Abstract
The generation of reactive oxygen species in mitochondria acts as a redox signal in triggering cellular events such as apoptosis, proliferation, and senescence. Overproduction of superoxide (O2·-) and O2·--derived oxidants change the redox status of the mitochondrial GSH pool. An electron transport protein, Mitochondrial Complex I, is the major host of reactive/regulatory protein thiols. An important response of protein thiols to oxidative stress is to reversibly form protein mixed disulfide via S-glutathiolation. Exposure of Complex I to oxidized GSH, GSSG, resulted in specific S-glutathiolation at the 51 kDa and 75 kDa subunits. Here, to investigate the molecular mechanism of S-glutathiolation of Complex I, we prepared isolated bovine Complex I under non-reducing conditions and employed the techniques of mass spectrometry and EPR spin trapping for analysis. LC/MS/MS analysis of tryptic digests of the 51 kDa and 75 kDa polypeptides from glutathiolated Complex I (GS-NQR) revealed that two specific cysteines (C206 and C187) of the 51 kDa subunit and one specific cysteine (C367) of the 75 kDa subunit were involved in redox modifications with GS binding. The electron transfer activity (ETA) of GS-NQR in catalyzing NADH oxidation by Q1 was significantly enhanced. However, O2·- generation activity (SGA) mediated by GS-NQR suffered a mild loss as measured by EPR spin trapping, suggesting the protective role of S-glutathiolation in the intact Complex I. Exposure of NADH dehydrogenase (NDH), the flavin subcomplex of Complex I, to GSSG resulted in specific S-glutathiolation on the 51 kDa subunit. Both ETA and SGA of S-glutathiolated NDH (GS-NDH) decreased in parallel as the dosage of GSSG increased. LC/MS/MS analysis of a tryptic digest of the 51 kDa subunit from GS-NDH revealed that C206, C187, and C425 were glutathiolated. C425 of the 51 kDa subunit is a ligand residue of the 4Fe-4S N3 center, suggesting that destruction of 4Fe-4S is the major mechanism involved in the inhibiton of NDH. The result also implies that S-glutathiolation of the 75 kDa subunit may play a role in protecting the 4Fe-4S cluster of the 51 kDa subunit from redox modification when Complex I is exposed to redox change in the GSH pool.
Mitochondrial Complex I (EC 1.6.5.3. NADH:ubiquinone oxidoreductase) is the first energy-conserving segment of the electron transport chain (ETC) (1-3). The enzyme catalyzes electron transfer from NADH to ubiquinone coupled with the translocation of four protons across the membrane. In addition to its functions of electron transfer and energy transduction, the catalysis of Complex I provides the major source of oxygen free radical generation in mitochondria (4-6). Two regions of the enzyme complex are hypothesized to be responsible for generating the superoxide anion radical (O2·-). One is located on the FMN cofactor and is modulated by its binding protein moiety (4, 5, 7), while the other is likely located on the ubiquinone-binding site and probably acts in the mediation of ubiquinone reduction (8, 9).
The generation of O2·- and the oxidants derived from it in mitochondria can act as a redox signal in triggering cellular events such as apoptosis, proliferation, and senescence. The redox pool in mitochondria is enriched in glutathione (GSH) with a physiological concentration of 5-10 mM (10). Overproduction of O2·- and O2·--derived oxidants increases the ratio of GSSG to GSH in mitochondria.
The proteins of mitochondrial ETC are rich in protein thiols (11, 12). It has been documented that Complex I is the major component of the ETC to host protein thiols, which comprise structural thiols involved in the ligands of iron sulfur clusters and the reactive/regulatory thiols which are thought to have biological functions of antioxidant defense and redox signaling (13, 14). The physiological roles of Complex I-derived regulatory thiols have been implicated in the regulation of the respiration, nitric oxide utilization (15, 16), and redox status of mitochondria (10-12).
An important response of protein thiols (PrSH) to oxidative stress is to reversibly form protein mixed disulfides (PrSSG) via S-glutathiolation (11-13). This post-translational modification has been suggested as a common mechanism regulating protein functions related to pathological changes such as disruption of the electron transfer activity and induction of membrane permeable transition pores through the cross-linking of membrane protein thiol(s) (10).
