Abstract
During apoptosis, cytochrome c (cyt c) is released from intermembrane space of mitochondria into the cytosol where it triggers caspase-dependent machinery. We discovered that cyt c plays another critical role in early apoptosis as a cardiolipin (CL)-specific oxygenase to produce CL hydroperoxides required for release of pro-apoptotic factors. We quantitatively characterized the activation of peroxidase activity of cyt c by CL and hydrogen peroxide. At low ionic strength and high CL/cyt c ratios, peroxidase activity of CL/cyt c complex was increased >50 times. This catalytic activity correlated with partial unfolding of cyt c monitored by Trp59 fluorescence and absorbance at 695 nm (Fe-S(Met80) band). The peroxidase activity increase preceded the loss of protein tertiary structure. Monounsaturated tetraoleoyl-CL (TOCL) induced peroxidase activity and unfolding of cyt c more effectively than saturated tetramyristoyl-CL (TMCL). TOCL/cyt c complex was found more resistant to dissociation by high salt concentration. These findings suggest that electrostatic CL/cyt c interactions are central to the initiation of the peroxidase activity, while hydrophobic interactions are involved when cyt c’s tertiary structure is lost. In the presence of CL, cyt c peroxidase activity is activated at lower H2O2 concentrations than for isolated cyt c molecules. This suggests that redistribution of CL in the mitochondrial membranes combined with increased production of H2O2 can switch on the peroxidase activity of cyt c and CL oxidation in mitochondria - a required step in execution of apoptosis.
Keywords: cytochrome c, cardiolipin, apoptosis, peroxidase activity
Studies of protein folding/unfolding are one of the central topics in contemporary structural biology. Theoretical considerations predict the potential heterogeneous nature of the process, however, the time resolution of commonly used biochemical techniques is often insufficient and masks the complexity inherent in their ensemble averaging (1). It also remains unclear whether protein folding/unfolding intermediates are functionally competent or not.
Cytochrome c (cyt c), an electron carrier between respiratory complexes III and IV, is a common “model protein” used in studies of folding-unfolding (reviewed in (2, 3)). Normally cyt c performs its functions by reversibly binding with its redox partners integrated into mitochondrial membranes and diffusing freely in the intermembrane space of mitochondria. A renewed interest in cyt c and its interactions with other cell components was brought up by the discovery of a vital role cyt c plays in the programmed cell death mechanism (apoptosis). The release of cyt c from mitochondria followed by its binding to Apaf-1 (apoptotic protease activating factor-1) constitutes a critical point of “no return” in the execution of the apoptotic program.
Cyt c is a positively charged protein (isoelectric point is near pH 10, net charge is +8e at neutral pH), while the inner mitochondrial membrane (IMM) contains a large fraction (up to 25%) of a negatively charged phospholipid, cardiolipin (CL) (4). Cyt c readily binds to anionic membranes and it was shown that this binding is accompanied by protein conformational transitions and changes in its chemical reactivity (5). It is believed that partial unfolding of cyt c upon its interactions with anionic phospholipids, including CL, confers peroxidase activity on the protein.
Recently, we demonstrated that at early stages of apoptosis cyt c bound to CL-containing mitochondrial membranes acts as a peroxidase that selectively catalyzes CL peroxidation; the latter contributes to the outer mitochondrial membrane permeation, release of cyt c into the cytosol and initiation of the apoptotic program (6). Numerous studies examined lipid/cyt c interactions (7-8). However, important qualitative and quantitative characteristics of cyt c binding with CL, resulting in protein conformational transitions and unfolding to yield the peroxidase activity, have not been sufficiently studied.
In the current study, we considered the interplay between peroxidase activity and lipid-induced conformational transitions of cyt c in simple biochemical model systems with the ultimate goal to understand better the specificity of mitochondrial lipid oxidation in apoptosis, the control mechanism of cyt c peroxidase activity in the cell and its connection to other apoptotic processes such Ca2+ release, mitochondria swelling and others.
First, we analyzed quantitatively binding of cyt c to CL-containing liposomes of different lipid composition at high and low ionic strength conditions using electrophoresis and competitive binding with a positively charged hydrophobic compound, nonyl acridine orange, NAO. Next, we described the results of electronic absorption and fluorescence measurements of structural transformations in cyt c upon binding to CL liposomes and contrasted them to CL/cyt c complex peroxidase activity data obtained using two oxidation assays with etoposide and 2′,7′-dichlorodihydrofluorescein (H2DCF). Finally, we present data showing that the activation and deactivation of the peroxidase activity of cyt c complexes is a combined result of binding to CL and chemical oxidative modification of amino acids and the heme moiety during the course of the peroxidase reaction.
Materials and Methods
Materials
Horse heart cytochrome c (cyt c, type C-7752, >95%), diethylenetriaminepentaacetic acid (DTPA), etoposide (Et, VP16, demethylepipodophyllotoxin-ethylediene-glucopyranoside), guanidine hydrochloride (GdmCl) and hydrogen peroxide (H2O2) were purchased from Sigma-Aldrich. 2′,7′-dichlorodihydrofluorescein diacetate (H2DCF), acridine orange 10-nonyl (nonyl acridine orange, NAO) were purchased from Molecular Probes Inc. (Eugene, OR). 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine (DOPC), 1,1′2,2′-Tetraoleoyl Cardiolipin (TOCL), 1,1′2,2′-tetralinoleoyl cardiolipin (TLCL), 1,1′,2,2′-Tetramyristoyl Cardiolipin (TMCL), bovine heart cardiolipin (BHCL) were purchased from Avanti Polar Lipids Inc.
Cytochrome c
Horse heart cyt c (type C-7752) used in our studies was prepared according to the method of Hagihara (9) with acetic acid without using TCA and had purity >95% (according to the manufacturer). The quality of cyt c preparation was evaluated using several tests. PAGE staining with Coomassie Brilliant blue revealed one band corresponding to monomeric form of the protein indicating that the content of cyt c polymers was negligible. The presence of denatured protein was assessed by: 1) accessibility of the heme site to NO, 2) initial and induced peroxidase activity, and 3) characteristic constitutive and induced fluorescence of Trp59. We observed a dramatic increase of the accessibility (> 15 times) of cyt c Fe-porphyrin to NO in the presence of CL-containing liposomes as indicated by the formation of EPR-detectable Fe-nitrosyl complexes. Moreover, the peroxidase activity of cyt c increased up to 50 times upon addition of CL-containing liposomes (see Results). Finally, characteristic fluorescence of Trp59 was observed from TOCL/cyt c complex, although this fluorescence response was essentially undetectable from cyt c alone (see Results). These results indicate that the presence of modified cyt c with lost distal Met80 ligand was negligible in our preparations of cyt c (as this would result in increased NO accessibility, peroxidase activity, and Trp59 fluorescence of cyt c samples in the absence of CL and reduce the observed stimulation by CL). Thus, a dominant fraction of cyt c used in our experiments (>∼95%) had Met80 ligand in place.
Small unilamellar liposomes were prepared by sonication in PBS buffer (10 mM plus 100 μM DTPA, pH=7.0), phosphate buffer (50 mM plus 100 μM DTPA, pH=7.0) or HEPES buffer (25 mM, pH=7.4). Liposomes were prepared from 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine (DOPC) and cardiolipin (CL) (1:1 ratio) or DOPC alone (2.5 mM/2.5 mM and 5 mM correspondingly). Cardiolipins used were: 1,1′2,2′-tetraoleoylcardiolipin (TOCL), 1,1′2,2′-tetramyristoylcardiolipin (TMCL), 1,1′2,2′-tetralinoleoylcardiolipin (TLCL) and bovine heart cardiolipin (BHCL).
EPR peroxidase activity measurements
Peroxidase activity of cyt c or CL/cyt c complex was quantitatively characterized monitoring the formation of etoposide phenoxyl radical at 25°C. Cyt c or liposomes/cyt c (40 μM or 400/40 μM) was mixed with H2O2 (100 μM) and etoposide (700 μM). Samples were placed in gas-permeable Teflon tubing (0.8 mm i.d., 0.013 mm thickness; Alpha Wire Corp. (Elizabeth, NJ)) folded into quarters. EPR instrument settings were: center field - 3350 G, scan range - 25 G, modulation amplitude 1 G, receiver gain - 1000, time constant - 0.03 s, scan time - 4 min, microwave power - 10 mW. The signal of etoposide radical have been recorded for 4 min every 20 sec, thus time-dependence of the EPR signal amplitude has been obtained. Through double integrating EPR signals of either etoposide radical or TEMPO as stable nitroxide radical of known concentration, we obtained the calibration coefficient (2.51/gain) connecting the amplitude of the observed etoposide radical EPR signal (AEPR) with its concentration:
Second order rate constant of peroxidase reaction of CL/cyt c has been derived fitting the observed time-dependence of etoposide radical concentration with results of numerical simulations using the following differential equations (assuming that for every hydrogen peroxide molecule reduced two etoposide molecules were oxidized to form Et· under conditions of the etoposide excess [Et] > 700 μM):
The value of the second order recombination rate of etoposide radicals was also obtained from fitting the time-dependence of etoposide radical concentration [Et·](t) :
Fluorometric determination of cyt c peroxidase activity
Peroxidase activity of cyt c and liposomes/cyt c was measured using Millipore Fluorescence Measurement System CytoFluor™ 2350. 2′,7′-dichlorofluorescein (DCF) fluorescence was detected using an excitation and emission filter B (485±20 nm, 530±25 nm). A Shimadzu spectrofluorimeter RF-5301 PC was employed for fluorescence measurements in a 0.2-ml quartz cuvette. DCF fluorescence spectra were detected using an excitation wavelength of 502 nm, emission 522 nm (excitation and emission slit of 1.5 nm). For measuring inactivation constant, the fluorescence spectra were recorded under the following conditions: 1 μM cyt c, 500 nM H2DCF, lipid:cyt c ratio 20:1, 50 mM phosphate buffer pH=7.0 with 100 μM DTPA. Peroxidase activity was estimated 30 sec after H2O2 treatment (10-100 μM) by monitoring fluorescence growth at 522 nm.
