Summary
The intestinal intraepithelial lymphocytes (IEL) represent multi-lineage T cell populations. In addition to a major γδTCR+ T cell subset, many IEL express αβTCRs and they can be separated into αβ sub-lineages. Some TCRαβ+IEL have characteristics in common with conventional TCRαβ+T cells whereas others share an unconventional phenotype with their TCRγδ+ counterparts. Because the latter are enriched for autoreactive TCRs and can be generated in the absence of a thymus, it has long been postulated that some IEL subsets develop locally in the intestine. Several new data however, indicate that under physiological conditions, IEL require a thymic education that directs lineage commitment and functional differentiation. This review will discuss the contributions of the thymus in shaping the various intestinal IEL sub-lineages.
Introduction
The intestinal epithelium is colonized by IEL that are numerous and diverse. Based on their phenotype and developmental origin, the IEL have been classified into two distinct groups [1]. One group consists of CD4+ or CD8αβ+TCRαβ+IEL, which are thought to have followed conventional thymic selection and reached the gut after antigenic stimulation in the periphery [2] (Figure 1). The other group of IEL encompasses gut resident T cells that either express a γδ- or αβTCR and that have adapted an antigen-experienced phenotype during their initial education in response to self-antigens [1,2] (Figure 1). IEL in the latter group do not express a TCR co-receptor but typically express homodimers of CD8α (CD8αα), which has been used as a hallmark to identify and distinguish these IEL from the conventional subset [3]. The frequent association of CD8αα expression with the gut microenvironment, led to the belief that CD8αα marks those IEL that developed locally within the intestine [3]. The identification of cryptopatches (CPs) which harbor ckit+, IL-7Rα+, Thy1+, CD44+ and lin− lymphoid precursor cells that under experimental conditions can differentiate to CD8αα+TCRγδ+ and TCRαβ+ T cells, further fueled the idea of extrathymic development for these cells [4-6]. Nevertheless, the appearance of CD8αα+TCRγδ+ and TCRαβ+IEL well before the initial development of CPs [4], together with the absence of CPs in any other organism other than mice, render the CPs as the exclusive differentiation niche for CD8αα+IEL unlikely. The most compelling evidence, that questions the intestine as the primary lymphoid tissue for CD8αα+ IEL, is the fact that congenitally athymic mice have much reduced γδTCR IEL and almost no αβTCR IEL [7,8]. Although the number of IEL in athymic mice does increase with age, it still remains far less than one would expect if CD8αα+ IEL developed extrathymically. Collectively these observations indicate that CD8αα expression is not consistent with extrathymic development and that the thymus serves an indispensable role in the differentiation of CD8αα+IEL. CD8αα on T cells is not a typical lineage marker but correlates more with an activated phenotype [9]. Even though CD8αα shares the same CD8α chain with CD8αβ TCR co-receptor, which marks MHC class I restricted T cells, CD8αα does not function as a stable co-receptor but rather negatively regulates TCR activation independently of MHC restriction [9, 10]. Furthermore, CD8αα can also be induced on CD4+ or CD8αβ+TCRαβ+T cells upon antigenic stimulation or as part of their adaptation to the gut environment [2,11,12] indicating that CD8αα cannot be used as a faithful lineage determinant. Therefore, we will refer in this review to the CD8αα+TCRαβ+IEL as the CD4−CD8αβ−TCRαβ+IEL instead.
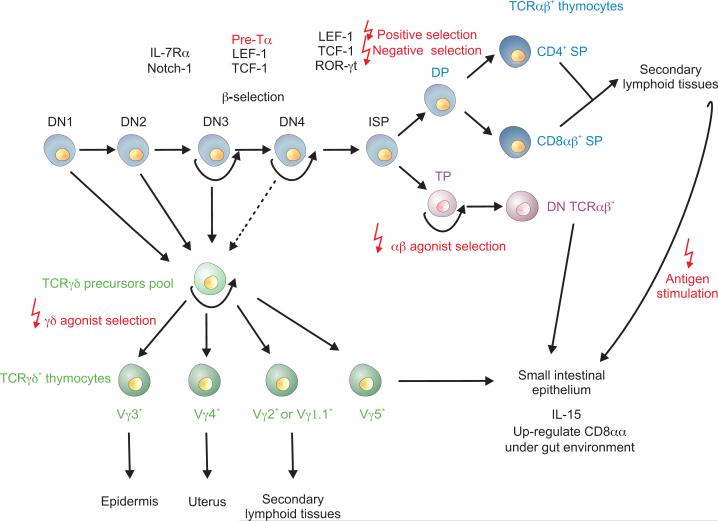
Figure 1. Differentiation pathways and checkpoints of TCRαβ and TcRγδ T cells.
DN thymocytes are subdivided in 4 populations (DN1 to DN4) that contain precursors belonging to the αβ- and γδ-lineage. DN TcRγδ precursors transition through agonist TCRγδ selection and give rise to waves of TCRγδ subsets with different homing capacity. Agonist selected TCRγδ IEL precursors migrate to the gut epithelium independently of functional S1P1. At the DN3 stage, precursors that successfully rearranged the β-chain will undergo pre-TCR driven β-selection and transition to the DP (CD4+CD8αβ+) or TP (CD4+CD8αβ+CD8αα+) stage. The DP population contains αβ lineage committed precursors that will undergo positive and negative conventional selection and commit to the CD4- or CD8αβ-SP sublineage. Positive selected SP thymocytes egress the thymus in a S1P1-dependent fashion and reside in the periphery as naïve T cells. Antigenic stimulation further differentiate these cells to effector cells that gain the capacity to migrate to the intestinal epithelium. TP thymocytes can proceed through agonist selection and give rise to mature DN TCRαβ thymocytes that home to the intestinal epithelium in a S1P1-independent way. Molecules that have important roles during T cell differentiation were added. Solid black arrows show established pathways whereas dashed arrows indicate uncertain transitions.