With the use of chaotropic anions such as perchlorate, Complex I can be resolved into three fractions: a flavoprotein fraction (Fp), an iron-sulfur (Fe-S) protein fraction (Ip), and a hydrophobic protein fraction (Hp) (17). The Fp fraction contains the enzymatic activity of NADH dehydrogenase and can be isolated as a three-subunit subcomplex from submitochondrial particles (SMP) (7). As demonstrated by EPR spin trapping with DEPMPO, the mechanism of O2·- generation by NDH is mainly controlled by FMN cofactor and its binding protein moiety at the 51 kDa subunit (7).
In previous studies we demonstrated that the biological relevance of C206 of the 51 kDa subunit in the oxidative damage of NADH dehydrogenase is to play the unique role involved in oxidative damage with protein radical formation based on the evidence of immunospin trapping with DMPO and mass spectrometry (7). Taylor et. al have employed a thiol-specific probe and proteomic approach to examine the redox biochemistry of mitochondria (14). Both the 51 kDa and 75 kDa subunits of Complex I have been implicated as hosts of the redox thiol(s) and are potentially involved in protein S-glutathiolation. This result was further verified by immunoblotting as reported by Beer et. al (13). However, the molecular mechanism of the above redox event remains unclear and needs to be defined.
The current study was undertaken to address the fundamental questions regarding the deep insights into the redox biochemistry of Complex I. Here we have identified the specific cysteine residues involved in the protein S-glutathiolation of Complex I. We have also functionally characterized NQR and NDH, including their electron transport and O2·- generation activities resulting from site-specific S-glutathiolation.
EXPERIMENTAL PROCEDURES
Reagents
Ammonium sulfate, diethylenetriaminepentaacetic acid (DTPA), Ubiquinone-1 (Q1), sodium cholate, deoxycholic acid, glutathione, oxidized glutathione, and β-nicotinamide adenine dinucleotide (reduced form, NADH) were purchased from Sigma Chemical Company (St. Louis, MO) and used as received. The 5-diethoxylphosphoryl-5-methyl-1-pyrroline N-oxide (DEPMPO) spin trap was purchased from ALEXIS Biochemicals (San Diego, CA).
Preparations of Mitochondrial Complex I and NDH subcomplex
Bovine heart mitochondrial Complex I was prepared under non-reducing conditions according to the published method with modifications (17). Submitochondrial particles were prepared as described and used as the starting material (18), starting with 2.5 pounds of trimmed bovine hearts with fat and connective tissues removed. The SMP preparation was suspended in 50 mM Tris-Cl buffer, pH 8.0, containing 1 mM histidine and 0.66 M sucrose (TSH), and then subjected to KCl fractionation (72 gm KCl was added to per liter of SMP) in the presence of deoxycholate (0.3 mg/mg protein). The supernatant thus obtained was mixed with an appropriate amount of cold water to precipitate trace amounts of cytochrome c oxidase, and then dialyzed against 10 mM Tris-Cl, pH 8.0, containing 1 mM EDTA for 6 h with one change of buffer. The dialysate was subjected to centrifugation (96,000 × g for 75 min). The pellet containing Complexes I, II, and III was homogenized in TSH buffer, and then subjected to repeated ammonium acetate fractionation in the presence of deoxycholate (0.5 mg/mg protein). Complex I was finally resolved (39% saturation of ammonium sulfate) and separated using ammonium sulfate precipitation (35.9% saturation) in the presence of potassium cholate (0.4 mg/mg protein).
The three-subunit subcomplex of Complex I containing NADH dehydrogenase was isolated from SMP under non-reducing conditions by following the established method described in a previous publication (7).
Analytical Methods
Optical spectra were measured on a Shimadzu 2401 UV/VIS recording spectrophotometer. The protein concentrations of SMP and Complex I were determined by the Biuret method using BSA as standard. The concentration of Q1 was determined by absorbance spectra from NaBH4 reduction using a millimolar extinction coefficient ε(275nm-290nm) = 12.25 mM-1cm-1 (19). The enzyme activity of NDH was assayed by measuring NADH oxidation by Q1 as described in a previous publication (7). The specific activity of NDH is about 140-150 μmol NADH oxidized min-1 mg-1. To measure the electron transfer activity of Complex I, an appropriate amount of Complex I was added to an assay mixture (1 ml) containing 20 mM potassium phosphate buffer, pH 8.0, 2mM NaN3, and 0.1 mM Q1, and 0.15 mM NADH as developed by Hatefi et al. (20). The Complex I activity was determined by measuring the decrease in absorbance at 340nm. The specific activity of Complex I was calculated using a molar extinction coefficient ε340nm = 6.22 mM-1cm-1. The purified Complex I exhibited a specific activity of ∼1.0 μmol NADH oxidized min-1 mg-1.
