Abstract
We previously reported the metabolic 15N labeling of a rat where enrichment ranged from 94% to 74%. We report here an improved labeling strategy which generates 94% 15N enrichment throughout all tissues of the rat. A high 15N enrichment of the internal standard is necessary for accurate quantitation, and thus, this approach will allow quantitative mass spectrometry analysis of animal models of disease targeting any tissue.
Introduction
The measurement of ion signals by mass spectrometers is precise and quantitative. Sampling handling and ionization are subject to variations that need to be controlled and measured, thus the use of internal standards to determine variations and errors is common. In addition, the measurement of relative changes in quantity requires an internal standard for every molecule to be compared. A general approach in quantitative mass spectrometry is to mix a protein sample containing only natural-abundance isotopes with an identical protein sample except the proteins are labeled with heavy stable isotopes (i.e. 2H, 13C, 15N, or 18O) 1, 2. The relative protein expression is calculated from the ion chromatograms of the labeled and unlabeled peptides 3-6.
Several strategies have been used to introduce labels into proteins including covalent attachment of stable isotope labeled reporter groups 7-10, proteolysis in the presence of 18O water 11-13, and metabolic incorporation of stable isotope labeled amino acids 1, 2, 7-9, 14-17. Compared to in vitro labeling methods, metabolic labeling ensures that every protein is enriched with a heavy stable isotope. Metabolic labeling is routinely performed with cultured cells ranging from bacteria, yeast to mammalian cells; and has been demonstrated in multi-cellular organisms such as C. elegans and D. melanogaster 18. Ishihama et al. took a creative approach by using a metabolically labeled 15N Neuro2A mouse cell line as an internal standard for mouse brain19. One drawback to this strategy is that the Neuro2A and brain proteome do not completely overlap. For example, neurons in vivo form synapses, while Neuro2A cultured cells do not. To pursue global quantification of complex mammalian tissues with mass spectrometry, we developed a technique to metabolically labeled a Sprague-Dawley rat by feeding it an 15N enriched protein diet for 44 days starting when the rat was 3 weeks old20. We demonstrated that the labeled rat was healthy and phenotypically identical to an unlabeled rat. Using the proteins from the 15N labeled rat liver as internal standards, we quantified global changes in the liver induced by a sub-lethal dose of cyclohexamide 20.
The introduction of stable isotope labels into mammalian organisms that serve as models for human physiology and disease creates an opportunity to study a wide range of diseases that effect tissues and organs. We observed, however, a striking difference in the levels of 15N enrichment in the various organs of the rat. While the average 15N enrichment of amino acids in liver proteins was 91%, the average 15N enrichment in brain was 74% 20. Although the nitrogen source is the same for all tissues, the amino acid precursor pools are not. Tissues with slower protein turnover rates will take longer for the 15N labeled amino acids to equilibrate with the normal amino acid precursor pool, which leads to a lower enrichment. Consistent with our measured levels of 15N enrichment, liver has a very a high protein turnover rate, while brain has a very low turnover rate 21, 22. The disadvantage of low 15N enrichment is that quantitation becomes less efficient and accurate.
In this report, we describe a method to more efficiently feed animals a 15N labeled protein diet and an improved metabolic labeling strategy to obtain high 15N protein enrichment of rat brain tissue. We designed and tested two different labeling protocols to determine the optimal procedure to ensure high 15N enrichment of brain. Furthermore, we show that 15N labeling of two generations of rats does not interfere with fetal development during gestation. In addition, we report this protocol is also applicable to other slow protein turnover tissues besides the brain. This method allows quantitative mass spectrometry analysis of animal models of neurological disorders or other types of diseases.
Materials and Methods
Materials
15N-Enriched (>99 atom % excess; ape) and unlabeled algal cells were purchased from Spectra Gases (Vista, CA). Digestion enzymes were purchased from Roche Applied Science (Indianapolis, IN). All Sprague-Dawley rats were purchased from Harlan (Indianapolis, IN). All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise noted. All methods involving animals were approved by the Institutional Animal Research Committee and accredited by the American Association for Accreditation of Laboratory Animal Care.