DN seeding from the thymus to the gut
Recent evidence suggested that the thymus may provide a critical source of immature T-committed precursors that egress the thymus before rearrangement of the Tcr genes and complete their further sublineage differentiation locally under the specific conditions of the gut microenvironment [13••]. Although this is a reasonable hypothesis that ties together the essential role of the thymus with the lymphopoietic capacity of the gut, it also raises new ambiguity. T cell differentiation in the thymus is a complex process that involves multiple checkpoints that direct T versus B, γδ versus αβ and among αβ progenitors, CD4 versus CD8 commitment in addition to conventional or specialized T cells differentiation [14]. It is not known whether under normal physiological conditions, undecided thymocytes can leave the thymus prematurely without transitioning through these different checkpoints. Furthermore, several developmentally more advanced and already lineage committed thymocytes have been identified that are direct progenitors of TCRγδ+ and TCRαβ+IEL subsets [15••,16] and the observation that immature TCRαβ thymocytes may act like LTi cells that can influence the gene expression profile of γδ thymic progenitors [16], all suggest that the thymic chapter for the development of IEL is likely to involve multiple stages and checkpoints.
γδ/αβIEL lineage checkpoints in the thymus
All T lineages develop from multipotent double negative (DN) (that express neither CD4 nor CD8), precursors. In the murine thymus these DN progenitors can be further subdivided into four maturation stages (DN1 to DN4) determined by the variable expression of CD44 and CD25 [17] (Figure 1). The γδ/αβ lineage decision occurs at these DN stages but the exact time point is debatable [18,19]. Furthermore, γδ T cell progenitors may not follow the same DN1 to DN4 pathway as their αβ counterparts and they basically branch off before Tcra gene recombination. Initial rearrangements of the TCRγ, TCRδ and TCRβ loci can be detected at the DN2 stage and are completed during the DN3 stage [20]. DN precursors that express a full γδTCR can commit to the γδ lineage, whereas DN3 cells that rearrange a functional TCRβ and form a pre-TCR together with the invariant pre-Tα chain and members of the CD3 complex, further mature along the αβ lineage [19] (Figure 1). Although successful Tcr gene rearrangements undoubtedly impact the eventual αβ/γδ-lineage commitment, several data indicate that DN2 cells may display a biased potential already before Tcr rearrangements [21]. Specific transcription factors might pre-set the stage and in that aspect various WNT signaling events have been identified that critically regulate expression of genes involved in lineage diversification. The WNT induced β-catenin controlled T cell factor-1 (TCF-1) and lymphoid enhancer factor-1 (LEF-1) were shown to play important albeit partially redundant roles in controlling development of αβ-lineage progenitors [22,23] (Figure 1). Nevertheless, in addition to TCRαβ+T cells also TCRγδ+IEL but not TCRγδ+splenocytes, were drastically reduced in Tcf1−/− mice pointing at a unique role for TCF-1 in the generation and/or maintenance of gut-specific TCRγδ+T cells [24]. T cell differentiation is also critically dependent on signals transduced through cytokine receptors belonging to the family of common γ-chain receptors. Especially IL-7R plays a non-redundant role during the initial survival and expansion of bipotent T cell progenitors [25] (Figure 1). Although differential expression of the IL-7R at the DN2 stage can be used to identify lineage bias [21], and although IL-7R signals promote TCRγ rearrangements [26], there is no direct evidence suggesting that IL7R signals play a decisive role in the TCRαβ/γδ lineage commitment. However, continuous IL7R signaling in IL-7Rα transgenic thymocytes does suppress the expression of the transcription factors TCF-1/LEF-1 and retinoic acid orphan nuclear receptors RORγ/RORγt, and prevents survival and further differentiation of TCRαβ progenitor cells [27] (Figure 1). In contrast transgene-driven local expression of IL-7 in the intestine of IL-7 deficient mice restored the TCRγδ+ IEL [28], suggesting that this IL-7 transgene-driven differentiation of progenitors in the gut might be different from the TCF-1 dependent TCRγδ+ IEL development.
Notch-ligand interactions have decisive roles for the initial T lineage commitment [29] and Notch-1−/− BM cells accumulate as immature B cells in the thymus of BM chimeric mice [30]. In sharp contrast however Notch-1−/− BM derived progenitor cells in the CPs of these animals remain arrested at the CD117+lin− stage, suggesting that either the intestine does not support development of immature B cells or alternatively that immature DN Notch−/− thymocytes do not migrate to the gut prematurely [30].
The defined lineage split of γδ versus αβ at the level of TCRγδ or pre-TCR expression respectively, has focused on TCR signal strength and duration for the instruction or confirmation of pre-determined lineage fate [31,32]. New evidence has indicated that in addition to phenotypic changes the quality of signals received through a full TCR versus a pre-TCR, has important implications for survival, migration and functional differentiation of thymocytes [33•, 34•, 35, 36]. The fact that in TCRγ transgenic mice few αβ TCR T cells develop [37] and in TCRβ transgenic mice most TCRγδ development is suppressed [38], indicate that signals generated by a full TCR or a pre-TCR are crucial elements in the αβ/γδ lineage choice. Several observations support the notion that strong signals from engaged full γδ- or αβTCRs expressed at the immature DN stages favor the γδ lineage, whereas reduced signaling through a γδTCR redirects development along the αβ lineage [33•, 34•]. The γδTCR-driven αβ lineage differentiation is insufficient and together with the constant association of pre-TCR with αβ lineage choice, they point to unique qualities of the pre-TCR for specific αβ lineage differentiation. Recently it was shown that some DN thymocytes express a full αβTCR [39]. It is possible that early expression of αβTCRs before the pre-TCR, may mimic γδTCR ligation signals rather than pre-TCR signaling and drive the precursor cells into γδ-like IEL [40]. However, the fact that non-transgenic CD8αα+TCRαβ+IEL strictly dependent on pre-TCR expression, indicate that early TCRαβ+ precursors are not major progenitor cells for CD4−CD8αβ−TCRαβ+IEL and that these TCRαβ+IEL are not part of the γδ lineage but represent genuine pre-TCR dependent αβ lineage cells.
