Abstract
Proper functioning of organelles necessitates efficient protein targeting to the appropriate subcellular locations. For example, degradation in the fungal vacuole relies on an array of targeting mechanisms for both resident hydrolases and their substrates. The particular processes that are used vary depending on the available nutrients. Under starvation conditions, macroautophagy is the primary method by which bulk cytosol is sequestered into autophagic vesicles (autophagosomes) destined for this organelle. Molecular genetic, morphological, and biochemical evidence indicates that macroautophagy shares much of the same cellular machinery as a biosynthetic pathway for the delivery of the vacuolar hydrolase, aminopeptidase I, via the cytoplasm-to-vacuole targeting (Cvt) pathway. The machinery required in both pathways includes a novel protein modification system involving the conjugation of two autophagy proteins, Apg12p and Apg5p. The conjugation reaction was demonstrated to be dependent on Apg7p, which shares homology with the E1 family of ubiquitin-activating enzymes. In this study, we demonstrate that Apg7p functions at the sequestration step in the formation of Cvt vesicles and autophagosomes. The subcellular localization of Apg7p fused to green fluorescent protein (GFP) indicates that a subpopulation of Apg7pGFP becomes membrane associated in an Apg12p-dependent manner. Subcellular fractionation experiments also indicate that a portion of the Apg7p pool is pelletable under starvation conditions. Finally, we demonstrate that the Pichia pastoris homologue Gsa7p that is required for peroxisome degradation is functionally similar to Apg7p, indicating that this novel conjugation system may represent a general nonclassical targeting mechanism that is conserved across species.
INTRODUCTION
The efficient turnover and recycling of proteins and redundant organelles are critical features of cell metabolism and physiology. In order to survive starvation conditions, nonessential cytosolic proteins and organelles must be delivered to the vacuole, where they are broken down and reused for essential cellular processes through the action of an array of vacuolar enzymes (reviewed by Klionsky et al., 1990; Klionsky, 1997). The secretory pathway and endocytosis define the classic paradigms for the vacuolar localization of both resident hydrolases and degradative substrates, respectively. For biosynthetic delivery, most characterized vacuolar enzymes transit through the early stages of the secretory pathway. They are diverted from being secreted at the trans-Golgi network and subsequently travel directly to the vacuole or arrive at the organelle via an endosomal intermediate (reviewed by Bryant and Stevens, 1998). Similarly, proteins destined for degradation transit from the cell surface or extracellular space by endocytosis to an endosomal compartment before their ultimate delivery to the vacuole (reviewed by Riezman et al., 1996). However, recent studies have demonstrated that alternate, nonclassical routes to the vacuole exist for both transport of the resident hydrolase aminopeptidase I (API)1 as well as delivery of cytoplasmic substrates (Klionsky et al., 1992; and reviewed by Scott and Klionsky, 1998).
Substrates destined for degradation are delivered to the vacuole through both specific and nonspecific recycling processes. The turnover of bulk cytosol is accomplished through macroautophagy, which is induced under conditions of nutrient deprivation (Takeshige et al., 1992; Baba et al., 1994). Cytosolic double-membrane autophagosomes sequester proteins and organelles for subsequent transport to the vacuole (Baba et al., 1994, 1995). In addition to the nonspecific process of autophagy, cells must specifically deliver targeted organelles for degradation under various nutrient conditions. For example, peroxisomes proliferate in response to certain growth conditions such as the presence of methanol or oleic acid as the sole carbon source. When glucose becomes available, peroxisomes, but not other organelles, are specifically sequestered and delivered to the vacuole for degradation by a process known as pexophagy (reviewed by Klionsky, 1997; and Scott and Klionsky, 1998). Peroxisome degradation is morphologically similar to nonspecific autophagy; however, the processes differ in the conditions for induction, the site of sequestration, and the kinetics of vacuolar delivery.
Molecular genetic analyses of both pexophagy and macroautophagy have begun to allow the isolation of mutants defective in these processes. The pdd, gsa, and pag mutants are blocked in various steps of peroxisome degradation (Titorenko et al., 1995; Tuttle and Dunn, 1995; Sakai et al., 1998). At present, however, only three of the corresponding genes have been identified (Yuan et al., 1997, 1999; Kiel et al., in press). Similar studies on autophagy have resulted in the identification of mutants, apg and aut, comprising ∼20 complementation groups that are defective in nonspecific protein delivery to the vacuole (Tsukada and Ohsumi, 1993; Thumm et al., 1994; Harding et al., 1996). While substantial progress has been made in cloning the apg and aut genes and characterizing the gene products (Kametaka et al., 1996; Funakoshi et al., 1997; Matsuura et al., 1997; Schlumpberger et al., 1997; Straub et al., 1997; Lang et al., 1998; Mizushima et al., 1998), key questions remain concerning the mechanism of sequestration and vacuolar delivery.
Simultaneous to these studies of macroautophagy, investigations into the transport of the resident vacuolar hydrolase API revealed that it is localized to the vacuole independent of the secretory pathway (Klionsky et al., 1992). Biochemical and morphological studies show that API is transported to the vacuole directly from the cytoplasm via a vesicle-mediated mechanism termed the cytoplasm-to-vacuole targeting (Cvt) pathway (Baba et al., 1997; Kim et al., 1997; Scott et al., 1997). Genetic studies resulted in the isolation of mutants, termed cvt, defective in API import and maturation (Harding et al., 1995, 1996). Of the 15 cvt complementation groups, 6 overlap with previously identified autophagy mutants (Harding et al., 1996; Scott et al., 1996). In addition, the majority, but not all, of the remaining cvt mutants have autophagy defects. Similarly, the majority of nonoverlapping autophagy mutants display an API import defect (Harding et al., 1996; Scott et al., 1996). Taken together, these studies suggest that the Cvt pathway and autophagy utilize many of the same molecular components.
Recent studies support the overlap between the Cvt and autophagy pathways by demonstrating that API is transported to the vacuole through two overlapping processes. During vegetative growth, API utilizes the Cvt pathway, while under starvation conditions the macroautophagy pathway transports API, as well as bulk cytosol, to the vacuole (Baba et al., 1997). However, in contrast to the slow, nonselective turnover of bulk cytosol, the kinetics of API import under both vegetative and starvation conditions remain relatively rapid (half-time of 30–45 min), suggesting that API maintains a specific mechanism for its delivery (Klionsky et al., 1992; Scott et al., 1996). The fact that API transport occurs under both vegetative and starvation conditions makes it a useful marker for the study of both the Cvt and macroautophagy pathways.
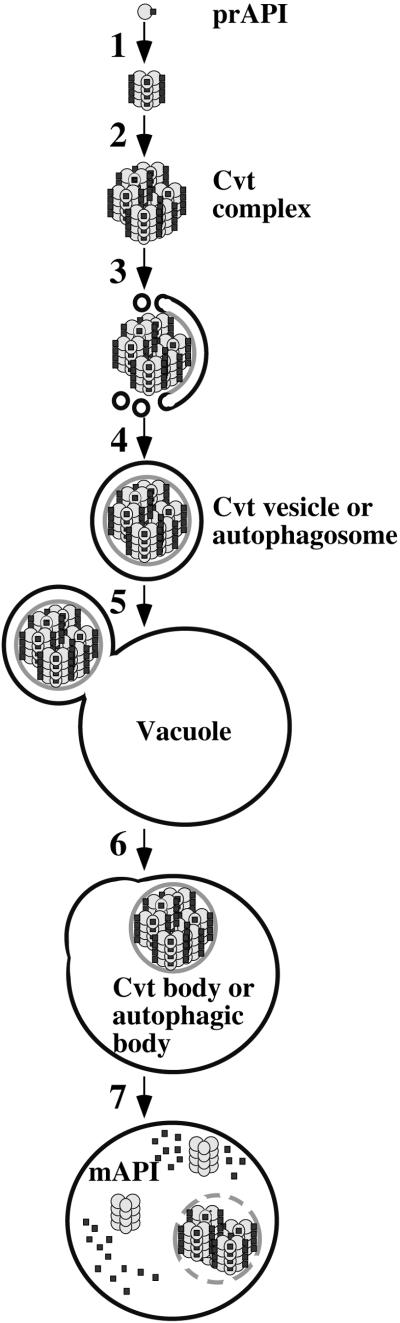
The study of API transport has begun to delineate discrete, sequential steps in the import process (Figure 1). API is synthesized in the cytosol as a 61-kDa precursor (prAPI) that includes an N-terminal 45-amino acid propeptide that is necessary for its vacuolar delivery (Oda et al., 1996). After synthesis, the 61-kDa prAPI rapidly oligomerizes in the cytosol into a dodecamer with a molecular mass of 732 kDa (Figure 1, step 1; Kim et al., 1997). Evidence from biochemical and electron microscopy studies indicates that prAPI subsequently forms a large protein complex in the cytosol defined as the Cvt complex (Figure 1, step 2; Baba et al., 1997; Scott et al., 1997). Subsequent to formation of the Cvt complex, a membrane recognition and sequestration event occurs in which the complex becomes enwrapped by a double membrane of unknown origin (Figure 1, step 3; Baba et al., 1997). In this step, prAPI in the Cvt complex remains accessible to the cytosol because of the incomplete formation of the surrounding vesicle. The completion of the sequestration event results in the formation of the double-membrane Cvt vesicle under vegetative growth conditions and the autophagosome in starvation conditions (Figure 1, step 4; Baba et al., 1997). The Cvt vesicles are ∼150 nm in diameter and contain an electron-dense core that appears distinct from cytosol. Autophagosomes are ∼300–900 nm in diameter and also contain the electron-dense Cvt complex, in addition to bulk cytosol (Baba et al., 1994, 1995, 1997; Scott et al., 1997).