Preparation of 15N enriched diet
10 g spirulina biomass (algae), unlabeled or 15N labeled, was mixed well with 30 g protein-free diet powder (Harlan TD 93328). Initially, rats were fed the powder mixture directly; however, we found this method to be a cumbersome and inefficient protocol to feed the rats. To improve our feeding protocol, we created 15N enriched food pellets. To make the pellets, we sprinkled 6 ml (for unlabeled) or 6.5 ml (for 15N labeled) of H2O in the powdered food, stirred and worked the mixture into a dough. The dough was shaped into a long cylinder about 2 cm thick. The cylinder was cut into 2 cm pieces lengthwise. The pellets were dried at 60 ° for 2-4 hours and then at 35 °C overnight using an Excalibur food dehydrator.
Metabolic Labeling with 15N-Labeled Algal Cells
Protocol 1
Two female Sprague-Dawley rats were delivered to the animal facility on pregnancy day 2. One rat was immediately fed a specialized diet consisting of a protein-free rodent diet (Harlan Teklad, TD93328) supplemented with algal cells containing only natural abundance isotopes (control diet) and the other rat was immediately placed on the 15N diet.
Protocol 2
Four female rats (littermates) arrived at the animal facility immediately after weaning. Two rats were immediately placed on the15N diet and housed in the same cage. The other two were placed on the control diet and housed in the same cage. Once the female rats reached sexual maturity, one male rat was placed in the female cage from 6 p.m. to 6 a.m. until pregnancy, which was determined by the observation of a vaginal plug. Before pregnancy, the female rats were offered the 15N diet or the control diet in powder form for 45 min every 6 hours (6a.m., 12p.m., 6p.m., and 12 a.m.). The male rats were removed from the cage, when the female rats were given the powdered food. The male rats were given standard Harlan rodent diet ad libitum when caged separately from the female rats.
For both protocols, pregnant female rats were caged separately and the food (in the form of dry pellets) was given to the rats ad libitum. After weaning, the pups were given food ad libitum. All rats had ad libitum access to water. All rats were maintained in temperature-controlled (23 °C) facility with a 12-h light/dark cycle. Taking the cost of the labeled algae into consideration, the resultant 15N labeled tissue usable as a quantitative internal standard is <$1/mg of protein.
Sample Preparation
Tissues were removed and stored as previously described23. Frozen tissues were homogenized in ice-cold homogenization buffer (4mM HEPES, pH 7.4, 0.32M sucrose) using 30 strokes in a tight-fitting Dounce homogenizer. Protein concentration was determined using the BCA protein assay (Pierce, Rockford, IL). 100 ug of the 15N tissue and the corresponding 14N tissue were mixed. For example, 15N labeled brain homogenate from a p1 pup was mixed with 14N labeled brain homogenate from a p1 pup. The sample was then centrifuged at 14,000 × g for 15 min, and the superntant was discarded. 120 microliters of 5X Invitrosol (Invitrogen, Carlsbad, CA) containing 8 M urea was added to the pellet, and tris(2-Carboxyethyl)-Phosphine Hydrochloride was added at a final concentration of 5 mM to reduce the proteins. This mixture was then vortexed for 1 hour at room temperature. Next, iodoacetamide was added to a final concentration of 10 mM and incubated in the dark at room temperature for 30 minutes to alkylate the sample. 100 mM Tris, pH 8.0, 2 mM CaCl2 was added to the mixture to dilute the 5X Invitrosol to 1X. The mixture was divided into two equal aliquots, and 4 ug of trypsin (1ug/ul) was added to each aliquot. The trypsin digestion was incubated at 37°C for 24 hours. The digestion was stored at -80°C until mass spectrometry analysis.
Multidimensional Protein Identification Technology (MudPIT)
MudPIT was performed as previously described using a LTQ linear ion trap mass spectrometer (ThermoFinnigan, San Jose, CA) and a quaternary HPLC pump (Agilent, Foster City, CA) except for the following modification in the acquisition of the mass spectra 24. A cycle of one full-scan mass spectrum (400-1400 m/z) followed by 6 data-dependent spectra was repeated continuously throughout each step of the multidimensional separation. The first data dependent event was a zoom scan from the most intense peak in order to achieve high resolution of the isotope distribution. The second data dependent event was an MS/MS scan of the most intense peak at a 35% normalized collision energy. The last four data dependent events consisted of zoom scans and MS/MS scans for the second and third most intense peaks. All zoom scans were acquired using 25 microscans to improve signal to noise. Application of mass spectrometer scan functions and HPLC solvent gradients were controlled by the Xcalibur data system (ThermoFinnigan, San Jose, CA).
