, De Lano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 32.Jones TA, Zou JY, Cowan SW, Kjeldjaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 33.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta. Crystallogr. D Biol. Crystallogr. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 34.DeLano WL. San Carlos; CA, USA: 2002. The PyMOL molecular graphics system, DeLano Scientific. [Google Scholar]
- 35.Hernick M, Fierke CA. Catalytic mechanism and molecular recognition of E. coli UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase probed by mutagenesis. submitted. [DOI] [PubMed]
- 36.Christianson DW. Structural biology and chemistry of the terpenoid cyclases. Chem. Rev. 2006;106:3412–3442. doi: 10.1021/cr050286w. [DOI] [PubMed] [Google Scholar]
- 37.Allen FH. The Cambridge Structural Database: a quarter of a million crystal structures and rising. Acta. Crystallogr. B. 2002;58:380–388. doi: 10.1107/s0108768102003890. [DOI] [PubMed] [Google Scholar]
- 38.Bock CW, Katz AK, Markham GD, Glusker JP. Manganese as a replacement for magnesium and zinc: functional comparison of divalent ions. J. Am. Chem. Soc. 1999;121:7360–7372. [Google Scholar]
- 39.Roy BC, Banerjee AL, Swanson M, Xiao GJ, Haldar MK, Mallik S, Srivastava DK. Two-prong inhibitors for Carbonic Anhydrase II. J. Am. Chem. Soc. 2004;126:13206–13207. doi: 10.1021/ja047271k. [DOI] [PubMed] [Google Scholar]
- 40.Banerjee AL, Eiler D, Roy BC, Jia X, Haldar MK, Mallik S, Srivastava DK. Spacer-based selectivity in the binding of two-prong ligands to recombinant human carbonic anhydrase I. Biochemistry. 2005;44:3211–3224. doi: 10.1021/bi047737b. [DOI] [PubMed] [Google Scholar]
- 41.Jude KM, Banerjee AL, Haldar MK, Manokaran S, Roy BC, Mallik S, Srivastava DK, Christianson DW. Ultrahigh resolution crystal structures of human carbonic anhydrases I and II complexed with “two-prong” inhibitors reveal the molecular basis of high affinity. J. Am. Chem. Soc. 2006;128:3011–3018. doi: 10.1021/ja057257n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clements JM, Coignard F, Johnson I, Chandler S, Palan S, Waller A, Wijkmans J, Hunter MG. Antibacterial activities and characterization of novel inhibitors of LpxC. Antimicrob, Agents Chemother. 2002;46:1793–1799. doi: 10.1128/AAC.46.6.1793-1799.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kline T, Andersen NH, Harwood EA, Bowman J, Malanda A, Endsley S, Erwin AL, Doyle M, Fong S, Harris AL, Mendelsohn B, Mdululi K, Raetz CRH, Stover CK, Witte PR, Yabannavar A, Zhu S. Potent, novel in vitro inhibitors of the Pseudomonas aeruginosa deacetylase LpxC. J. Med. Chem. 2002;45:3112–3129. doi: 10.1021/jm010579r. [DOI] [PubMed] [Google Scholar]
- 44.Pirrung MC, Tumey LN, Raetz CRH, Jackman JE, Snehalatha K, McClerren AL, Fierke CA, Gantt SL, Rusche KM. Inhibition of the antibacterial target UDP-(3-O-acyl)-N-acetylglucosamine deacetylase (LpxC): isoxazoline zinc amidase inhibitors bearing diverse metal binding groups. J. Med. Chem. 2002;45:4359–4370. doi: 10.1021/jm020183v. [DOI] [PubMed] [Google Scholar]
- 45.McClerren AL, Endsley S, Bowman JL, Andersen NH, Guan Z, Rudolph J, Raetz CRH. A slow, tight-binding inhibitor of the zinc-dependent deacetylase LpxC of lipid A biosynthesis with antibiotic activity comparable to ciproflaxin. Biochemistry. 2005;44:16574–16583. doi: 10.1021/bi0518186. [DOI] [PMC free article] [PubMed] [Google Scholar]