CL/cyt c binding constant assessments
DOPC/CL-liposomes (2 mM total lipids, 1:1) were incubated with cyt c (40 μM) for 30 min. The mixture was then incubated in the presence of acridine 10-nonyl bromide (NAO, 2-12 mM) in 50 mM phosphate buffer (pH=7.0) for 1 h at room temperature. Samples (5 μl) were applied to 7.5% PAGE and the reverse mode electrophoresis was performed. After that, the fluorescence of NAO was monitored at 520 nm band (excitation by UV) on a BioRad MultiImager (Hercules, CA) to confirm that band signals from NAO and cyt c do not interfere. Then cyt c in the gel was stained with 0.25% Coomassie Brilliant Blue R-250 and quantitated by optical density measurements. CL/cyt c binding constants were calculated using the following equation (10):
where [NAO] - concentration of NAO that displaces 90% of cyt c from the complex with CL as estimated from the observed optical density of the cyt c band in the gel (proportional to the concentration of unbound cyt c). Klipid-NAO = 2 × 106 M-1 for cardiolipin (11).
Agarose gel electrophoresis was run for 15 min h at 50 V in non-denaturing 35 mM HEPES buffer (pH 7.4) containing imidazole (43 mM) using horizontal gel system “Mupid-21” (Cosmo Bio). Samples with cyt c and CL at different ratios were incubated in HEPES buffer (25 mM, pH=7.4) and applied onto 0.8 % agarose gel (120 pmol protein per sample). Gels were stained for protein with 0.25% Coomassie Brilliant Blue R-250 in 45% methanol, 10% acetic acid.
Absorption spectroscopy
Optical spectra were recorded on UV160U spectrophotometer (Shimadzu, Japan) in 1 ml cuvettes. To increase sensitivity and obtain cyt c spectra in the 650-750 nm range, the following procedure of “sample exchange” was used. Two cuvettes containing (1) reference and (2) sample with cyt c, were placed in the spectrophotometer with cuvette 2 as a reference sample and cuvette 1 as a measured sample; then the baseline was established and recorded. After exchanging the cuvettes in the compartments the spectrum was recorded again. The amplitude was twice as high as that of a usual spectrum. Liposomes were used as a reference for liposomes/cyt c sample. Conditions were: cyt c 50 μM, liposomes/cyt c 10:1, H2O2 1 mM. The absorption in the 650-750 nm area is strongly affected by a broad shoulder of the strong 550 nm peak. We approximated this by slowly changing the absorption shoulder with a linear function and subtracted the linear fit from the total spectrum to emphasize the 695 nm charge-transfer band. The absorption at 410 nm was determined in a 20-fold buffer-diluted 50 μl sample.
Tryptophan fluorescence
The tryptophan fluorescence of cyt c in the presence and in the absence of liposomes was measured on RF-5301 PC spectrofluorimeter (Shimadzu, Japan) using standard quartz cuvettes with an optical path length of 1 cm. The excitation wavelength was 293 nm; the emission spectra were recorded at 300-500 nm regions. Cyt c 5 μM. Complex liposomes/cyt c incubation was 15 min.
Statistical Analyses
Data are expressed as means ±S.D. of at least triplicate determinations. Changes in variables were analyzed by one-way ANOVA for multiple comparisons. Difference were considered significant at p<0.05.
Results
Binding of cyt c to CL/PC liposomes: effects of CL with different acyl chains
Ferric horse heart cytochrome c (cyt c) has a net charge +8e at neutral pH. Therefore, cyt c strongly interacts electrostatically with membranes containing acidic phospholipids (such as CL) and the interaction is very sensitive to ionic strength (12). Hydrophobic interactions and hydrogen bonding are also important for lipid/cyt c binding although their role is much more difficult to characterize (13).
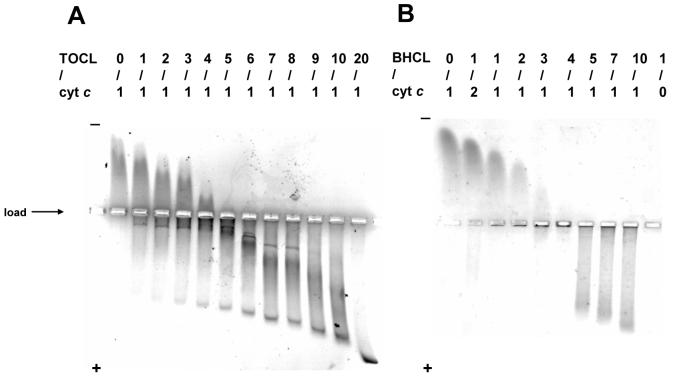
We studied the binding stoichiometry of cyt c and CL-containing liposomes probing electrophoretic mobility of CL/cyt c complexes in native agarose gels (Figure 1). Four types of cardiolipin differing by the degree of saturation of their acyl chains have been used (fully saturated tetramyristoyl-CL (TMCL), mono-unsaturated tetraoleoyl-CL (TOCL), polyunsaturated tetralinoleoyl-CL (TLCL), and polyunsaturated bovine heart CL (BHCL) predominantly containing TLCL). Cyt c was incubated with different amounts of CL (methanol-chloroform, ∼4:1 mixture, less than 2%) in 50 mM phosphate buffer for 1 hr. The gel density (0.8 %) allows cyt c, CL or CL/cyt c complexes to enter the gel matrix, Coomassie Blue staining was been used for detection of free and CL-bound cyt c. As expected, positively charged cyt c molecules moved to the cathode (Figure 1, first lane), while anionic CL liposomes migrated to the anode (last lane, not seen in the stained for protein gels). The apparent mobility of cyt c decreased with increased lipid/protein ratio indicating that, at a low ionic strength, the protein tightly binds to CL-containing membranes and its positive charge is neutralized by the negative charge of CL.
Figure 1. Native agarose gels of CL/cyt c complexes at different CL/cyt c ratios.
A: TOCL: B: Bovine heart CL (BHCL).
TOCL or BHCL dissolved in methanol-chloroform (∼4:1 mixture, less than 2%) were added to cyt c solution in HEPES buffer (25 mM, pH=7.4 with 100 μM DTPA) and incubated for 15 min. Then, the samples were loaded onto 0.8% aragose gel (as indicated on the figure) and the electrophoresis was run for 15 min in HEPES-imidazole buffer (35 mM HEPES, 43 mM imidazole, pH = 7.4). The gels were stained with Coomassie Brilliant Blue R-250. Cyt c - 720 pmol (120 μM, 6 μl), CL concentration was changed to achieve CL/cyt c ratios from 0:1 up to 20:1 molar ratios.
CL/cyt c ratios that corresponded to the reversal in direction of the electrophoretic mobility of the CL/cyt c complex (neutral charge of CL/cyt c complex) were the same for TOCL and TLCL. CL has two phosphate groups and thus, its formal electric charge in the deprotonated state is -2 and CL should neutralize cyt c molecules with a stoichiometry of 4:1. Electrostatic interactions between phosphate groups within the same CL molecule and between neighboring molecules increase the pKa values of the phosphate groups and thus, in CL-containing membranes, only a fraction of the CL molecules are ionized (the proportion of deprotonated CL molecules is lower for membranes with higher CL-content). Electrostatic interactions with positively charged cyt c molecules should shift the pKa values of phosphate groups to smaller values and increase the absolute value of the effective negative charge of CL.
The electrophoretic mobility of the BHCL/cyt c complex becomes zero at a CL/cyt c ratio of 4:1 (Figure 1B) corresponding to the ratio at which the cyt c and CL formal charge cancel. Also, the mobility of the TOCL/cyt c complex becomes zero only at a 4:1 lipid/protein ratio. Similar experiments were also performed with liposomes (data not shown). The agarose gel with low density (0.8%) allows small liposomes to enter the gel matrix and migrate in electric field. Gels with liposome-cyt c samples looked essentially very similar to those obtained with CL dissolved in methanol-chloroform (∼4:1 mixture, less than 2%) except that the isoelectric point was shifted. While isoelectric point was observed at a 4:1 ratio (CL:cyt c) when CL was added in a solvent (as expected for non-charged CL/cyt c complex), in the presence of liposomes the isoelectric point was found at a 5:1 (CL:cyt c) ratio. This difference probably indicates a partial neutralization of CL in liposomes due to electrostatic repulsion of CL molecules.
Incubation of cyt c with CL in high-ionic strength buffer (50 mM phosphate buffer with 1 M KCl) abolishes the complex formation, as the apparent electrophoretic mobility of cyt c does not change even at a 25:1 lipid/protein molar ratio (data not shown). Saturated TMCL did not reveal any significant binding with cyt c as evidenced by a lack of changes in cyt c migration after incubation with TMCL-containing liposomes.
