SUMMARY
To efficiently transcribe genes, RNA Polymerase II (Pol II) must overcome the barrier imposed by nucleosomes and higher order chromatin structure. Many genes, including Drosophila melanogaster Hsp70, undergo changes in chromatin structure upon activation. To characterize these changes, we mapped the nucleosome landscape of Hsp70 following an instantaneous heat shock at high spatial and unprecedented temporal resolution. Surprisingly, we find an initial disruption of nucleosomes across the entire gene within 30 seconds following activation, faster than the rate of Pol II transcription, followed by a second further disruption within 2 minutes. This initial change occurs independently of Pol II transcription. Furthermore, the rapid loss of nucleosomes extends beyond Hsp70 and halts at the scs and scs’ insulating elements. An RNAi screen of 28 transcription and chromatin-related factors reveal that depletion of heat shock factor, GAGA Factor, or Poly(ADP)-Ribose Polymerase or its activity abolishes the loss of nucleosomes upon Hsp70 activation.
INTRODUCTION
In eukaryotic cells, DNA is packaged into chromatin providing a natural barrier to factors requiring access to DNA (Rando, and Ahmad, 2007). Many essential cellular processes, including gene expression, rely on the ability of the cell to regulate and alleviate the restrictive properties of chromatin. In vitro studies have shown that the transcriptional rate and processivity of RNA Polymerase II (Pol II), which is responsible for expressing all mRNA-encoding genes, is severely inhibited by nucleosomes (Knezetic, and Luse, 1986; Lorch, LaPointe, and Kornberg, 1987). However, in vivo studies show that Pol II, and even the first ‘pioneer’ Pol II to transcribe an induced gene, is able to transcribe at rates (~1.5 kb/min) that suggest it is not inhibited by the presence of nucleosomes (O'Brien, and Lis, 1993; Thummel, Burtis, and Hogness, 1990). These results indicate that eukaryotic cells have mechanisms that modulate nucleosome position and structure at active genes.
The diversity of factors that act on chromatin indicate that eukaryotic cells use multiple general mechanisms to alter the position or composition of the nucleosome, allowing critical factors such as Pol II access to DNA (Li, Carey, and Workman, 2007). Chromatin remodeling complexes, such as SWI/SNF and ISWI, provide the cell with the ability to remove, transfer, or slide a nucleosome along a DNA template (Saha, Wittmeyer, and Cairns, 2006). Additionally, many transcription elongation factors and histone chaperones, such as FACT, Spt6, and Asf1 aid in the disassembly of nucleosomes and their reassembly in the wake of transcribing Pol II (Bortvin, and Winston, 1996; Orphanides, LeRoy, et al, 1998; Schwabish, and Struhl, 2006). Finally, histone modifying enzymes can acetylate, methylate, phosphorylate, monoubiquitinate, sumoylate, or ADP-ribosylate histones, or carry out the reverse of each reaction (Li, Carey, and Workman, 2007). Many of these modifications can modulate inter and intranucleosomal interactions (Reinke, and Horz, 2003; Shogren-Knaak, and Peterson, 2006) and most are likely to serve as specific targets for effectors (Jenuwein, and Allis, 2001; Wysocka, Swigut, et al, 2006), which can act locally to alleviate or reinforce the repressive structure of chromatin.
Understanding where nucleosomes are positioned and how these positions change during transcription is critical in deciphering how the changes occur. Early studies with micrococcal nuclease (MNase) or DNase1 on chromatin isolated from cells produced the first views of the chromatin structure of Drosophila melanogaster heat shock (HS) genes in vivo. These analyses showed that both the promoter and the 3’ end of the Hsp70 gene contained large hypersensitive regions and that nucleosomes on the coding regions before HS were disrupted after an extended HS (Wu, Wong, and Elgin, 1979; Wu, 1980). Recently, whole genome ChIP-chip and ChIP-seq studies have shown that many genes exhibit similar patterns of nucleosome occupancy (Johnson, Tan, et al, 2006; Lee, Tillo, et al, 2007; Schones, Cui, et al, 2008). In particular, nucleosomes are often absent in promoter regions just upstream of the transcriptional start site, allowing the transcriptional machinery easy access to DNA elements (Crawford, Holt, et al, 2006; Mito, Henikoff, and Henikoff, 2005; Yuan, Liu, et al, 2005). Moreover, genes in Saccharomyces cerevisiae on average contain 1 well-positioned nucleosome on each side of the promoter region and nucleosomes become progressively less positioned beyond this point (Albert, Mavrich, et al, 2007; Lee, Tillo, et al, 2007; Yuan, Liu, et al, 2005). Thus, while many promoters are open and accessible, and even occupied by Pol II (Lis, 2007), Pol II still must overcome nucleosomes occluding its downstream path into the gene.
D. melanogaster Hsp70 is a model gene used to study general mechanisms by which Pol II is able to transcribe through chromatin. Hsp70 is rapidly activated within seconds after HS, and concomitantly its chromatin structure decondenses, evident by the formation of large puffs on polytene chromosomes (Boehm, Saunders, et al, 2003). Not surprisingly, an entire battery of transcription factors is recruited to Hsp70 upon activation, under the direction of the master heat shock factor (HSF) activator (Saunders, Core, and Lis, 2006). Curiously, although puff size at HS induced genes increases with transcript length and promoter strength (Simon, Sutton, et al, 1985), previous studies also show that puffing of HS loci can be decoupled from active transcription of the gene using a chemical stimulus (Winegarden, Wong, et al, 1996). Although informative, these studies do not demonstrate at the molecular level in living cells that changes are indeed happening to the nucleosomes occupying Hsp70.
The dramatic change in the chromatin structure upon gene activation begs several questions. Does this puffing entirely represent changes in nucleosomes? Are the nucleosomes evicted, or are their positions or configuration altered? How rapidly can changes in the chromatin structure be detected at the molecular level? And finally, which factors are responsible? Previous studies have shown that deletion of either Heat Shock Elements (HSEs) or GAGA factor (GAF) binding sites in the Hsp70 promoter region result in reduced HSF binding and a loss in puff formation upon HS (Shopland, Hirayoshi, et al, 1995). GAF, encoded by the Trithorax-like gene, binds to repeating (GA)n sequences and is present at the Hsp70 promoter before HS (O'Brien, Wilkins, et al, 1995). GAF itself has been shown to play important roles in gene activation presumably by regulating chromatin structure by itself or through its interactions with the NURF remodeling complex (Tsukiyama, Becker, and Wu, 1994; Tsukiyama, and Wu, 1995).
In addition to HSF and GAF, recent evidence indicates that Poly(ADP-)Ribose Polymerase (PARP) is important in puff formation at many loci including Hsp70 (Tulin, and Spradling, 2003). PARP is an enzyme that catalyzes the polymerization of ADP ribose units from NAD+ onto target proteins (primarily itself) and interacts with DNA and nucleosomes (Kim, Mauro, et al, 2004; Pinnola, Naumova, et al, 2007). PARP proteins have roles in several nuclear processes, including DNA damage responses (D'Amours, Desnoyers, et al, 1999), but have only recently been examined with respect to transcription (Kraus, and Lis, 2003). A P-element insertion that disrupts PARP expression, or inhibition of PARP’s catalytic activity, displays decreased puff sizes in polytene chromosomes and reduced Hsp70 protein levels upon HS (Tulin, and Spradling, 2003). To understand puff formation and the roles of HSF, GAF, and PARP, examination of chromatin at the nucleosomal level is required. Moreover, the potential role of other factors needs to be evaluated.
In this paper, we map the nucleosome architecture of the D. melanogaster Hsp70 gene at a 30 bp resolution and track its changes seconds after an instantaneous HS. We find that before HS, the chromatin structure of Hsp70 has characteristics similar to general genome-wide features at most genes. After HS, we find that the chromatin architecture at Hsp70 has an initial dramatic change throughout the gene that is so rapid that it occurs even before the first wave of transcribing polymerase reaches the corresponding regions of the gene. Furthermore, we find this initial loss of nucleosomes is independent of transcription and extends to natural insulating elements flanking the HS genes. Through an RNAi screen of known coactivators, chromatin remodeling enzymes, histone modifiers, nucleosome assembly and disassembly factors, modifiers of DNA topology, and elongation factors, we indentify HSF, GAF, and PARP as each necessary for the rapid loss of nucleosomes at the Hsp70 gene following HS. The effect of PARP requires its activity and a specific inhibitor added only 10 minutes before HS is sufficient to block the nucleosome loss.