Figure 1.
Steps in API import via the Cvt and autophagy pathways. Details are discussed in INTRODUCTION. Step 1: prAPI dodecamerization; step 2: Cvt complex formation; step 3: recognition and membrane sequestration; step 4: membrane expansion and vesicle completion; step 5: targeting, docking, and fusion of the Cvt vesicle or autophagosome with the vacuole; step 6: release of the Cvt body or autophagic body into the vacuole lumen; step 7: breakdown of the Cvt body or autophagic body and maturation of API.
Upon completion of formation, the Cvt vesicle/autophagosome is specifically targeted to the vacuole, where the outer membrane of the vesicle docks and fuses with the vacuolar membrane (Figure 1, step 5; Baba et al., 1995, 1997). This fusion reaction is followed by the release of the inner membrane of the vesicle, the Cvt body, or the autophagic body into the vacuolar lumen (Figure 1, step 6; Baba et al., 1994, 1995, 1997; Scott et al., 1997). In the final step of the Cvt and autophagy pathways, the vesicle membrane is degraded in a vacuolar protease-dependent manner. Vesicle breakdown also requires the proper vacuolar pH (Nakamura et al., 1997). The breakdown of the inner membrane vesicle allows the content of the Cvt body and the bulk cytosol inside the autophagic body to be accessible to vacuolar proteases, resulting in the processing of prAPI to its mature form as well as the degradation and recycling of bulk cytosol transported inside the autophagic bodies (Figure 1, step 7; Takeshige et al., 1992; Scott et al., 1997).
The apg7/cvt2 mutants were isolated in two independent screens based on defects in autophagy and API import, respectively (Tsukada and Ohsumi, 1993; Harding et al., 1995). A mutation in a homologous gene, GSA7, was also identified in a screen for defects in peroxisomal degradation (Yuan et al., 1999). A recent study indicated that Apg7p contains considerable homology to E1 ubiquitin-activating enzymes and was required in a novel conjugation reaction of two other autophagy proteins, Apg5p and Apg12p (Mizushima et al., 1998). These reactions were discovered to be essential for autophagy as well as API import via the Cvt pathway. Therefore, unlike the analogous ubiquitin-mediated, proteasome-dependent proteolysis system, the Apg7p-dependent covalent modification system is not restricted to a degradation function, but may function as part of a novel, general targeting mechanism. In this study, we sought to determine the site of function of Apg7p in the Cvt/autophagy pathways. We demonstrate that Apg7p is required for completion of the sequestration event and show that its interaction with membranes is dependent on its substrate Apg12p. Complementation of the apg7 defect by the GSA7 gene suggests that this protein functions in three overlapping but discrete pathways, autophagy, Cvt, and pexophagy.
MATERIALS AND METHODS
Strains and Media
Wild-type haploid Saccharomyces cerevisiae yeast strains used in this study were: SEY6210 (MATα leu2–3112 ura3–52 his3-Δ200 trp1-Δ901 lys2–801 suc2-Δ9 GAL) and SEY6211 (MATa leu2–3112 ura3–52 his3-Δ200 trp1-Δ901 ade2–101 suc2-Δ9 GAL) (Robinson et al., 1988). Mutant strains THY193a (cvt2–1) and THY226a (cvt2–2), YNM101 (MATα trp1 leu2 his3 ura3 apg12Δ::HIS3), and WSY99 (MATα leu2–3112 ura3–52 his3-Δ200 trp1-Δ901 lys2–801 suc2-Δ9 GAL ypt7Δ::HIS3) were isolated and characterized previously (Harding et al., 1995; Mizushima et al., 1998; Wurmser and Emr, 1998, respectively).
The apg7Δ disruption strain VDY101 (MATα leu2–3112 ura3–52 his3-Δ200 trp1-Δ901 lys2–801 suc2-Δ9 GAL apg7Δ::LEU2) was generated by one-step disruption of strain SEY6210 in this study. The apg5Δ strain, MGY101 (MATα leu2–3112 ura3–52 his3-Δ200 trp1-Δ901 lys2–801 suc2-Δ9 GAL apg5Δ::LEU2) was provided by Michael George (George and Klionsky, manuscript in preparation).
Yeast strains were grown in synthetic minimal medium (SMD; 0.067% yeast nitrogen base, 2% glucose, and auxotrophic amino acids and vitamins as needed) for immunoblotting, radiolabeling, and immunoprecipitation. YPD (containing 1% yeast extract, 2% peptone, and 2% glucose) was used for cell growth before transformation. Synthetic minimal medium containing 2% glucose without ammonium sulfate or amino acids was used for nitrogen starvation experiments (SD-N).
Materials
To prepare antiserum to Apg7p, synthetic peptides corresponding to amino acids 427–443 and 606–630 of Apg7p were synthesized and conjugated individually to keyhole limpet hemocyanin (Multiple Peptide Systems; San Diego, CA). Standard procedures were used to generate antiserum in a New Zealand white rabbit. Antisera against CPY and API were prepared as described previously (Klionsky et al., 1992, 1988, respectively). Antiserum against phosphoglycerate kinase was provided by Dr. Jeremy Thorner (Baum et al., 1978).
All restriction enzymes and Vent DNA polymerase were from New England Biolabs (Beverly, MA). Expre35S35S Protein Labeling Mix was from Dupont-NEN Research Products (Boston, MA). Immobilon-P (polyvinylidene fluoride) was from Millipore (Bedford, MA). Con A Sepharose was from Pharmacia (Piscataway, NJ). Complete EDTA-free protease inhibitor cocktail was from Boehringer Mannheim Biochemicals (Indianapolis, IN). Oxalyticase was from Enzogenetics (Corvallis, OR). The Vistra ECF Western Blotting System was obtained from Amersham (Arlington Heights, IL). YNB was from Difco (Detroit, MI). YNB-copper was from BIO 101 (Vista, CA). Oligonucleotides to pBR322 were from Promega (Madison, WI); all other oligonucleotides were synthesized by Operon Technologies (Alameda, CA). Vectors for expressing GFP fusions were a gift from Dr. Jodi Nunnari (University of California, Davis). The copper-inducible promoter-based vectors, pCu416 and pCu426, were gifts from Dr. Dennis J. Thiele (Labbe and Thiele, in press). The Pichia pastoris plasmid pYM8 containing the GSA7 open reading frame (ORF) was a gift from Dr. William A. Dunn, Jr. (University of Florida College of Medicine, Gainesville). All other reagents were from Sigma (St. Louis, MO).
Cloning APG7 by Nitrogen Starvation
The cvt2/apg7 strain (Harding et al., 1995) was transformed with a yeast genomic plasmid library (Rose et al., 1987), and the transformants were selected on SMD-ura plates. The transformants were then pooled, resuspended in SMD-ura at a concentration of 0.5 OD600/ml, and allowed to double before being transferred to SD-N media for 17 d. Survivors were plated onto SMD-ura plates and then screened by Western blot for the complementation of API maturation defect as described previously (Harding et al., 1995). The complementing plasmid was recovered and sequencing (University of California, Davis, Division of Biological Sciences Automated DNA Sequencing Facility) was performed using pBR322 oligonucleotides. Partial sequences were entered in the Saccharomyces Genome Database (SGD; http://genome-www.stanford.edu/Saccharomyces/), which identified a region of DNA on chromosome VIII. Subsequent subclonings identified a 4.3-kilobase (kb) SacII/KpnI fragment that complemented the API maturation defect. This fragment contained one continuous ORF, YHR171W, corresponding to the recently identified gene APG7 (Mizushima et al., 1998). The pRS414 plasmid containing the APG7 sequence was named pAPG7(414). The multicopy plasmid used to overexpress Apg7p, pAPG7(424), was constructed by subcloning a 3.1-kb SacI/BamHI fragment from pAPG7(414) into the SacI/BamHI sites of pRS424. The SacI/BamHI APG7-containing fragment was also subcloned into pRS416 (pAPG7(416)) and pRS426 (pAPG7(426)). All of the APG7 plasmids complemented the API import defect by immunoblot analysis.
Disruption of the APG7 Chromosomal Locus
To disrupt the APG7 chromosomal locus, pAPG7(414) was digested with XhoI/SpeI to remove a 694-base pair (bp) fragment from the APG7 gene. A XhoI/XbaI fragment containing the LEU2 gene was isolated from the plasmid pJLS2 and ligated with the XhoI/SpeI-digested pAPG7(414) to generate pAPG7Δ::LEU2. Primers to the flanking sequences of the APG7 gene (5′-CACCCGCGGAATCTCAGCAG-3′, 5′-CGAACTTAAAACGTATTGATTGAGGGCCCG-3′) were synthesized, and the linear knockout construct was amplified by PCR. The PCR product was used to transform the yeast strain SEY6210. Transformants were selected on SMD-leu plates, and the apg7Δ knockout strain was identified by the accumulation of prAPI. Tetrad analysis was performed to confirm that the deleted APG7 gene maps to the same locus as the original cvt2/apg7 mutant: the apg7Δ strain was transformed with the pAPG7(416) plasmid and crossed to both alleles of cvt2/apg7 (THY193a cvt2–1 and THY226a cvt2–2). Diploids were sporulated, and the resulting 31 tetrads were dissected. Isolated tetrads were plated onto 5-Fluoro-orotic acid plates to cure the germinants of the pAPG7(416) plasmid. Colonies were then screened for the API maturation defect. All cured germinants showed a precursor API phenotype indicating that the APG7 gene maps to the correct locus.