γδTCR IEL precursor checkpoints in the thymus
The TCR recombination system creates the potential to generate a repertoire of almost unlimited diversity. Despite this, TCRγδ+T cells display a very restricted repertoire that is tissue specific and strongly correlates with particular variable gene segments of the TCRγ (Vγ) [41]. Furthermore, various γδ subsets develop with different time kinetics that are in part programmed by stem cell subpopulations [42]. In mice, TCRγδ+IEL populate the intestine around birth and sometime thereafter and they almost all express Vγ5* [43*, 44]. Because Vγ5 is rarely expressed by DN4 thymocytes and productive Vg5 gene segments are underrepresented at the DN3 stage in C57BL/6 mice, it suggests that TCRγδ+IEL precursors might exit the thymus on or before the DN3 stage [45]. Although genetically programming is involved in γδ gene rearrangements, selective TCRγδ-associated signals may control in part the migration ability of the TCRγδ+thymocytes. For example, G8-TCRγδ+thymocytes specific for the MHC class Ib molecules, T10b/T22b, differentiate in the presence of their cognate antigen and readily migrate to the gut but not elsewhere [46]. In that aspect it was shown that unlike the conventional selected TCRαβ precursors, functional S1P1 was not required for thymic egress of γδ-selected progenitors of small intestine TCRγδ+IEL [47••]. The existence of γδ-selection was also demonstrated using GFP-Tcrd reporter mice [48••], which indicated that DN3 thymocytes require a full γδTCR together with CD3ε and functional linker for activation of T cells (LAT) for their differentiation to mature γδTCR+ T cells. A gene expression analysis of TCRγδ+IEL identified a γδ-biased profile that distinguishes them from conventional selected TCRαβ+T cells [49]. Interestingly, rather than typical for TCRγδ expressing IEL this profile was also shared in part by the CD4−CD8αβ−TCRαβ+IEL [49, 50•, 51•], which also require agonistic selection [35] and which also egress from the thymus in a S1P1 independent manner [47].
The specific loss of TCRVγ5+IEL in mice with defects in IL-15R mediated signals [52] and the increase of Vγ5+IEL in IL-15 transgenic mice [53•], suggest a central role for IL-15 in the generation and/or maintenance of TCRγδ+IEL. In support of this, enforced expression of Bcl-2 partially restored the numbers of Vγ5+IEL in IL-15 deficient mice [54]. Nevertheless, Bcl-2-rescued Vγ5+IEL did not display normal effector's functions indicating that IL-15 may also serve important roles for the functional differentiation of Vγ5+IEL. In addition, the selective reduction of Vγ5+thymocytes in IL-15−/− or IL-15 signal transducer stat5−/− newborn mice [53•], indicated that IL-15 specifically influences development and/or survival of Vγ5+thymocyte precursors. Consistent with this, it was shown that, prior to Tcrg rearrangement, IL-15 is able to direct biased Vg5 transcription by specifically regulating chromatin domain modifications and accessibility in the exclusive vicinity of the Vg5 gene [53•]. Nevertheless, the absence of Vγ5 transcripts among IL-7r−/− thymocytes whilst they were detectable among IL-7r−/− precursors in the gut [53•], suggests that Vg5 rearrangements controlled by IL-15 are not a major mechanism during thymic differentiation but it might operate in the small intestine to allow for specific Vγ5 differentiation when thymic selected IEL are limiting.
Thymic checkpoints for TCRαβ+ IEL precursors
Whereas co-receptor expressing TCRαβ+IEL mainly differentiate from conventional positively selected CD4+ and CD8αβ+ T cells that have encountered a cognate non-self antigen in the periphery [1,2], CD4−CD8αβ−TCRαβ+IEL have acquired their antigen experienced phenotype during agonist selection in the thymus [35, 55] (Figure 1). In contrast however to TCRγδ+ thymocytes, which commit to the γδ lineage upon ligation of their mature γδTCR, αβ progenitors pass through additional checkpoints that involve a pre-TCR stage followed by a full αβTCR-based selection process (Figure 1). Although CD4−CD8αβ−TCRαβ+IEL display phenotypic and functional similarities with the γδ subset, this is likely not due to a shared lineage commitment but rather it coincides with the full TCR-based agonist selection process in the thymus. Some DN thymocytes can commit to the γδ-lineage upon expression/ligation of a full αβTCR, as is the case in male H-YTCR transgenic mice [40, 56], however the nearly absence of CD4−CD8αβ−TCRαβ+IEL in pre-Tcr−/− animals [57,58], makes it unlikely that this pre-TCR independent pathway is contributing significantly to the CD4−CD8αβ−TCRαβ+IEL sublineage in normal mice. Consistent with this, H-YTCR transgenic mice in which the transgenic TCR was driven by the CD4 promoter, do not generate DN H-YTCRαβ+ thymocytes and male mice do not generate significant numbers of CD4−CD8αβ−H-YTCR+IEL [59•]. Furthermore, the functional capacity of the transgenic CD4−CD8αβ−H-YTCRαβ+IEL are distinct from their natural occurring counterparts and suggests that true CD4−CD8αβ−TCRαβ+IEL may depend on a pre-TCR pathway for their lineage and functional differentiation.