RESULTS
The Chromatin Structure at Hsp70 is Rapidly and Dramatically Altered Following Heat Shock
We employed a previously developed method (Sekinger, Moqtaderi, and Struhl, 2005) to track in vivo changes in the chromatin structure of the Hsp70 gene at the mononucleosome level following activation. In our assay, Hsp70 is induced in Drosophila S2 cells by an instantaneous HS for various times. The cells are cross-linked with formaldehyde and chromatin is then isolated and split into mock and MNase treated samples (Figure 1A). The DNA is then probed with over 100 separate primer sets that on average are spaced 30 bp apart and amplify 100 bp regions along the Hsp70Ab copy (Figure 1B and Table S1). The ability of a primer pair to amplify depends on the amount of contiguous DNA between the primers that remains after MNase digest; MNase cleaves linker DNA between nucleosomes and cleaves nucleosome-free DNA. To determine the amount of digestion in a particular region at the Hsp70 gene, we calculated the relative ratio of the amount of digested DNA to the undigested control using quantitative PCR (qPCR). Efficient amplification of 100 bp regions ensures that amplifiable DNA in the digested sample is mononucleosome or subnucleosome in size but larger than tetrameric or hexameric structures. The amount of MNase was titrated to produce mononucleosomes from the bulk chromatin of nuclei (Figure S1A), and to observe protection under uninduced conditions at Hsp70 with amplification of 100 bp sized fragments but not with 190 bp sized fragments (Figure S1B). This ensures that our assay has mononucleosome resolution, and provides a precise way to measure and track the positions of nucleosomes along Hsp70 following a HS.
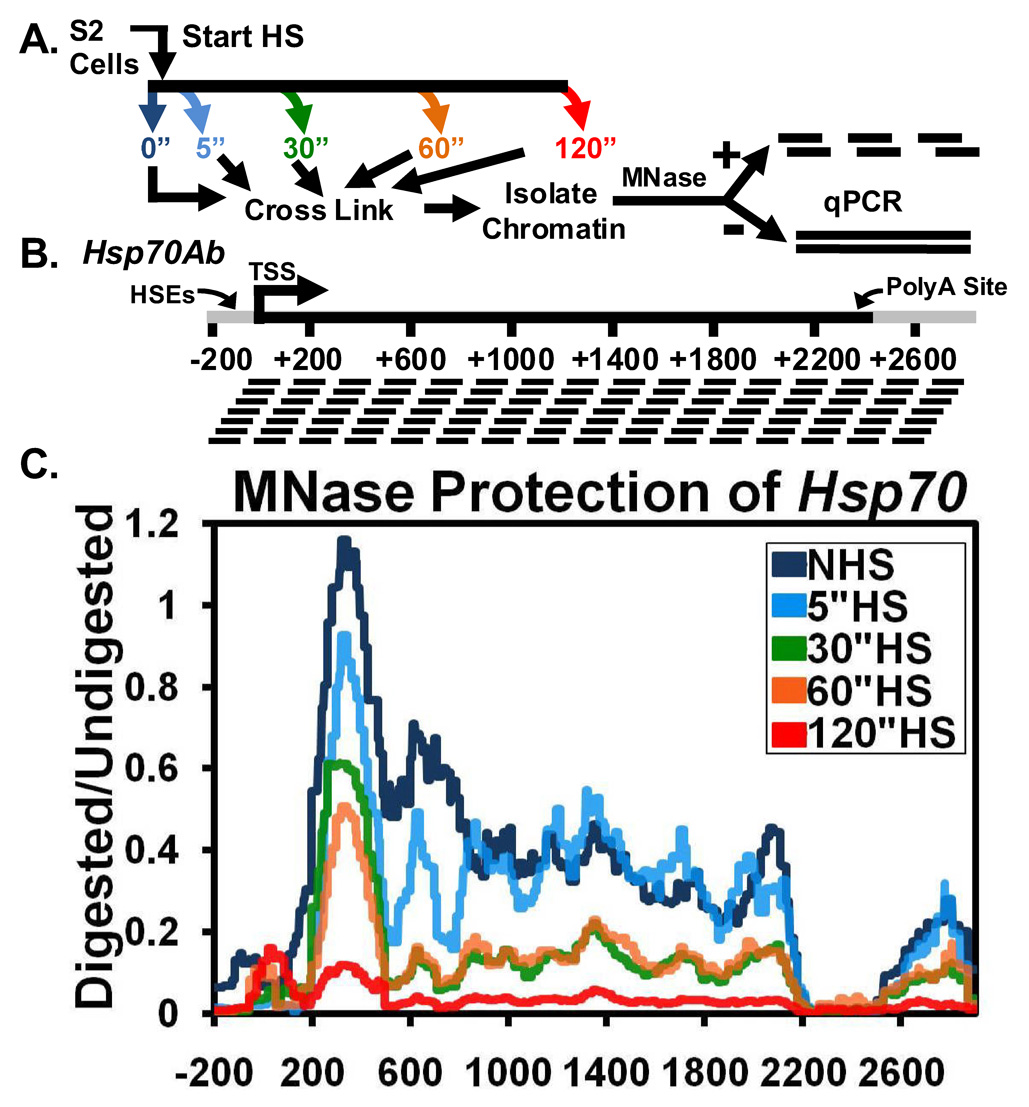
Figure 1. Rapid Loss of Chromatin Structure at Hsp70 upon Heat Shock Detected by a High Resolution MNase Scanning Assay.
(A) Diagram depicting the procedure followed for the high resolution MNase assay. S2 cells are heat shocked for 0 (dark blue), 5 (light blue), 30 (green), 60 (orange), or 120 seconds (red) (colors refer to 1C), immediately cooled to room temperature and crosslinked, and their chromatin is isolated. Purified DNA products from samples are treated with 0 or 500 U of MNase and used for qPCR.
(B) Diagram showing the PCR amplicons used at the Hsp70Ab gene. PCR products are 100±5 bp in size and are spaced 30±6 bp apart. The mRNA-encoding unit is shown in black. The gene nucleotide location, corresponding to panel C, is numbered below with the HSEs, TSS, and PolyA site (at +2343) indicated.
(C) The HS time course chromatin profile of Hsp70 is determined by normalizing the amount of the MNase digested PCR product to that of the undigested product using the ΔC(t) method (y-axis), which is plotted against the gene nucleotide location (x-axis). Values from overlapping primer sets are averaged. The x-axis represents base pair units with 0 being the TSS. Lines represent the average of 3 separate experiments with error bars omitted for clarity.
Under non heat shock (NHS) conditions, Hsp70 contains 4 distinct chromatin regions: a nucleosome free promoter region, a well-positioned nucleosome centered approximately 330 nucleotides after the transcription start site (TSS), poorly positioned nucleosomes in the body of the gene, and a second nucleosome free region at the 3’ end of the gene (Figure 1C, dark blue). (Each individual time point can be seen as a separate graph with error bars in Figures S2A–E.) The nucleosome free region at the 5’ and 3’ ends of Hsp70 agree with earlier studies that show these regions are hypersensitive to nucleases (Wu, 1980). The 5’ hypersensitive region extends further into the transcription unit than where Pol II is known to be transcriptionally engaged and paused 20 to 40 base pairs downstream of its initiation site (Giardina, Perez-Riba, and Lis, 1992; Rasmussen, and Lis, 1993; Rougvie, and Lis, 1990). Beyond the first nucleosome, centered at +330 and well downstream of the paused Pol II, the body of the gene contains nucleosomes that gradually lose their positioning. This is seen from the relatively even distribution of protection along the gene. A nucleosome just upstream of the polyadenylation (PolyA) site of Hsp70 bookends the nucleosomes on the 3’ end of the gene. Overall, under NHS conditions, the chromatin structure of the Hsp70 gene accommodates paused polymerase but still provides an impending barrier to transcription elongation.
To determine how the chromatin structure of Hsp70 changes upon an instantaneous HS, we used the high-resolution MNase assay following 5, 30, 60, and 120 seconds of HS. Within 5 seconds of HS, the protection of DNA in the immediate 5’ region of the gene decreases (Figure 1C, light blue). By 30 seconds (Figure 1C, green), further loss in the 5’ region is observed, but now losses are also observed extending past the 3’ region of the gene. Surprisingly, these initial changes in the 3’ end of the gene occur before RNA polymerase reaches the 3’ end, which takes approximately 2 minutes (Boehm, Saunders, et al, 2003; O'Brien, and Lis, 1993). No significant changes in nucleosome protection are seen between 30 and 60 seconds (Figure 1C, orange). However, by 120 seconds of HS (Figure 1C, red) another broad loss in the protection of nucleosomes occurs along the entire gene. In contrast to NHS conditions, the nucleosome protection pattern of a 120 second HS is decimated. This 2 minute protection pattern remains even after 20 minutes of HS (data not shown). Similar changes in nucleosomes were also observed on the same time scale for the shorter, HS inducible Hsp26 gene (Figures S3A and S3B). During HS, the positions of nucleosomes neither move into nucleosome free regions nor do they increase in their relative level of protection.