Electron Paramagnetic Resonance Experiments
EPR measurements were performed using the EPR Core Facilities at the Ohio State University’s Davis Heart and Lung Research Institute. Experiments were carried out on a Bruker EMX spectrometer operating at 9.86 GHz with 100 kHz modulation frequency at room temperature. The reaction mixture was transferred to a 50 μl capillary, which was then positioned into the HS cavity (Bruker Instrument, Billerica, MA). The sample was scanned using the following parameters: center field, 3510 G; sweep width, 140 G; power, 20 mW; receiver gain, 2×105; modulation amplitude, 1G; time of conversion, 163.84 ms; time constant, 163.84 ms; number of scans, 1 scan. The spectral simulations were performed using the WinSim program developed at NIEHS by Duling (21). The hyperfine coupling constants used to simulate the spin adduct of DEPMPO/·OOH were isomer 1: aN = 13.14 G, aHß = 11.04 G, aHγ = 0.96 G, aP= 49.96 G (80% relative concentration); isomer 2: aN = 13.18 G, aHß = 12.59 G, aHγ = 3.46 G, aP= 48.2 G (20% relative concentration) (22). The correlation coefficient of simulated spectrum is typically more than 0.950. Therefore, the simulated spectrum is suitable for spin quantitation (7, 23).
Immunoblotting Analysis
The reaction mixture was mixed with the Laemmli sample buffer at a ratio 4:1 (v/v), incubated at 70 °C for 10 min, and then immediately loaded onto a 4-20 % Tris-glycine polyacrylamide gradient gel. Samples were run at room temperature for 2 h at 100 V [current 30-40 mA/gel (start); 12-13 mA/gel (end)] Protein bands were electrophoretically transferred to nitrocellulose membrane in 25 mM Tris, 192 mM glycine, and 10% methanol. Membranes were blocked for 1 h at room temperature (R.T.) in Tris-buffered saline (TBS) containing 0.1% Tween-20 (TTBS) and 5% dry milk (BioRad). The blots were then incubated overnight with anti-GSH monoclonal antibody (glutathione as the specific epitope of the antibody, ViroGen Coorporation, Watertown, MA) at 4 °C. Blots were then washed 3 times in TTBS, and incubated for 1 h with horseradish peroxidase-conjugated anti-mouse IgG in TTBS at R.T. The blots were again washed twice in TTBS and twice in TBS, and then visualized using ECL Western Blotting Detection Reagents (Amersham Biosciences).
Mass Spectrometry
The sample of protein was subjected to SDS-PAGE using 4-20% gradient polyacrylamde for Complex I and 10% acrylamide for NDH. Protein bands on the gel were then stained with Coomassie blue and subjected to MS measurement.
(a) In -Gel Digestion
Gels were digested with sequencing grade trypsin (Promega, Madison WI) and chymotrypsin (Roche Diagnostics, Indianapolis, IN) using the Montage In-Gel Digestion Kit from Millipore (Bedford, MA) following the manufacturer’s recommended protocols with minor changes for optimization of peptide extraction. Briefly, the bands of interest were trimmed as closely as possible to minimize background protein material. After being washed twice in 50% methanol/5% acetic acid for several hours, the gel bands were dehydrated with acetonitrile and washed again with cycles of acetonitrile and 100 mM ammonium bicarbonate buffer. The gels were then dried using a speed vac. A 50μl aliquot of trypsin (20 ng/μl) or chymotrypsin (25 ng/μl) in 50mM ammonium bicarbonate buffer was added to the dehydrated gel. The gel was set on ice for 10 min for rehydration before the addition of another 20 μl of 50mM ammonium bicarbonate buffer. The mixture was then incubated at room temperature overnight. The peptides were extracted from the gel using 50% acetonitrile with 5% formic acid several times and pooled together. The extracted pools were concentrated in a speed vac to ∼25 μl.