Pre-TCR also promotes the transition of the DN4 to the double positive CD4CD8αβ+ (DP) stage together with a bust of proliferation (Figure 1). At the DNA level productive pre-TCR signals coincide with β-selection and cessation of further TCRβ rearrangements together with initiation of TCRα rearrangements. Upon expression of a full TCR, DP thymocytes pass through a second checkpoint where αβTCR-based selection events eliminate self-reactive TCRs from the conventional repertoire and allow for functional differentiation of positively selected thymocytes. The observation that the TCR repertoire of CD4−CD8αβ−TCRαβ+IEL is greatly enriched for self-reactive TCRs is inconsistent with negative selection of CD4−CD8αβ−TCRαβ+IEL precursor cells in the thymus. Instead, the enhanced accumulation of CD4−CD8αβ−TCRαβ+IEL in the presence of cognate antigen suggests the existence of an alternative pathway for thymic differentiation that operates in parallel with negative selection and that preserves and differentiates some TCRαβ+thymocytes with high affinity for self-antigens [35]. In support of this, when DP thymocytes from H-YTCR female mice were exposed to H-Y antigen in vitro, a large number of them survived and differentiated to CD8αα+TCRαβ+T cells with the typical gene transcription signature of CD4−CD8αβ−TCRαβ+IEL [36]. This indicated that precursors of CD4−CD8αβ−TCRαβ+IEL might be positively selected by self-agonist at the DP stage. Direct evidence for this was provided by genetic cell-fate mapping using expression of green fluorescent protein (GFP) driven by the CD4 promoter, which confirmed that CD4−CD8αβ−TCRαβ+IEL thymic progenitors transition through the DP checkpoint [60]. These observations imply that DP thymocytes might be heterogenic and contain precursors for conventional T cells as well as CD4−CD8αβ-TCRαβ+IEL. We recently identified a new subset of DP thymocytes that co-express CD8αα [15••]. Using TCR transgenic cells, we further demonstrated that, in contrast to DP thymocytes, CD8αα expressing, triple positive (TP) thymocytes, survived, expanded and adapted to the CD4−CD8αβ-TCRαβ+IEL phenotype in response to in vitro stimulation with their cognate antigen [15••]. Intrathymic injection of TP but not DP thymocytes generated a significant population of CD4−CD8αβ−TCRαβ+IEL in vivo, indicating that TP thymocytes are indeed immature thymic precursors of CD4−CD8αβ-TCRαβ+IEL (Figure 1). Consistent with this, thymocytes that survived agonist selection conditions in vitro acquired an antigen experienced phenotype as well as innate-like features and gut-specific homing receptors [36,51•]. A gene array analysis of ex vivo CD8αα+TCRαβ+IEL indicated an almost identical transcription signature [50•] and further underscores the importance of the thymic agonist selection process as a central mechanism for the unique differentiation and migration of CD4−CD8αβ-TCRαβ+IEL.
The initial induction of CD8αα is consistent with a pre-TCR signal since it does not depend on a full αβTCR or MHC ligation [15••]. Not all pre-TCR triggered thymocytes, however, induce CD8αα indicating that diversity in thymocyte precursors may in part be determined at the pre-TCR level before full αβTCR expression. It is not known how TP thymocytes survive agonist selection conditions but it is likely that CD8αα may function as a repressor to reduce the signal strength and allow for survival [9].
We identified mature TCRαβ+DN thymocytes as post agonist selected precursors and showed that these cells differentiate to CD8αα+CD5−TCRαβ+IEL in vivo without the need for further checkpoints in the thymus [15••]. The co-receptor negative phenotype of agonist selected thymocytes is consistent with the accumulation of co-receptor independent high affinity TCRs among CD4−CD8αβ−TCRαβ+IEL. The acquisition of CD8αα and the downregulation of CD5 are post thymic events that occur locally in the intestine and that are actively promoted by IL-15 [15••].
Conclusions
T cells do not absolutely require a thymus for their development and many tissues with lymphoietic capacity, including BM, spleen, LNs and without a doubt, the intestinal epithelium can effectively support T cell poiesis. The thymus however has evolved as a specific T cell organ equipped with a three dimensional network of stromal and epithelial cells that interact closely with the developing thymocytes. It is this exclusive microenvironment that provides the unique combination of cellular interactions together with cytokines and chemokines that guide thymocytes through defined TCR-dependent “self”-based selection processes that lead to the functional differentiation of self- and nonself-reactive T cells. The gene profile that is uniquely shared by agonist-selected γδ- and αβ-lineage IEL as opposed to their conventional selected counterparts, importantly underscores that the functional diversity and homing capacity of the various IEL subsets is directly linked to the specific selection process of their thymic precursor cells.
This however does not exclude lymphopoiesis in the gut and it is possible that RORγt+ lymphoid tissue inducer (LTi)-like cells may transform CPs to lymphoid follicles [61] that together with cytokines such as IL-15, promote a transient burst of non-selective T cell poiesis in the gut in response to challenges imposed by infections or local tissue damage.