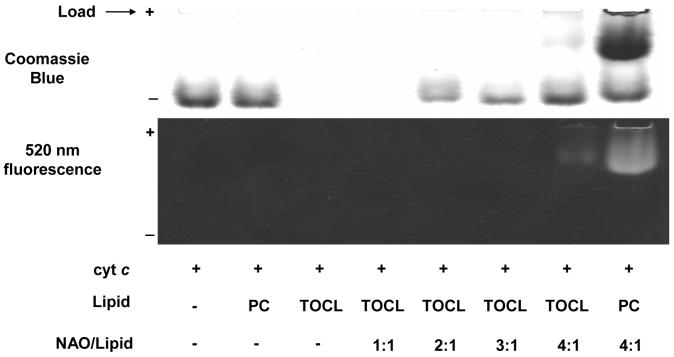
We determined the strength of CL/cyt c interactions using the competitive binding technique. Acridine 10-nonyl bromide (nonyl acridine orange, NAO) has been shown to specifically interact with CL (both electrostatically and hydrophobically) and is widely used as a probe for CL in cells (14-15). We used NAO as a competitor of cyt c for binding to CL-containing liposomes. Based on the known value of the binding constant CL-NAO (11), we calculated the CL/cyt c binding constant (Figure 2). At a 2:1 ratio (NAO:TOCL) approximately 25% of the cyt c was displaced from liposomes, and at a 4:1 ratio almost all of the cyt c (90%) was forced out liposomes. DOPC did not bind cyt c at all, and NAO did not interact with DOPC or cyt c. The data on Kb for different CLs are shown in Table 1. The values of the binding constants obtained are in agreement with previous measurements of cyt c/anionic membrane affinities at low ionic strength as well as with the theoretical estimates that only take into account electrostatic interactions (8, 16). Although consideration of only electrostatic interactions between cyt c and CL-containing membranes is sufficient to get a crude estimate of the strength of cyt c/membrane binding, substantial variations of the binding constants for different types of cardiolipin presented in Table 1 indicate that acyl chain chemical composition and CL/cyt c hydrophobic interactions are important for cyt c/membrane binding. Binding of cyt c is increased in the same progression TMCL < TOCL = TLCL < BHCL as the increase in the number of double bonds in CL acyl chains.
Figure 2. Native PAGE of CL/cyt c complexes in the presence of NAO.
Typical PAGE stained for protein (upper panel) and for NAO fluorescence (lower panel). Before staining of gels with Coomassie Brilliant Blue R-250 for protein, fluorescence of NAO was measured using a 520 nm band pass filter. Note that NAO and cyt c have different mobility in the gel.
Table 1.
Cyt c binding to CL-containing membranes at low ionic strength
| Lipid | K binding constant (Kb), × 109 M-1 |
|---|---|
| Dioleoyl phosphatidylcholine | Not detectable |
| Tetraoleoyl cardiolipin | 1.7 ± 1.0 |
| Tetralinoleoyl cardiolipin | 1.6± 0.2 |
| Bovine heart cardiolipin | 6.6 ± 3.1 |
| Tetramyristoyl cardiolipin | Not detectable |
To confirm that competition between cyt c and NAO for binding with CL affected the peroxidase activity of CL/cyt c complexes, the peroxidase activity was measured by an ESR assay. Incubation of CL-containing liposomes with cyt c, and subsequent addition of NAO lead to a complete inhibition of peroxidase activity at 1:1 ratio of NAO to CL (data not shown). Addition of an unfolding reagent, guanidine hydrochloride (GdmCl, 6 M) to cyt c induced peroxidase activity by its partially denaturing effect. Because thus triggered peroxidase activity is produced in the absence of CL, any effect of NAO would be independent of CL. We found that NAO exerted only a slight (less than 25%) inhibition of the peroxidase activity as assessed by the etoposide phenoxyl radical formation. Moreover, we established that NAO was completely ineffective in inhibiting the activity of horseradish peroxidase assessed by the etoposide phenoxyl radical generation. These results suggest that the inhibitory action of NAO on the peroxidase complex CL/cyt c is likely realized through interactions of NAO with CL.
Changes of cyt c structure and peroxidase activity upon binding to CL/PC liposomes
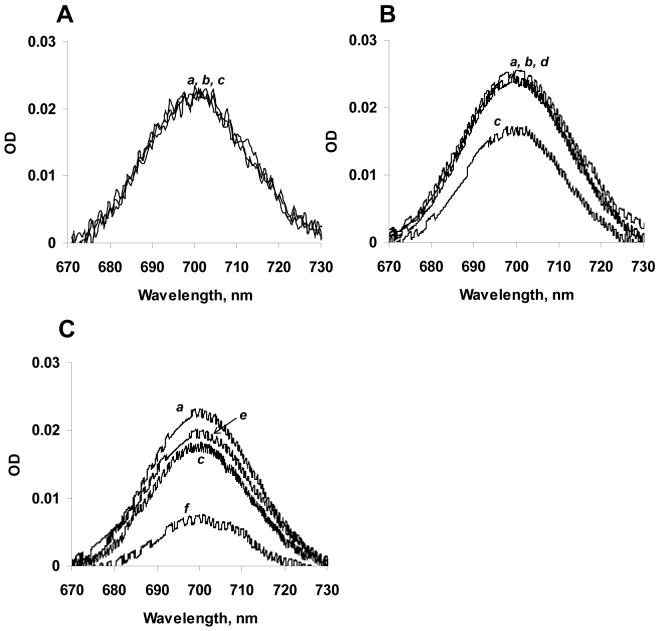
When cyt c binds to anionic liposomes, its tertiary structure changes (17, 18). The heme of cyt c is covalently attached to the polypeptide chain by residues Cys14 and Cys17. The proximal and distal axial ligand positions of the cyt c heme iron are normally occupied by His18 and Met80 respectively. A weak absorption band at 695 nm in the electronic spectrum of cyt c - associated with the Fe-S(Met80) bond - has been commonly used for assessments of conformational transitions in cyt c associated with the disruption of Fe-S(Met80) bond. Incubation of cyt c with excess of CL/PC liposomes in a low ionic strength buffer resulted in weakening of the 695 nm band absorbance at CL/cyt c molar ratio 1:10 (Figure 3B, c) with no observable effect in 50 mM phosphate buffer (Figure 3A). At high ionic strength conditions (25 mM HEPES with 1 M KCl) or in the absence of CL, PC-containing liposomes did not cause changes of absorbance at 695 nm (Figure 3B, b and d). This suggests that interactions of CL with cyt c in a low ionic strength environment apparently induce disruption of the Fe-S(Met80) bond as has been argued previously for cyt c interacting with anionic membranes (19). Incubation of TOCL/DOPC liposomes with cyt c (10:1 ratio, 15 min) caused a decrease of the absorbance peak at 695 nm by ∼25%. Addition of KCN (10-fold excess over cyt c) to cyt c with a “weakened” Met80-heme bond resulted in a decrease of 695 nm peak by 70% (Figure 3C, f). In the control experiment, the same concentration of KCN caused only a 10% loss of the 695 nm absorption in intact cyt c (Figure 3C, e).
Figure 3. Optical spectra of CL/cyt c complexes corresponding to absorbance of Fe-S(Met80) bond.
Adjusted 695 nm spectra of CL/cyt c complexes in 50 mM phosphate buffer (A) and 25 mM HEPES buffer (B, C): (a) cyt c, (b) DOPC liposomes with cyt c after 15 min incubation, (c) TOCL/DOPC liposomes with cyt c after 15 min incubation, (d) TOCL/DOPC with cyt c after 15 min incubation in high-ionic buffer (25 mM HEPES plus 1 M KCl), (e) cyt c incubated with KCN for 15 min, (f) TOCL/DOPC liposomes with cyt c and KCN after 15 min incubation. Liposomes 1 mM total lipids, cyt c 50 μM, KCN 500 μM.
To further elucidate relationships between any disruption of the Fe-S(Met80) bond and the resulting peroxidase activity upon interaction of cyt c with CL, we performed measurements of the peroxidase activity of CL/cyt c complexes using the production of etoposide radicals. We found that the activity increased dramatically in the presence of CL/PC liposomes as a function of CL/cyt c molar ratio (Figure 4). On the one hand, this supports the notion that the peroxidase activity of CL/cyt c in low ionic strength conditions is closely associated with the loss of the Met80 heme ligand. However, a careful examination of the 695 nm absorbance band at 5:1 lipid/cyt c ratio indicated that no changes in the 695 nm absorbance occurred within 30 min of incubation of cyt c with the liposomes compared to free cyt c (within 2-3 % error) (Figure 3A). Under the same conditions, the peroxidase activity increased several times and reached 10% of the “plateau” value (Figure 4A). Thus, the CL/cyt c complex peroxidase activity is not directly proportional to any Fe-S(Met80) bond disruption. The catalytic activity can be strongly increased even when the Fe-S(Met80) bond is still clearly intact.