Construction of Apg7 Fusions with GFP
To construct the pAPG7GFP centromeric and multicopy plasmids, a cassette containing the GFP ORF and actin termination sequences was first removed from pRS305Mip1-GFP using a BamHI/HindIII digest and subcloned into pRS416 and pRS426, resulting in pCGFP(416) and pCGFP(426). The BamHI site was in frame with the GFP ORF. The APG7 gene, including the 363-bp upstream sequence, was then PCR amplified from the pAPG7(414) template using oligonucleotides that incorporated a NotI site on the 5′-primer and an in-frame BamHI site on the 3′-primer (5′-GGAGTCGAGAACGCGGCCGCTGAATCTCAG-3′, 5′-GCAATCTCATCGGATCCATCATCTTCCC-3′). The PCR product was subcloned into pCGFP(416) and pCGFP(426), resulting in pAPG7GFP(416) and pAPG7GFP(426). To test for function, the fusion plasmids were transformed into the apg7Δ strain and examined for the rescue of the API import defect.
Construction of APG7 and GSA7 under the CUP1 Copper-inducible Promoter
The APG7 ORF was PCR amplified from pAPG7(414) using oligonucleotides that incorporated a BamHI site on the 5′-primer and a SalI site on the 3′-primer (5′-GAGGATCCAGAATAAAATGTCGTCAG-3′,5′-GTGAGTAAAGTCAAGAATTTGTCGACTTG-3′). The PCR product was digested with BamHI/SalI and subcloned into the BamHI/SalI sites in pCu416 and pCu426 (centromeric and multicopy-pRS–based vectors containing the CUP1 copper-inducible promoter; Labbe and Thiele, in press), resulting in pCu416APG7 and pCu426APG7, respectively.
The GSA7 ORF was PCR amplified from P. pastoris plasmid pYM8 containing the GSA7 ORF (Yuan et al., 1999). The oligonucleotides used in the PCR contained an engineered upstream BamHI site on the 5′-primer and a downstream SalI site on the 3′-primer (5′-GTCTAACCTTTAGAAGGATCCTTCCCCCAC-3′, 5′-GAGAGAAGGAGAGCAGTCGACCACTAATAAAGAG-3′). The PCR product was digested with BamHI/SalI and subcloned into the BamHI/SalI sites in pCu416 and pCu426, resulting in pCu416GSA7 and pCu426GSA7.
Cell Viability under Nitrogen Starvation Conditions
To examine the survival of various yeast strains under nitrogen starvation conditions, cells were grown to an OD600 = 1 in SMD, washed in SD-N, and then resuspended in SD-N to an OD600 = 1. For the analysis of the apg7Δ strain transformed with the APG7 and GSA7 genes under the regulable control of the CUP1 copper-inducible promoter, cells were grown in SMD containing 100 μM CuSO4 to an OD600 = 1 before shift into SD-N medium. At the indicated times, aliquots were removed and plated onto YPD plates in triplicate. Colonies that survived the nitrogen starvation regimen were counted after 2–3 d.
Cell Labeling and Subcellular Fractionations
For cell-labeling experiments, cells were grown to an OD600 = 1 and resuspended in SMD at 20–30 OD600/ml. The resuspended cells were labeled with 10–20 μCi of 35S Express label/OD600 for the indicated times followed by a chase reaction in SMD supplemented with 0.2% yeast extract, 4 mM methionine, and 2 mM cysteine at a final cell density of 1 OD600/ml. The labeled cells were precipitated with 10% trichloroacetic acid (TCA) on ice, followed by two acetone washes. Crude extracts were prepared by glass-bead lysis, as described previously (Harding et al., 1995).
To biochemically examine the subcellular distribution of Apg7p under vegetative growth and nitrogen starvation conditions, appropriate yeast strains were transformed with the multicopy plasmid of APG7 [pAPG7(426)] and grown to OD600 = 1 in SMD or subsequently transferred into SD-N medium for 15 h. The cells were then converted into spheroplasts and subjected to subcellular fractionation procedures. Spheroplasts were prepared using a modification of a previously described protocol (Kim et al., 1997). In brief, cells were incubated for 15 min at 30°C in a buffer of 0.1 M Tris-SO4, pH 9.4, and 10–20 mM DTT. After a 5000 × g spin for 5 min, the cells were resuspended in an osmotically supportive spheroplasting medium (1 M sorbitol, 50 mM sodium phosphate, pH 7.4, in SMD or SD-N, depending on the growth media used) containing 1–5 μg/OD600 of oxalyticase and incubated for 30 min at 30°C. The spheroplasts were then pelleted at 1500 × g for 5 min and lysed by pipeting in a physiological salts buffer (100 mM KOAc, 50 mM KCl, 5 mM MgCl2, 20 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), pH 6.8) containing a cocktail of protease inhibitors at a spheroplast density of 20 OD600/ml in lysis buffer. Unlysed spheroplasts were removed by a centrifugation step at 500 × g for 5 min at 4°C. The precleared total lysate (T) was subjected to centrifugation at 13,000 × g for 10 min at 4°C and separated into supernatant and pellet fractions (S13 and P13, respectively). The S13 fraction was then subjected to centrifugation at 100,000 × g for 30 min at 4°C and separated into high-speed supernatant and pellet fractions (S100 and P100, respectively). Aliquots of the T, S13, P13, S100, and P100 fractions were TCA precipitated, washed in acetone, and subjected to SDS-PAGE and immunoblotting analysis.
The immunoblot procedure has been described previously (Oda et al., 1996). For quantitation analysis, the immunoblotting procedure was modified, and immunodetection was executed using a Vistra ECF chemifluorescent substrate as described previously (Kim et al., 1997). The procedures to test for the presence of carbohydrate using tunicamycin treatment and Con A Sepharose precipitations were followed exactly as previously described (Klionsky et al., 1992). All samples from radiolabeling and quantitative immunoblot experiments were examined using the Molecular Dynamics Storm System equipped with both phosphorscreen and blue fluorescence/chemifluorescence scanners (Molecular Dynamics, Sunnyvale, CA).
Membrane Flotation Analysis
The procedure for the membrane flotation experiments is a modification of a previously described protocol (Babst et al., 1997). Spheroplasts from the apg7Δ strain were lysed in an osmotic lysis buffer (20 mM PIPES, pH 6.8, 200 mM sorbitol, 5 mM MgCl2, Complete EDTA-free protease inhibitor cocktail) on ice at a spheroplast density of 20 OD600/ml. The lysate from 16 OD600 cell equivalents was subjected to a 5000 × g centrifugation for 5 min at 4°C, resulting in low-speed supernatant (S5) and pellet (P5) fractions. The P5 fractions, which contain all of the precursor API, were resuspended in 300 μl of 60% sucrose (wt/vol) in gradient buffer (GB; 20 mM PIPES, pH 6.8, 5 mM MgCl2, Complete EDTA-free protease inhibitor cocktail) with or without the addition of 1% Triton X-100. The resuspended P5 fractions were overlaid with 900 μl of 55% sucrose in GB and then 900 μl of 35% sucrose in GB. The resulting step gradients were subjected to centrifugation at 100,000 × g for 60 min at 4°C. Fractions were collected from the top. The first 1.2 ml was the float fraction, the remaining 900 μl was the nonfloat fraction, and the gradient pellet was considered the pellet fraction. The resulting fractions were TCA precipitated and washed twice with acetone before being subjected to immunoblot analysis with anti-API antiserum.
Protease Sensitivity Analysis
To examine the protease sensitivity of API in the apg7Δ and ypt7Δ strains, spheroplasts were lysed in the osmotic lysis buffer at a spheroplast density of 20 OD600/ml. The resulting lysate (20 OD600 cell equivalents per incubation condition) was separated into S5 supernatant and P5 pellet fractions by centrifugation at 5000 × g for 5 min at 4°C. The P5 pellet was resuspended in osmotic lysis buffer in the presence or absence of proteinase K (50 μg/ml) and 0.2% Triton X-100. The resuspended P5 pellets were incubated on ice for 30 min, followed by TCA precipitation, acetone wash, and immunoblot analysis with anti-API antiserum.
Fluorescence Microscopy
Strains with Apg7pGFP fusion protein were grown to midlog stage in SMD medium and shifted to SD-N for 8–20 h. Cells were examined on a Leica DM IRB confocal microscope (Leica, Deerfield, IL) utilizing a 410- to 425-nm band pass filter. The images captured were the result of an average of eight scans of a single focal plane. For analysis of fluorescent cell populations (Table 1), a Nikon Axiophot epifluorescence microscope (Nikon, Garden City, NY) was used.
Table 1.
Apg7pGFP displays an Apg12p-dependent punctate staining pattern upon shift to nitrogen starvation conditions
| Time in SD-N (h) | Strain | Total cells counted | Fluorescent cells detected | % Fluorescent cellsa | Cells with punctate staining | % Cells with punctate stainingb |
|---|---|---|---|---|---|---|
| 0 | Wild-type | 291; 406 | 130; 160 | 42 ± 4 | 0; 0 | 0 ± 0 |
| 4 | Wild-type | 368; 451 | 136; 203 | 41 ± 6 | 30; 49 | 23 ± 1 |
| 15 | Wild-type | 633; 597 | 293; 225 | 42 ± 6 | 122; 78 | 38 ± 5 |
| 0 | apg5Δ | 211; 436 | 138; 272 | 64 ± 2 | 0; 0 | 0 ± 0 |
| 4 | apg5Δ | 233; 327 | 139; 184 | 58 ± 2 | 33; 41 | 23 ± 1 |
| 15 | apg5Δ | 465; 670 | 311; 421 | 65 ± 3 | 220; 310 | 72 ± 2 |
| 0 | apg12Δ | 293; 603 | 154; 362 | 56 ± 5 | 0; 0 | 0 ± 0 |
| 4 | apg12Δ | 183; 247 | 120; 127 | 58 ± 10 | 3; 0 | 1 ± 2 |
| 15 | apg12Δ | 548; 479 | 337; 297 | 62 ± 1 | 0; 0 | 0 ± 0 |
The percent of total cells counted that were fluorescent.