Analysis of Tandem Mass Spectra
Tandem mass spectra were analyzed using the following software analysis protocol. Poor quality spectra were removed from the dataset using an automated spectral quality assessment algorithm25. Tandem mass spectra remaining after filtering were searched with the SEQUEST™ algorithm version 27 26 against the EBI-IPI_rat_3.05_04-04-2005 database concatenated to a decoy database in which the sequence for each entry in the original database was reversed 27. All searches were parallelized and performed on a Beowulf computer cluster consisting of 100 1.2 GHz Athlon CPUs 28. No enzyme specificity was considered for any search. SEQUEST results were assembled and filtered using the DTASelect (version 2.0) program29. DTASelect 2.0 uses a quadratic discriminant analysis to dynamically set XCorr and DeltaCN thresholds for the entire dataset to achieve a user-specified false positive rate (5% in this analysis). The false positive rates are estimated by the program from the number and quality of spectral matches to the decoy database.
The percent atomic enrichment was estimated using a previously published procedure 30. Briefly, the high resolution zoom scans were employed to determine the 15N peptide isotopic distributions, which are characteristic of the 15N atomic percent enrichment of the peptide. The 15N percent enrichment for a tissue was calculated from one MudPIT analysis of 100 ug of tissue. The spectra were searched for 14N peptides, an algorithm was used to find the corresponding 15N peptide in the MS1 zoom scan. The algorithm predicts isotope distributions over a range of enrichments and compares the predicted distributions to experimental peptide isotope distributions. In addition, the algorithm generates a correlation factor between the matching 14N and 15N peptide with zero being no correlation and 1.0 being a perfect correlation. We used only 15N peptides with a correlation factor greater than 0.7 to obtain the average 15N enrichment for a given tissue. In addition, we determined that at least 100 15N peptides are required to determined an accurate 15N average percent enrichment of a tissue sample.
Statistical Analysis
We performed unpaired t-tests (two-tailed) to determine significant differences between enrichment protocols. The sample size equaled the number of peptides that we were able to determine a confident 15N enrichment percentage (correlation value > 0.7). All statistical analyses were performed using Prism 4.0 software (GraphPad Software, San Diego, CA)
Results and Discussion
Enrichment Protocols
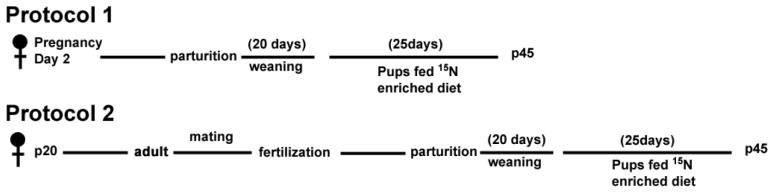
We designed two protocols for which rats were fed a 15N enriched diet for two generations (Figure 1). This diet was formulated by supplementing a protein-free Harlan rodent diet with 15N-enriched algal cells included as the sole protein source. Control rats were fed the same diet formulated with algae containing only natural abundance isotopes. In protocol 1, a pregnant rat was placed on the 15N enriched diet on its second day of pregnancy (E2). This rat remained on the 15N enriched diet after parturition and until it finished weaning its pups after 20 days. In protocol 2, a female rat was placed on the 15N diet immediately after it was weaned. The rat remained on the diet during mating and pregnancy. For both protocols, the mother was sacrificed after it weaned its pups and the pups remained on the 15N diet for an additional 25 days (Figure 1).
Figure 1. Diagram of the enrichment protocols.
A, In protocol 1, a female rat on pregnancy day 2 (E2) was placed on the 15N enriched diet. It remained on the 15N diet during pregnancy and the weaning of its pups. After weaning, the pups were placed on the 15N diet for 25 days. B, In protocol 2, a weaned female rat was placed on a 15N diet and remained on the 15N diet during mating, pregnancy and the weaning of its pups. After weaning, the pups were placed on the 15N diet for 25 days.