Acknowledgements
We thank Marieke Cheroutre for her contribution. This work was supported by a grant from the NIH (NIH RO1 DK054451-09). This is manuscript # 992 of the La Jolla Institute for Allergy and Immunology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Cheroutre H. Starting at the beginning: new perspectives on the biology of mucosal T cells. Annu Rev Immunol. 2004;22:217–246. doi: 10.1146/annurev.immunol.22.012703.104522. [DOI] [PubMed] [Google Scholar]
- 2.Cheroutre H, Madakamutil L. Acquired and natural memory T cells join forces at the mucosal front line. Nat Rev Immunol. 2004;4:290–300. doi: 10.1038/nri1333. [DOI] [PubMed] [Google Scholar]
- 3.Rocha B. The extrathymic T-cell differentiation in the murine gut. Immunol Rev. 2007;215:166–177. doi: 10.1111/j.1600-065X.2006.00467.x. [DOI] [PubMed] [Google Scholar]
- 4.Kanamori Y, Ishimaru K, Nanno M, Maki K, Ikuta K, Nariuchi H, Ishikawa H. Identification of novel lymphoid tissues in murine intestinal mucosa where clusters of c-kit+ IL-7R+ Thy1+ lympho-hemopoietic progenitors develop. J Exp Med. 1996;184:1449–1459. doi: 10.1084/jem.184.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saito H, Kanamori Y, Takemori T, Nariuchi H, Kubota E, Takahashi-Iwanaga H, Iwanaga T, Ishikawa H. Generation of intestinal T cells from progenitors residing in gut cryptopatches. Science. 1998;280:275–278. doi: 10.1126/science.280.5361.275. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki K, Oida T, Hamada H, Hitotsumatsu O, Watanabe M, Hibi T, Yamamoto H, Kubota E, Kaminogawa S, Ishikawa H. Gut cryptopatches: direct evidence of extrathymic anatomical sites for intestinal T lymphopoiesis. Immunity. 2000;13:691–702. doi: 10.1016/s1074-7613(00)00068-6. [DOI] [PubMed] [Google Scholar]
- 7.Bandeira A, Itohara S, Bonneville M, Burlen-Defranoux O, Mota-Santos T, Coutinho A, Tonegawa S. Extrathymic origin of intestinal intraepithelial lymphocytes bearing T-cell antigen receptor gamma delta. Proc Natl Acad Sci U S A. 1991;88:43–47. doi: 10.1073/pnas.88.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guy-Grand D, Cerf-Bensussan N, Malissen B, Malassis-Seris M, Briottet C, Vassalli P. Two gut intraepithelial CD8+ lymphocyte populations with different T cell receptors: a role for the gut epithelium in T cell differentiation. J Exp Med. 1991;173:471–481. doi: 10.1084/jem.173.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheroutre H, Lambolez F. Doubting the TCR Coreceptor Function of CD8alphaalpha. Immunity. 2008;28:149–159. doi: 10.1016/j.immuni.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 10.Gangadharan D, Cheroutre H. The CD8 isoform CD8alphaalpha is not a functional homologue of the TCR co-receptor CD8alphabeta. Curr Opin Immunol. 2004;16:264–270. doi: 10.1016/j.coi.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 11.Madakamutil LT, Christen U, Lena CJ, Wang-Zhu Y, Attinger A, Sundarrajan M, Ellmeier W, von Herrath MG, Jensen P, Littman DR, et al. CD8alphaalpha-mediated survival and differentiation of CD8 memory T cell precursors. Science. 2004;304:590–593. doi: 10.1126/science.1092316. [DOI] [PubMed] [Google Scholar]
- 12.Morrissey PJ, Charrier K, Horovitz DA, Fletcher FA, Watson JD. Analysis of the intra-epithelial lymphocyte compartment in SCID mice that received coisogenic CD4+ T cells. Evidence that mature post-thymic CD4+ T cells can be induced to express CD8 alpha in vivo. J Immunol. 1995;154:2678–2686. [PubMed] [Google Scholar]
- •• 13.Lambolez F, Arcangeli ML, Joret AM, Pasqualetto V, Cordier C, Di Santo JP, Rocha B, Ezine S. The thymus exports long-lived fully committed T cell precursors that can colonize primary lymphoid organs. Nat Immunol. 2006;7:76–82. doi: 10.1038/ni1293. [This paper shows that precursor cells can exite a thymus graft prematurely before T cell receptor rearrangements and that immature thymocytes can colonize lymphoid organs such as the thymus and the gut in recipient mice. Migration of the thymic T cell-committed precursors led to permanent colonization of the gut precursor compartment and further differentiate into T cells. Based on these observations the authors suggest here a new function for the thymus in peripheral seeding with T cell precursors that become long lived after thymus export.] [DOI] [PubMed] [Google Scholar]
- 14.Baldwin TA, Hogquist KA, Jameson SC. The fourth way? Harnessing aggressive tendencies in the thymus. J Immunol. 2004;173:6515–6520. doi: 10.4049/jimmunol.173.11.6515. [DOI] [PubMed] [Google Scholar]
- •• 15.Gangadharan D, Lambolez F, Attinger A, Wang-Zhu Y, Sullivan BA, Cheroutre H. Identification of pre- and postselection TCRalphabeta+ intraepithelial lymphocyte precursors in the thymus. Immunity. 2006;25:631–641. doi: 10.1016/j.immuni.2006.08.018. [This study shows that CD4−CD8αβ−CD8αα+IEL, arose from a unique subset of DP thymocytes that co-express CD8αα and are referred to as TP thymocytes. These TP precursors are precommitted to preferentially give rise to CD4−CD8αβ−CD8αα+IEL, but required self-agonist-based selection in the thymus for their initial differentiation. Agonist-selected thymocytes are DN and their final maturation, including the induction of CD8αα, appear to occur after thymus export in the IL-15-rich environment of the gut. These developmental steps, including precommitment of immature thymocytes, TCR-mediated agonist selection, and postthymic differentiation promoted by cytokines, define novel pathways for the generation of CD4−CD8αβ−CD8αα+IEL.] [DOI] [PubMed] [Google Scholar]
- 16.Silva-Santos B, Pennington DJ, Hayday AC. Lymphotoxin-mediated regulation of gammadelta cell differentiation by alphabeta T cell progenitors. Science. 2005;307:925–928. doi: 10.1126/science.1103978. [DOI] [PubMed] [Google Scholar]