The percent of fluorescent cells that exhibited punctate structures.
Copper-induced Expression Analysis
To examine the expression of Apg7p and Gsa7p under the regulable control of the CUP1 copper-inducible promoter, the appropriate strains were grown to 0.2 OD600/ml in SMD lacking copper and then induced with 100 mM CuSO4 for 7 h. In the 0 μM CuSO4 cultures, bathocuproinedisulfonic acid copper chelator was added to a final concentration of 100 μM. Cells were then harvested and examined by immunoblot analysis.
RESULTS
Apg7p Is a Component of Both the Macroautophagy and Cvt Pathways
Genetic mutants in the Cvt pathway were originally isolated on the basis of their precursor API accumulation phenotype. Further analysis revealed that the majority of these cvt mutants share a genetic overlap with mutants defective in macroautophagy (Harding et al., 1996; Scott et al., 1996). For example, cvt2 was found to be allelic to apg7 and demonstrated both sensitivity to nitrogen starvation conditions as well as a defect in API transport. Therefore, survival in nitrogen-poor medium offered a convenient initial selection strategy to clone the complementing gene for the cvt2/apg7 mutant. The sequence of the APG7 gene was recently published (Mizushima et al., 1998). Accordingly, we will use the APG7 nomenclature hereafter in this study.
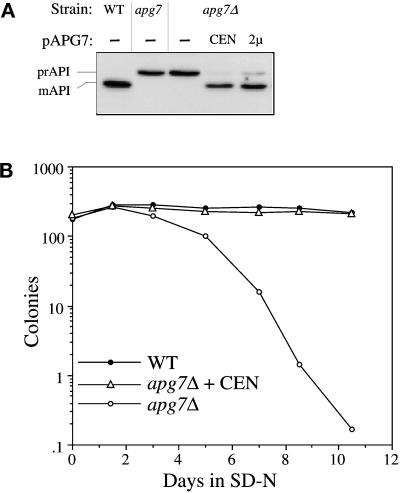
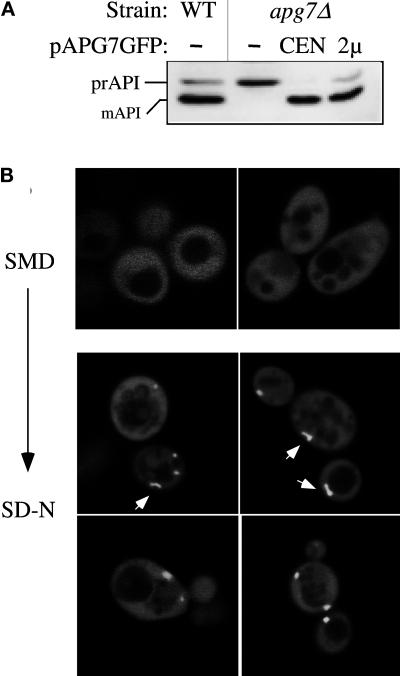
The apg7 mutant was transformed with a yeast genomic library (Rose et al., 1987), grown to early-log phase, and transferred to synthetic minimal medium lacking nitrogen (SD-N) for 17 d. As a secondary screen, transformants that survived the nitrogen starvation regimen were examined by immunoblot analysis for complementation of the precursor API mutant phenotype. The genomic library plasmids from transformants that passed both screenings were isolated. Partial sequences from these complementing genomic plasmids were entered into the Saccharomyces Genome Database (SGD) to obtain full-length sequences, which contained overlapping fragments of 7–8 kb. Subcloning allowed the identification of an ORF that rescued the API-processing defect in the apg7 mutant. A strain in which 37% of the apg7-complementing ORF was deleted and replaced by the LEU2 auxotrophic marker showed the precursor API accumulation phenotype (Figure 2A, lane 3), confirming that Apg7p is required for API import. Subsequent tetrad analysis revealed that the deleted gene mapped to the same locus as the original apg7 mutant gene (our unpublished results). The API-processing defect in apg7Δ was rescued by the APG7 gene on centromeric or multicopy plasmids (Figure 2A). In addition, survival in nitrogen starvation was also restored in the apg7Δ strain transformed with the APG7 centromeric plasmid (Figure 2B), suggesting that Apg7p is a shared component of both macroautophagy and API import by the Cvt pathway.
Figure 2.
Cloning and characterization of APG7. (A) WT (wild-type, SEY6210), apg7 (THY193), apg7Δ (VDY1), and the apg7Δ strain transformed with single copy (CEN, pAPG7(414)) or multicopy (2 μ, pAPG7(424)) plasmids encoding APG7 were grown to log phase in SMD. Protein extracts were prepared and analyzed by immunoblot using antiserum to API as described in MATERIALS AND METHODS. The positions of precursor and mature API are indicated. The APG7 gene complements the precursor API accumulation phenotype of the apg7Δ mutant. (B) WT (wild-type, SEY6210), apg7Δ (VDY1), and the apg7Δ strain transformed with the APG7 centromeric plasmid pAPG7(414) were grown in SMD and transferred to SD-N as described in MATERIALS AND METHODS. Aliquots were removed at the indicated times and spread onto YPD plates in triplicate. Numbers of viable colonies were determined after 2–3 d. The APG7 gene complements the starvation-sensitive phenotype of the apg7Δ mutant.
Precursor API Is Membrane Associated in the apg7Δ Strain
Once synthesized in the cytoplasm, precursor API rapidly forms a dodecamer and assembles into a pelletable Cvt complex (Baba et al., 1997; Kim et al., 1997). Biochemical and electron microscopy studies have demonstrated that the Cvt complex is subsequently enwrapped by a membrane to form the Cvt vesicle (Baba et al., 1997; Scott et al., 1997). Membrane flotation experiments were performed to assess the stage of API import in which accumulated prAPI is blocked in the apg7Δ strain. In this analysis, membrane-associated proteins would be recovered in the float (F) fraction, while soluble proteins would remain in the nonfloat (NF) fraction, and proteins associated with large, protein complexes would be recovered as a gradient pellet (P2).
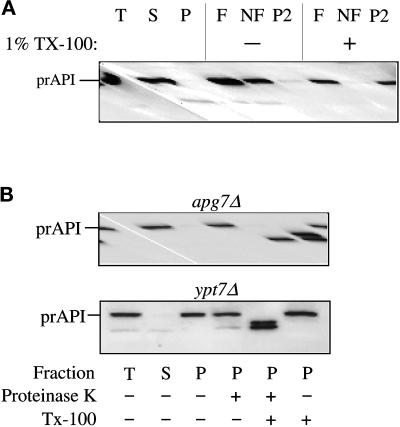
Spheroplasts were prepared from the apg7Δ strain and subjected to differential osmotic lysis, which preserves the membrane integrity of the vacuole while disrupting the plasma membrane. After a 5000 × g low-speed centrifugation step, prAPI was recovered entirely in a pelletable (P) fraction (Figure 3A). This pellet fraction was then subjected to the membrane flotation step gradient. In the apg7Δ strain, the majority of prAPI was found in the float fraction (F) in the absence of detergent, suggesting that it is directly or indirectly membrane-associated (Figure 3). In addition, a large fraction was also found in the P2 pellet fraction, indicating that a significant portion of prAPI is part of a large pelletable complex (Figure 3A). In the presence of detergent, the majority of prAPI was recovered from the P2 pellet with only a minor amount recovered in the soluble protein pool of the nonfloat fraction. These findings suggest that prAPI in apg7Δ is part of a large protein complex that, in part, associates with a membraneous compartment. In addition, prAPI maintains its binding to the protein complex even after the membrane association has been disrupted by detergent treatment.
Figure 3.
The apg7Δ mutant accumulates precursor API in a membrane-associated and protease-sensitive form that is part of a large complex. (A) The apg7Δ strain (VDY1) was grown in SMD to midlog phase and converted to spheroplasts. The spheroplasts were lysed in osmotic lysis buffer (see MATERIALS AND METHODS). An aliquot was removed for a total lysate control (T). The remainder of the lysed spheroplasts were separated into supernatant (S) and pellet (P) fractions by centrifugation at 5000 × g. The pellet fraction was resuspended in 60% sucrose in GB in the presence or absence of Triton X-100 and overlaid with 55 and 35% sucrose in GB. The step gradients were centrifuged at 100,000 × g for 60 min. Membrane-containing float (F), nonfloat (NF), and pellet (P2) fractions were collected and subjected to immunoblot analysis with antiserum to API as described in MATERIALS AND METHODS. The position of precursor API is indicated. (B) Precursor API in apg7Δ is protease accessible. Spheroplasts isolated from apg7Δ (VDY1) and ypt7Δ (WSY99) cells were lysed in osmotic lysis buffer. Supernatant (S) and pellet (P) fractions after a 5000 × g centrifugation were collected, and the pellet fractions were subjected to protease treatment in the absence or presence of 0.2% Triton X-100 as described in MATERIALS AND METHODS. The resulting samples were subjected to immunoblot analysis with antibody against API.
Apg7p Acts at the Step of Vesicle Formation
In the apg7Δ strain, the precursor form of API accumulates in a large pelletable complex that associates with a membrane. We next wanted to examine whether the accumulated prAPI was accessible to protease treatment. If Apg7p is required for the completion of vesicle formation (Figure 1, step 4), then prAPI would be sensitive to exogenously added proteases and processed to the mature form in the apg7Δ strain. Alternatively, if Apg7p functions at a downstream event after the completion of the enwrapping membrane, then prAPI would remain in a protease-protected precursor state. This protease-protected phenotype is observed in mutants blocked in fusion of vesicles with the vacuole, such as vps18 ts (Scott et al., 1997), vam3 ts (Darsow et al., 1997) and ypt7Δ. Ypt7p is a rab guanosine triphosphatase required for homotypic vacuole fusion (Haas et al., 1995). In addition, the ypt7Δ strain displays a strong block in the maturation of API in both rich and starvation conditions, suggesting that it is required for both the delivery of Cvt and macroautophagic vesicles to the vacuole (our unpublished results).
