Abstract
The triarylpyridines are potent G-quadruplex ligands that are highly discriminating against duplex DNA and show promising selectivity between intramolecular quadruplexes.
G-Quadruplexes are nucleic acid secondary structures formed from guanine-rich sequences, and comprise a planar arrangement of four guanines (G-tetrad) stabilised by Hoogsteen hydrogen bonding and monovalent cations.1 Putative quadruplex forming sequences have been identified throughout the genome, raising the possibility of function.2 The best-studied example is that of the human telomeric quadruplex, whose stabilisation by a small molecule leads to the inhibition of telomerase activity3 and interference with telomere biology.4 Quadruplex motifs are enriched in gene promoters,2b which is suggestive of their involvement in gene transcription. Promoter quadruplexes have been studied for several proto-oncogenes that include c-myc,5 VEGF,6 bcl-2,7 KRAS8 and c-kit.9
Quadruplex ligands have potential as anti-cancer agents10 that act by interference with telomere maintenance11 or by alteration of oncogene expression levels.5,12 The modification of flat, aromatic molecules has led to G-quadruplex ligands that bind by a mode presumed to involve stacking with the terminal G-tetrad(s).13 However, the G-tetrads are common to all quadruplexes, making discrimination between quadruplexes challenging. Given the potential for a large number of quadruplexes to exist in the genome,2 it is an important goal to create quadruplex ligands that show specificity between different quadruplex structures.
We considered that the triarylpyridines (TAPs, Fig. 1) offer an attractive template for ligand design. The TAPs possess adaptive structural features arising from three rotatable bonds that provide potential for different conformations while retaining a degree of rigidity, somewhat akin to α-helix mimics that have been based on linked aryl groups.14 The TAP scaffold could allow recognition of the quadruplex whereby the central core orients side chains to target the hypervariable loops that distinguish each quadruplex, thereby providing the potential for G-quadruplex discrimination.
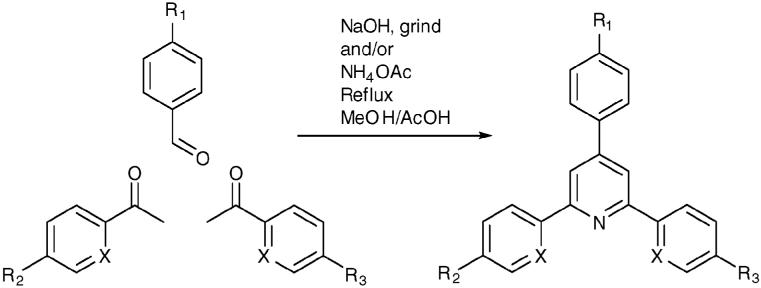
Fig. 1.
Synthesis of triarylpyridines, X = CH or N.
The TAPs were synthesized by straightforward 1-4 step procedures15,16 from commercially available starting materials (Fig. 1 and ESI†). Side chains R1-3 were introduced to provide interactions with distinct quadruplex features (Table 1). In particular, the amine substituents provide potential for hydrogen bonding and cation dipole interactions with the sugar-phosphate backbone and the loops. The heteroatom X was varied (X = CH, N) to explore the effects arising from altered rotamer preferences and potential cation coordination.
Table 1.
Structures of the triarylpyridines 1-20, dissociation constants (Kd) determined by SPR and stabilization potentials (ΔTm) determined by FRET melting
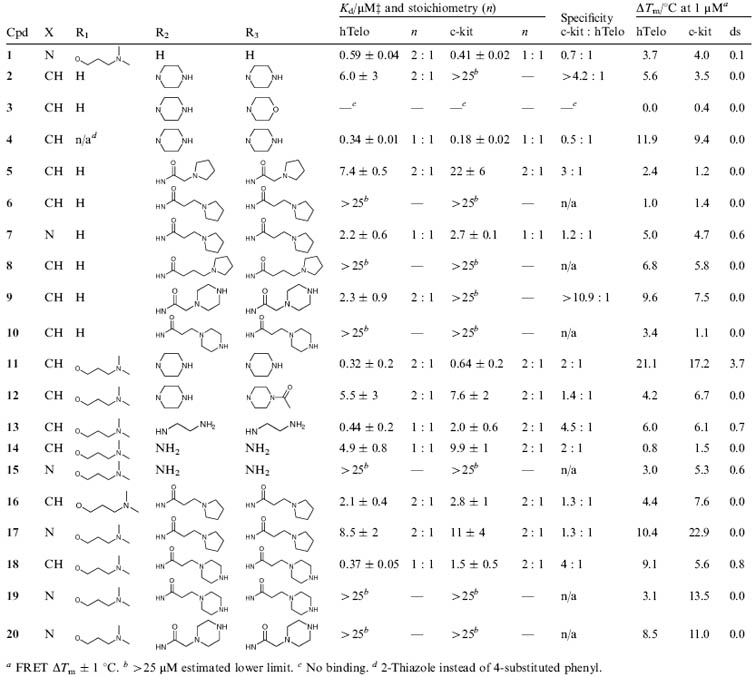 |
FRET ΔTm ± 1 °C.