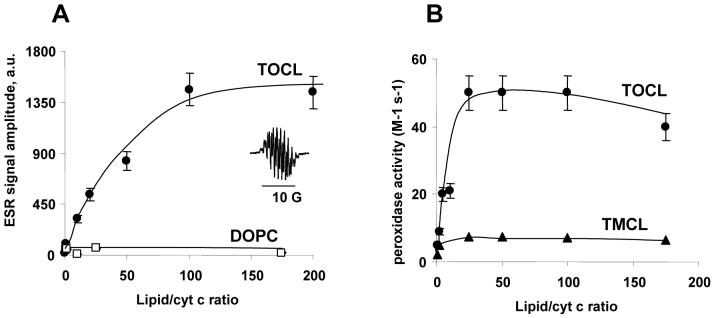
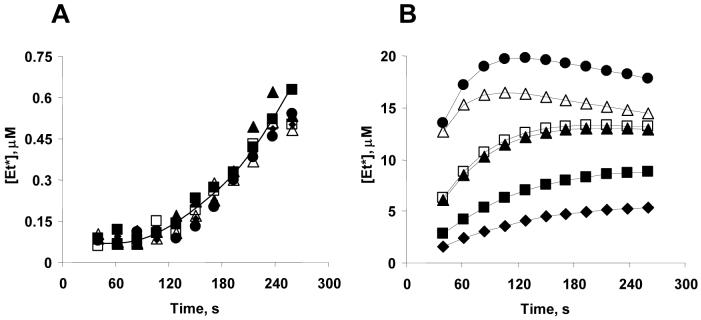
Figure 4. Peroxidase activity of lipid/cyt c complexes at different ratios as assessed by the etoposide phenoxyl radical ESR assay.
A - Formation of phenoxyl radical as a result a CL/cyt c peroxidase activity measured by ESR. Cyt c 40 μM, TOCL/DOPC liposomes 1:1, etoposide 700 μM, H2O2 1 mM. Inset: typical ESR signal of etoposide phenoxyl radical. (●) - TOCL, (▲) - TMCL, (□) DOPC. B - Second order rate constant of interaction of cyt c and H2O2 and second order rate constant of recombination of etoposide radicals were used as adjustable parameters in the fit (see Methods for details). (●) - TOCL, (▲) - TMCL.
Tryptophan fluorescence and peroxidase activity induced by cardiolipin liposomes with saturated and unsaturated acyl chains
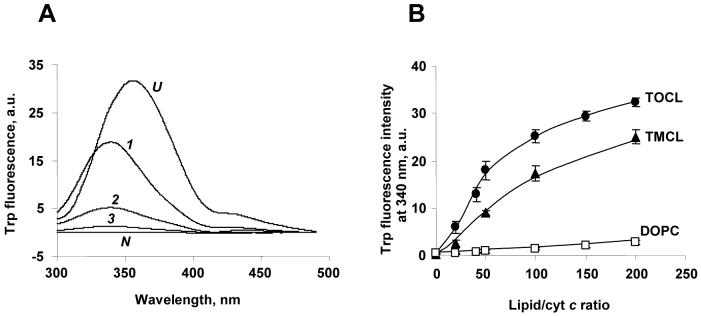
Another common approach to study conformational changes of cyt c is monitoring of its tryptophan fluorescence. Trp59 is the only tryptophan residue in horse heart cyt c (3). In the native protein, Trp59 fluorescence is efficiently quenched by the closely located heme group (Figure 5A, N). Addition of an unfolding reagent, guanidine hydrochloride (GdmCl, 6 M) (20), induced a strong Trp59 fluorescence increase (fully denatured state, Figure 5A, U) and also shifted the maximum of its emission to longer wavelengths (more polar tryptophan environment). Interaction with CL/PC liposomes at low ionic strength conditions and high lipid/protein molar ratio (50:1) also increased Trp59 fluorescence (Figure 5A, 1), although the fluorescence was weaker than under GdmCl conditions, indicating a more compact “molten globule”-like state of cyt c unfolded by CL. In this case, almost no shift in the fluorescent emission occurred, indicating that Trp59 was still in the hydrophobic environment in the unfolded CL/cyt c complex.
Figure 5. Cyt c tryptophan (Trp) fluorescence induced upon its binding with CL.
A. Typical cyt c Trp59 fluorescence emission spectra obtained after excitation at 293 nm. (N, native state cyt c) cyt c, 5 μM; (1) TOCL/DOPC liposomes incubated for 15 min with cyt c (at 50:1 molar ratio) in 25 mM HEPES buffer, pH=7.4; (2) same as 1 plus 150 mM KCl after the incubation, (3) same as 1, plus 1 M KCl after the incubation; (U, unfolded state of cyt c) cyt c after addition of 6.0 M GdmCl. B. Dependence of the emission of cyt c Trp59 fluorescence (340 nm) on the lipids:cyt c ratio (25 mM HEPES, pH=7.4), (●) - TOCL, (▲) - TMCL, (□) DOPC.
We analyzed the tryptophan fluorescence of cyt c and its peroxidase activity as a function of lipid/protein ratio at low ionic strength conditions for two types of CLs of different acyl chain composition - mono-unsaturated TOCL and saturated TMCL. Tryptophan fluorescence data presented in Figure 5B show that TOCL is a more effective inducer of unfolding of cyt c than TMCL. About two-fold higher concentration of TMCL liposomes was required to achieve the same level of Trp59 fluorescence as the one induced by TOCL liposomes. Cyt c Trp59 fluorescence increased steadily as a function of lipid/cyt c ratio and began to slowly flatten out of a lipid/protein molar ratio of about 200:1. Half maximal Trp59 fluorescence was observed at a ratio of 50:1 for both TOCL liposomes and TMCL liposomes.
We next assessed the peroxidase activity of CL/cyt c complexes by modeling the kinetics of accumulation of etoposide radical and comparing it to the actual time dependence of etoposide radical concentration from EPR measurements. The second order rate constant of interaction of cyt c and hydrogen peroxide as well as the second order rate constant of recombination of etoposide radicals were used as adjustable parameters in the fit. Figure 4B shows the computed peroxidase activity of CL/cyt c complexes as a function of lipid/cyt c molar ratios. The peroxidase activity of cyt c increases dramatically even at low lipid/cyt c ratio, reaching half-maximal value at a ratio of 10:1 and flattening out above 25:1, both for TLCL and TOCL lyposomes (Note that tryptophan fluorescence continued to increase with increasing lipid/cyt c ratio, see Figure 5B). These observations are similar to our above described comparative findings on the peroxidase activity of the CL/cyt c complex and a loss of the electronic absorption band associated with Fe-S(Met80) at 695 nm. Apparently, the free energy barrier for a hydrogen peroxide molecule to reach the cyt c heme and replace the Met80 ligand is much less then a the free energy needed for a partial unfolding of cyt c accompanied by the breaking of the Fe-S(Met80) bond and consequent heme-tryptophan distance increase. Therefore, the peroxidase activity of cyt c is activated already in the “electrostatically” bound cyt c conformation, when the tertiary structure of cyt c is still essentially intact.
The peroxidase activity of TOCL/cyt c complex at high lipid/cyt c molar ratios is about 5 times greater than that of the TMCL/cyt c complex, indicating that CL acyl chain composition is an important factor controlling the peroxidase activity of a bound cyt c.
Effects of ionic strength on cyt c peroxidase activity and cyt c binding with CL
To further characterize the importance of electrostatic and hydrophobic interactions in inducing the peroxidase activity of cyt c by CL-liposomes, we studied the effects of different ionic strength conditions. When cyt c and CL-containing liposomes were mixed in high-ionic strength buffer (25 mM HEPES with 1 M KCl), the Fe-S(Met80)-associated 695 nm band of cyt c remained unchanged, and the peroxidase activity stayed at essentially the same low level as that of soluble cyt c (Figure 6). Tryptophan fluorescence was very low (Figure 5A, (3)) and electrophoretic measurements did not detect changes of cyt c mobility in the presence of CL-liposomes in high ionic strength buffers. These data show that electrostatic interactions are necessary to initiate interactions between cyt c and CL. If, however, 1M KCl was added after the CL/cyt c complex was formed, the tryptophan fluorescence decreased only slightly and the peroxidase activity was similar to that observed in the low ionic strength TOCL/cyt c complex. The effect of 1M KCl on the TMCL/cyt c complex was much more pronounced: the enhancement of peroxidase activity was almost completely blocked after the addition of the high salt concentration. This indicates that hydrophobic interactions are important in stabilizing unfolded conformation of cyt c on the CL membrane surface. It is probable that reduced mobility of saturated acyl chains of TMCL precluded its close interactions with cyt c hydrophobic residues. As a result, cyt c and TMCL interacted only electrostatically and cyt c could be easily washed out by 1M KCL. While some of our data indicate that interactions between cyt c and TMCL take place, binding of cyt c to TMCL was not detectable in the PAGE experiment (25:1 ratio of TMCL to cyt c). Apparently a weak complex between cyt c and TMCL is much more dynamic than the complex between cyt c and unsaturated species of CL, thus cyt c can escape into the gel in the strong electric field. This result agrees with a complete blocking of the peroxidase activity of TMCL/cyt c by salt solutions and relative resistance of TOCL/cyt c complex and its peroxidase activity to the effect of high ionic strength. Thus TMCL/cyt c and TOCL/cyt c binding differ not only thermodynamically but also kinetically.
Figure 6. Peroxidase activity of CL/cyt c complexes in buffers with different ionic strengths.