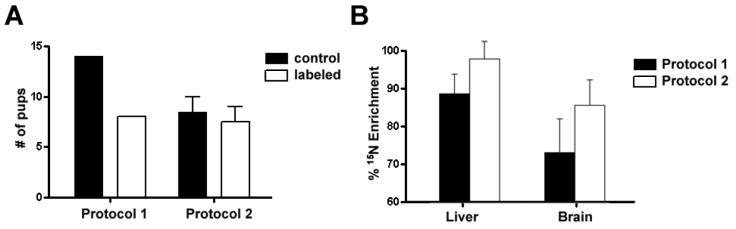
We previously demonstrated that a rat on a 15N diet lasting 44 days had no adverse physiological or histological consequences on a three week old rat. One potential concern, however, is the effect of 15N labeling on fertilization, fetal development, gestation or parturition. All the litters of rats on the 15N diet were within the range of the average litter size for a rat, which is 7-11 pups (data obtained from Harlan, our rodent supplier) (Figure 2A). In protocol 1, the mother was on the 15N diet for 50 days while the mother from protocol 2 was on the 15N diet for 107 days. Accordingly, we observed a lower enrichment of 15N in tissues from mother1 (liver 86.0% +/- 4.69; brain 71.9% +/- 8.13) compared to mother2 (liver 94.4% +/- 4.24; brain 83.3% +/- 6.09) (Figure 2B). As might be expected, 15N enrichment in the liver and brain increased with the longer labeling times and consistent with our previous report 20, the 15N enrichment in the brain was lower than the enrichment in the liver in both protocols.
Figure 2. A, The number of pups in a litter was unaffected by the metabolic labeling of the mothers.
The white bars represent litters from 15N labeled rats and the black bars represent litters from rats on the control diet. In protocol 1, one pregnant rat was placed on the 15N diet and one pregnant rat on the control diet. In protocol 2, two female rats were placed on the 15N diet and two on the control diet before mating. B, The percent 15N enrichment of liver and brain tissues from maternal rats. The black bars represent tissues from a maternal rat from protocol 1 and the white boxes represent tissues from a maternal rat from protocol 2. Values represent averages +/- SD.
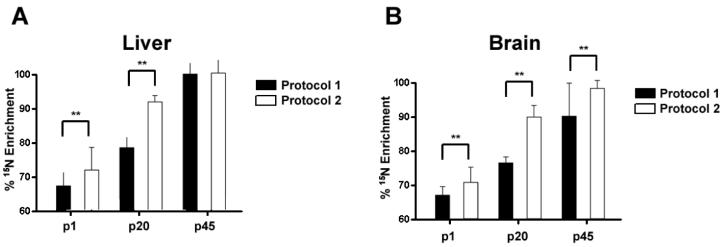
We analyzed the 15N enrichment of tissues from the pups. We determined the enrichment levels of 15N for the pups at p1 (postnatal day 1), p20, and p45. In liver, protocol 2 had an enrichment of 70.8% +/- 6.48 and 88.8% +/- 1.37 at p1 and p20, respectively. This is significantly higher 15N enrichment compared to the liver in the p1 (66.3% +/- 2.77) and p20 (76.6% +/- 2.68) pups of protocol 1 (Figure 3A). This is likely due to the differences in amount of 15N labeled protein obtained while breast feeding. At p1 and p20, the sole source of protein for these pups had been breast milk from the mother, and the enrichment of 15N in proteins was lower in the mother from protocol 1 (liver 86.0% +/- 4.69; brain 71.9% +/- 8.13) compared to the mother from protocol 2 (liver 94.4% +/- 4.24; brain 83.3% +/- 6.09) (Figure 2B). Since the enrichment in the pups cannot exceed their 15N source, the pups from protocol 1 would be expected to have less 15N enrichment than the pups from protocol 2. After weaning, however, the pups from both protocols were placed on the same diet enriched with 99% 15N. After 25 days, the 15N enrichment of pups from protocol 1 (96.2% +/- 2.34) reached the level of the pups from protocol 2 (96.5% +/- 5.07). These data suggest that 96% is the maximum tissue enrichment for two generations of rats on a diet consisting of 99% 15N.
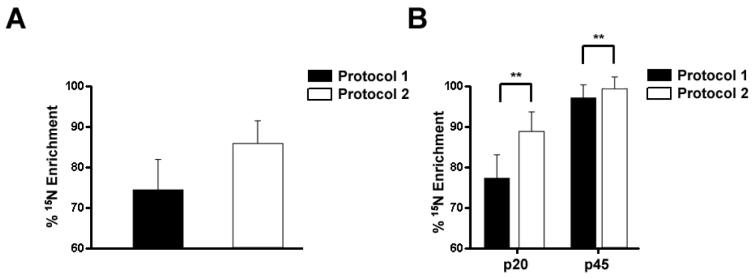
Figure 3. The percent 15N enrichment of liver and brain tissues from rat pups during development.