To investigate whether Apg7p functions in the vesicle formation step of API targeting, a protease-protection assay was performed. Protease-accessible precursor API is usually digested to the mature form by exogenous protease. This presumably reflects the protease-resistant nature of mature API as a resident vacuolar hydrolase. Spheroplasts from both the apg7Δ and ypt7Δ strains were lysed osmotically and separated into low-speed supernatant and pellet fractions. In both strains, prAPI was recovered in the membrane-associated pellet fraction (Figure 3B). When the pellet fraction was subjected to proteinase K digestion in the apg7Δ strain, prAPI was completely sensitive to proteolysis (Figure 3B), independent of detergent addition, suggesting that the surrounding membrane had not completely formed. However, prAPI in the ypt7Δ strain was protected from exogenously added proteinase K and was only protease accessible after the addition of detergent (Figure 3B). In both strains, detergent treatment in the absence of protease did not alter the prAPI pattern (Figure 3B). These findings suggest that Apg7p is required for the complete formation of the vesicle around the prAPI protein complex (Figure 1, step 4), while Ypt7p acts at a downstream event after vesicle formation has been completed (Figure 1, step 5).
Apg7p Biosynthesis
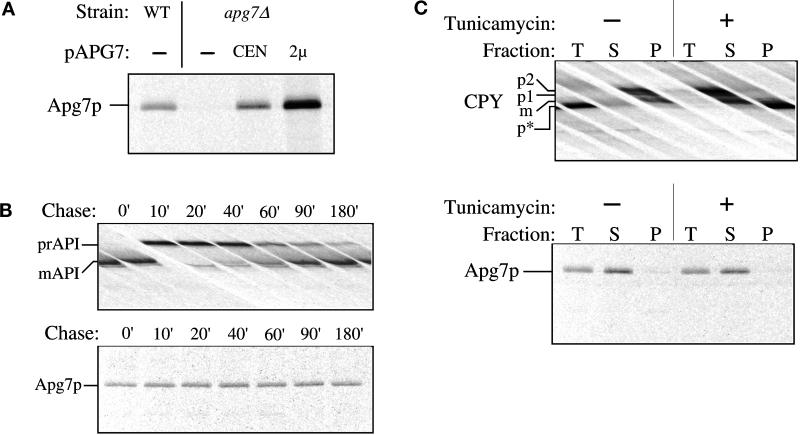
A previous study revealed that Apg7p shows significant homology with E1 ubiquitin-activating enzymes in S. cerevisiae as well as in other species (Mizushima et al., 1998; Tanida et al., 1999). The deduced amino acid sequence of Apg7p predicts a 630-amino acid protein with a molecular mass of 71.4 kDa. To further characterize Apg7p, antiserum to the protein was raised against two synthetic peptides. To examine the expression of Apg7p, wild-type and apg7Δ strains harboring the centromeric or multicopy APG7 plasmid were radiolabeled for 10 min. After cell lysis, the extracts were subjected to immunoprecipitation with the anti-Apg7p antibody (Figure 4A). Immunoprecipitated Apg7p migrates at the predicted size of 71 kDa by SDS-PAGE. The 71-kDa band is absent in the apg7Δ strain (Figure 4A) and shows a dose-dependent increase in level in strains bearing plasmids that encode the APG7 gene (Figure 4A). These data suggest that the 71-kDa band corresponds to Apg7p.
Figure 4.
Apg7p is not glycosylated or proteolytically modified. (A) WT (wild-type, SEY6210), apg7Δ (VDY1) and the apg7Δ strain transformed with single-copy (CEN) or multicopy (2 μ) plasmids encoding APG7 were grown to log phase in SMD. Cells were labeled for 10 min and immunoprecipitated with antiserum to Apg7p and analyzed by SDS-PAGE as described in MATERIALS AND METHODS. Apg7p is detected as a 71-kDa protein. (B) Wild-type (SEY6210) cells were radiolabeled for 10 min and subjected to a nonradioactive chase. At the indicated time points an aliquot was removed and precipitated with TCA. Protein extracts were prepared and successively immunoprecipitated with antiserum to API and Apg7p and analyzed as above. The positions of precursor and mature API and of Apg7p are indicated. The absence of a molecular mass shift indicates that Apg7p is not proteolytically modified. (C) Wild-type (SEY6210) cells were converted to spheroplasts and treated with tunicamycin (final concentration 20 μg/ml) to inhibit glycosylation 15 min before the addition of radioactive label as indicated. Labeling was allowed to continue for 20 min. Samples were TCA precipitated and divided in half. One-half was immunoprecipitated immediately with antiserum to CPY or Apg7p for a total (T) control. The remaining half was precipitated with Con A-Sepharose and separated into supernatant (S, not bound to Con A) and pellet (P, bound to Con A) fractions as referenced in MATERIALS AND METHODS. The separate fractions were then immunoprecipitated with antiserum to CPY or Apg7p. The positions of precursor and mature forms of glycosylated CPY (p1, p2, m) and unglycosylated precursor CPY (p*) and of Apg7p are shown. Apg7p does not bind Con A, suggesting that it is not glycosylated.
To determine whether Apg7p undergoes proteolytic processing, a detailed pulse/chase analysis was performed (Figure 4B). Wild-type cells were radiolabeled for 10 min, followed by a nonradioactive chase. Immunoprecipitation analysis under reducing conditions indicated that Apg7p does not undergo any apparent proteolytic processing after synthesis (Figure 4B, bottom panel). As a control, we examined API, which was shown to be processed from the 61-kDa precursor form to the 50-kDa mature enzyme with a half-time of 30 min (Figure 4B, top panel).
Sequence analysis of Apg7p revealed several potential sites for the addition of N-linked oligosaccharides. Addition of carbohydrate moieties would result in an increase in molecular mass that should be detectable by SDS-PAGE. However, Apg7p did not show a glycosylation-dependent modification over a 3-h time course based on this criterion (Figure 4B). To confirm that Apg7p is not glycosylated, we tested for the presence of N-linked and O-linked oligosaccharides through the use of tunicamycin and Con A (concanavalin A) (Figure 4C). Tunicamycin-treated N-linked glycoproteins migrate as a lower-molecular-mass species by SDS-PAGE while Con A lectin binds to both N- and O-linked mannose-containing oligosaccharides.
Wild-type cells were incubated for 15 min in the presence or absence of tunicamycin before radiolabeling for 20 min. Half of the labeled samples were directly immunoprecipitated (total) with anti-Apg7p and carboxypeptidase Y (CPY) antibodies while the remaining sample was treated with Con A Sepharose and separated into Con A-precipitable (pellet, P) and nonprecipitable (supernatant, S) fractions before immunoprecipitation reactions. The wild-type vacuolar hydrolase CPY binds Con A in the absence of tunicamycin, consistent with its being glycosylated. However, in the presence of tunicamycin, CPY is not glycosylated and migrates as a lower molecular mass species that does not bind Con A and appears in the Con A supernatant fraction (Figure 4C, top panel). In contrast, the molecular mass of Apg7p remains unchanged in the presence or absence of tunicamycin. Similarly, under both conditions, Apg7p fails to bind Con A and remains in the Con A supernatant fraction (Figure 4C, bottom panel). These results suggest that Apg7p does not contain N- or O-linked oligosaccharides.
In Vivo Localization of an Apg7pGFP Fusion Protein Reveals Association with a Membrane Structure
A recent study by Ohsumi and colleagues demonstrated that Apg7p function is required for the conjugation of Apg12p to Apg5p, a reaction that is essential for macroautophagy to occur (Mizushima et al., 1998). In addition, most of the Apg5p and Apg5p-Apg12p conjugate and more than half of the free Apg12p were present in 100,000 × g pellet fractions, suggesting that they associate with some membrane compartment. Therefore, in order for Apg7p to function in the conjugation reaction of Apg5p and Apg12p, we postulated that the site of its function would be on a membrane compartment. Alternatively, the Apg7p-mediated conjugation reaction may occur in the cytosol before membrane binding.
To investigate the subcellular location of Apg7p, we constructed a fusion of GFP to the C terminus of Apg7p and examined the localization by fluorescence microscopy. The advantage of this approach was that the localization of Apg7p could be examined in vivo. The plasmids expressing the Apg7pGFP fusion, pAPG7GFP(416) and pAPG7GFP(426), complement the API- processing defect in the apg7Δ strain, suggesting that the fusion proteins are functional (Figure 5A). Unfortunately, the level of fluorescence from the centromeric plasmid was below practical levels of detection. apg7Δ cells harboring the multicopy pAPG7GFP plasmid were grown to midlog in SMD and then shifted to SD-N medium for 15 h and visualized by confocal microscopy. Under vegetative growth conditions, the Apg7pGFP fusion protein is distributed uniformly in the cytoplasm (Figure 5B). In contrast, the Apg7pGFP fusion in cells shifted to SD-N showed both a diffuse cytoplasmic population as well as an intense punctate localization (Figure 5B). Generally found in, but not restricted to, the perivacuolar region, these punctate structures were generally circular in shape, but rod-like structures were also detected as indicated by arrows in Figure 5B. The vacuoles in cells grown in both SMD and SD-N can be seen as areas in which fluorescence is excluded. The staining pattern of Apg7p in the wild-type background appeared to be identical to the pattern in the apg7Δ background (our unpublished results). The shift in the Apg7pGFP localization to punctate structures under nitrogen-starvation conditions suggests that a detectable portion of Apg7p is associated with membrane structures when macroautophagy is induced.