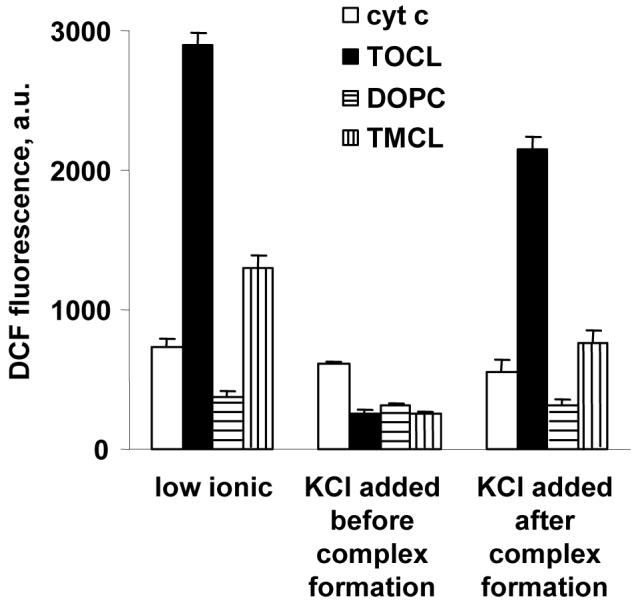
Fluorescence intensity of DCF, the product of H2DCF oxidation, with the excitation at 502 nm and emission at 522 nm is shown. Cyt c (1.25 μM), liposomes (125 μM, DOPC/CL 1:1 ratio), H2DCF 4 μM, H2O2 50 μM, spectra obtained 30 min after addition H2O2. First set of bars: cyt c was incubated for 15 min with CL/DOPC liposomes in 25 mM HEPES with 100 μM DTPA. Second set of bars: same plus 1 M KCl. Third set of bars: 1 M KCl was added after 15 min incubation of cyt c and CL/DOPC liposomes in 25 mM HEPES).
The importance of hydrophobic interactions between cyt c and CL is also emphasized by a strong dependence of the peroxidase activity of CL/cyt c complex on the type of oxidizable substrate. With a natural hydrophobic substrate, polyunsaturated TLCL, the peroxidase activity was equally high in both the low and high ionic-strength environments. Indeed, incubation of cyt c (20 μM) with 2 mM TLCL/DOPC liposomes at low ionic strength conditions (25 mM HEPES buffer with 100 μM DTPA in the presence of 100 μM H2O2 for 1 hr) lead to accumulation of 186±16 pmol TLCL hydroperoxides/nmol lipid. When the CL/cyt c complex was formed in low-ionic strength buffer after which 1 M KCl was added, the oxidation level was essentially the same: 172±19 pmol TLCL hydroperoxides/nmol lipid.
Self-activation and deactivation of CL/cyt c peroxidase activity
Characterization of the peroxidase activity of cyt c is complicated by oxidative chemical modifications of the protein itself by reactive intermediates of the reaction. Oxidative transformations of amino acids and the heme cofactor can both increase and decrease the peroxidase activity of cyt c (21-22).
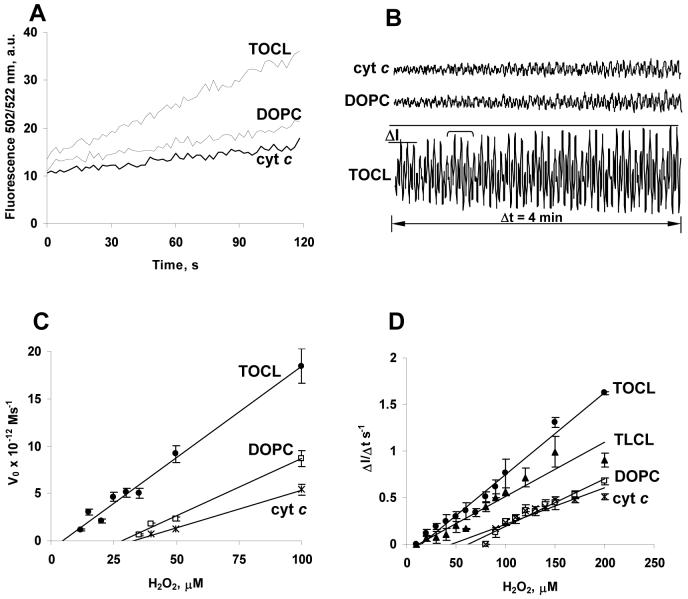
The time course of the etoposide radical accumulation showed that DOPC/cyt c peroxidase activity (similar to cyt c alone) markedly increased 2 minutes after the addition of H2O2 (Figure 7A) (from ∼0.1 M-1s-1 to ∼1 M-1s-1). Similar self-activation of the peroxidase activity of solubilized cyt c has been noted in (21, 23). The peroxidase activity of the CL/cyt c complex was much higher and detectable immediately after the addition of H2O2 (Figure 7B). However, excess H2O2 caused destruction of the cyt c heme and ceased the peroxidase activity. Figure 8A shows the time course of H2O2-induced changes in absorbance of cyt c monitored at two wavelengths. The loss of absorbance at 695 nm is associated with disruption of the Fe-S(Met80) bond (likely due to oxidation of the Met80 residue) while a loss of absorbance at 410 nm (Soret band) indicates heme destruction. We found that disruption of the Fe-S(Met80) bond preceded heme degradation. Five minutes after the addition of H2O2, more than 70% of the cyt c lost its characteristic Fe-S(Met80) bond absorbance whereas more than 90% of the heme still remained intact. Upon the addition of H2O2 to the CL/cyt c complex, the absorbance associated with the 695 nm band disappeared immediately. EPR measurements at this time point, showed a 5-fold increase in the peroxidase activity of cyt c, confirming that Fe-S(Met80) bond disruption was sufficient to promote cyt c reactivity. The increase of peroxidase activity of cyt c - coincident with (likely caused by) Fe-S(Met80) bond disruption - was much smaller than that achieved by the interaction of cyt c with CL-containing liposomes (Figure 8B). This suggests that the peroxidase activity of cyt c is controlled not only by the strength of Fe-S(Met80) bond but by the protein matrix creating a free energy barrier for permeability of H2O2 molecules to the heme crevice.
Figure 7. Time-course of etoposide phenoxyl radicals generated by the peroxidase activity of cyt c in the presence of DOPC (A) or TOCL (B).
Incubation conditions: cyt c - 40 μM, TOCL/DOPC liposomes 1:1, etoposide 700 μM, H2O2 1 mM. Ratios of lipids to cyt c: (◆) - 1:1, (■) - 2:1, (▲) - 5:1, (□) - 10:1, (△) - 25:1, (●) - 50:1. Concentrations of etoposide radicals were calculated based on the magnitudes of the ESR signals as detailed in Materials and Methods.
Figure 8. Effects of H2O2 pretreatment on the peroxidase activity of cyt c and its complexes with TOCL and DOPC.
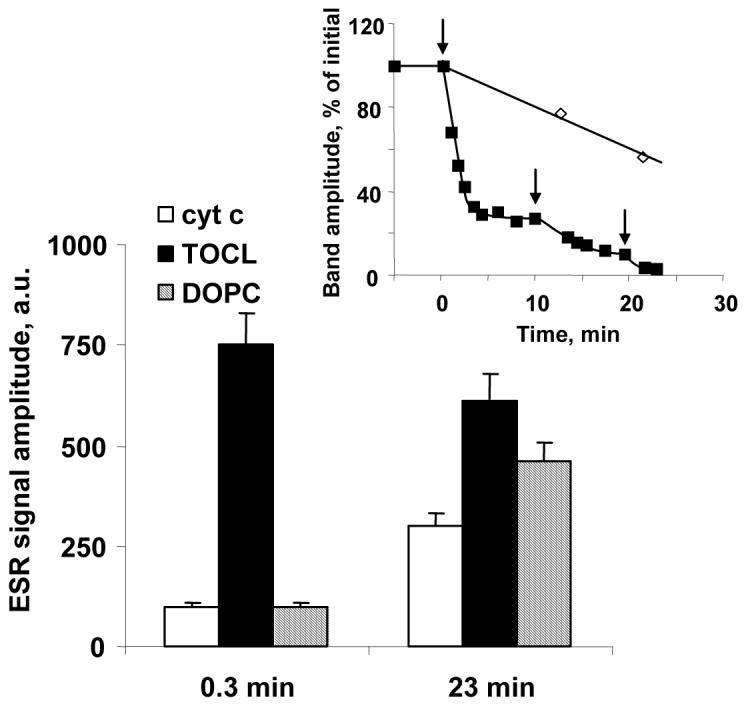
Peroxidase activity was evaluated by ESR-detectable generation of etoposide phenoxyl radicals. Conditions: cyt c (1 mM) in 10 mM PBS (pH 7.0) was incubated with 5 mM H2O2 (added three times as shown by the arrows on the inset). Aliquots of cyt c (containing 40 μM of cyt c) were taken at indicated time intervals (0.3 min, 23 min) and mixed with liposomes (10:1 lipid/cyt c ratio) to perform measurements of peroxidase activity by ESR.
Inset: Changes of cyt c absorbance in the Soret band ((◇), 410 nm) and Fe-S(Met80) band ((■), 695nm) induced by H2O2. Arrows show additions of H2O2. Absorbance at 695 nm was determined under the following conditions: cyt c (1 mM), H2O2 (5 mM). Absorbance at 410 nm was determined in the diluted samples: cyt c (10 μM), H2O2 (50 μM).