(b) Nano-LC MS/MS (LC/MS/MS)
Capillary-liquid chromatography tandem mass spectrometry (Nano-LC MS/MS) was performed on a Micromass hybrid quadrupole time-of-flight Q-TOF II (Micromass, Wythenshawe, UK) mass spectrometer equipped with an orthogonal nanospray source (New Objective, Woburn, MA) operated in positive ion mode. The capillary LC system was a Dionex UltiMate system (Dionex, Sunnyvale, CA). Solvent A was water containing 50mM acetic acid and Solvent B was acetonitrile. A 5 cm 75 μM ID BioBasic C18 column (New Objective, Woburn, MA) packed directly in the nanospray tip was used for chromatographic separations. 2.5 μl aliquots of each sample were injected onto the column for analysis. Peptides were eluted directly off the column into the Q-TOF system using a gradient of 2-80% Solvent B over 48 minutes, with a flow rate of ∼300 nl/min. The total run time was 55 minutes. The nanospray capillary voltage was set at 3.0 kV and the cone voltage at 40 V. The source temperature was maintained at 100 °C. Mass spectra were acquired from m/z 400-2,000 every 0.9 seconds with a resolution of 8,000 (FWHM) and recorded using MassLynx 4.0 with automatic switching functions. When the desired peak was detected at a minimum of 15 ion counts, the mass spectrometer automatically switched to acquire a CID (Collision Induced Dissociation) spectrum of the individual peptide; mass spectra were acquired from m/z 75 - 2000 to detect immonium ions. Collision energy was set depending on charge state recognition properties. Sequence information from the MS/MS data was processed using the Mascot Distiller software with standard data processing parameters. Database searches were performed using the MASCOT (Matrix Science, Boston, MA) and PEAKS (Bioinformatics Solutions, Waterloo, ON Canada) programs.
RESULTS
Protein S-glutathiolation of Mitochondrial Complex I
One commonly hypothesized pathway leading to the formation of protein-GSH mixed disulfide (PrSSG) via S-glutathiolation is thiol-disulfide exchange between protein thiols and GSSG (13) (equation 1). Therefore, to induce protein S-glutathiolation of Complex I, the protein (2.8 mg/ml, ∼2.8 μM) was incubated with various amounts of GSSG (0-3 mM) in phosphate buffered saline (PBS) at room temperature for 1 h.
| (Eq. 1) |
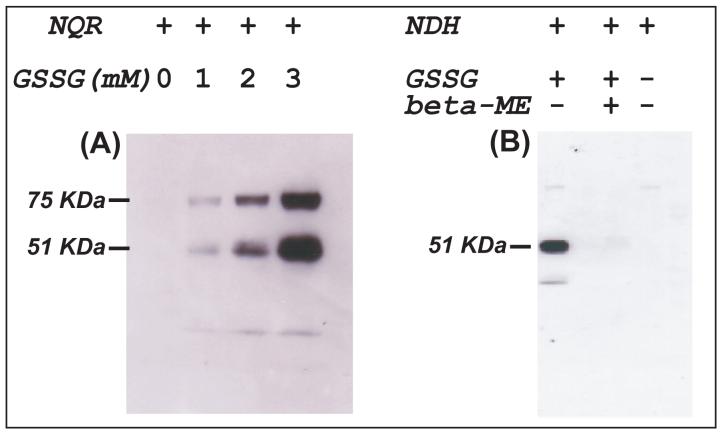
When the reaction mixture was subjected to SDS-PAGE and immunoblotted with the monoclonal antibody against GSH, Complex I-derived PrSSG was detected as indicated in Fig. 1A. The detected Western signal was diminished in the presence of the reducing agents beta-mercaptoethanol (β-ME), suggesting a reversible process (data not shown). The subunit involved in the protein S-glutathiolation was specifically located on the 51 kDa and 75 kDa subunits of Complex I. The intensity of the Western signal was enhanced in proportion to the dose of GSSG (Fig. 1A). The equilibrium constant of thiol exchange between free cysteine and GSSG was reported to be 0.71 (24, 25). The equilibrium constant between GSSG and thiol agent, dithiothreitol, was reported to be 2.00-2.20 × 102(26). Presumably, protein conformation and localized environment of cysteine residue in the NQR limited the extent of protein S-glutathiolation even though the ratio of GSSG/NQR is high (350-1050). The addition of NADH (0.15 mM) and Q1 (0.1 mM) to the mixture did not significantly affect the intensity of detected Western signal, suggesting that protein S-glutathiolation of Complex I can take place under the conditions of enzyme turnover (data not shown).