Figure 5.
Apg7pGFP membrane association in vivo. (A) WT (wild-type, SEY6210), apg7Δ (VDY1), and the apg7Δ strain transformed with single-copy (CEN) or multicopy (2μ) plasmids encoding Apg7p-GFP were grown to log phase in SMD. Protein extracts were prepared and analyzed by Western blot using antiserum to API as described in MATERIALS AND METHODS. The positions of precursor and mature API are indicated. The Apg7pGFP hybrid protein complements the precursor API accumulation phenotype of the apg7Δ strain, indicating that the protein retains Apg7p function. (B) The apg7Δ strain transformed with the multicopy pAPG7GFP plasmid was grown in SMD to log phase and shifted to SD-N for 15 h. Cells from the SMD and SD-N cultures were examined by fluorescence microscopy as described in MATERIALS AND METHODS. Apg7pGFP displays primarily a diffuse cytosolic staining, but punctate structures are detected in SD-N medium. Some of the punctate structures appear rod-like in shape as indicated by arrows.
Subcellular Fractionation of Apg7p
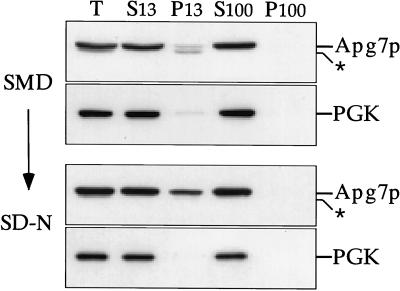
The analysis of Apg7pGFP suggested that we could detect a population of Apg7p associated with a membrane fraction under starvation conditions. We next performed subcellular fractionation analyses under both vegetative growth and nitrogen-starvation conditions to determine whether this observation could be confirmed biochemically. Wild-type cells transformed with the APG7 multicopy plasmid were grown to OD600 = 1 in SMD medium and subsequently shifted to nitrogen-lacking SD-N medium for 15 h. Cells from both SMD and SD-N cultures were converted into spheroplasts and subjected to differential osmotic lysis as described in MATERIALS AND METHODS.
Immunoblot analysis of the subcellular fractions indicated that under vegetative growth conditions, Apg7p was predominantly localized in the S13 fraction with only a minor Apg7p band detected in the P13 fraction (Figure 6). When the S13 sample was further separated into high-speed supernatant and pellet fractions, Apg7p remained in the soluble S100 pool. This observation is consistent with the diffuse cytosolic staining seen with the Apg7pGFP fusion protein in vegetatively growing cells (Figure 5), and the findings of Tanida et al. (1999), indicating that Apg7p is a cytosolic protein under vegetative conditions. In contrast, the fractionation pattern of cells shifted to SD-N for 15 h indicated that a significant portion of the Apg7p cytosolic pool now accumulated in the P13 pellet (Figure 6). Quantitation of the S13 and P13 fractions revealed that there was an increase, from 5.3% ± 0.4% to 25.2% ± 0.6%, in the amount of Apg7p found in the P13 pellet when cells were shifted from vegetative growth to nitrogen starvation conditions. The accumulation of Apg7p in the P13 fraction under SD-N conditions is consistent with the Apg7pGFP punctate localization pattern observed when cells were starved for nitrogen.
Figure 6.
Apg7p subcellular fractionation pattern in SMD and SD-N. Cells from the wild-type strain (SEY6210) transformed with the multicopy plasmid encoding APG7 were grown in SMD to midlog phase and shifted to SD-N medium for 15 h before converting them to spheroplasts. The spheroplasts were then lysed osmotically in a physiological salts buffer. After a preclearing centrifugation step at 500 × g for 5 min to remove unlysed spheroplasts, the total lysate (T) was separated into 13,000 × g supernatant (S13) and pellet (P13) fractions. The S13 fraction was further separated into 100,000 × g supernatant (S100) and pellet (P100) fractions. The T, S13, P13, S100, and P100 subcellular fractions were subjected to immunoblot analysis using antiserum to Apg7p and phosphoglycerate kinase (a cytosolic marker protein). A background band (*) appears below the signal for Apg7p.
Apg7pGFP Localization to Punctate Structures Is Dependent on Apg12p
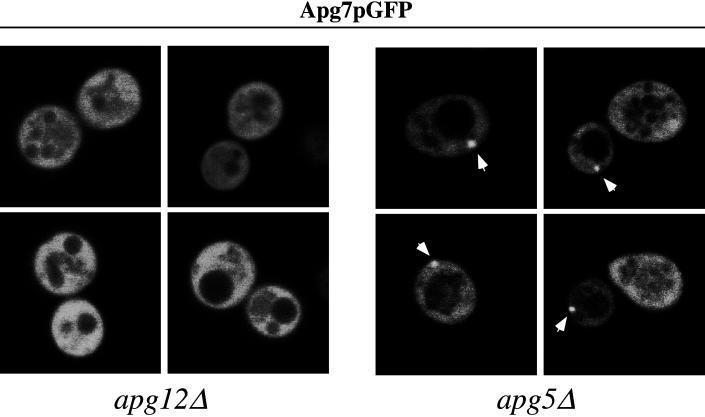
We decided to extend our analysis of Apg7p localization by examining the fluorescence pattern of the Apg7pGFP fusion protein in various mutant strains. Previous data (Mizushima et al., 1998) and the studies by Yuan et al. (1999) and Tanida et al. (1999) indicate that Apg12p is conjugated to Apg7p via a thioester bond in a reaction analogous to the conjugation of ubiquitin to E1 enzymes. Furthermore, because more than half of Apg12p can be found as a membrane-associated species that pellets after a 100,000 × g centrifugation (Mizushima et al., 1998), we hypothesized that the membrane association of Apg7pGFP may be dependent, in part, on Apg12p, its transient substrate for the conjugation reaction. To examine this possibility, Apg7pGFP was expressed in the apg12Δ strain and examined under vegetative and nitrogen starvation conditions. As controls, Apg7pGFP was also introduced into the wild-type and apg5Δ backgrounds.
The wild-type, apg12Δ, and apg5Δ strains harboring the multicopy pAPG7GFP plasmid were grown to midlog phase in SMD and then examined in SD-N over a period of 15 h (Table 1 and Figure 7). Under vegetative growth conditions, Apg7pGFP was uniformly distributed in the cytoplasm in both the apg12Δ and apg5Δ strains (our unpublished results), consistent with its localization pattern in the wild-type strain background. However, in contrast to the punctate localization of Apg7pGFP observed under nitrogen starvation conditions in the wild-type background, Apg7pGFP was unable to form these punctate structures in the apg12Δ strain even after shifting to SD-N. Instead, Apg7pGFP retained its uniform, cytosolic pattern (Figure 7, left panels; and Table 1), reminiscent of the Apg7pGFP distribution pattern under vegetative growth conditions. Apg7pGFP in the apg5Δ background maintained its punctate localization pattern in SD-N, similar to that seen in the wild-type strain. There was a qualitative difference, however, in the staining pattern of Apg7pGFP in the wild-type and apg5Δ backgrounds. Whereas the wild-type punctate pattern exhibited both rod-like and circular structures (Figure 5), only the latter punctate pattern could be detected in the apg5Δ background (Figure 7). The number of fluorescent cells exhibiting punctate structures in both the wild-type and apg5Δ background strains increased over time under starvation conditions while the total number of fluorescent cells remained constant over 15 h in SD-N (Table 1). These results suggest that the punctate localization of Apg7pGFP, which presumably represents its membrane associated form in vivo, is dependent on Apg12p but not Apg5p. Furthermore, a higher percentage of apg5Δ cells relative to wild-type displayed a punctate staining pattern after 15 h in SD-N (Table 1). This may reflect an accumulation of the Apg7p-Apg12p conjugate in the absence of Apg5p.
Figure 7.
Membrane association of Apg7pGFP is dependent on Apg12p. The apg5Δ (MGY101) and apg12Δ (YNM101) strains transformed with the multicopy pAPG7GFP(426) plasmid were analyzed by fluorescence microscopy following transfer to SD-N as described in the legend to Figure 5. The punctate staining pattern of Apg7pGFP, marked by arrows, is dependent on the Apg12 protein.
GSA7, the P. pastoris Homologue to APG7, Partially Complements the API-Import Defect and the Starvation-sensitive Phenotype of apg7Δ
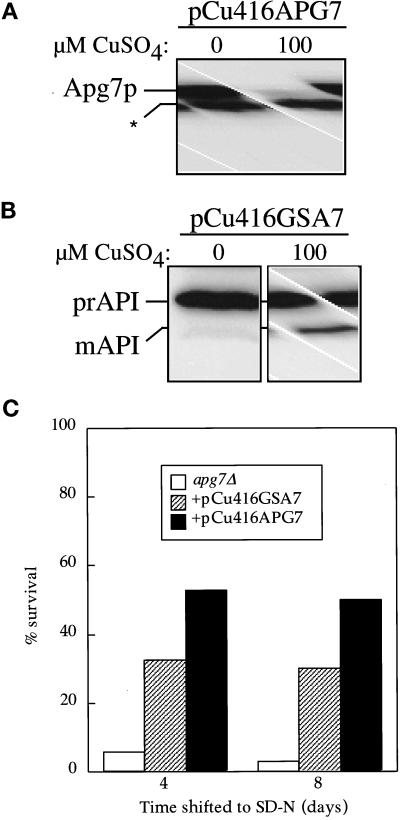
In media containing methanol, the methylotrophic yeast P. pastoris proliferates peroxisomes and peroxisomal proteins. Upon shift of the cells to media containing glucose, excess peroxisomes are specifically degraded in the vacuole via micropexophagy (Tuttle et al., 1993). The GSA7 gene is essential to this glucose-stimulated micropexophagy pathway (Yuan et al., 1999). Sequence analysis indicated that GSA7 was the P. pastoris homologue of APG7. To determine whether Gsa7p could functionally substitute for Apg7p, we transformed S. cerevisiae apg7Δ cells with the P. pastoris GSA7 gene under the control of a regulable promoter.