To describe the effect of substrate concentration on the enzyme kinetics Michaelis-Menten constant (24) is normally used. It has been shown, that for cyt c at pH 6.0 to 7.5 Km=65 mM (25). The same millimolar range has been obtained also in (26). For carboxymethylated cyt c at pH 7.0 Km is 25 mM (27). However, Michaelis-Menten kinetics is not strictly valid for the description of the behavior of a suicidal enzyme, like cyt c. In this case more proper is Kitz-Wilson plot (28), which give the inactivation constant represents the inversed affinity of the inhibitor for the enzyme active site. Given a likely interference due to the suicidal nature of the catalytic process, we chose to characterize the peroxidase efficacy by detecting the initial rate of peroxidase reaction in a wide range of substrate concentrations and determining the minimal substrate concentration sufficient for the enzyme activation. The time-dependent development of the peroxidase activity was monitored at several concentrations of H2O2 (Figure 9A present data of H2DCF oxidation by cyt c at different H2O2 concentrations, Figure 9B represents a typical plot of phenoxyl radical formation assessed by ESR). The slope of each line was determined and plotted as a concentration dependence (Figure 9C and D). Our estimates showed that the peroxidase activity was readily detectable at H2O2 levels as low as 5-10 μM (Figure 9C and D). The fact that this minimal concentration was not zero may indicates that peroxidase activity of cyt c increased in the course of the reaction. This minimal H2O2 concentration for activation is likely dependent on the incubation time (several minutes in our model experiments). In physiological processes such as apoptosis where the relevant time scale is about 30-60 minutes, the minimal H2O2 concentrations sufficient to activate the peroxidase activity of CL/cyt c complex are likely to be lower.
Figure 9. Kinetic parameters of peroxidase reaction of CL/cyt c complexes.
A. Time course of H2DCF oxidation. H2O2 (50 μM) was added to start peroxidase reaction. Time-courses of peroxidase activity of cyt c in the absence and presence of liposomes at different H2O2 concentrations were obtained. The slopes of each line were calculated to yield the concentration dependence shown on Figure 9C. Cyt c 1 μM, liposomes 20 μM, H2DCF 0.5 μM, H2O2 50 μM.
B. Typical ESR recordings of etoposide phenoxyl radicals over 4 min period of time. The rate of growth of this signal magnitude at different H2O2 concentrations was measured and replotted as shown on Figure 9D. Cyt c 25 μM, liposomes 500 μM, etoposide 350 μM, H2O2 100 μM, record time 4 min.
C. Initial reaction rates of H2DCF oxidation by cyt c (*), DOPC/cyt c (20:1) (□), TOCL/cyt c (10:10:1) (●) in 10mM PBS with 100 μM DTPA (pH=7.4). Reaction mixture contained 1 μM cyt c, 20 μM liposomes and 0.5 μM H2DCF. Extrapolation of linear dependences of the rates for cyt c, DOPC/cyt c and TOCL/cyt c interception with the X-axis provides an estimate of the initial H2O2 concentrations capable of activating the peroxidase activity: H2O2cyt c∼ 35 μM; H2O2DOPC/cyt c∼ 30 μM; H2O2TOCL/cyt c∼ 5 μM.
D. Initial rate of phenoxyl radical formation, measured by ESR assay. Cyt c (*), DOPC/cyt c (20:1) (□), TOCL/cyt c (10:1) (●), TLCL/cyt c (10:1) (○) in 10mM PBS with 100 μM DTPA (pH=7.4). Reaction mixture contained 25 μM cyt c, 500 μM lipid liposomes and 350 μM etoposide. Extrapolation of linear dependences of the rates for cyt c, DOPC/cyt c and TOCL/cyt c interception with the X-axis provides an estimate of the initial H2O2 concentrations capable of activating the peroxidase activity: H2O2cyt c∼ 60 μM; H2O2DOPC/cyt c∼ 40 μM; H2O2TOCL/cyt c∼ 10 μM, H2O2TLCL/cyt c∼ 12 μM.
Discussion
Cytochrome c (cyt c) is an electron carrier between mitochondrial respiratory complexes III and IV, but under certain conditions it can be released into the cytosol and initiate the caspases cascade of proteolytic reactions designating a point of no return in programmed cell death or apoptosis [reviewed in (29)]. The precise mechanisms regulating release of cyt c and other pro-apoptotic factors remain not fully understood. Recently, we reported a new redox function of cyt c in apoptosis (6) whereby cyt c, through its specific binding with CL, serves as a catalyst of CL peroxidation in mitochondria occurring upstream of cyt c release into the cytosol. Importantly, accumulation of CL oxidation products is required for the release of cyt c and other pro-apoptotic factors from mitochondria during permeabilization of the outer mitochondrial membrane (6, 30). These findings encouraged us to conduct more detailed studies of mechanisms involved in the activation of the peroxidase activity of cyt c by CL-containing membranes.
The ability to catalytically reduce H2O2 with the intermediate formation of highly active oxo-ferryl compounds is prototypical of many, if not all, hemoproteins (31). However, their respective peroxidase activities vary drastically. For example, horseradish peroxidase has a second order rate constant of interaction with hydrogen peroxide of 1.4 107 M-1s-1, while horse heart cyt c peroxidase activity is less than 1 M-1s-1 (32). One of the most important factors that control peroxidase activity of hemoproteins is accessibility of their iron-site for H2O2 binding. Hemoproteins with the vacant sixth iron ligand positions, such as myoglobin or hemoglobin, and isolated heme groups display peroxidase activity of about 103 M-1s-1 (32) which is three orders of magnitude higher than that of six-coordinated cyt c. An additional gain of four orders of magnitude in the peroxidase activity in horseradish and some other peroxidases is explained by the specific structure of peroxidase catalytic sites active, in particular by the presence of a histidine (general base) residue located in a precise position to facilitate proton transfer from one oxygen atom of H2O2 molecule to another (33-34).
The folded structure of cyt c, optimal for maintenance of its hexacoordinate arrangement, is essential for its function as a mobile electron-carrier. One can assume, however, that unfolding and destabilization of cyt c would confer the peroxidase activity on the protein. Indeed, cyt c completely unfolded by guanidine hydrochloride has the peroxidase activity on the order of 103 M-1s-1 (32) - similar to that of an isolated heme groups. While several types of different anionic molecules, including anionic lipids (35) have been found to activate cyt c into a peroxidase, the physiological relevance of this transformation remained elusive. Our data have now clearly demonstrated that CL is a very potent activator of cyt c’s peroxidase activity. The underlying mechanisms and the extent to which binding of cyt c to anionic phospholipid membranes causes formation of their complex, perturbation of the cyt c structure and appearance of the peroxidase activity, depends on many factors such as ionic strength, protein/lipid ratio, membrane phospholipid composition, pH, etc.
At low-ionic strength and high lipid/protein ratios, peroxidase activity of CL/cyt c complexes may be as high as 50 M-1s-1 as compared with less than 1 M-1s-1 for a solubilized lipid-free cyt c. This suggests that membrane-bound cyt c may function as a competitive peroxidase in mitochondria. The maximal peroxidase activity of the CL/cyt c complex is achieved in a tightly bound state achieved through a combination of electrostatic and hydrophobic interactions with CL-containing liposomes that has a molten-globule like structure (36). A relatively lower value of the maximal peroxidase activity of CL-bound cyt c compared to that of fully unfolded protein can be attributed to a compactness of a molten globule-like conformation of the CL/cyt c complex and higher local concentrations of imidazole and amino groups that compete with H2O2 for binding to heme iron.
The tightly-bound membrane state of cyt c is strongly stabilized by hydrophobic interactions as demonstrated by a strong effect of the CL acyl chain composition on cyt c peroxidase activity. TMCL with fully saturated acyl chains was shown to be five times less efficient than TOCL in inducing peroxidase activity of cyt c. High ionic strength conditions were sufficient to disassemble TMCL/cyt c complexes and eliminate the associated peroxidase activity; however, 1M KCl was much less effective in disrupting TOCL/cyt c complexes which retain their peroxidase activity after treatment with the salt. Strong stabilization of tightly membrane bound state cyt c by hydrophobic interactions with unsaturated acyl chains of CL was also evident from a greater ability of TOCL vs TMCL to unfold cyt c (as monitored by tryptophan fluorescence ) and increased binding energy of cyt c to CL in the row TMCL < TOCL = TLCL < BHCL. Importantly, the critical role of hydrophobic interactions for the peroxidase activity of CL/cyt c complexes towards their natural substrate, polyunsaturated CL, is emphasized by the same level of TLCL peroxidation detected in experiments at either low or high ionic strength. Thus, apoptosis-related CL-oxygenase function of cyt c requires hydrophobic interactions suggesting that changes in ionic environment, occurring during mitochondrial membrane permeabilization, are not likely to affect cyt c’s role as a specific catalyst of CL peroxidation.