>25 μM estimated lower limit.
No binding.
2-Thiazole instead of 4-substituted phenyl.
Ligand-quadruplex interactions were evaluated by two complementary methods: Surface Plasmon Resonance17 (SPR) and Fluorescence Resonance Energy Transfer18 (FRET) melting. SPR measures equilibrium binding, while FRET melting analysis provides a measure of the ligand-induced stabilisation of a folded quadruplex.
SPR experiments were performed using three different immobilized DNA targets: the human telomeric sequence d(biotin-[GT2A(G3T2A)4G2]) (hTelo), a c-kit promoter quadruplex9b d(biotin-[C3G3CG3CGCGAG3AG4AG2]) (c-kit) and double-stranded DNA (ds) comprising the oligonucleotide d(biotin-[G2CATAGTGCGTG3CGT2AGC]) hybridized with its complementary strand. FRET experiments were performed using dual-labeled hTelo and c-kit quadruplexes, in addition to a double stranded DNA (see ESI†). The two quadruplexes differ in their conformational heterogeneity and dynamics.19,20 The results are summarized in Table 1 and example data for ligand 1 are given in Fig. 2.
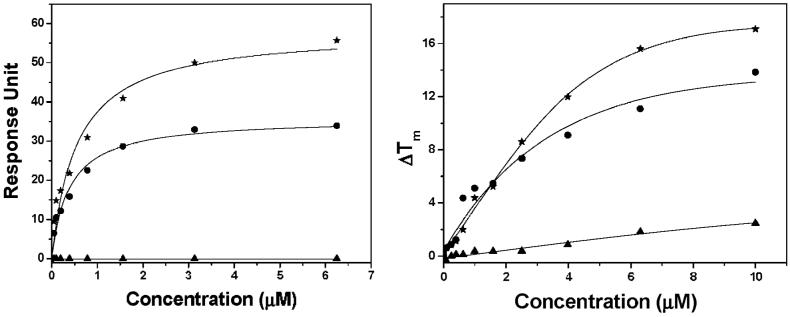
Fig. 2.
(Left) SPR binding curves for 1 binding to hTelo (*), c-kit (•) and ds (▲) DNA (running buffer: 50 mM Tris-HCl pH 7.4, 100 mM KCl). (Right) FRET assay for hTelo (*), c-kit (•) and ds (▲) DNA in the presence of 1 (buffer: 60 mM potassium cacodylate pH 7.4).
In support of our reasoning, we found that relatively small changes in the structure of TAPs significantly altered binding and selectivity with respect to DNA quadruplexes (Table 1). It is noteworthy that none of the TAPs showed detectable interaction with double-stranded DNA by SPR‡ or by FRET melting. This is indicative of a general preference of TAPs for quadruplex, rather than duplex DNA, a pre-requisite for selective chemical intervention of cell biology.
There was gratifyingly wide variation in quadruplex binding affinity observed. Ligand 4 showed the highest affinity for a quadruplex with a preference for c-kit (Kd = 180 nM). Ligands 11, 13 and 18 show a preference for hTelo with submicromolar affinities (Kd values of 320, 440 and 370 nM, respectively). In contrast, TAP 3 showed no detectable interaction with either DNA quadruplex or duplex by SPR or FRET melting. Overall, the ligands exhibited a wide range of quadruplex binding affinities spanning greater than 2 orders of magnitude. Our design hypothesis presumed that side chains are the key to quadruplex discrimination. Ligand 1 has only one side chain and displays good affinity for quadruplexes hTelo and c-kit (Kd values of 590 nM and 410 nM, respectively), but without discrimination between them. While several ligands exhibited a preference for either c-kit or hTelo in the 2-5 fold range (5, 11, 13, 18), ligand 9 showed a preference for hTelo vs. c-kit of at least an order of magnitude.‡
The relationship between stabilisation (ΔTm) and equilibrium binding (Kd) is not straightforward, and thus a simple correlation for ΔTm and Kd was neither observed nor expected. However, for cases where no significant binding was detected by SPR the associated ΔTm was always found to be near zero (e.g. most cases for duplex). Ligand 11 is an example that showed good binding affinity to hTelo and c-kit (Kd = 320 nM and 640 nM, respectively) and stabilisation (ΔTm = 21 °C and 17 °C, respectively). However, some molecules showed strong stabilisation of DNA without a correspondingly high binding affinity. For example, 17 exhibits a large ΔTm for c-kit (23 °C)§ but a modest Kd (11 μM for c-kit). Molecules that induce a large ΔTm may be able to influence DNA topology by stabilisation of the quadruplex form, whereas a molecule with low Kd (strong binding) but low ΔTm (weak stabilisation) may be better suited to interfere with protein-quadruplex recognition.21,¶