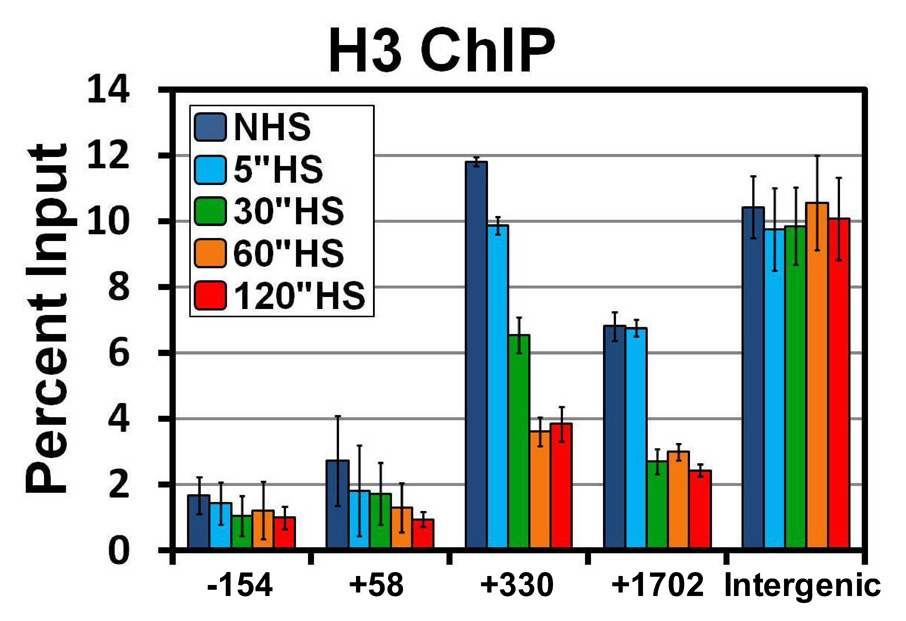
To address whether or not these changes in the chromatin landscape are due to either a loss of histones or just an increased accessibility of the DNA at this locus, we performed traditional sonication ChIP with an antibody that recognizes histone H3 and its variants. The histone NHS landscape determined by ChIP matches that of the MNase protection assay (Figure 2, dark blue). Additionally, the same changes in the MNase protection pattern are seen with a histone H3 ChIP at 5, 30, and 60 seconds of HS (Figure 2, light blue, green, orange). The combination of increased MNase accessibility and decreased histone density indicate disruption of nucleosome structure on the gene, or a loss in nucleosomes. The only significant difference observed between the 2 methods is that there is not a change in histone levels between 60 and 120 seconds of HS. One likely explanation for these differences is that Pol II accumulates across the body of the gene by this point, resulting in further disruption of canonical nucleosomal structures. This could lead to an increased accessibility of this DNA to the MNase without a further decrease in the amount of histone H3 present. Again, similar results were found for Hsp26 (Figure S3C). Figure 1 and Figure 2 indicate that changes in nucleosomal structures at Hsp70 start at the 5’ end of the gene and move more rapidly than Pol II towards the 3’ end.
Figure 2. Rapid loss of Histone H3 from Hsp70 upon Heat Shock.
Histone density across the Hsp70 gene detected by ChIP using an H3 antibody. S2 cells are heat shocked for 0 (dark blue), 5 (light blue), 30 (green), 60 (orange), or 120 seconds (red). The y-axis represents the percent of input material immunoprecipitated. Error bars represent the SEM of 3 independent experiments. The x-axis represents base pair units along the Hsp70 gene with 0 as the TSS; each number represents the center of the PCR amplicon. The intergenic region represents a region 32 kb downstream of Hsp70Bc that does not change upon HS.
Nucleosomes at Hsp70 can be Lost Independently of Transcription
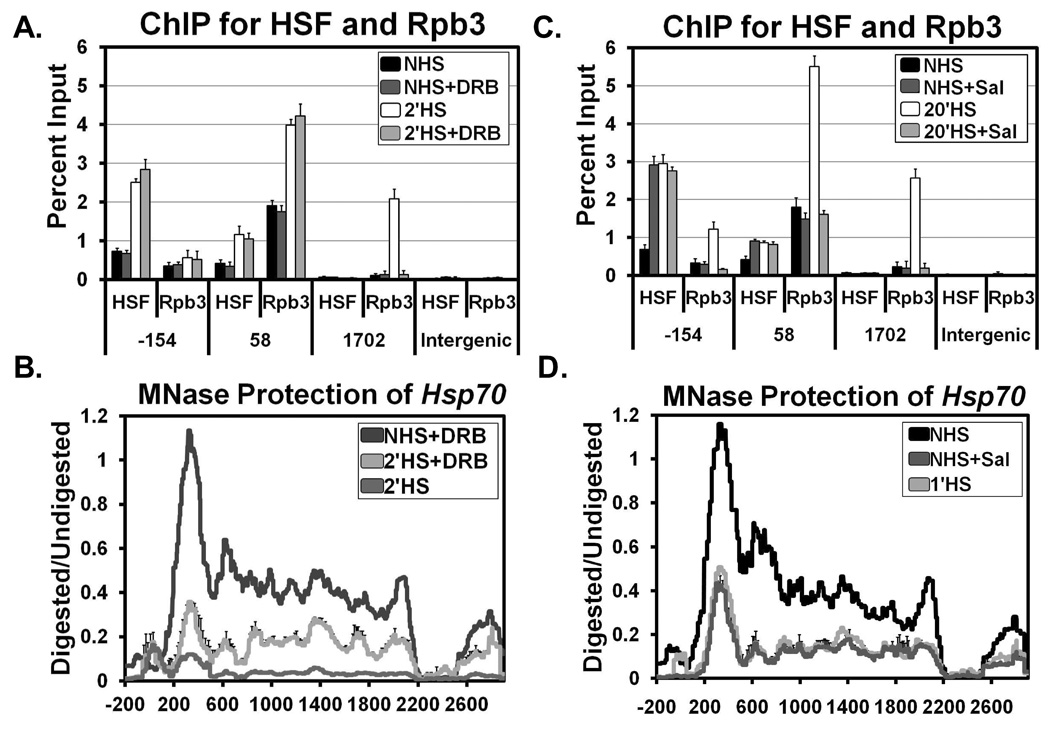
The loss of nucleosomes at Hsp70 occurring prior to Pol II’s occupancy of these regions suggests that the rapid changes in chromatin, which occur well before maximal puff formation at 20 min HS (Lewis, Helmsing, and Ashburner, 1975), might also occur independently of transcription. To catalogue at high-resolution what chromatin changes occur independently of transcription, we used sodium salicylate or the nucleotide analog 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB). Both of these chemicals reduce the level of HS induced Hsp70 transcription (Giardina, and Lis, 1993; Winegarden, Wong, et al, 1996). Treatment of S2 cells with DRB followed by a 2 minute HS resulted in normal recruitment of HSF to the promoter (−154) (Figure 3A). Pol II was also normally recruited to the pause site (+58); however, it was not detected in regions further downstream (+1702) (Figure 3A), indicative of transcription elongation being inhibited. The nucleosome protection profile of a 2 minute (or 30 second) HS with DRB revealed that the initial loss of nucleosome structure occurred even though Pol II never moved into the body of the gene (Figure 3B and Figure S15A).
Figure 3. Initial Loss of Nucleosomes at Hsp70 is Independent of Transcription.
(A) ChIP at Hsp70 for HSF and Rpb3 (Pol II) with and without 125 µM of the transcription inhibitor DRB before (NHS, black bars and NHS+DRB, dark gray bars) and during HS (2’HS, white bars and 2’HS+DRB, light gray bars). The y-axis represents the percent of input material immunoprecipitated. x-axis values represent base pair units centered on the HSEs (−154), the Pol II pause site (+58), a downstream region (+1702), and an intergenic region outside the scs and scs’ regions. Error bars represent the SEM of 3 independent experiments.
(B) MNase protection profile of NHS with DRB (dark gray), 2’HS with DRB (light gray), and 2’HS (medium gray) as in Figure 1C. Lines represent the average of 3 separate experiments. Error bars representing the SEM are plotted just for the 2’HS+DRB line for clarity.