We also found that the peroxidase activity of cyt c is strongly activated in conditions when the protein tertiary structure, as measured by optical absorption at 695 nm and tryptophan fluorescence, is essentially intact. This suggests that a loosely (electrostatically) bound state of cyt c can be responsible for the initiation of the peroxidase reaction. The free energy barrier for the initiation of peroxidase activity of cyt c (3.8 kcal/mol as estimated by a 1,000-fold increase of the peroxidase activity of cyt c upon complete unfolding) is substantially less than the free energy of partial unfolding of the protein (5-6 kcal/mol) and is likely determined by a probability to form a “channel” in the cyt c globule facilitating access of H2O2 to the heme iron. Indeed, analysis of the cyt c structure reveals a narrow cavity formed by Lys79, Met80, Ile81 and the heme that can potentially provide access of H2O2 to the heme iron. Destabilization of cyt c by its interactions with anionic CL-membranes facilitates this and allows H2O2 molecule to reach the heme-iron catalytic site, substitute the Met80 ligand and initiate the peroxidase reaction. This interpretation is consistent with a recent identification of several hierarchically denatured domains in cyt c, whereby unfolding of each of the domains sequentially occurs only after denaturation of the previous domain (37). Notably, Met80 was found to be a part of the second domain whose unfolding was preceded by structural changes in domain 1 (37). Thus, detectable conformational changes occur in the cyt c molecule before any involvement of Met80 takes place.
Similar to other peroxidases, the protein backbone of cyt c is modified during the peroxidase reaction. These chemical modifications of the protein can both increase the peroxidase activity and inhibit it. We found that the peroxidase activity of solubilized lipid-free cyt c is activated after several minutes of incubation with H2O2 (100 μM). Notably, ten-fold lower H2O2 concentrations were needed to activate cyt c bound to CL-containing membranes. DOPC/cyt c peroxidase activity (similar to cyt c alone) markedly increased 2 minutes after the addition of H2O2 - from ∼0.1 M-1s-1 to ∼1 M-1s-1 (see Figure 7A). This data is in agreement with the results of Radi et al (25) who reported an increase in catalytic activity of ferricytochrome c after a brief preincubation with H2O2.
Reduction of H2O2 to water catalyzed by cyt c heme probably results in the formation of Compounds I and II (oxo-ferryl heme intermediates) (23), although none of these species were observed spectroscopically - in contrast to other heme proteins with peroxidase activity (38). One possible explanation for a short-life time of oxo-ferryl heme intermediates of cyt c can be their fast interactions with aromatic amino-acid residues of the protein. Cyt c contains 4 tyrosine and 1 tryptophan residues that can be potentially oxidized by compounds I and II (31). Some of these residues Tyr67, Trp59, Tyr48 are within 5.0 Å of the heme porphyrin ring and can readily reduce heme oxo-ferryl reactive intermediates. ESR spin-trapping studies of the cyt c/H2O2 system have shown the formation of cyt c tyrosyl radicals (39). Oxidation of Tyr and Trp residues should “loosen” the cyt c structure, increase its peroxidase activity and stabilize the tightly (hydrophobically) bound state of cyt c.
Under normal physiological conditions, low availability of CL in the outer leaflet of the inner mitochondria membrane (IMM) and its absence from the inner leaflet of the outer mitochondria membrane (OMM) as well as relatively high ionic strength (150 mM), are not conducive of efficient interactions between CL and cyt c and its unfolding. This is not surprising given that the main function of cyt c is that of an electron shuttle between complexes III and IV and interactions with cytochrome b5 in the OMM. Clearly, this electron-transport function would be impeded if all or a significant fraction of the cyt c was tightly bound and unfolded on the membrane surface. Recent findings, however, indicate that significant amounts of cyt c are loosely (electrostatically) bound to the IMM (40-41). Our results show that the peroxidase activity of cyt c is already activated in the loosely membrane bound conformation. After the reaction with H2O2 and oxidation of cyt c’s amino-acid residues, stability of its tertiary structure decreases while its peroxidase activity increases. This activation of peroxidase activity of cyt c in the complex with CL is achieved (as we showed above) at low micromolar concentrations of H2O2 corresponding to the elevated levels of reactive oxygen species (ROS) production during the early stages of apoptosis.
The oxidatively modified form of the protein is likely associated with its increased propensity to convert into a tightly (hydrophobically)-bound form with an even more distorted tertiary structure and still greater peroxidase activity. Because of a higher reactivity of the tightly-membrane-bound form of cyt c, H2O2 will be interacting preferentially with these unfolded cyt c molecules. It is tempting to speculate that the fraction of tightly bound cyt c present in mitochondria under physiological conditions consists mostly of cyt c molecules that reacted at least once with H2O2 and thus obtained a decreased structural stability. Because this fraction of cyt c molecules has the highest peroxidase activity, it is likely that CL oxidation is predominantly catalyzed by tightly bound cyt c.
The combined effect of CL redistribution in the mitochondrial membranes and increased ROS production may increase the cyt c peroxidase activity many-fold: from ∼0.1 M-1s-1 for unmodified free cyt c to ∼50 M-1s-1 for cyt c in complex with CL. Thus, the peroxidase activity of cyt c can effectively integrate signals due to changes of mitochondrial membrane composition and H2O2 concentration and convert them into another signal - CL oxidation products.
It should be noted that CL is not the only physiologically relevant anionic phospholipid capable of triggering unfolding of cyt c and activating its peroxidase catalytic function. Phosphatidylserine (PS), an anionic phospholipid lacking from the mitochondria but common in membranes of other organelles and the cytosolic leaflet of the plasma membrane, is also a good “peroxidatic” activator of cyt c. It has been postulated that peroxidase activity of PS/cyt c complexes is a significant contributor to another signaling pathway in apoptosis, namely, PS externalization essential for recognition and clearance of apoptotic cells by macrophages. Specific mechanisms of PS-induced unfolding and associated peroxidase activity of cyt c remain to be elucidated.
In summary, cyt c fulfills several different roles in mitochondria. Its participation in long range electron-transfer reactions between complexes III and IV, occurring via transient protein-protein interfaces, is dependent on electron tunneling and conformational dynamics (42). This function is effectively accommodated by the cyt c’s hexa-coordinate structure with His18 and Met80 as the two axial ligands. In contrast, the catalytic competence of cyt c as a peroxidase requires direct interaction of its heme moiety with H2O2, hence, disruption of the Met80-heme iron bond. Thus, the employment of Met80, is a very effective strategy that effectively serves both functions of cyt in mitochondria. Not surprisingly, Met80 is highly conserved in cyt c from different species along with Lys72 and Lys73 which are essential for the electrostatic and hydrophobic interactions of the protein with anionic phospholipids respectively.
Acknowledgments
This work was supported by NIH U19 AI068021, NIH HL61411, The Human Frontier Science Program, RFFI (Russian Foundation of Fundamental Investigations) 05-04-49765-a.
Abbreviations
- cyt c
cytochrome c
- CL
cardiolipin
- TOCL
1,1′2,2′-Tetraoleoyl cardiolipin
- TLCL
1,1′2,2′-tetralinoleoyl cardiolipin
- TMCL
1,1′,2,2′-Tetramyristoyl cardiolipin
- BHCL
bovine heart cardiolipin
- DOPC, PC
1,2-Dioleoyl-sn-Glycero-3-Phosphocholine
- NAO
acridine orange 10-nonyl (nonyl acridine orange)
- H2DCF
2′,7′-dichlorodihydrofluorescein
- GdmCl
guanidine hydrochloride
- IMM
inner mitochondrial membrane
- OMM
outer mitochondria membrane
- ROS
reactive oxygen species
References
- (1).Cecconi C, Shank EA, Bustamante C, Marqusee S. Direct observation of the three-state folding of a single protein molecule. Science. 2005;309:2057–60. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- (2).Pettigrew GW, Moore GR. Cytochrome c - Biological aspects. Springer-Verlaf; Berlin. Heidelberg: 1987. [Google Scholar]
- (3).Bushnell GW, Louie GV, Brayer GD. High-resolution three-dimensional structure of horse heart cytochrome c. J Mol Biol. 1990;214:585–95. doi: 10.1016/0022-2836(90)90200-6. [DOI] [PubMed] [Google Scholar]