- 17.Godfrey DI, Kennedy J, Suda T, Zlotnik A. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3-CD4-CD8- triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J Immunol. 1993;150:4244–4252. [PubMed] [Google Scholar]
- 18.Joachims ML, Chain JL, Hooker SW, Knott-Craig CJ, Thompson LF. Human alpha beta and gamma delta thymocyte development: TCR gene rearrangements, intracellular TCR beta expression, and gamma delta developmental potential--differences between men and mice. J Immunol. 2006;176:1543–1552. doi: 10.4049/jimmunol.176.3.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narayan K, Kang J. Molecular events that regulate alphabeta versus gammadelta T cell lineage commitment: old suspects, new players and different game plans. Curr Opin Immunol. 2007;19:169–175. doi: 10.1016/j.coi.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 20.Capone M, Hockett RD, Jr., Zlotnik A. Kinetics of T cell receptor beta, gamma, and delta rearrangements during adult thymic development: T cell receptor rearrangements are present in CD44(+)CD25(+) Pro-T thymocytes. Proc Natl Acad Sci U S A. 1998;95:12522–12527. doi: 10.1073/pnas.95.21.12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang J, Volkmann A, Raulet DH. Evidence that gammadelta versus alphabeta T cell fate determination is initiated independently of T cell receptor signaling. J Exp Med. 2001;193:689–698. doi: 10.1084/jem.193.6.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ioannidis V, Beermann F, Clevers H, Held W. The beta-catenin--TCF-1 pathway ensures CD4(+)CD8(+) thymocyte survival. Nat Immunol. 2001;2:691–697. doi: 10.1038/90623. [DOI] [PubMed] [Google Scholar]
- 23.Okamura RM, Sigvardsson M, Galceran J, Verbeek S, Clevers H, Grosschedl R. Redundant regulation of T cell differentiation and TCRalpha gene expression by the transcription factors LEF-1 and TCF-1. Immunity. 1998;8:11–20. doi: 10.1016/s1074-7613(00)80454-9. [DOI] [PubMed] [Google Scholar]
- 24.Ohteki T, Wilson A, Verbeek S, MacDonald HR, Clevers H. Selectively impaired development of intestinal T cell receptor gamma delta+ cells and liver CD4+ NK1+ T cell receptor alpha beta+ cells in T cell factor-1-deficient mice. Eur J Immunol. 1996;26:351–355. doi: 10.1002/eji.1830260213. [DOI] [PubMed] [Google Scholar]
- 25.von Freeden-Jeffry U, Solvason N, Howard M, Murray R. The earliest T lineage-committed cells depend on IL-7 for Bcl-2 expression and normal cell cycle progression. Immunity. 1997;7:147–154. doi: 10.1016/s1074-7613(00)80517-8. [DOI] [PubMed] [Google Scholar]
- 26.Schlissel MS, Durum SD, Muegge K. The interleukin 7 receptor is required for T cell receptor gamma locus accessibility to the V(D)J recombinase. J Exp Med. 2000;191:1045–1050. doi: 10.1084/jem.191.6.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu Q, Erman B, Park JH, Feigenbaum L, Singer A. IL-7 receptor signals inhibit expression of transcription factors TCF-1, LEF-1, and RORgammat: impact on thymocyte development. J Exp Med. 2004;200:797–803. doi: 10.1084/jem.20032183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laky K, Lefrancois L, Lingenheld EG, Ishikawa H, Lewis JM, Olson S, Suzuki K, Tigelaar RE, Puddington L. Enterocyte expression of interleukin 7 induces development of gammadelta T cells and Peyer's patches. J Exp Med. 2000;191:1569–1580. doi: 10.1084/jem.191.9.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garbe AI, Krueger A, Gounari F, Zuniga-Pflucker JC, von Boehmer H. Differential synergy of Notch and T cell receptor signaling determines alphabeta versus gammadelta lineage fate. J Exp Med. 2006;203:1579–1590. doi: 10.1084/jem.20060474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson A, Ferrero I, MacDonald HR, Radtke F. Cutting edge: an essential role for Notch-1 in the development of both thymus-independent and -dependent T cells in the gut. J Immunol. 2000;165:5397–5400. doi: 10.4049/jimmunol.165.10.5397. [DOI] [PubMed] [Google Scholar]
- 31.Hayes SM, Love PE. Strength of signal: a fundamental mechanism for cell fate specification. Immunol Rev. 2006;209:170–175. doi: 10.1111/j.0105-2896.2006.00356.x. [DOI] [PubMed] [Google Scholar]
- 32.Lauritsen JP, Haks MC, Lefebvre JM, Kappes DJ, Wiest DL. Recent insights into the signals that control alphabeta/gammadelta-lineage fate. Immunol Rev. 2006;209:176–190. doi: 10.1111/j.0105-2896.2006.00349.x. [DOI] [PubMed] [Google Scholar]
- • 33.Haks MC, Lefebvre JM, Lauritsen JP, Carleton M, Rhodes M, Miyazaki T, Kappes DJ, Wiest DL. Attenuation of gammadeltaTCR signaling efficiently diverts thymocytes to the alphabeta lineage. Immunity. 2005;22:595–606. doi: 10.1016/j.immuni.2005.04.003. [This paper demonstrates that impairing the signaling capacity of a γδTCR complex enables it to efficiently direct thymocytes to the αβ-lineage. Engaging a transgenic γδTCR with its cognate ligand efficiently drives differentiation into the γδ-lineage, while in the absence of ligand, the same γδTCR promotes αβ-lineage development with an efficiency comparable to the pre-TCR. In contrast, attenuating γδTCR signaling through Lck deficiency diverts thymocytes to the αβ-lineage even in the presence of the cognate ligand. Conversely, increased γδTCR signaling favored γδ-lineage development. The data support a model whereby γδ-versus αβ-lineage commitment is in part controlled by TCR signal strength.] [DOI] [PubMed] [Google Scholar]