(C) ChIP as described in (A) except with and without 10 mM of sodium salicylate before (NHS, black bars and NHS+salicylate, dark gray bars) and during HS (20’HS, white bars and 20’HS+salicylate, light gray bars). Error bars represent the SEM of 3 independent experiments.
(D) MNase protection profile as in (B) with NHS (black), NHS+salicylate (dark gray), and 1’HS (light gray). Lines represent the average of 3 separate experiments. Error bars representing the SEM are plotted just for the NHS+salicylate line for clarity.
Treatment of cells with sodium salicylate under NHS conditions resulted in the recruitment of HSF levels equivalent to that of a 20 minute HS (Figure 3C). The amount of Pol II at the pause site or in downstream regions, however, did not change from NHS levels (Figure 3C). These results are consistent with previous studies showing that sodium salicylate induces HSF binding to its HSEs, but does not result in additional promoter melting by the paused Pol II (Giardina, and Lis, 1995). This NHS treatment with sodium salicylate, like HS in the presence of DRB, resulted in the loss of nucleosomes throughout the gene similar to the initial loss found by 30 or 60 seconds of HS (Figure 3D). Similar results for both experiments were found for Hsp26 as well (Figures S4A–D). These experiments demonstrate that the initial loss of nucleosomes is due to a mechanism that is independent of transcription and the subsequent loss in protection is due to a transcription-dependent mechanism.
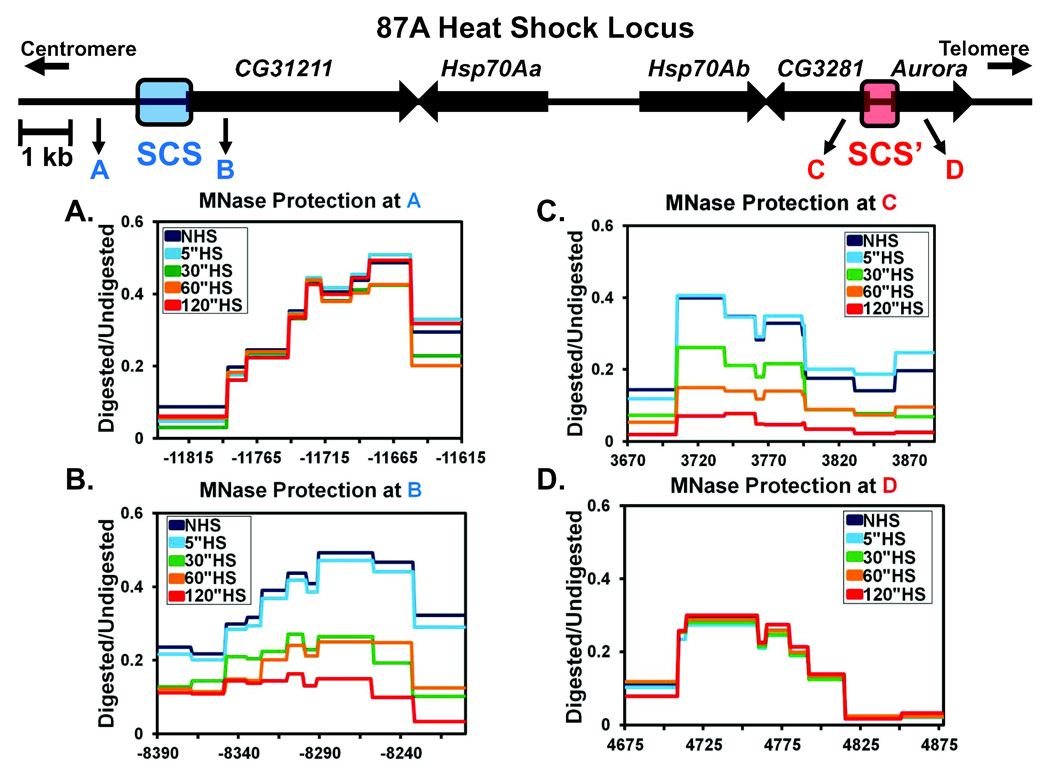
The Loss of Nucleosomes at Hsp70 Halts at the Drosophila scs and scs’ Boundary Elements
In our initial assay, we observed chromatin changes across the entire Hsp70 region that we surveyed (Figure 1C). Next, we chose to determine where the loss in nucleosomal protection ceases. Decondensed polytene chromosome puffs do not spread indefinitely, suggesting that a relatively defined border of nucleosome loss might exist. To identify where this border occurs, we progressively walked downstream of the Hsp70 genes at 87A to the scs and scs’ boundary elements (Figure 4) (Udvardy, Maine, and Schedl, 1985). As insulators, the scs and scs’ regions protect against positive and negative chromatin position effects, and can block cis-acting, promoter-enhancer interactions (Kellum, and Schedl, 1991; Kuhn, Hart, and Geyer, 2004). The scs and scs’ regions are known to contain nuclease hypersensitive regions as well as DNA binding elements for the zeste-white 5 (Zw5) and boundary element associated factor 32 (BEAF-32) DNA binding proteins, respectively (Gaszner, Vazquez, and Schedl, 1999; Vazquez, and Schedl, 1994; Zhao, Hart, and Laemmli, 1995). Surprisingly, protection of nucleosomes anywhere between the Hsp70 copies and either the scs or scs’ regions was lost upon HS activation with similar kinetics to the loss observed at Hsp70 (Figures 4B, 4C, and data not shown). However, nucleosomes outside the region enclosed by the scs and scs’ elements were unaffected (Figures 4A and 4D). These results indicate that the scs and scs’ regions also act as barriers to the spread of severely decondensed chromatin and that rapid nucleosome loss upon transcription activation is not confined to the transcription unit, but can extend several kb upstream and downstream of the activated gene. Interestingly, even though changes in nucleosomes occur over multiple genes within the region, only the Hsp70 genes are induced following a HS (Figure S5A). Moreover, the loss of nucleosomes across this broad domain, corresponding to the 87A polytene puff, occurs well before maximal puff formation; therefore, puffing does not merely represent nucleosomal changes, but must also denote higher order structural changes at the locus.
Figure 4. The scs and scs’ Regions Insulate the Heat Shock Locus from the Spread of Nucleosome Loss.
(A –D) MNase HS time course protection as in Figure 1C flanking the scs and scs’ insulators. The 87A HS locus shown above depicts the 4 sites analyzed flanking scs and scs’. Regions A and C are outside and B and D are inside of the scs and scs’ insulators respectively. Regions A–D also correspond to A–D in the graphs. The x-axis of all 4 graphs uses the TSS of the Hsp70Ab copy as the 0 point. Each line represented is the average of 3 independent experiments. Error bars are omitted for clarity but the SEM from 3 independent experiments is less than ±0.06.