Fig. 1. Immunoblotting of the protein GSH mixed disulfide (PrSSG) of NQR (A) and NDH (B) using an anti-GSH monoclonal antibody.
A, The NQR-derived PrSSG was induced by a thiol disulfide exchange reaction (Equation 1) from NQR (2.8 mg/ml) in PBS with various amounts (0-3 mM) of GSSG at room temperature for 1 h. B, NDH-derived PrSSG from the reaction (Equation 1) of NDH (0.23 mg/ml) in PBS with GSSG (1 mM) at room temperature for 1 h.
Involvement of C367 at the 75 kDa subunit in the site-specific glutathiolation of NQR as determined by mass spectrometry
To provide further direct evidence for the molecular mechanism of Complex I-derived protein S-glutathiolation induced by GSSG, it was imperative to determine the location of GS binding. Complex I-derived PrSSG was obtained from incubation of NQR (2.8 mg/ml) with GSSG (2 mM, ratio of GSSG/NQR ∼ 700) at RT for 1 h (Fig. 1A, lane 3) and the reaction mixture subjected to SDS-PAGE under non-reducing conditions. The protein band at 75 kDa was cut out and subjected to in-gel digestion with trypsin and chymotrypsin respectively as described in “Experimental Procedures”, followed by LC/MS/MS analysis. The resulting mass spectra acquired from the tryptic and chymotryptic digests contained ions that corresponded in mass to tryptic and chymotryptic peptides of NQR and accounted for over 70.45% of the amino acid sequence of the 75 kDa subunit (Fig. 2).
Fig. 2. Amino acid sequence of the 75 kDa subunit precursor of NQR.
The region labeled with bold represents the amino acid residues identified with LC/MS/MS. The underlined regions are the proposed sequence motif of 4Fe-4S binding (aa residues 64-92, 124-137, 176-226). The residues highlighted with gray color are involved in GS-binding (C367). The region labeled with a dotted underline is the signal peptide (residues 1-23), which acts as an import sequence and does not exist in the mature protein.
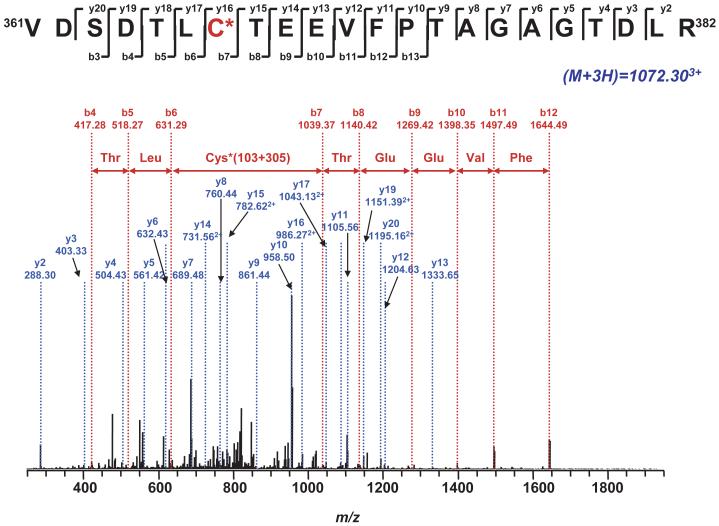
The addition of one glutathione to native protein will increase the molecular weight of the protein by 305 Da. Therefore, the mass spectra from the proteolytic digest of PrSSG of the NQR-75 kDa polypeptide were examined for the addition of 305 Da to the tryptic or chymotryptic peptides. This mass difference was observed in both tryptic and chymotryptic digest. MS/MS results indicated that the mass shift of 305 Da occurred on the same residue C367 in both digests. The tryptic peptide was identified as fragment 361VDSDTLC367TEEVFPTAGAGTDLR382, named GSC367 (aa 361-382, Fig. 3) and the chymotryptic peptide was identified as fragment C367TEEVFPTAGAGTDLRSNY385.,(data not shown)
Fig. 3. Tandem mass spectrum (MS/MS) of the triply protonated molecular ion of the GS-binding peptide (361VDSDTLC367TEEVFPTAAGTDLR382) of the 75 kDa subunit from GS-NQR.