The GSA7 and APG7 genes were cloned behind the CUP1 promoter as described in MATERIALS AND METHODS. The apg7Δ strain harboring either plasmid was grown in SMD lacking copper to 0.2 OD600/ml and then induced for 7 h with 100 μM CuSO4. Cells were then harvested and examined by immunoblot analysis. Regulation of expression was monitored by immunodetection of Apg7p (Figure 8A). The CUP1 promoter is leaky so that a low level of Apg7p is synthesized even in the presence of the copper chelator. This amount of Apg7p is similar to the level from expression at the chromosomal locus and complements the prAPI phenotype of the apg7Δ strain (our unpublished results). Induction in the presence of copper results in a dramatic increase in the level of Apg7p from the pCu416APG7 plasmid (Figure 8A).
Figure 8.
The Pichia pastoris GSA7 gene partially complements the apg7 defect. (A) The apg7Δ strain (VDY1) was transformed with a plasmid encoding the APG7 gene cloned behind the regulable CUP1 promoter (pCu416APG7). Cells were grown in YNB minus copper containing either 100 μM BCS copper chelator or 100 μM copper sulfate. After 7 h, cells were TCA precipitated, and the protein extract was analyzed by immunoblot using antiserum to Apg7p. Induction by copper results in a significant increase in Apg7p synthesis. A background band has been indicated by an asterisk (*). (B) The apg7Δ strain was transformed with a plasmid encoding the GSA7 gene cloned behind the regulable CUP1 promoter (pCu416GSA7). Cells were grown as above and analyzed by immunoblot with antiserum to API. The GSA7 gene partially complements the apg7 defect. (C) The apg7Δ strain transformed with the pCu416GSA7 or the pCu416APG7 plasmid was grown in YNB minus copper containing 100 μM coppper sulfate and transferred to SD-N as described in MATERIALS AND METHODS. Aliquots were removed at the indicated times and spread onto YPD plates in triplicate. Numbers of viable colonies were determined after 2–3 d. The pCu416GSA7 plasmid partially complements the starvation-sensitive phenotype of the apg7Δ mutant.
The Gsa7 protein was not detectable with antiserum directed against Apg7p, preventing a determination of expression levels. In the absence of copper, apg7Δ cells harboring the pCu416GSA7 plasmid accumulated prAPI. In contrast, upon copper induction this strain showed a low but reproducible level of mature API (Figure 8B). These results indicate that at high levels, the Gsa7 protein can functionally complement the API-import defect of the apg7Δ strain, although only weakly. This is similar to the result seen with expression of APG7 in the P. pastoris gsa7 mutant (Yuan et al., 1999), where the Apg7 protein is able to complement the micropexophagy defect.
Since Gsa7p partially complements the API-import defect of the apg7Δ strain, we further investigated whether Gsa7p would also partially rescue the nitrogen starvation defect of the apg7Δ strain. The apg7Δ strain, transformed with either pCu416GSA7 or pCu416APG7, was grown to OD600=1 in SMD medium containing 100 μM CuSO4 and subsequently shifted to SD-N medium. Survival under nitrogen starvation conditions was measured at the indicated times by plating aliquots onto YPD plates and counting viable colonies after 2–3 d. The results indicated that Gsa7p partially complemented the nitrogen-sensitive phenotype of the apg7Δ strain and demonstrates that Gsa7p can replace Apg7p in Cvt and macroautophagy pathways in S. cerevisiae.
DISCUSSION
In this study we have demonstrated that Apg7p is a shared component of three nonclassical pathways to the yeast vacuole in S. cerevisiae. The APG7 gene complemented the API import defect of the apg7Δ strain under vegetative growth conditions, indicating that it is involved in the Cvt pathway (Figure 2A). In addition, APG7 rescued the nitrogen-starvation defect in the apg7Δ strain, demonstrating its role in macroautophagy (Figure 2B). The APG7 homologue, GSA7, in the methylotrophic yeast P. pastoris, is involved in the specific degradation of peroxisomes by micropexophagy (Yuan et al., 1999). Expression of GSA7 under a regulable promoter in S. cerevisiae partially complemented the API import defect as well as conferring greater viability during starvation conditions in the apg7Δ background (Figure 8). A corresponding role for Apg7p in pexophagy in S. cerevisiae is suggested by the finding that the apg7Δ strain is also defective in peroxisome degradation (Hutchins and Klionsky, manuscript in preparation). Taken together, we have demonstrated for the first time that a component of the recently characterized novel conjugation system functions in three distinct targeting pathways of Cvt, macroautophagy, and micropexophagy.
A small amount of precursor API was reproducibly detected when APG7 was expressed on a multicopy plasmid (Figure 2A). The precursor API accumulation phenotype increased when APG7 was expressed at higher levels under the control of the regulable CUP1 promoter on a multicopy plasmid (our unpublished results). These findings suggest that overexpression of Apg7p may have a saturating effect on other components in the API-import pathway.
The site of accumulated precursor API in the apg7Δ strain was found to be part of a membrane-associated, large protein complex, presumably the Cvt complex, that was pelletable upon detergent treatment (Figure 3A). In addition, prAPI was accessible to exogenously added protease, suggesting that the vesicle around the Cvt complex had not formed completely. These experiments place the action of Apg7p at a step of vesicle formation around the Cvt complex in vegetative conditions or around cytosol during starvation (Figure 1, step 4). Experiments on the biogenesis of Apg7p indicated that this 71-kDa protein was not posttranslationally modified by proteolytic processing or glycosylation (Figure 4). Tanida et al. (1999) demonstrate that Apg12p is conjugated to Apg7p via a thioester modification and that the two proteins can be coimmunoprecipitated. Due to the reducing conditions of our pulse/chase labeling conditions, we did not detect this event.
The subcellular localization of Apg7p was determined biochemically by subcellular fractionations and in vivo through the use of a fusion of Apg7p with GFP. The biochemical fractionations localized Apg7p predominantly to a soluble S100 fraction under vegetative growth conditions, suggesting a cytosolic localization. The shift to starvation conditions resulted in a small but significant amount of Apg7p to be localized to a low-speed P13 pellet (Figure 6). The association of Apg7p with the P13 pellet fractionation was sensitive to extended lysis times; reproducible results were obtained only when the lysis and subsequent centrifugation steps were performed immediately after lysis. We believe this is indicative of a weak interaction of Apg7p with the P13 pellet fraction. The in vivo examination of the Apg7pGFP fusion protein indicated that a portion of the Apg7pGFP was localized to strongly staining punctate structures that could be detected under nitrogen starvation conditions (Figure 5). This punctate localization of Apg7pGFP was dependent on the presence of Apg12p but not Apg5p (Figure 7). Apg12p shows a partially pelletable distribution in the cell (Mizushima et al., 1998). One interpretation of the fluorescence data is that the punctate pattern for Apg7p represents the intermediate Apg7p-Apg12p conjugate. The formation of punctate Apg7pGFP structures under starvation conditions is consistent with the accumulation of a small amount of Apg7p to the P13 pellet fraction in our biochemical fractionation experiments.
The conjugation of Apg12p to Apg7p has been demonstrated to be required for both API import and autophagy by Tanida et al. (1999). Therefore, if the site of action of Apg7p is on the membrane of the punctate structures seen under nitrogen starvation conditions, why aren’t these structures also seen under vegetative growth conditions? This may be due to the limitations of fluorescence microscopy in detecting the significantly smaller size of the vesicles being formed during vegetative growth (Cvt vesicles, 150 nm diameter) versus those that are formed under starvation conditions (autophagosomes, 300–900 nm diameter). In addition, electron microscopy data suggest that the number of Cvt vesicles formed during vegetative growth is substantially lower than the number of autophagosomes that are generated during starvation. Because association of Apg7p with Apg12p is transient, there is a greater likelihood of detecting this interaction in starvation conditions.
The requirement for Apg7p-dependent conjugation in both rich and starvation conditions suggests that conjugation is not a regulatory step. The reactions that occur during conjugation, however, may be regulated by other factors. For example, in micropexophagy, invaginations of the vacuolar membrane or finger-like protrusions from the vacuole are thought to capture the targeted peroxisomes and sequester them inside the vacuole for degradation. In the study by Yuan et al. (1999), electron micrographs show that the micropexophagy defect in the gsa7 strain appears to be in the completion of vacuole membrane fusion around peroxisomes. Accordingly, the site of action of the Gsa7p in P. pastoris appears to be at the vacuolar membrane. In addition, Yuan et al. (1999), indicate that the gsa7 mutant is defective in protein turnover under starvation conditions, suggesting that Gsa7p is also required for macroautophagy in the yeast P. pastoris. Similarly, the proposed site of Apg7p function in S. cerevisiae is at the formation or completion of the autophagosome or Cvt vesicle. This dual function in micropexophagy and macroautophagy may reflect the role of accessory proteins that differentially target Apg7p/Gsa7p and, by extension, the conjugation system, depending on the nutrient conditions. Alternatively, the vacuole membrane may serve as the donor membrane compartment for the formation of the autophagosome and Cvt vesicle via retrograde movement from this organelle. In this case, the apg7 and gsa7 mutants would both be blocked in events involving sequestration by vacuolar membrane.