HSF and GAF are Necessary for Nucleosome Loss at Drosophila Hsp70
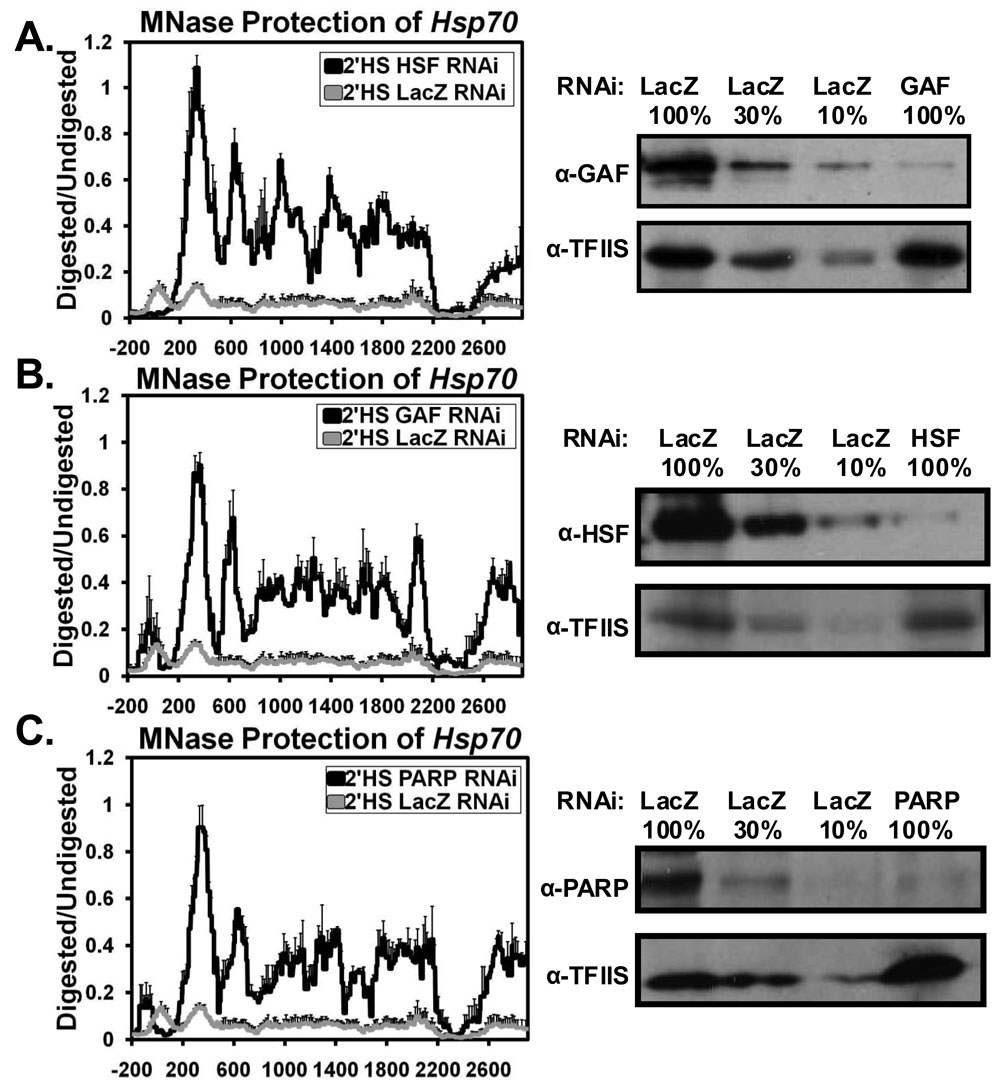
The above results defined the location and rate of nucleosome loss at Hsp70 but did not identify what factors are required for the nucleosome loss. To address this question we coupled the high resolution MNase assay to transient depletions of candidate factors by RNAi. Given that a mutation in HSF or deletion or mutation of the HS or GAGA elements greatly inhibits the formation of HS puffs (Jedlicka, Mortin, and Wu, 1997; Shopland, Hirayoshi, et al, 1995), we first targeted HSF and GAF for depletion. Depletion of HSF or GAF to less than 10% of LacZ control cells (Figures 5A and 5B), abolished the changes in the chromatin structure of Hsp70 after a 2 minute HS (Figures 5A and 5B) and with a shorter 30 second HS (Figures S15B and 15C). Therefore, both HSF and GAF are critical for bringing about the loss of nucleosomes upon HS.
Figure 5. HSF, GAF, and PARP are Essential for the Loss of Nucleosomes at Hsp70.
MNase protection profile as in Figure 1C comparing the chromatin architecture following an RNAi depletion of either (A) HSF, (B) GAF, or (C) PARP (black lines) in comparison to a control RNAi depletion of LacZ (gray lines) after a 2’HS at Hsp70. Each line represents the average of 3 independent experiments with error bars representing the SEM. Western blots showing corresponding knockdown of HSF, GAF, or PARP are shown to the right of each figure with TFIIS used as a loading control and a serial dilution of LacZ RNAi to quantify each knockdown.
Poly(ADP)-Ribose Polymerase is Necessary for Nucleosomal Loss at Hsp70
To determine if other factors are required for the rapid changes in nucleosome structure upon HS, we targeted many additional factors by RNAi depletion: known players in the HS response, nucleosome remodelers, histone-interacting proteins, elongation factors, factors affecting DNA topology, and known boundary element factors. The results of these experiments are summarized in Table 1 and in Figures S7–13. Although not all knock-downs were confirmed, due to the unavailability of antibodies, those that were checked had protein levels decreased to 5–20% of control cells (Figure S14). Under NHS conditions, the ISWI remodeling complex containing Nurf301 and Chd1 had the largest effects, as nucleosomes on the body of the gene were better positioned in ISWI, Nurf301, and Chd1 RNAi treated cells (Figures S10B, S10C, and S10E). Additionally, RNAi depletion of HDAC3 reduced the protection of DNA corresponding to the positions of the first 2 nucleosomes (Figure S11D). After HS, some transcription related factors including Med15, P-TEFb, Spt6, and ERCC3 (Figures S7D, S9A, S9D, and S12C respectively) showed nucleosome profiles more closely resembling a 1 minute rather than a 2 minute HS, which may be a consequence of a transcription defect at Hsp70 (Ni, Saunders, et al, 2007) or possibly a direct role in the disassembly of nucleosomes associated with elongating Pol II. Regardless, depletion of these 4 factors reinforces the finding that the second loss in MNase protection (occurring between 1 to 2 minutes of HS) is a transcription-dependent event. Depletion of other factors resulted in slightly more protection at the site of the first nucleosome after the TSS upon HS (e.g. Figures S8D, S9B, S9C, etc). Additionally, a subset of factors, including Zw5 and BEAF-32, were chosen to determine if RNAi depletion of any factor allowed disruption of nucleosomes outside scs and scs’, and none were found (Figure S6). Overall, most factors targeted for depletion did not show any difference in the nucleosome profile in comparison to control LacZ RNAi treatments before or after a 2 minute HS (e.g. Figures S7C, S7E, S8A, etc).
Table 1.
Affect of RNAi Depletion of Different Factors on the Chromatin Architecture at Hsp70
| Factor | Change from NHS | Change from 2’HS | |
|---|---|---|---|
| Upstream Activators | HSF1 | None | NHS4 |
| GAF1 | None | NHS4 | |
| CBP | None | None | |
| Med15 | None | 1’HS5 | |
| Med23 | None | None | |
| SAGA Subunits | Gcn5 | None | None |
| Tra1 | None | None | |
| Spt3 | None | None | |
| Ubp8 | None | NMP6 | |
| Elongation Factors | PTEF-b (Cyclin T1) | None | 1’HS5 |
| Paf11 | None | NMP6 | |
| FACT (Spt161) | None | NMP6 | |
| Spt61 | None | 1’HS5 | |
| Chromatin Remodelers | Swi/Snf (Brm) | None | None |
| ISWI | NWP2 | None | |
| Nurf301 | NWP2 | None | |
| MI-2 | None | None | |
| Chd1 | NWP2 | NMP6 | |
| Kismet | None | None | |
| Nucleosome Interacting Proteins | HIRA | None | None |
| Asf1 | None | None | |
| PARP1 | None | NHS4 | |
| HDAC3 | NLP3 | NMP6 | |
| DNA Topology | Topo11 | None | None |
| Topo2 | None | NMP6 | |
| TFIIH (ERCC31) | None | 1’HS5 | |
| Boundary Elements | BEAF- | None | NMP6 |
| Zw5 | None | NMP6 |
Knockdown confirmed by Western to be 5–20% of LacZ RNAi levels.
Nucleosomes on the body of the gene are better positioned.
DNA at the 1st and 2nd nucleosomes after the TSS is less protected.
Resembles a NHS profile.
Resembles a 1’ HS profile.
DNA at the 1st nucleosome after the TSS is more protected.
Beyond HSF and GAF only one additional factor, Poly(ADP)-Ribose Polymerase (PARP), was found to be essential for the dramatic changes in chromatin structure upon HS. In comparison to LacZ depleted cells which were heat shocked for 2 minutes, depletion of PARP to ~10% of control levels, resulted in a nucleosome profile more closely resembling NHS (Figure 5C), indicating PARP’s importance in bringing about changes in chromatin structure upon HS. Similar results were found using a shorter 30 second HS (Figure S15D).