The sequence-specific ions are labeled as y and b ions on the spectrum. The amino acid residues involved in GS binding are identified by asterisks.
The triply protonated molecular ion, (M+3H)3+, of tryptic peptide GSC367 was observed at m/z 1072.30, which has a mass shift of 305 Da if compared with the parent ion [m/z 2908.40 for (M+H)+]. These data suggest that one GSH is covalently bound to one of the residues of the GSC367 peptide.
To determine which amino acid(s) was covalently linked with the GSH for the GSC367 peptide, the MS/MS spectrum of the (GSC367)3+ ion at m/z 1072.303+ was obtained. As shown in Fig. 3, under the conditions of low energy CID, both y and b product ions were observed, corresponding to cleavages along the peptide backbone (27, 28). The y series ions result from C-terminal peptide cleavages, while the b series ions result from cleavages at the N-terminal.
In the spectrum of the (GSC367)3+ ion ( Fig. 3), some of the structurally informative fragment ions including b7-b13 and y16-y20 were observed with a mass shift of 305 Da compared to the native fragment ions, thus allowing unequivocal assignment of the glutathiolated adduct to the cysteine-367 residue of the tryptic peptide, 361VDSDTLC367TEEVFPTAGAGTDLR382. Other sequence informative ions including b3-b6 and y2-y15 provided the evidence to ensure that the sequence of (GSC367)3+ was matched to the sequence of aa residues 361-382 of the 75 kDa subunit.
Involvement of C206 and C187 at the 51 kDa subunit of NQR in site-specific S-glutathiolation
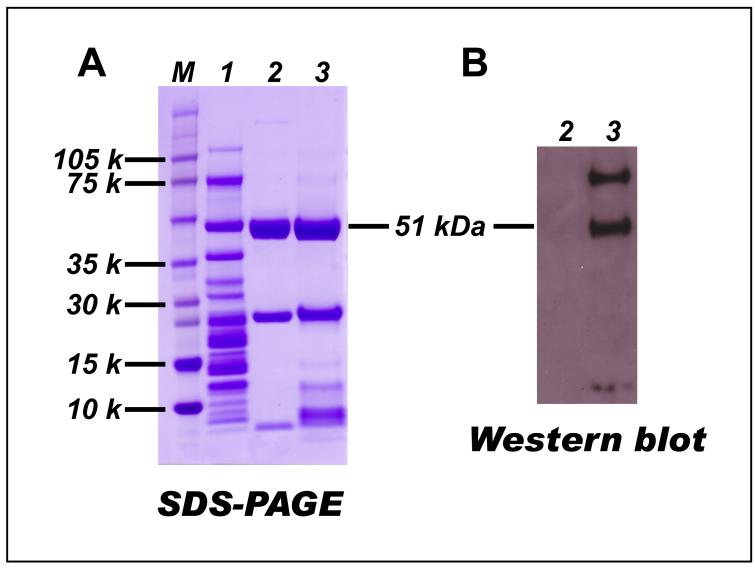
It was observed that the protein band of the 51 kDa subunit in the SDS-PAGE of NQR overlapped with the bands from the subunits of 49 kDa (Ip) and ND5 (Hp) (Fig. 4A, lane 1) as verified by MS (data not shown). To facilitate the identification of specific cysteine residue(s) in the 51 kDa subunit of glutathiolated NQR (GS-NQR), the Fp fraction of GS-NQR was partially purified by a procedure involving ethanol (9% v/v) extraction at 40 °C, which removed the Hp fraction and most of the Ip fraction of NQR (Fig. 4A) (7). This partially purified Fp fraction (namely crude NADH dehydrogenase) containing multiple subunits was subjected to SDS-PAGE (Fig. 4A, lane 3). Both the 51 kDa and 75 kDa subunits were recognized by the monoclonal antibody against GSH as verified by Western blotting (Fig. 4B, lane 3).
Fig. 4. Preparation of the crude NADH dehydrogenase (NDH) from GS-NQR.