A, The percent 15N enrichment was determined for liver tissue at 3 developmental ages, p1, p20, and p45. There were significant differences (** p < .001) detected between the two enrichment protocol at p1 and p20, but no significant differences were detected at p45. B, The percent 15N enrichment was determined for brain tissue at 3 developmental ages, p1, p20, and p45. There were significant differences (** p < .001) detected between the two enrichment protocol at p1 and p20. In contrast to the liver tissue, there was a significant difference detected at p45 between the two enrichment protocols. Values represent averages +/- SD.
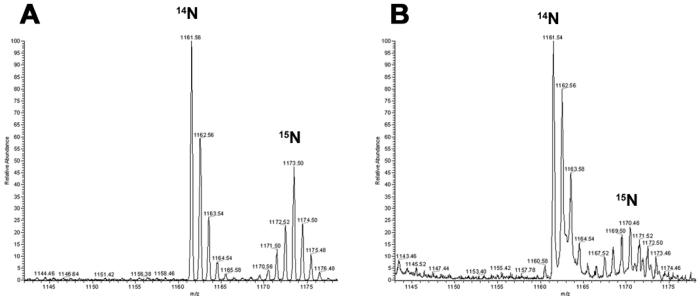
Determination of 15N enrichment in the brain revealed p1 (69.6% +/- 4.08) and p20 (87.3% +/- 2.40) pups of protocol 2 had significantly higher 15N enrichment in proteins compared to the p1 (66.4% +/- 2.27) and p20 (74.9% +/- 2.34) pups of protocol 1 (Figure 3B). This is consistent with the observation from the liver. Furthermore, the 15N enrichment of liver and brain is comparable in both protocols in the p1 and p20 pups. After approximately 40 days of metabolic labeling starting at fertilization, p20 pups in protocol 2 demonstrated a greater 15N enrichment in brain tissue (>85%) compared to our previous attempt (74%) of labeling a 3 week old rat for 44 days 20. Thus, beginning the labeling process earlier increases 15N enrichment in the brain. This is consistent with reports demonstrating protein turnover in muscle tissue decreases with age 31, 32. We did observe, however, a difference in 15N enrichment between liver and brain in the p45 pups. In protocol 1, the 15N brain enrichment was 87.0% +/- 8.95 at p45, while in protocol 2, the 15N brain enrichment was 95% +/- 2.25 at p45, but in liver there was no significant difference between the 15N enrichment of the two protocols at p45. Consistent with reported changes in mass spectra as the enrichment decreases30, the mass spectra of the same peptide from the p45 and p1 brain samples are quite different (Figure 4). As a result, the signal to noise ratio will decrease, which causes two problems. First, identification of 15N labeled peptides becomes difficult. Since the labeled and unlabeled protein samples are mixed 1:1, low enrichment will result in fewer proteins identified and then fewer proteins quantified. Second, low signal intensity hinders the precision of extracting the ion chromatogram of the 15N peptide, which leads to inaccurate quantification. Therefore, labeling for two generations is preferred to adequately enrich brain tissue with 15N labeled proteins for high accuracy quantitative measurements.
Figure 4. The 15N enrichment affects the isotopic distribution.
The peptide, K.EITALAPSTMK.I, was identified in both p45 (A) and p1 (B) brain samples from protocol 2. The peptide was measured to be 95% enriched in the p45 sample and 68% enriched in the p1 sample. The isotopic distribution of the 14N peptide is identical in both mass spectra with the major peak at 1161.5 m/z, but the isotopic distribution for the 15N peptide is different between the two samples. The major isotopic peak of the 15N peptide in p45 is at 1173.5 m/z, while it is at 1170.5 m/z in p1. In addition, the 15N isotopic distribution is broader, less intense in p1 compared to p45. The y-axis is relative abundance and the x-axis is mass/charge.