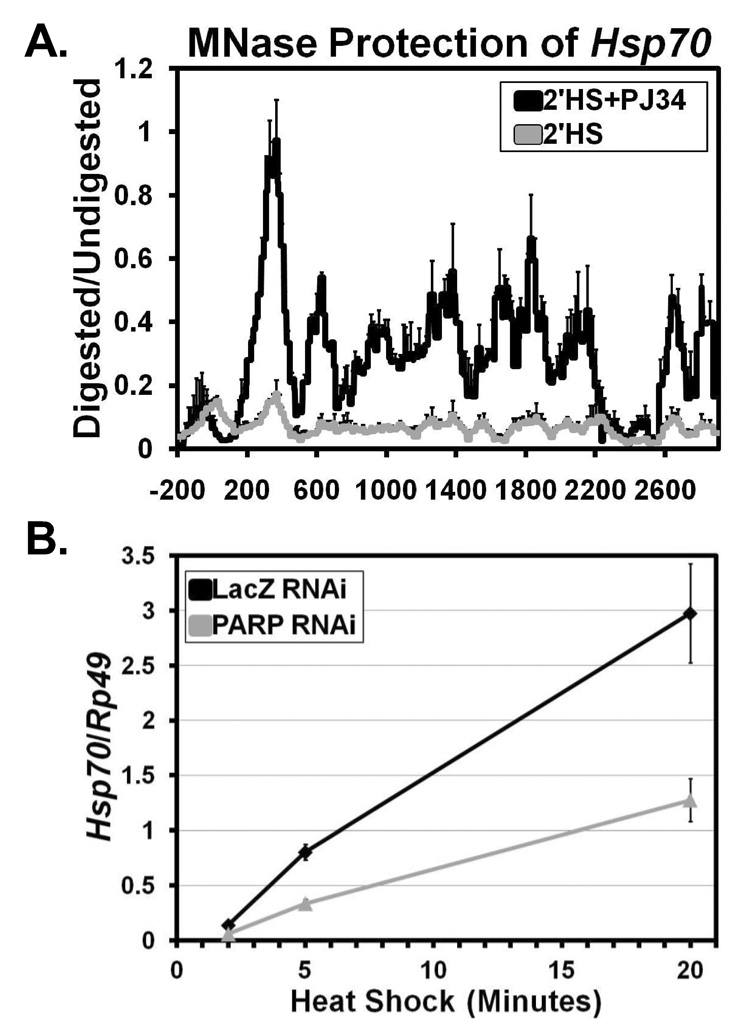
To determine if PARP’s ability to mediate changes in chromatin structure were dependent on its catalytic activity, we used a specific PARP catalytic inhibitor, PJ34 (Virag, and Szabo, 2002). Treatment of S2 cells with 300 nM PJ34 for 10 minutes did not affect the NHS chromatin profile (data not shown). However, treatment of cells with 300 nM PJ34 followed by 2 minutes of HS (Figure 6A) or 30 seconds of HS (Figure S15E) resulted in retention of the NHS nucleosome profile in comparison to untreated cells. These results confirm the RNAi knockdown data, arguing against an off target effect of the transient knockdown, but also show that PARP’s enzymatic activity is required for the loss of nucleosomes at Hsp70 following HS.
Figure 6. The Enzymatic Activity of PARP is Needed for Nucleosome Loss at Hsp70 and PARP is Required for full Hsp70 Expression.
(A) MNase protection profile of Hsp70 as in Figure 1C comparing the chromatin architecture of S2 cells treated (black line) or not treated (gray line) with the PARP enzymatic inhibitor PJ34 after a 2’HS. Each line represents the average of 3 independent experiments with error bars representing the SEM.
(B) Hsp70 mRNA levels following a 2, 5, and 20 minute HS were measured for S2 cells RNAi depleted of LacZ (black) or PARP (gray). Hsp70 expression levels were measured by oligo dT primed reverse transcription followed by qPCR using specific Hsp70Ab primers. Hsp70 mRNA levels are normalized to the Rp49 gene with error bars representing the SEM of 3 replicates.
We then sought to determine if depletion of PARP has a functional consequence beyond retention of chromatin structure following HS. To address this, we measured Hsp70 mRNA levels following 2, 5, and 20 minutes of HS. The result was almost a 3-fold reduction in the amount of transcript following each of the time points (Figure 6B), comparable to a previously estimated 5-fold reduction in Hsp70 protein levels (Tulin, and Spradling, 2003). Collectively, our results show that upon HS, PARP is necessary for rapid nucleosome loss at Hsp70 and consequently, full transcriptional activation of the gene.
DISCUSSION
Using a high resolution in vivo approach to map changes in the chromatin structure of the rapidly induced Hsp70 gene, we observed a broad disruption of nucleosome structure that occurred at a rate faster than transcribing Pol II and broader than a single transcription unit, ceasing at the natural insulating elements. Furthermore, we found that the initial changes in chromatin architecture at Hsp70 can be decoupled from transcription of the gene, whereas the second disruption by 2 minutes is transcription-dependent. A selective RNAi screen identified HSF, GAF, and PARP as each being necessary for the changes in chromatin landscape at Hsp70.
Before HS, the Hsp70 gene contains a chromatin landscape that has many general, as well as some distinct features. Like many other TATA containing genes (Albert, Mavrich, et al, 2007), a highly positioned nucleosome exists downstream of the promoter region and the adjacent nucleosomes on the body of Hsp70 gradually lose their positioning. Likewise, as seen with many genome-wide studies, the promoter, and a region at the 3’ end of the gene, is relatively nucleosome free (Mito, Henikoff, and Henikoff, 2005; Trinklein, Karaoz, et al, 2007; Yuan, Liu, et al, 2005). It is yet to be determined why 3’ ends of genes are hypersensitive to nucleases. However, while many genes in yeast contain a positioned nucleosome starting within the first 100 bp of the transcription unit, Hsp70 contains a nucleosome free region that extends further, with the first nucleosome centered 330 bp following the TSS. This extended nucleosome-free region may be a more general feature of genes containing a paused polymerase.
Our HS time course shows that within 2 minutes following HS, the chromatin landscape of Hsp70 drastically changes. By our assay, following 2 minutes of HS, there no longer exists appreciable protection of a contiguous 100 bp piece of DNA that would normally be provided from a histone octamer. However, there are still detectable levels of histone H3 on the body of the gene, albeit three-fold less than NHS levels. Although these results differ from early observations that histone levels on Hsp70 do not change following HS (Nacheva, Guschin, et al, 1989; Solomon, Larsen, and Varshavsky, 1988), our 3-fold decrease measured by qPCR agrees with more recent quantifications of histone levels following HS (Adelman, Wei, et al, 2006) and may have gone undetected in the qualitative analysis of these early experiments. Early electron microscopy spreads of native transcribing Pol II complexes with a growing RNA chain from D. melanogaster indicate that the bulk of transcribing Pol II in vivo appears to have nucleosomes flanking its path (McKnight, and Miller, 1979). Our results, however, suggest that at least for the rapidly induced Hsp70 gene, the nucleosomal structure present before HS no longer exists following activation of the gene.
We also found that changes in chromatin upon Hsp70 induction extend well beyond the transcription unit of Hsp70 and halt at the scs and scs’ insulating elements. Previous studies of scs and scs’ have shown that these insulators are capable of blocking enhancer functions and establishing chromatin domains that are resistant to position effects (Kellum, and Schedl, 1991; Kuhn, Hart, and Geyer, 2004). However, the scs and scs’ regions have been located by DNA FISH on squashed polytene chromosomes to be within a HS puff at the endogenous 87A HS locus (Kuhn, Hart, and Geyer, 2004). This indicates that the scs and scs’ regions by themselves are not absolute boundaries to changes in chromosome architecture, and supports our observation that puffing is maximal at a time well after nucleosome disruption and therefore denotes additional structural alterations beyond those observed here. Although transcription of CG31211, CG3281, and Aurora did not change following HS, and no factor targeted for RNAi permitted the disruption of nucleosomes beyond scs or scs’, both of these regions include a TSS with detectable amounts of Pol II (Figure S5B), consistent with previous results (Muse, Gilchrist, et al, 2007). It is therefore possible that the promoter architecture with Pol II present at these genes may be responsible for establishing a barrier at these sites. Overall, our results show that scs and scs’ provide a primary barrier to the spread of chromatin decondensation, at least at the nucleosomal level, and add to our limited knowledge of the chromatin architecture of a puff.
Previous results, combined with ours, indicate that transcription-independent chromatin decondensation may prove more general. Changes in chromatin structure independent of transcription have been implicated at Hsp70 in humans (Brown, and Kingston, 1997) and also at developmentally regulated puffs in Drosophila (Crowley, Mathers, and Meyerowitz, 1984). Furthermore, our results indicate that the changes in chromatin at D. melanogaster Hsp70 do not depend on many different transcription factors. In Saccharomyces cerevisiae, many HS genes also lose histone density within the body of the gene by 2 minutes of HS (Zhao, Herrera-Diaz, and Gross, 2005), and as in our study, these changes are independent of SWI/SNF, Gcn5, and Paf1. Overall, transcription-independent chromatin decondensation might allow cells to rapidly activate genes by clearing the obstacles in the path of Pol II prior to its movement, together with its entourage of elongation factors, through the gene.