GS-NQR was prepared according to the procedure (using 3 mM GSSG here) described in the legend of Fig. 1. 100 % ethanol was added to a final concentration of 9% (v/v), and the mixture was incubated at 40 °C for 10 min. The sample was then chilled in an ice-salt water bath for 10 min and subjected to centrifugation at 25,000 rpm for 30 min. The supernatant containing crude NDH was collected and concentrated with Centricon 30. A, SDS-PAGE: lane 1, isolated NQR (32 μg); lane 2, isolated native NADH dehydrogenase (7.5 μg); lane3, crude NADH dehydrogenase (12 μg). M represents a molecular weight marker. B, Western blot using a monoclonal antibody against GSH: lane 2, isolated native NADH dehydrogenase; lane 3, crude NADH dehydrogenase obtained from GS-NQR.
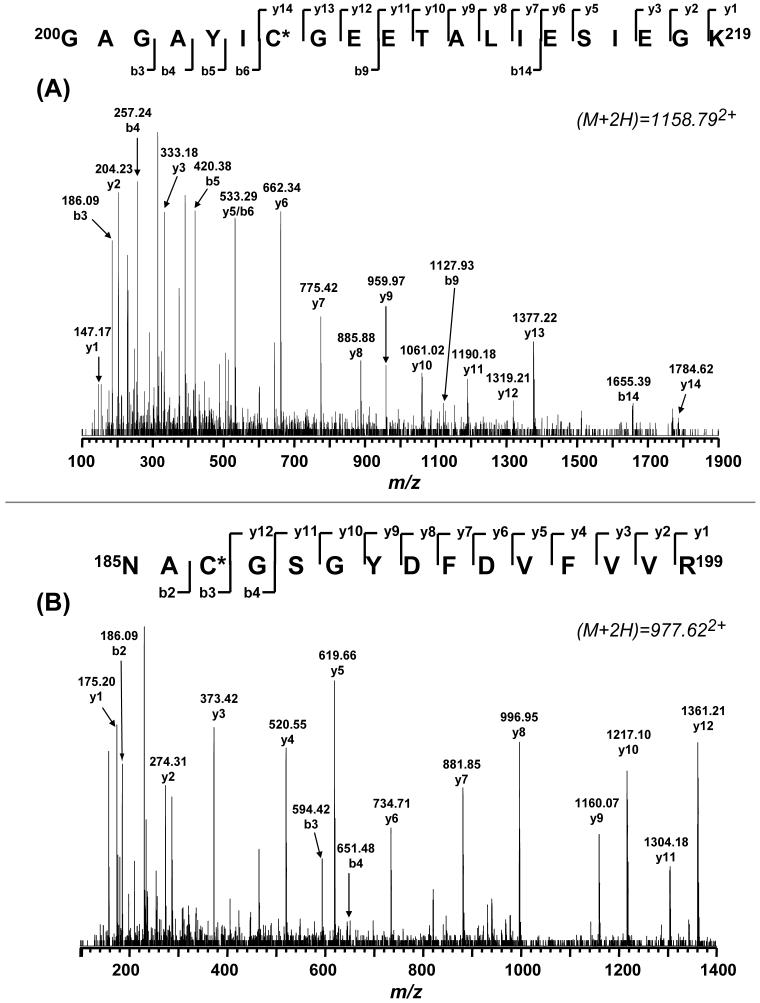
The protein band at 51 kDa was cut out and subjected to in-gel digestion with trypsin and chymotrypsin respectively as described previously, and followed by LC/MS/MS analysis. The mass spectra from the proteolytic digests of the 51 kDa polypeptide were investigated for the addition of 305 Da for S-glutathiolation. This mass difference was observed for two specific tryptic peptides: GSC206, 200GAGAYIC206GEETALIESIEGK219 (aa 200-219) and GSC187, 185NAC187ACGSGYDFDVFVVR199 (aa 185-199).
The doubly and triply protonated molecular ions (M+2H)2+ and (M+3H)3+ of the tryptic peptide GSC206 were observed at m/z 1158.79 and 772.93, where each ion corresponded in mass to the parent ion [m/z 2010.97 for (M+H)+] with an additional 305 Da. MS/MS spectra of the (GSC206)2+ ion of m/z 1158.792+ (Fig. 5A) and the (GSC206)3+ ion of m/z 772.933+ (data not shown) further revealed C206 to be glutathiolated.