- (4).McMillin JB, Dowhan W. Cardiolipin and apoptosis. Biochim Biophys Acta. 2002;1585:97–107. doi: 10.1016/s1388-1981(02)00329-3. [DOI] [PubMed] [Google Scholar]
- (5).Jori G, Tamburro AM, Azzi A. Photooxidative and spectral studies on cytochrome c. Conformational changes induced by binding of cardiolipin. Photochem Photobiol. 1974;19:337–45. doi: 10.1111/j.1751-1097.1974.tb06521.x. [DOI] [PubMed] [Google Scholar]
- (6).Kagan VE, Tyrin VA, Jiang J, Tyrina YY, Ritov VB, Amoscato AA, Osipov AN, Belikova NA, Kapralov AA, Kini V, Vlasova II, Zhao Q, Zou M, Di P, Svistunenko DA, Kurnikov IV, Borisenko GG. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nature Chem Biol. 2005;1:223–232. doi: 10.1038/nchembio727. [DOI] [PubMed] [Google Scholar]
- (7).Pinheiro TJ. The interaction of horse heart cytochrome c with phospholipid bilayers. Structural and dynamic effects. Biochimie. 1994;76:489–500. doi: 10.1016/0300-9084(94)90173-2. [DOI] [PubMed] [Google Scholar]
- (8).Oellerich S, Lecomte S, Paternostre M, Heimburg T, Hildebrandt P. Peripheral and Integral Binding of Cytochrome c to Phospholipids Vesicles. J. Phys. Chem. B. 2004;108:3871–3878. [Google Scholar]
- (9).Hagihara B, Sekuzu I, Tagawa K, Yoneda M, Okunuki K. Crystallization of cytochrome c from pigeon breast muscle. Nature. 1958;181:1588–9. [PubMed] [Google Scholar]
- (10).Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- (11).Petit JM, Maftah A, Ratinaud MH, Julien R. 10N-nonyl acridine orange interacts with cardiolipin and allows the quantification of this phospholipid in isolated mitochondria. Eur J Biochem. 1992;209:267–73. doi: 10.1111/j.1432-1033.1992.tb17285.x. [DOI] [PubMed] [Google Scholar]
- (12).Nicholls P. Cytochrome c binding to enzymes and membranes. Biochim Biophys Acta. 1974;346:261–310. doi: 10.1016/0304-4173(74)90003-2. [DOI] [PubMed] [Google Scholar]
- (13).Tuominen EK, Wallace CJ, Kinnunen PK. Phospholipid-cytochrome c interaction: evidence for the extended lipid anchorage. J Biol Chem. 2002;277:8822–6. doi: 10.1074/jbc.M200056200. [DOI] [PubMed] [Google Scholar]
- (14).Gallet PF, Maftah A, Petit JM, Denis-Gay M, Julien R. Direct cardiolipin assay in yeast using the red fluorescence emission of 10-N-nonyl acridine orange. Eur J Biochem. 1995;228:113–9. doi: 10.1111/j.1432-1033.1995.tb20238.x. [DOI] [PubMed] [Google Scholar]
- (15).Kirkland RA, Adibhatla RM, Hatcher JF, Franklin JL. Loss of cardiolipin and mitochondria during programmed neuronal death: evidence of a role for lipid peroxidation and autophagy. Neuroscience. 2002;115:587–602. doi: 10.1016/s0306-4522(02)00512-2. [DOI] [PubMed] [Google Scholar]
- (16).Heimburg T, Marsh D. Protein surface-distribution and protein-protein interactions in the binding of peripheral proteins to charged lipid membranes. Biophys J. 1995;68:536–46. doi: 10.1016/S0006-3495(95)80215-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Sinibaldi F, Mei G, Polticelli F, Piro MC, Howes BD, Smulevich G, Santucci R, Ascoli F, Fiorucci L. ATP specifically drives refolding of non-native conformations of cytochrome c. Protein Sci. 2005;14:1049–58. doi: 10.1110/ps.041069405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Pinheiro TJ, Elove GA, Watts A, Roder H. Structural and kinetic description of cytochrome c unfolding induced by the interaction with lipid vesicles. Biochemistry. 1997;36:13122–32. doi: 10.1021/bi971235z. [DOI] [PubMed] [Google Scholar]
- (19).de Jongh HH, Ritsema T, Killian JA. Lipid specificity for membrane mediated partial unfolding of cytochrome c. FEBS Lett. 1995;360:255–60. doi: 10.1016/0014-5793(95)00115-p. [DOI] [PubMed] [Google Scholar]
- (20).Kita N, Kuwajima K, Nitta K, Sugai S. Equilbrium and kinetics of the unfolding of alpha-lactalbumin by guanidine hydrochloride (II) Biochim Biophys Acta. 1976;427:350–8. doi: 10.1016/0005-2795(76)90310-x. [DOI] [PubMed] [Google Scholar]
- (21).Radi R, Turrens JF, Freeman BA. Cytochrome c-catalyzed membrane lipid peroxidation by hydrogen peroxide. Arch Biochem Biophys. 1991;288:118–25. doi: 10.1016/0003-9861(91)90172-f. [DOI] [PubMed] [Google Scholar]
- (22).Chen YR, Deterding LJ, Sturgeon BE, Tomer KB, Mason RP. Protein oxidation of cytochrome C by reactive halogen species enhances its peroxidase activity. J Biol Chem. 2002;277:29781–91. doi: 10.1074/jbc.M200709200. [DOI] [PubMed] [Google Scholar]
- (23).Burkitt MJ, Wardman P. Cytochrome C is a potent catalyst of dichlorofluorescin oxidation: implications for the role of reactive oxygen species in apoptosis. Biochem Biophys Res Commun. 2001;282:329–33. doi: 10.1006/bbrc.2001.4578. [DOI] [PubMed] [Google Scholar]
- (24).Pryse KM, Bruckman TG, Maxfield BW, Elson EL. Kinetics and mechanism of the folding of cytochrome c. Biochemistry. 1992;31:5127–36. doi: 10.1021/bi00137a006. [DOI] [PubMed] [Google Scholar]
- (25).Radi R, Thomson L, Rubbo H, Prodanov E. Cytochrome c-catalyzed oxidation of organic molecules by hydrogen peroxide. Arch Biochem Biophys. 1991;288:112–7. doi: 10.1016/0003-9861(91)90171-e. [DOI] [PubMed] [Google Scholar]
- (26).Kim NH, Jeong MS, Choi SY, Kang JH. Peroxidase activity of Cytochrome c. Bull. Korean Chem. Soc. 2004;25:1889–1892. [Google Scholar]
- (27).Prasad S, Maiti NC, Mazumdar S, Mitra S. Reaction of hydrogen peroxide and peroxidase activity in carboxymethylated cytochrome c: spectroscopic and kinetic studies. Biochim Biophys Acta. 2002;1596:63–75. doi: 10.1016/s0167-4838(02)00205-4. [DOI] [PubMed] [Google Scholar]
- (28).Kitz R, Wilson IB. Esters of methanesulfonic acid as irreversible inhibitors of acetylcholinesterase. J Biol Chem. 1962;237:3245–9. [PubMed] [Google Scholar]
- (29).Green DR, Amarante-Mendes GP. The point of no return: mitochondria, caspases, and the commitment to cell death. Results Probl Cell Differ. 1998;24:45–61. doi: 10.1007/978-3-540-69185-3_3. [DOI] [PubMed] [Google Scholar]
- (30).Nakagawa Y. Initiation of apoptotic signal by the peroxidation of cardiolipin of mitochondria. Ann N Y Acad Sci. 2004;1011:177–84. doi: 10.1007/978-3-662-41088-2_18. [DOI] [PubMed] [Google Scholar]
- (31).Svistunenko DA. Reaction of haem containing proteins and enzymes with hydroperoxides: the radical view. Biochim Biophys Acta. 2005;1707:127–55. doi: 10.1016/j.bbabio.2005.01.004. [DOI] [PubMed] [Google Scholar]
- (32).Diederix RE, Ubbink M, Canters GW. Peroxidase activity as a tool for studying the folding of c-type cytochromes. Biochemistry. 2002;41:13067–77. doi: 10.1021/bi0260841. [DOI] [PubMed] [Google Scholar]
- (33).Poulos TL, Kraut J. The stereochemistry of peroxidase catalysis. J Biol Chem. 1980;255:8199–205. [PubMed] [Google Scholar]
- (34).Finzel BC, Poulos TL, Kraut J. Crystal structure of yeast cytochrome c peroxidase refined at 1.7-A resolution. J Biol Chem. 1984;259:13027–36. [PubMed] [Google Scholar]
- (35).Jiang J, Kini V, Belikova N, Serinkan BF, Borisenko GG, Tyurina YY, Tyurin VA, Kagan VE. Cytochrome c release is required for phosphatidylserine peroxidation during Fas-triggered apoptosis in lung epithelial A549 cells. Lipids. 2004;39:1133–42. doi: 10.1007/s11745-004-1340-1. [DOI] [PubMed] [Google Scholar]
- (36).Sanghera N, Pinheiro TJ. Unfolding and refolding of cytochrome c driven by the interaction with lipid micelles. Protein Sci. 2000;9:1194–202. doi: 10.1110/ps.9.6.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Maity H, Maity M, Krishna MM, Mayne L, Englander SW. Protein folding: the stepwise assembly of foldon units. Proc Natl Acad Sci U S A. 2005;102:4741–6. doi: 10.1073/pnas.0501043102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Makris TM, Denisov IG, Sligar SG. Haem-oxygen reactive intermediates: catalysis by the two-step. Biochem Soc Trans. 2003;31:516–9. doi: 10.1042/bst0310516. [DOI] [PubMed] [Google Scholar]
- (39).Barr DP, Gunther MR, Deterding LJ, Tomer KB, Mason RP. ESR spin-trapping of a protein-derived tyrosyl radical from the reaction of cytochrome c with hydrogen peroxide. J Biol Chem. 1996;271:15498–503. doi: 10.1074/jbc.271.26.15498. [DOI] [PubMed] [Google Scholar]
- (40).Ott M, Robertson JD, Gogvadze V, Zhivotovsky B, Orrenius S. Cytochrome c release from mitochondria proceeds by a two-step process. Proc Natl Acad Sci U S A. 2002;99:1259–63. doi: 10.1073/pnas.241655498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Berezhna S, Wohlrab H, Champion PM. Resonance Raman investigations of cytochrome c conformational change upon interaction with the membranes of intact and Ca2+-exposed mitochondria. Biochemistry. 2003;42:6149–58. doi: 10.1021/bi027387y. [DOI] [PubMed] [Google Scholar]
- (42).Kang SA, Crane BR. Effects of interface mutations on association modes and electron-transfer rates between proteins. Proc Natl Acad Sci U S A. 2005;102:15465–70. doi: 10.1073/pnas.0505176102. [DOI] [PMC free article] [PubMed] [Google Scholar]