Our results show that in addition to HSF and GAF, which have previously been implicated in the decondensation at Hsp70 loci (Shopland, Hirayoshi, et al, 1995), PARP is also necessary for rapid changes in the nucleosome architecture of Hsp70. This is consistent with the finding that reduction of PARP expression results in decreased HS puff sizes (Tulin, and Spradling, 2003). Our results go further in demonstrating that PARP aids the rapid removal of nucleosomes within 2 minutes of HS. Poly(ADP-)Ribose (PAR) polymers are the enzymatic product of PARP and have similar chemical and structural features as a nucleic acid (D'Amours, Desnoyers, et al, 1999). Upon activation, PARP polyribosylates itself, which results in PARP’s release from chromatin (Wacker, Ruhl, et al, 2007). The result of this could be two fold. First, since PARP binds to nucleosomes in a similarly repressive manner as linker histone H1 (Kim, Mauro, et al, 2004), the activation of PARP could result in its release from chromatin to reverse any repressive effects on the chromatin structure at Hsp70. Second, the ADP-ribosylation of histones may destabilize the nucleosome, and the creation of these PAR polymers could act locally as a nucleic acid that attracts and removes histones from the body of the Hsp70 gene. Alternatively, PARP could covalently modify another protein to activate its role in removal of nucleosomes.
In addition to histones, PAR could also attract transcription factors that bind nucleic acids. This could explain the rapid recruitment of Pol II and other important transcription factors to the site of active HS transcription (Boehm, Saunders, et al, 2003). Likewise, PAR could also provide a means through which transcription factors recruited to the gene are then retained locally (Yao, Ardehali, et al, 2007). The activation of PARP could thus provide a rapid, transcription-independent method to deplete histones and promote transcription of the Hsp70 gene.
EXPERIMENTAL PROCEDURES
ChIP
Heat shocks were performed as in (Boehm, Saunders, et al, 2003) and ChIP was performed as in (Adelman, Wei, et al, 2006) with the following exceptions. Extracts were sonicated at 4°C on the high sonication setting of the Bioruptor (Diagenode) using one 15’ followed by three 5’ intervals of 20” bursts followed by 1’ of inactivity (corresponding to 24 total bursts) to achieve average fragment sizes of 400bp. The amount of antibody per IP used was: 4 µL rabbit anti-Rpb3 (Adelman, Wei, et al, 2006), 2 µL rabbit anti-HSF (Boehm, Saunders, et al, 2003), and 2 µL rabbit anti-Histone H3-ChIP grade (Abcam ab1791).
High-resolution MNase Mapping
The same procedure for ChIP was used up until resuspension in sonication buffer. At this point, cross-linked cells were resuspended in hypertonic buffer A (300 mM sucrose, 2 mM Mg Acetate, 3 mM CaCl2, 10 mM Tris pH 8.0, 0.1% Triton X-100, and 0.5 mM DTT) to 1 × 108 cells/mL, incubated on ice for 5’, and dounced 20 times with a 2 mL dounce homogenizer (tight pestle, Wheaton). Nuclei were collected by centrifuging at 4 °C for 5’ at 720 g. The pellets were washed twice in buffer A, and then resuspended in buffer D (25% glycerol, 5 mM Mg Acetate, 50 mM Tris pH 8.0, 0.1 mM EDTA, 5 mM DTT) at 1 × 108 nuclei/mL. Chromatin was collected by centrifuging at 4 °C for 5’ at 720 g. The pellets were washed twice in buffer D and then resuspended in buffer MN (60 mM KCl, 15 mM NaCl, 15 mM Tris pH 7.4, 0.5 mM DTT, 0.25 M sucrose, 1.0 mM CaCl2) at 1 × 108nuclei/mL. The equivalent of 2 × 107 nuclei was used per MNase reaction. MNase (USB), diluted in buffer MN, was added so that 0, 0.5, 5, 50, and 500 total units were used per reaction and timed for 30’ at room temperature. Reactions were stopped with the addition of EDTA and SDS to final concentrations of 12.5 mM and 0.5% respectively. The equivalent of 5 × 106 nuclei was removed and processed like ChIP samples from the point of elution from the beads.
Quantitative Real-Time PCR Analysis
ChIP and RT-qPCR primer sets have previously been characterized as in (Adelman, Wei, et al, 2006). Real-Time PCR was performed as in (Ni, Saunders, et al, 2007). For ChIP samples, a standard curve was generated by serially diluting input samples to quantify IP samples. For MNase digests, a fold difference was calculated (Schmittgen, Zakrajsek, et al, 2000) between MNase treated and untreated samples. All values used were collected from the linear range of amplification.
Chemical Treatments
All chemicals were added to S2 cells in media, at final concentrations of 125 µM DRB, 10 mM Sodium Salicylate, and 300 nM PJ34 and allowed to mix for 10’ at room temperature. Cells were then collected following NHS or 2’ HS conditions outlined in the ChIP section.
RNAi Treatments
All RNAi treatments were preformed as in (Adelman, Marr, et al, 2005) except knockdown was allowed to proceed for 96 hours. RNAi primers and RefSeq DNA Identifiers for all knockdowns are provided in Table S1. Briefly, S2 cells were treated with double stranded RNA, designed using the Ambion MEGAscript manual, targeting either the coding sequence of the listed factor or β-galactosidase (LacZ, as a negative control). Cells were collected and split into NHS and 2’ HS samples to be processed using the high resolution MNase assay.
mRNA Expression Analysis
All mRNA expression analyses were as performed in (Adelman, Wei, et al, 2006). Briefly, total RNA was isolated (Qiagen RNeasy) from PARP RNAi and LacZ RNAi S2 cells following 2, 5, and 20 minutes of HS. Hsp70 levels were determined from oligo dT mediated quantitative real-time reverse transcription-PCR. The stable ribosomal protein RpL32 gene (Rp49) was used to internally standardize for the amount of RNA.
Western Blots
Western blots were performed using standard conditions, and input dilutions were used as a quantitative indication of signal linearity. Antibody lab stocks of HSF (Shopland, Hirayoshi, et al, 1995), GAF (O'Brien, Wilkins, et al, 1995), and TFIIS (Adelman, Marr, et al, 2005) were used at dilutions of 1:2000, 1:1000, and 1:3000 respectively. Rabbit anti-Parp serum raised to recognize the C-terminus (Kim, Mauro, et al, 2004) was a gift of W. Lee Kraus and used at a 1:1000 dilution.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank all members of the Lis lab for comments on the manuscript, particularly Abbie Saunders, and Abbie Saunders for ERCC3 antibody production. This work was supported by an NIH grant GM25232 to J.T.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adelman K, Marr MT, Werner J, Saunders A, Ni Z, Andrulis ED, Lis JT. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol. Cell. 2005;17:103–112. doi: 10.1016/j.molcel.2004.11.028. [DOI] [PubMed] [Google Scholar]
- Adelman K, Wei W, Ardehali MB, Werner J, Zhu B, Reinberg D, Lis JT. Drosophila Paf1 modulates chromatin structure at actively transcribed genes. Mol. Cell. Biol. 2006;26:250–260. doi: 10.1128/MCB.26.1.250-260.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- Boehm AK, Saunders A, Werner J, Lis JT. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol. Cell. Biol. 2003;23:7628–7637. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortvin A, Winston F. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science. 1996;272:1473–1476. doi: 10.1126/science.272.5267.1473. [DOI] [PubMed] [Google Scholar]
- Brown SA, Kingston RE. Disruption of downstream chromatin directed by a transcriptional activator. Genes Dev. 1997;11:3116–3121. doi: 10.1101/gad.11.23.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford GE, Holt IE, Whittle J, Webb BD, Tai D, Davis S, Margulies EH, Chen Y, Bernat JA, Ginsburg D, et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS) Genome Res. 2006;16:123–131. doi: 10.1101/gr.4074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley TE, Mathers PH, Meyerowitz EM. A trans-acting regulatory product necessary for expression of the Drosophila melanogaster 68C glue gene cluster. Cell. 1984;39:149–156. doi: 10.1016/0092-8674(84)90200-9. [DOI] [PubMed] [Google Scholar]
- D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999;342(Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- Gaszner M, Vazquez J, Schedl P. The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev. 1999;13:2098–2107. doi: 10.1101/gad.13.16.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardina C, Lis JT. Sodium salicylate and yeast heat shock gene transcription. J. Biol. Chem. 1995;270:10369–10372. doi: 10.1074/jbc.270.18.10369. [DOI] [PubMed] [Google Scholar]
- Giardina C, Lis JT. Polymerase processivity and termination on Drosophila heat shock genes. J. Biol. Chem. 1993;268:23806–23811. [PubMed] [Google Scholar]
- Giardina C, Perez-Riba M, Lis JT. Promoter melting and TFIID complexes on Drosophila genes in vivo. Genes Dev. 1992;6:2190–2200. doi: 10.1101/gad.6.11.2190. [DOI] [PubMed] [Google Scholar]