We have demonstrated that Apg7p functions in three nonclassical targeting pathways to the yeast vacuole. The fact that APG7 homologues exist in higher eukaryotes including humans indicates the function of this novel conjugation system, which includes Apg7p, Apg12p, and Apg5p, may be as a conserved, general targeting mechanism for a variety of nonclassical pathways to the vacuole/lysosome.
ACKNOWLEDGMENTS
We thank Dr. Heiner Matthies and Dawn Signor for assistance with confocal microscopy. This work was supported by a National Institutes of Health Molecular and Cellular Biology training grant to J.K. and K.P.E., a National Science Foundation Plant Cell Biology training grant to V.M.D., an American Cancer Society, California Division Senior Postdoctoral Fellowship to S.V.S., and by Public Health Service grant GM-53396 from the National Institutes of Health to D.J.K.
Abbreviations used:
- API
aminopeptidase I
- Con A
concanavalin A
- CPY
carboxypeptidase Y
- Cvt
cytoplasm-to-vacuole targeting
- GFP
green fluorescent protein
- PIPES
piperazine-N,N′-bis(2-ethanesulfonic acid)
- SMD
synthetic minimal medium containing 2% glucose, essential amino acids, and ammonium sulfate
- SD-N
synthetic minimal medium containing 2% glucose but lacking essential amino acids and ammonium sulfate
- TCA
trichloroacetic acid
REFERENCES
- Baba M, Osumi M, Ohsumi Y. Analysis of the membrane structures involved in autophagy in yeast by freeze-replica method. Cell Struct Funct. 1995;20:465–471. doi: 10.1247/csf.20.465. [DOI] [PubMed] [Google Scholar]
- Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Takeshige K, Baba N, Ohsumi Y. Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J Cell Biol. 1994;124:903–913. doi: 10.1083/jcb.124.6.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Sato TK, Banta LM, Emr SD. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 1997;16:1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum P, Thorner J, Honig L. Identification of tubulin from the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1978;75:4962–4966. doi: 10.1073/pnas.75.10.4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant NJ, Stevens TH. Vacuole biogenesis in Saccharomyces cerevisiae: protein transport pathways to the yeast vacuole. Microbiol Mol Biol Rev. 1998;62:230–247. doi: 10.1128/mmbr.62.1.230-247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsow T, Rieder SE, Emr SD. A multispecificity syntaxin homologue, Vam3p, esential for autophagic and biosynthetic protein transport to the vacuole. J Cell Biol. 1997;138:517–529. doi: 10.1083/jcb.138.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi T, Matsuura A, Noda T, Ohsumi Y. Analyses of APG13 gene involved in autophagy in yeast. Saccharomyces cerevisiae. Gene. 1997;192:207–213. doi: 10.1016/s0378-1119(97)00031-0. [DOI] [PubMed] [Google Scholar]
- Haas A, Scheglmann D, Lazar T, Gallwitz D, Wickner W. The GTPase Ypt7p of Saccharomyces cerevisiae is required on both partner vacuoles for the homotypic fusion step of vacuole inheritance. EMBO J. 1995;14:5258–5270. doi: 10.1002/j.1460-2075.1995.tb00210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding TM, Hefner-Gravink A, Thumm M, Klionsky DJ. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein targeting pathway. J Biol Chem. 1996;271:17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kametaka S, Matsuura A, Wada Y, Ohsumi Y. Structural and functional analyses of APG5, a gene involved in autophagy in yeast. Gene. 1996;178:139–143. doi: 10.1016/0378-1119(96)00354-x. [DOI] [PubMed] [Google Scholar]
- Kiel, J.A.K.W., Rechinger, K.B., van der Klei, I.J., Salomons, F.A., Titorenko, V.I., and Veenhuis, M. (1999). The Hansenula polymorpha PDD1 gene, essential for selective degradation of peroxisomes, is a homologue of Saccharomyces cerevisiae Vps34. Yeast (in press). [DOI] [PubMed]
- Kim J, Scott SV, Oda M, Klionsky DJ. Transport of a large oligomeric protein by the cytoplasm to vacuole protein targeting pathway. J Cell Biol, 1997;137:609–618. doi: 10.1083/jcb.137.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ. Protein transport from the cytoplasm into the vacuole. J Membr Biol. 1997;157:105–115. doi: 10.1007/s002329900220. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ. Nonclassical protein sorting to the yeast vacuole. J Biol Chem. 1998;273:10807–10810. doi: 10.1074/jbc.273.18.10807. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Banta LM, Emr SD. Intracellular sorting and processing of a yeast vacuolar hydrolase: proteinase A propeptide contains vacuolar targeting information. Mol Cell Biol. 1988;8:2105–2116. doi: 10.1128/mcb.8.5.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Cueva R, Yaver DS. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Herman PK, Emr SD. The fungal vacuole: composition, function, and biogenesis. Microbiol Rev. 1990;54:266–292. doi: 10.1128/mr.54.3.266-292.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbe, S., and Thiele, D.J. (1999). Copper ion inducible and repressible promoter systems in yeast. Methods Enzymol. (in press). [DOI] [PubMed]
- Lang T, Schaeffeler E, Bernreuther D, Bredschneider M, Wolf DH, Thumm M. Aut2p and Aut7p, two novel microtubule-associated proteins are essential for delivery of autophagic vesicles to the vacuole. EMBO J. 1998;17:3597–3607. doi: 10.1093/emboj/17.13.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–50. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation sytem essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Matsuura A, Wada Y, Ohsumi Y. Acidification of vacuole is required for the autophagic degradation in the yeast Saccharomyces cerevisiae. J Biochem. 1997;121:338–344. doi: 10.1093/oxfordjournals.jbchem.a021592. [DOI] [PubMed] [Google Scholar]
- Oda MN, Scott SV, Hefner-Gravink A, Caffarelli AD, Klionsky DJ. Identification of a cytoplasm to vacuole targeting determinant in aminopeptidase I. J Cell Biol. 1996;132:999–1010. doi: 10.1083/jcb.132.6.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riezman H, Munn A, Geli MI, Hicke L. Actin-, myosin- and ubiquitin-dependent endocytosis. Experientia. 1996;52:1033–1041. doi: 10.1007/BF01952099. [DOI] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Novick P, Thomas JH, Botstein D, Fink GR. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Sakai Y, Koller A, Rangell LK, Keller GA, Subramani S. Peroxisome degradation by microautophagy in Pichia pastoris: identification of specific steps and morphological intermediates. J Cell Biol. 1998;141:625–636. doi: 10.1083/jcb.141.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlumpberger M, Schaeffeler E, Straub M, Bredschneider M, Wolf DH, Thumm M. AUT1, a gene essential for autophagocytosis in the yeast Saccharomyces cerevisiae. J Bacteriol. 1997;179:1068–1076. doi: 10.1128/jb.179.4.1068-1076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Baba M, Ohsumi Y, Klionsky DJ. Aminopeptidase I is targeted to the vacuole by a nonclassical vesicular mechanism. J Cell Biol. 1997;138:37–44. doi: 10.1083/jcb.138.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Hefner-Gravink A, Morano KA, Noda T, Ohsumi Y, Klionsky DJ. Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc Natl Acad Sci USA. 1996;93:12304–12308. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Klionsky DJ. Delivery of proteins and organelles to the vacuole from the cytoplasm. Curr Opin Cell Biol. 1998;10:523–529. doi: 10.1016/s0955-0674(98)80068-9. [DOI] [PubMed] [Google Scholar]
- Straub M, Bredschneider M, Thumm M. AUT3, a serine/threonine kinase gene, is essential for autophagocytosis in Saccharomyces cerevisiae. J Bacteriol. 1997;179:3875–3883. doi: 10.1128/jb.179.12.3875-3883.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119:301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Mizushima N, Kiyooka M, Ohsumi M, Ueno T, Ohsumi Y, Kominami E. Apg7p/Cvt2p: a novel protein activating enzyme essential for autophagy. Mol Biol Cell. 1999;10:1367–1379. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumm M, Egner R, Koch M, Schlumpberger M, Straub M, Veenhuis M, Wolf DH. Isolation of autophagocytosis mutants of Saccharoymces cerevisiae. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- Titorenko VI, Keizer I, Harder W, Veenhuis M. Isolation and characterization of mutants impaired in the selective degradation of peroxisomes in the yeast Hansenula polymorpha. J Bacteriol. 1995;177:357–363. doi: 10.1128/jb.177.2.357-363.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- Tuttle DL, Dunn WA., Jr Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris. J Cell Sci. 1995;108:25–35. doi: 10.1242/jcs.108.1.25. [DOI] [PubMed] [Google Scholar]
- Tuttle DL, Lewin AS, Dunn WA., Jr Selective autophagy of peroxisomes in methylotrophic yeasts. Eur J Cell Biol. 1993;60:283–290. [PubMed] [Google Scholar]
- Wurmser AE, Emr SD. Phosphoinositide signaling and turnover: PtdIns(3)P, a regulator of membrane traffic, is transported to the vacuole and degraded by a process that requires lumenal vacuolar hydrolase activities. EMBO J. 1998;17:4930–4942. doi: 10.1093/emboj/17.17.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Strombaug PE, Dunn WA., Jr Glucose-induced autophagy of peroxisomes in Pichia pastoris requires a unique E1-like protein. Mol Biol Cell. 1999;10:1353–1366. doi: 10.1091/mbc.10.5.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan WY, Tuttle DL, Shi Y-J, Ralph GS, Dunn WA., Jr Glucose-induced microautophagy in Pichia pastoris requires the α-subunit of phosphofructokinase. J Cell Sci. 1997;110:1935–1945. doi: 10.1242/jcs.110.16.1935. [DOI] [PubMed] [Google Scholar]