Like brain tissue, muscle tissue also possesses a slow protein turnover rate 22. In our previous report, skeletal muscle had the lowest 15N enrichment (79%) of any tissue with the exception of brain tissue 20. We analyzed skeletal muscle from our two protocols to determine if 15N enrichment improves in muscle in addition to brain. Similar to the other tissues examined, the mother from protocol 2 (83.4% +/- 5.04) had a higher 15N enrichment in skeletal muscle compared to protocol 1 (72.9%+/- 6.88) (Figure 5A). Furthermore, the p20 pups from protocol 2 (86.7% +/- 2.59) had a higher 15N enrichment in skeletal muscle compared to protocol 1 (76.1% +/- 4.7) as expected. Similar to brain, there was a significant difference in 15N enrichment in skeletal muscle between the two protocols (protocol 1 93.6% +/- 2.37; protocol 2 95.9% +/- 2.62) in p45 pups (Figure 5B). Thus, metabolic labeling of a rat for two generations produces a uniformly higher level of 15N enrichment in all tissues known to have slow protein turnover.
Figure 5. The 15N enrichment of skeletal muscle.
A, The percent 15N enrichment was determined for skeletal muscle from the maternal rats. Consistent with other maternal tissues, the percent enrichment of skeletal muscle was greater from the maternal rat from protocol 2 compared to protocol 1. B, The percent 15N enrichment was determined for skeletal muscle at p20, and p45. There were significant differences (** p < .001) detected between the two enrichment protocol both at p20 and p45. Values represent averages +/- SD.
Conclusions
We developed a metabolic labeling strategy to generate a rat with high levels of 15N enrichment in proteins. The laboratory rat (Rattus norvegicus) is an extensively studied model organism for a range of aspects of human health and disease, physiology, toxicology, neurobiology and drug development. For several human diseases, some clinical and molecular aspects are best studied in the rat, particularly neurodegenerative diseases and disorders affecting higher brain function, such as schizophrenia, anxiety, depression and addiction. In addition, the mouse is another important disease animal model and initial experiments have demonstrated this protocol is also applicable to this species33. Quantitative mass spectrometry has the potential to provide novel insight into these complex neurological disease models, and the ability to metabolically label rats with 15N to a high degree of enrichment will allow more accurate and precise measurement of protein expression to better understand these diseases. We conclude here that labeling rats for two generations generates high 15N enrichment of all tissues regardless of their intrinsic protein turnover rate. This labeling strategy will be important in applying quantitative mass spectrometry to more complex systems such as tissues and organs with low protein turnover rates such as the brain.
Supplementary Material
Supplementary Data 1 A list of all the peptides and their calculated percentage 15Nenrichment for liver, brain, and muscle tissues of the rat pups from both protocols. In addition, the table for brain tissue from protocol 1 includes the protein from which the peptides where derived from and the spectrum count of that protein Since spectrum count has previously been demonstrated to correspond to protein abundance, we demonstrate that high enrichment of both high and low abundant proteins.
Supplementary Data 2 The distribution of the percentage 15N enrichment measurements for liver, brain, and muscle tissues of the rat pups from both protocols.
Supplementary Data 3 Three additional spectra of 15N enriched peptides The y-axis is relative abundance and the x-axis is mass/charge.
Acknowledgements
The authors would like to acknowledge financial support from National Institutes of Health grants 5R01 MH067880-02 and P41 RR11823-10.