There are noteworthy correlations between structural features of TAPs and their binding properties. The terpyridines (X = N) always show higher stabilisation temperatures compared to their benzene (X = CH) counterparts. For example, 16 shows moderate stabilisation of hTelo and c-kit (ΔTm = 4 °C and 8 °C, respectively), whereas 17 shows significantly higher stabilisation (ΔTm = 10 °C and 23 °C, respectively). The simple replacement of an amine in 2 with oxygen (3) leads to loss of detectable binding and stabilizing properties, suggestive that protonation of the amine of 2 may be critical. Replacement of the benzene ring in the 4-position of the central pyridine of 2 with a thiazole (i.e. 4) results in > 100-fold stronger binding to c-kit (Kd = 180 nM for 4) compared to 2 (Kd > 25 μM for c-kit). The origin of this may be steric interactions between the hydrogens on the phenyl ring and the central pyridine core of 2, which could cause the ring to reside slightly out of plane, in contrast with 4 being able to adopt a more planar structure owing to reduced steric interactions of the heteroatoms (N and S) and smaller ring size. It was also notable that some ligands were found to bind with 2 : 1 stoichiometry and others with 1 : 1 binding, suggestive of more than one mode of binding. Overall it was found that the ligand 4 is the tightest binder and has preference for c-kit quadruplex, while ligands 11, 13 and 18 show preference for hTelo quadruplex.
The TAPs are a new class of quadruplex binding ligands that show versatility in their specificities, affinities and stabilisation potential. These ligands do not bind to duplex DNA and have provided proof of concept for the discrimination between different genomic DNA quadruplexes by a small molecule. Investigations into the biological activities of these molecules are currently underway.
Acknowledgments
We thank the BBSRC for project funding and CRUK for program funding. We also thank the BBSRC for a studentship (Z.A.E.W.) and the EPSRC Mass Spectrometry Service for MS analysis. S.B. is a BBSRC Career Development Research Fellow. We thank James Redman and Colin Raston for discussions prior to this project.
Footnotes
Electronic supplementary information (ESI) available: Experimental procedures for synthesis of the TAPs, SPR and FRET.
Above 25 μM, the TAPs were generally observed to adhere to the SA Biacore sensor chip leading to data that could not be fitted; thus 25 μM has been quoted as a lower limit for weak binding ligands.
The Tm of c-kit by FRET melting (in 60 mM cacodylate buffer) was found to be 71 ± 1 °C and ΔTm 24 °C (i.e. Tm = 95 °C) is the maximum ΔTm that can be confidently measured.
We cannot rule out the possibility that molecules in the FRET melting experiments stabilise a non-quadruplex secondary structure.
Notes and references
- 1.Neidle S, Balasubramanian S, editors. Quadruplex Nucleic Acids. Cambridge, UK: Royal Society of Chemistry; 2006. [Google Scholar]
- 2.(a) Huppert JL, Balasubramanian S. Nucleic Acids Res. 2007;35:406. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Huppert JL, Balasubramanian S. Nucleic Acids Res. 2005;33:2908. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Todd AK, Johnston M, Neidle S. Nucleic Acids Res. 2005;33:2901. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun DY, Thompson B, Cathers BE, Salazar M, Kerwin SM, Trent JO, Jenkins TC, Neidle S, Hurley LH. J. Med. Chem. 1997;40:2113. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 4.Gomez D, Paterski R, Lemarteleur T, Shin-ya K, Mergny J-L, Riou J-F. J. Biol. Chem. 2004;279:41487. doi: 10.1074/jbc.M406123200. [DOI] [PubMed] [Google Scholar]