- • 34.Hayes SM, Li L, Love PE. TCR signal strength influences alphabeta/gammadelta • lineage fate. Immunity. 2005;22:583–593. doi: 10.1016/j.immuni.2005.03.014. [Impact of TCR signal strength on αβ-/γδ-lineage development was analyzed by altering the surface expression or signalling potential of the γδTCR. The data indicate that increased γδTCR signal strength favores γδ-lineage development, whereas reduced γδTCR signalling favors αβ-lineage development. These data support a model in which the strength of the TCR signal controls in part the lineage fate decision.] [DOI] [PubMed] [Google Scholar]
- 35.Leishman AJ, Gapin L, Capone M, Palmer E, MacDonald HR, Kronenberg M, Cheroutre H. Precursors of functional MHC class I- or class II-restricted CD8alphaalpha(+) T cells are positively selected in the thymus by agonist self-peptides. Immunity. 2002;16:355–364. doi: 10.1016/s1074-7613(02)00284-4. [DOI] [PubMed] [Google Scholar]
- 36.Yamagata T, Mathis D, Benoist C. Self-reactivity in thymic double-positive cells commits cells to a CD8 alpha alpha lineage with characteristics of innate immune cells. Nat Immunol. 2004;5:597–605. doi: 10.1038/ni1070. [DOI] [PubMed] [Google Scholar]
- 37.Kang J, Coles M, Cado D, Raulet DH. The developmental fate of T cells is critically influenced by TCRgammadelta expression. Immunity. 1998;8:427–438. doi: 10.1016/s1074-7613(00)80548-8. [DOI] [PubMed] [Google Scholar]
- 38.von Boehmer H, Bonneville M, Ishida I, Ryser S, Lincoln G, Smith RT, Kishi H, Scott B, Kisielow P, Tonegawa S. Early expression of a T-cell receptor beta-chain transgene suppresses rearrangement of the V gamma 4 gene segment. Proc Natl Acad Sci U S A. 1988;85:9729–9732. doi: 10.1073/pnas.85.24.9729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aifantis I, Bassing CH, Garbe AI, Sawai K, Alt FW, von Boehmer H. The E delta enhancer controls the generation of CD4- CD8- alphabetaTCR-expressing T cells that can give rise to different lineages of alphabeta T cells. J Exp Med. 2006;203:1543–1550. doi: 10.1084/jem.20051711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Egawa T, Kreslavsky T, Littman DR, von Boehmer H. Lineage diversion of T cell receptor transgenic thymocytes revealed by lineage fate mapping. PLoS ONE. 2008;3:e1512. doi: 10.1371/journal.pone.0001512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiong N, Raulet DH. Development and selection of gammadelta T cells. Immunol Rev. 2007;215:15–31. doi: 10.1111/j.1600-065X.2006.00478.x. [DOI] [PubMed] [Google Scholar]
- 42.Ikuta K, Kina T, MacNeil I, Uchida N, Peault B, Chien YH, Weissman IL. A developmental switch in thymic lymphocyte maturation potential occurs at the level of hematopoietic stem cells. Cell. 1990;62:863–874. doi: 10.1016/0092-8674(90)90262-d. [DOI] [PubMed] [Google Scholar]
- 43.Garman RD, Doherty PJ, Raulet DH. Diversity, rearrangement, and expression of murine T cell gamma genes. Cell. 1986;45:733–742. doi: 10.1016/0092-8674(86)90787-7. [DOI] [PubMed] [Google Scholar]
- 44.Goodman T, Lefrancois L. Intraepithelial lymphocytes. Anatomical site, not T cell receptor form, dictates phenotype and function. J Exp Med. 1989;170:1569–1581. doi: 10.1084/jem.170.5.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krotkova A, Smith E, Nerz G, Falk I, Eichmann K. Delayed and restricted expression limits putative instructional opportunities of Vgamma1.1/Vgamma2 gammadelta TCR in alphabeta/gammadelta lineage choice in the thymus. J Immunol. 2004;173:25–32. doi: 10.4049/jimmunol.173.1.25. [DOI] [PubMed] [Google Scholar]
- 46.Lin T, Yoshida H, Matsuzaki G, Guehler SR, Nomoto K, Barrett TA, Green DR. Autospecific gammadelta thymocytes that escape negative selection find sanctuary in the intestine. J Clin Invest. 1999;104:1297–1305. doi: 10.1172/JCI7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •• 47.Kunisawa J, Kurashima Y, Higuchi M, Gohda M, Ishikawa I, Ogahara I, Kim N, Shimizu M, Kiyono H. Sphingosine 1-phosphate dependence in the regulation of lymphocyte trafficking to the gut epithelium. J Exp Med. 2007;204:2335–2348. doi: 10.1084/jem.20062446. [Here the data show that IELs can be divided into two groups according to their dependency on sphingosine 1-phosphate (S1P) for trafficking into the intestines. Conventional T cells originating from SP thymocytes express high levels of S1P1 and their migration into the intestine is regulated by S1P. In contrast, whereas DN thymic IEL precursors expressing either an αβ- or γδTCR migrate to the intestine. in a S1P1 independent fashion and down-regulation of S1P1 or disruption of the S1P gradient prohibited conventional T cells to migrate to the gut, but did not affect the trafficking of DN thymic IEL precursors. These data are the first to demonstrate that thymic precursors of IEL subsets have different gut homing capacity independent from their lineage commitment but consistent with their specific thymic selection requirement.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- •• 48.Prinz I, Sansoni A, Kissenpfennig A, Ardouin L, Malissen M, Malissen B. Visualization of the earliest steps of gammadelta T cell development in the adult thymus. Nat Immunol. 2006;7:995–1003. doi: 10.1038/ni1371. [Using reporter mice that express a reporter gene ‘knocked into’ the Tcrd constant region gene, it was shown that a single checkpoint controls the γδ-switch during the immature DN stage. This checkpoint requires ligand triggering and signaling through a full γδTCR which induces a burst of sustained proliferation similar to that induced by the pre-TCR.] [DOI] [PubMed] [Google Scholar]
- 49.Pennington DJ, Silva-Santos B, Shires J, Theodoridis E, Pollitt C, Wise EL, Tigelaar RE, Owen MJ, Hayday AC. The inter-relatedness and interdependence of mouse T cell receptor gammadelta+ and alphabeta+ cells. Nat Immunol. 2003;4:991–998. doi: 10.1038/ni979. [DOI] [PubMed] [Google Scholar]