References
- 1.Oda Y, Huang K, Cross FR, Cowburn D, Chait BT. Accurate quantitation of protein expression and site specific phosphorylation. Proc. Natl. Acad. Sci. 1999;96:6591–6596. doi: 10.1073/pnas.96.12.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conrads TP, Alving K, Veenstra TD, Belov ME, Anderson GA, Anderson DJ, Lipton MS, Pasa-Tolic L, Udseth HR, Chrisler WB, Thrall BD, Smith RD. Quantitative analysis of bacterial and mammalian proteomes using a combination of cysteine affinity tags and 15N-metabolic labeling. Anal Chem. 2001;73(9):2132–9. doi: 10.1021/ac001487x. [DOI] [PubMed] [Google Scholar]
- 3.MacCoss MJ, Wu CC, Liu H, Sadygov R, Yates JR., 3rd A correlation algorithm for the automated quantitative analysis of shotgun proteomics data. Anal Chem. 2003;75(24):6912–21. doi: 10.1021/ac034790h. [DOI] [PubMed] [Google Scholar]
- 4.Pan C, Kora G, McDonald WH, Tabb DL, Verberkmoes NC, Hurst GB, Pelletier DA, Samatova NF, Hettich RL. ProRata: A Quantitative Proteomics Program for Accurate Protein Abundance Ratio Estimation with Confidence Interval Evaluation. Anal Chem. 2006;78(20):7121–7131. doi: 10.1021/ac060654b. [DOI] [PubMed] [Google Scholar]
- 5.Gruhler A, Olsen JV, Mohammed S, Mortensen P, Faergeman NJ, Mann M, Jensen ON. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol Cell Proteomics. 2005;4(3):310–27. doi: 10.1074/mcp.M400219-MCP200. [DOI] [PubMed] [Google Scholar]
- 6.Everley PA, Bakalarski CE, Elias JE, Waghorne CG, Beausoleil SA, Gerber SA, Faherty BK, Zetter BR, Gygi SP. Enhanced analysis of metastatic prostate cancer using stable isotopes and high mass accuracy instrumentation. J Proteome Res. 2006;5(5):1224–31. doi: 10.1021/pr0504891. [DOI] [PubMed] [Google Scholar]
- 7.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17(10):994–9. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 8.Goodlett DR, Keller A, Watts JD, Newitt R, Yi EC, Purvine S, Eng JK, von Haller P, Aebersold R, Kolker E. Differential stable isotope labeling of peptides for quantitation and de novo sequence derivation. Rapid Commun Mass Spectrom. 2001;15(14):1214–21. doi: 10.1002/rcm.362. [DOI] [PubMed] [Google Scholar]
- 9.Chakraborty A, Regnier FE. Global internal standard technology for comparative proteomics. J Chromatogr A. 2002;949(12):173–84. doi: 10.1016/s0021-9673(02)00047-x. [DOI] [PubMed] [Google Scholar]
- 10.Munchbach M, Quadroni M, Miotto G, James P. Quantitation and facilitated de novo sequencing of proteins by isotopic N-terminal labeling of peptides with a fragmentation-directing moiety. Anal Chem. 2000;72(17):4047–57. doi: 10.1021/ac000265w. [DOI] [PubMed] [Google Scholar]
- 11.Reynolds KJ, Yao X, Fenselau C. Proteolytic 18O Labeling for Comparative Proteomics: Evaluation of Endoprotease Glu-C as the Catalytic Agent. Journal of Proteome Research. 2002;1:27–33. doi: 10.1021/pr0100016. [DOI] [PubMed] [Google Scholar]
- 12.Yao X, Freas A, Ramirez J, Demirev PA, Fenselau C. Proteolytic 18O Labeling for Comparative Proteomics: Model Studies with Two Serotypes of Adenovirus. Anal. Chem. 2001;73(13):2836–42. doi: 10.1021/ac001404c. [DOI] [PubMed] [Google Scholar]
- 13.Johnson KL, Muddiman DC. A method for calculating 16O/18O peptide ion ratios for the relative quantification of proteomes. J Am Soc Mass Spectrom. 2004;15(4):437–45. doi: 10.1016/j.jasms.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 14.Hunter TC, Yang L, Zhu H, Majidi V, Bradbury EM, Chen X. Peptide mass mapping constrained with stable isotope-tagged peptides for identification of protein mixtures. Anal Chem. 2001;73(20):4891–902. doi: 10.1021/ac0103322. [DOI] [PubMed] [Google Scholar]
- 15.Zhu H, Pan S, Gu S, Bradbury EM, Chen X. Amino acid residue specific stable isotope labeling for quantitative proteomics. Rapid Communications in Mass Spectrometry. 2002;16(22):2115–23. doi: 10.1002/rcm.831. [DOI] [PubMed] [Google Scholar]
- 16.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1(5):376–86. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 17.Washburn MP, Ulaszek R, Deciu C, Schieltz DM, Yates JR., III Analysis of quantitative proteomic data generated via multidimensional protein identification technology. Analytical Chemistry. 2002;74(7):1650–1657. doi: 10.1021/ac015704l. [DOI] [PubMed] [Google Scholar]