- 5.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11593. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun D, Guo K, Rusche JJ, Hurley LH. Nucleic Acids Res. 2005;33:6070. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dexheimer TS, Sun D, Hurley LH. J. Am. Chem. Soc. 2006;128:5404. doi: 10.1021/ja0563861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cogoi S, Xodo LE. Nucleic Acids Res. 2006;34:2536. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Fernando H, Reszka AP, Huppert JL, Ladame S, Rankin S, Venkitaraman AR, Neidle S, Balasubramanian S. Biochemistry. 2006;45:7854. doi: 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Rankin S, Reszka AP, Huppert J, Zloh M, Parkinson GN, Todd AK, Ladame S, Balasubramanian S, Neidle S. J. Am. Chem. Soc. 2005;127:10584. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Burger AM, Dai F, Schultes CM, Reszka AP, Moore MJB, Double JA, Neidle S. Cancer Res. 2005;65:1489. doi: 10.1158/0008-5472.CAN-04-2910. [DOI] [PubMed] [Google Scholar]; (b) Pennarun G, Granotier C, Gauthier LR, Gomez D, Hoffschir F, Mandine E, Riou J-F, Mergny J-L, Mailliet P, Boussin F. Oncogene. 2005;24:2917. doi: 10.1038/sj.onc.1208468. [DOI] [PubMed] [Google Scholar]; (c) Kim MY, Vankayalapati H, Shin-Ya K, Wierzba K, Hurley LH. J. Am. Chem. Soc. 2002;124:2098. doi: 10.1021/ja017308q. [DOI] [PubMed] [Google Scholar]
- 11.Zhou J, Zhu X, Lu Y, Deng R, Huang Z, Mei Y, Wang Y, Huang W, Liu Z, Gu L, Zeng Y. Oncogene. 2006;25:503. doi: 10.1038/sj.onc.1209067. [DOI] [PubMed] [Google Scholar]
- 12.(a) Bejugam M, Sewitz S, Shirude PS, Rodriguez R, Shahid R, Balasubramanian S. J. Am. Chem. Soc. 2007;129:12926. doi: 10.1021/ja075881p. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ou T, Lu Y, Zhang C, Huang Z, Wang X, Tan J, Chen Y, Ma D, Wong K, Tang JC, Chan AS, Gu L. J. Med. Chem. 2007;50:1465. doi: 10.1021/jm0610088. [DOI] [PubMed] [Google Scholar]
- 13.(a) Shirude PS, Gilles ER, Ladame S, Godde F, Shin-ya K, Huc I, Balasubramanian S. J. Am. Chem. Soc. 2007;129:11890. doi: 10.1021/ja073775h. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) White EW, Tanious F, Ismail MA, Reszka AP, Neidle S, Boykin DW, Wilson DW. Biophys. Chem. 2007;126:140. doi: 10.1016/j.bpc.2006.06.006. [DOI] [PubMed] [Google Scholar]; (c) Seenisamy J, Bashyan S, Gokhale V, Vankayalapati H, Sun D, Siddiqui-Jain A, Steiner AN, Shin-ya K, White E, Wilson DW, Hurley LH. J. Am. Chem. Soc. 2005;127:2944. doi: 10.1021/ja0444482. [DOI] [PubMed] [Google Scholar]
- 14.Orner BP, Ernst JT, Hamilton AD. J. Am. Chem. Soc. 2001;123:5382. doi: 10.1021/ja0025548. [DOI] [PubMed] [Google Scholar]
- 15.Cave GWV, Raston CL. J. Chem. Soc., Perkin Trans. 1. 2001:3258. [Google Scholar]
- 16.Tamami B, Yeganeh H. Polymer. 2000;42:415. [Google Scholar]
- 17.(a) Teulade-Fichou M-P, Carrasco C, Guittat L, Bailly C, Alberti P, Mergny J-L, David A, Lehn J-M, Wilson WD. J. Am. Chem. Soc. 2003;125:4732. doi: 10.1021/ja021299j. [DOI] [PubMed] [Google Scholar]; (b) Schouten JA, Ladame S, Mason SJ, Cooper MA, Balasubramanian S. J. Am. Chem. Soc. 2003;125:5594. doi: 10.1021/ja029356w. [DOI] [PubMed] [Google Scholar]
- 18.(a) De Cian A, Guittat L, Kaiser M, Sacca B, Amrane S, Bourdoncle A, Alberti P, Teulade-Fichou M-P, Lacroix L, Mergny J-L. Methods. 2007;42:183. doi: 10.1016/j.ymeth.2006.10.004. [DOI] [PubMed] [Google Scholar]; (b) Mergny J-L, Maurizot J-L. ChemBioChem. 2001;2:124. doi: 10.1002/1439-7633(20010202)2:2<124::AID-CBIC124>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 19.(a) Ying L, Green JJ, Li H, Klenerman D, Balasubramanian S. Proc. Natl. Acad. Sci. U. S. A. 2003;100:14629. doi: 10.1073/pnas.2433350100. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lee JY, Okumus B, Kim DS, Ha T. Proc. Natl. Acad. Sci. U. S. A. 2003;102:18938. doi: 10.1073/pnas.0506144102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shirude PS, Okumus B, Ying L, Ha T, Balasubramanian S. J. Am. Chem. Soc. 2007;129:7485. doi: 10.1021/ja070497d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fry M. Front. Biosci. 2007;12:4336. doi: 10.2741/2391. [DOI] [PubMed] [Google Scholar]