- Jedlicka P, Mortin MA, Wu C. Multiple functions of Drosophila heat shock transcription factor in vivo. EMBO J. 1997;16:2452–2462. doi: 10.1093/emboj/16.9.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Tan FJ, McCullough HL, Riordan DP, Fire AZ. Flexibility and constraint in the nucleosome core landscape of Caenorhabditis elegans chromatin. Genome Res. 2006;16:1505–1516. doi: 10.1101/gr.5560806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum R, Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991;64:941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- Kim MY, Mauro S, Gevry N, Lis JT, Kraus WL. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 2004;119:803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Knezetic JA, Luse DS. The presence of nucleosomes on a DNA template prevents initiation by RNA polymerase II in vitro. Cell. 1986;45:95–104. doi: 10.1016/0092-8674(86)90541-6. [DOI] [PubMed] [Google Scholar]
- Kraus WL, Lis JT. PARP goes transcription. Cell. 2003;113:677–683. doi: 10.1016/s0092-8674(03)00433-1. [DOI] [PubMed] [Google Scholar]
- Kuhn EJ, Hart CM, Geyer PK. Studies of the role of the Drosophila scs and scs' insulators in defining boundaries of a chromosome puff. Mol. Cell. Biol. 2004;24:1470–1480. doi: 10.1128/MCB.24.4.1470-1480.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- Lewis M, Helmsing PJ, Ashburner M. Parallel changes in puffing activity and patterns of protein synthesis in salivary glands of Drosophila. Proc. Natl. Acad. Sci. U. S. A. 1975;72:3604–3608. doi: 10.1073/pnas.72.9.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Lis JT. Imaging Drosophila gene activation and polymerase pausing in vivo. Nature. 2007;450:198–202. doi: 10.1038/nature06324. [DOI] [PubMed] [Google Scholar]
- Lorch Y, LaPointe JW, Kornberg RD. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987;49:203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- McKnight SL, Miller OL., Jr. Post-replicative nonribosomal transcription units in D. melanogaster embryos. Cell. 1979;17:551–563. doi: 10.1016/0092-8674(79)90263-0. [DOI] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat. Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacheva GA, Guschin DY, Preobrazhenskaya OV, Karpov VL, Ebralidse KK, Mirzabekov AD. Change in the pattern of histone binding to DNA upon transcriptional activation. Cell. 1989;58:27–36. doi: 10.1016/0092-8674(89)90399-1. [DOI] [PubMed] [Google Scholar]
- Ni Z, Saunders A, Fuda NJ, Yao J, Suarez JR, Webb WW, Lis JT. PTEFb is critical for the maturation of RNA Polymerase II into productive elongation in vivo. Mol. Cell. Biol. 2007 doi: 10.1128/MCB.01859-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien T, Lis JT. Rapid changes in Drosophila transcription after an instantaneous heat shock. Mol. Cell. Biol. 1993;13:3456–3463. doi: 10.1128/mcb.13.6.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien T, Wilkins RC, Giardina C, Lis JT. Distribution of GAGA protein on Drosophila genes in vivo. Genes Dev. 1995;9:1098–1110. doi: 10.1101/gad.9.9.1098. [DOI] [PubMed] [Google Scholar]
- Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- Pinnola A, Naumova N, Shah M, Tulin AV. Nucleosomal core histones mediate dynamic regulation of poly (ADP-Ribose) polymerase 1 protein binding to chromatin and induction of its enzymatic activity. J. Biol. Chem. 2007 doi: 10.1074/jbc.M705989200. [DOI] [PubMed] [Google Scholar]
- Rando OJ, Ahmad K. Rules and regulation in the primary structure of chromatin. Curr. Opin. Cell Biol. 2007;19:250–256. doi: 10.1016/j.ceb.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Rasmussen EB, Lis JT. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc. Natl. Acad. Sci. U. S. A. 1993;90:7923–7927. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke H, Horz W. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell. 2003;11:1599–1607. doi: 10.1016/s1097-2765(03)00186-2. [DOI] [PubMed] [Google Scholar]
- Rougvie AE, Lis JT. Postinitiation transcriptional control in Drosophila melanogaster. Mol. Cell. Biol. 1990;10:6041–6045. doi: 10.1128/mcb.10.11.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR. Chromatin remodelling: the industrial revolution of DNA around histones. Nat. Rev. Mol. Cell Biol. 2006;7:437–447. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Zakrajsek BA, Mills AG, Gorn V, Singer MJ, Reed MW. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal. Biochem. 2000;285:194–204. doi: 10.1006/abio.2000.4753. [DOI] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol. Cell. 2006;22:415–422. doi: 10.1016/j.molcel.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Sekinger EA, Moqtaderi Z, Struhl K. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol. Cell. 2005;18:735–748. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Peterson CL. Switching on chromatin: mechanistic role of histone H4-K16 acetylation. Cell Cycle. 2006;5:1361–1365. doi: 10.4161/cc.5.13.2891. [DOI] [PubMed] [Google Scholar]
- Shopland LS, Hirayoshi K, Fernandes M, Lis JT. HSF access to heat shock elements in vivo depends critically on promoter architecture defined by GAGA factor, TFIID, and RNA polymerase II binding sites. Genes Dev. 1995;9:2756–2769. doi: 10.1101/gad.9.22.2756. [DOI] [PubMed] [Google Scholar]
- Simon JA, Sutton CA, Lobell RB, Glaser RL, Lis JT. Determinants of heat shock-induced chromosome puffing. Cell. 1985;40:805–817. doi: 10.1016/0092-8674(85)90340-x. [DOI] [PubMed] [Google Scholar]
- Solomon MJ, Larsen PL, Varshavsky A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988;53:937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- Thummel CS, Burtis KC, Hogness DS. Spatial and temporal patterns of E74 transcription during Drosophila development. Cell. 1990;61:101–111. doi: 10.1016/0092-8674(90)90218-4. [DOI] [PubMed] [Google Scholar]
- Trinklein ND, Karaoz U, Wu J, Halees A, Force Aldred S, Collins PJ, Zheng D, Zhang ZD, Gerstein MB, Snyder M, Myers RM, Weng Z. Integrated analysis of experimental data sets reveals many novel promoters in 1% of the human genome. Genome Res. 2007;17:720–731. doi: 10.1101/gr.5716607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T, Becker PB, Wu C. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature. 1994;367:525–532. doi: 10.1038/367525a0. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- Tulin A, Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299:560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- Udvardy A, Maine E, Schedl P. The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J. Mol. Biol. 1985;185:341–358. doi: 10.1016/0022-2836(85)90408-5. [DOI] [PubMed] [Google Scholar]
- Vazquez J, Schedl P. Sequences required for enhancer blocking activity of scs are located within two nuclease-hypersensitive regions. EMBO J. 1994;13:5984–5993. doi: 10.1002/j.1460-2075.1994.tb06944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virag L, Szabo C. The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol. Rev. 2002;54:375–429. doi: 10.1124/pr.54.3.375. [DOI] [PubMed] [Google Scholar]
- Wacker DA, Ruhl DD, Balagamwala EH, Hope KM, Zhang T, Kraus WL. The DNA Binding and Catalytic Domains of Poly(ADP-ribose) Polymerase-1 Cooperate in the Regulation of Chromatin Structure and Transcription. Mol. Cell. Biol. 2007 doi: 10.1128/MCB.01314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winegarden NA, Wong KS, Sopta M, Westwood JT. Sodium salicylate decreases intracellular ATP, induces both heat shock factor binding and chromosomal puffing, but does not induce hsp 70 gene transcription in Drosophila. J. Biol. Chem. 1996;271:26971–26980. doi: 10.1074/jbc.271.43.26971. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980;286:854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Wu C, Wong YC, Elgin SC. The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell. 1979;16:807–814. doi: 10.1016/0092-8674(79)90096-5. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- Yao J, Ardehali MB, Fecko CJ, Webb WW, Lis JT. Intranuclear Distribution and Local Dynamics of RNA Polymerase II during Transcription Activation. Mol. Cell. 2007;28:978–990. doi: 10.1016/j.molcel.2007.10.017. [DOI] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- Zhao J, Herrera-Diaz J, Gross DS. Domain-wide displacement of histones by activated heat shock factor occurs independently of Swi/Snf and is not correlated with RNA polymerase II density. Mol. Cell. Biol. 2005;25:8985–8999. doi: 10.1128/MCB.25.20.8985-8999.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Hart CM, Laemmli UK. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell. 1995;81:879–889. doi: 10.1016/0092-8674(95)90008-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.