- 18.Krijgsveld J. Metabolic labeling of C.elegans and D.melanogaster for quantitative proteomics. Nat. Biotechnology. 2003 doi: 10.1038/nbt848. advanced online publication. [DOI] [PubMed] [Google Scholar]
- 19.Ishihama Y, Sato T, Tabata T, Miyamoto N, Sagane K, Nagasu T, Oda Y. Quantitative mouse brain proteomics using culture-derived isotope tags as internal standards. Nat Biotechnol. 2005;23(5):617–21. doi: 10.1038/nbt1086. [DOI] [PubMed] [Google Scholar]
- 20.Wu C, MacCoss M, III JRY. Metabolic labeling of mammalian organisms with stable isotopes for quantitative proteomic analysis. Anal. Chem. 2004 doi: 10.1021/ac049208j. In press. [DOI] [PubMed] [Google Scholar]
- 21.Bark TH, McNurlan MA, Lang CH, Garlick PJ. Increased protein synthesis after acute IGF-I or insulin infusion is localized to muscle in mice. Am J Physiol. 1998;275(1 Pt 1):E118–23. doi: 10.1152/ajpendo.1998.275.1.E118. [DOI] [PubMed] [Google Scholar]
- 22.McNurlan MA, Pain VM, Garlick PJ. Conditions that alter rates of tissue protein synthesis in vivo. Biochem Soc Trans. 1980;8(3):283–5. doi: 10.1042/bst0080283. [DOI] [PubMed] [Google Scholar]
- 23.Wu CC, MacCoss MJ, Howell KE, Matthews DE, Yates JR., 3rd Metabolic labeling of mammalian organisms with stable isotopes for quantitative proteomic analysis. Anal Chem. 2004;76(17):4951–9. doi: 10.1021/ac049208j. [DOI] [PubMed] [Google Scholar]
- 24.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19(3):242–7. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 25.Bern M, Goldberg D, McDonald WH, Yates JR., 3rd Automatic quality assessment of Peptide tandem mass spectra. Bioinformatics. 2004;20(Suppl 1):I49–I54. doi: 10.1093/bioinformatics/bth947. [DOI] [PubMed] [Google Scholar]
- 26.Eng J, McCormack A, Yates JR., 3rd An Approach to Correlate Tandem Mass Spectral Data of Peptides with Amino Acid Sequences in a Protein Database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 27.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003;2(1):43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 28.Sadygov RG, Eng J, Durr E, Saraf A, McDonald H, MacCoss MJ, Yates JR., III Code developments to improve the efficiency of automated MS/MS spectra interpretation. Journal of Proteome Research. 2002;1(3):211–215. doi: 10.1021/pr015514r. [DOI] [PubMed] [Google Scholar]
- 29.Tabb DL, McDonald WH, Yates JR., III DTASelect and Contrast: Tools for Assembling and Comparing Protein Identifications from Shotgun Proteomics. Journal of Proteome Research. 2002;1(1):21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacCoss MJ, Wu CC, Matthews DE, Yates JR., 3rd Measurement of the isotope enrichment of stable isotope-labeled proteins using high-resolution mass spectra of peptides. Anal Chem. 2005;77(23):7646–53. doi: 10.1021/ac0508393. [DOI] [PubMed] [Google Scholar]
- 31.Millward DJ. The regulation of muscle-protein turnover in growth and development. Biochem Soc Trans. 1978;6(3):494–9. doi: 10.1042/bst0060494. [DOI] [PubMed] [Google Scholar]
- 32.Lavie L, Reznick AZ, Gershon D. Decreased protein and puromycinyl-peptide degradation in livers of senescent mice. Biochem J. 1982;202(1):47–51. doi: 10.1042/bj2020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sikela JM, Maclaren EJ, Kim Y, Karimpour-Fard A, Cai WW, Pollack J, Hitzemann R, Belkap J, McWeeney S, Kerns RT, Downing C, Johnson TE, Grant KJ, Tabakoff B, Hoffman P, Wu CC, Miles MF. DNA microarray and proteomic strategies for understanding alcohol action. Alcohol Clin Exp Res. 2006;30(4):700–8. doi: 10.1111/j.1530-0277.2006.00081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data 1 A list of all the peptides and their calculated percentage 15Nenrichment for liver, brain, and muscle tissues of the rat pups from both protocols. In addition, the table for brain tissue from protocol 1 includes the protein from which the peptides where derived from and the spectrum count of that protein Since spectrum count has previously been demonstrated to correspond to protein abundance, we demonstrate that high enrichment of both high and low abundant proteins.
Supplementary Data 2 The distribution of the percentage 15N enrichment measurements for liver, brain, and muscle tissues of the rat pups from both protocols.
Supplementary Data 3 Three additional spectra of 15N enriched peptides The y-axis is relative abundance and the x-axis is mass/charge.