- • 50.Denning TL, Granger SW, Mucida D, Graddy R, Leclercq G, Zhang W, Honey K, Rasmussen JP, Cheroutre H, Rudensky AY, et al. Mouse TCRalphabeta+CD8alphaalpha intraepithelial lymphocytes express genes that down-regulate their antigen reactivity and suppress immune responses. J Immunol. 2007;178:4230–4239. doi: 10.4049/jimmunol.178.7.4230. [Using microarray analysis together with real-time quantitative PCR and flow cytometryit was shown thatsimilar to γδTCR IEL, CD4−CD8αβ−CD8αα+IEL preferentially express genes that suppress activation including members of the Ly49 family of NK receptors and other inhibitory receptors, along with some activating receptors. The signaling molecules expressed by both γδTCR IEL and CD4−CD8αβ−CD8αα+IEL are similar but differ with the conventional selected IEL subset. The CD4−CD8αβ−CD8αα+IEL also show increased expression of genes that are related to immune regulation, including TGF-β and lymphocyte activation gene-3. These data show that γδ-lineage IEL and CD4-CD8αβ−CD8αα+IEL have partially overlapping signatures but also unique features that clearly separate them as individual but related subsets.] [DOI] [PubMed] [Google Scholar]
- • 51.Yamagata T, Benoist C, Mathis D. A shared gene-expression signature in innate-• like lymphocytes. Immunol Rev. 2006;210:52–66. doi: 10.1111/j.0105-2896.2006.00371.x. [This paper brings forward several data that indicate that innate-like lymphocytes might share a common gene transcription signature. The signature observed in agonist-triggered CD4−CD8αβ−CD8αα+ T cells derived from fetal or neonatal thymocytes was compared with gene expression profiles of innate-like lymphocytes and closely paired adaptive system counterparts. A statistically significant ‘innate signature’ indeed was distilled that can be designated as an innate signature among lymphocytes that require self-agonist selection for their development.] [DOI] [PubMed] [Google Scholar]
- 52.Schluns KS, Nowak EC, Cabrera-Hernandez A, Puddington L, Lefrancois L, Aguila HL. Distinct cell types control lymphoid subset development by means of IL-15 and IL-15 receptor alpha expression. Proc Natl Acad Sci U S A. 2004;101:5616–5621. doi: 10.1073/pnas.0307442101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • 53.Zhao H, Nguyen H, Kang J. Interleukin 15 controls the generation of the • restricted T cell receptor repertoire of gamma delta intestinal intraepithelial lymphocytes. Nat Immunol. 2005;6:1263–1271. doi: 10.1038/ni1267. [The data presented in this paper show that the generation of the restricted TCRVγ5 gene repertoire of IEL can be regulated by interleukin 15, which induced local chromatin modifications specific for the Vγ5 gene segment and enhanced accessibility conducive to subsequent targeted gene rearrangement. This cytokine-directed tissue-specific TCR repertoire formation is distinct from γδ- and αβTCR repertoires generated under specific selection conditions in the thymus and indicates that thymus independent development pathways for T cells exist that lead to the generation of T cells with limited specificity and functional capacity.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakazato K, Yamada H, Yajima T, Kagimoto Y, Kuwano H, Yoshikai Y. Enforced expression of Bcl-2 partially restores cell numbers but not functions of TCRgammadelta intestinal intraepithelial T lymphocytes in IL-15-deficient mice. J Immunol. 2007;178:757–764. doi: 10.4049/jimmunol.178.2.757. [DOI] [PubMed] [Google Scholar]
- 55.Cheroutre H, Madakamutil L. Mucosal effector memory T cells: the other side of the coin. Cell Mol Life Sci. 2005;62:2853–2866. doi: 10.1007/s00018-005-5232-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Croxford AL, Akilli-Ozturk O, Rieux-Laucat F, Forster I, Waisman A, Buch T. MHC-restricted T cell receptor signaling is required for alphabeta TCR replacement of the pre T cell receptor. Eur J Immunol. 2008;38:391–399. doi: 10.1002/eji.200737054. [DOI] [PubMed] [Google Scholar]
- 57.Lambolez F, Azogui O, Joret AM, Garcia C, von Boehmer H, Di Santo J, Ezine S, Rocha B. Characterization of T cell differentiation in the murine gut. J Exp Med. 2002;195:437–449. doi: 10.1084/jem.20010798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pennington DJ, Silva-Santos B, Silberzahn T, Escorcio-Correia M, Woodward MJ, Roberts SJ, Smith AL, Dyson PJ, Hayday AC. Early events in the thymus affect the balance of effector and regulatory T cells. Nature. 2006;444:1073–1077. doi: 10.1038/nature06051. [DOI] [PubMed] [Google Scholar]
- • 59.Baldwin TA, Sandau MM, Jameson SC, Hogquist KA. The timing of TCR{alpha} • expression critically influences T cell development and selection. J Exp Med. 2005;202:111–121. doi: 10.1084/jem.20050359. [This paper shows that expression of the H-YTCR at the physiological DP stage does not result in reduced DP and increased DN cellularity typically seen in male H-YTCR transgenic mice and H-YTCR CD4−CD8αβ−CD8αα+ do not expand in these “on time” transgenic male mice. These data indicate that the abundant generation of CD4−CD8αβ-CD8αα+ IEL in male H-YTCR transgenic mice is due to premature expression of the transgenic H-YTCR at the immature DN stage which favors the γδ-lineage differentiation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eberl G, Littman DR. Thymic origin of intestinal alphabeta T cells revealed by fate mapping of RORgammat+ cells. Science. 2004;305:248–251. doi: 10.1126/science.1096472. [DOI] [PubMed] [Google Scholar]
- 61.Eberl G. Inducible lymphoid tissues in the adult gut: recapitulation of a fetal developmental pathway? Nat Rev Immunol. 2005;5:413–420. doi: 10.1038/nri1600. [DOI] [PubMed] [Google Scholar]