Fig. 5. Tandem mass spectra (MS/MS) of the doubly protonated molecular ions of the GS-binding peptides (A) 200GAGAYIC206GEETALIESIEGK219 and (B) 185NAC187GSGYDFDVFVVR199 of the 51 kDa subunit from GS-NDH.
The sequence-specific ions are labeled as y and b ions on the spectra. The amino acid residues involved in GS-binding are identified by asterisks.
In the spectrum of the (GSC206)2+ ion of m/z 1158.79 (Fig. 5A), some of the structurally informative fragment ions including b9, b14, and y14 were observed with a mass shift of 305 Da, thus allowing unequivocal assignment of the glutathiolation to the cysteine-206 residue of the tryptic peptide 200GAGAYIC206GEETALIESIEGK219. Other sequence informative ions including b3-b6 and y1-y13 provided the evidence to ensure that the sequence of (GSC206)2+ was matched to the sequence of aa residues 200-219.
In the spectrum of the (GSC206)3+ ion (m/z 772.933+), sequence informative ions including b2-b6, and y1-y13 were matched to the sequence of aa residues 200-219 (see Supporting Information Figure 1). However, two structurally informative ions with weak intensity were identified. They were b9 (m/z 1127.93) and b10 (m/z 1256.91), which corresponded to glutathiolated adducts of the peptides 200GAGAYICGE208 and 200GAGAYICGEE209 respectively.
Likewise, the doubly and triply protonated molecular ions of the tryptic peptide GSC187 (aa 185-199) were detected at m/z 977.622+ and m/z 652.433+. As indicated in Figure 5B, the structurally informative fragment ions in the spectrum of (GSC187)2+ revealed an increase of 305 Da at b3 (peptide 185NAC187) and b4 (peptide 185NACG188), suggesting the addition of glutathione to the residue cysteine-187. In the spectrum of the (GSC187)3+ ion of m/z 652.433+ (see Supporting Information Figure 2), one structurally informative ion with weak intensity was identified as b8 (m/z 1073.79), corresponding to the glutathiolated adduct of the peptide 185NACGSGYD192. Both data confirmed C187 to be glutathiolated.
Involvement of C206, C187, and C425 at the 51 kDa subunit of NADH dehydrogenase in site-specific glutathiolation as determined by mass spectrometry
To gain a deeper insight into the S-glutathiolation of the 51 kDa subunit, the flavin protein subcomplex of NQR was isolated by the established method (7). Purified NDH (0.23 mg/ml, ∼2.8 μM) was incubated with GSSG (1 mM) in PBS at room temperature for 1 h. The reaction mixture was then subjected to SDS-PAGE and immunoblotted with the monoclonal antibody against GSH. This resulted in detection of NDH-derived PrSSG specifically at the 51 kDa subunit as indicated in Fig. 1B (lane 1). The detected Western signal was diminished in the presence of the reducing agent β-ME. The intensity of the detected Western signal was enhanced in proportion to the dose of GSSG (data not shown).
To further provide direct evidence for the molecular mechanism of NDH-derived S-glutathiolation induced by GSSG, the NDH-derived PrSSG was obtained from incubation of NDH with GSSG (1 mM) at RT for 1 h, and the reaction mixture was subjected to SDS-PAGE under non-reducing conditions. The protein band at 51 kDa was cut out and subjected to in-gel digestion with trypsin and chymotrypsin respectively as described previously, and followed by LC/MS/MS analysis. The resulting mass spectra acquired from the tryptic and chymotryptic digests contained ions corresponding in mass to tryptic and chymotryptic peptides of NDH and accounted for over 87.61% of the amino acid sequence of NDH (Fig. 6).
Fig. 6. Amino acid sequence of the precursor of NADH dehydrogenase 51 kDa subunit.
The regions labeled with bold represents the amino acid residues identified with LC/MS/MS. The underlined regions are the sequence motif of 4Fe-4S binding (aa 379-425). The residues involved in GS-binding are highlighted with gray. The region labeled with a dotted underline is the signal peptide (aa 1-20), which acts as an import sequence and does not exist in the mature protein.
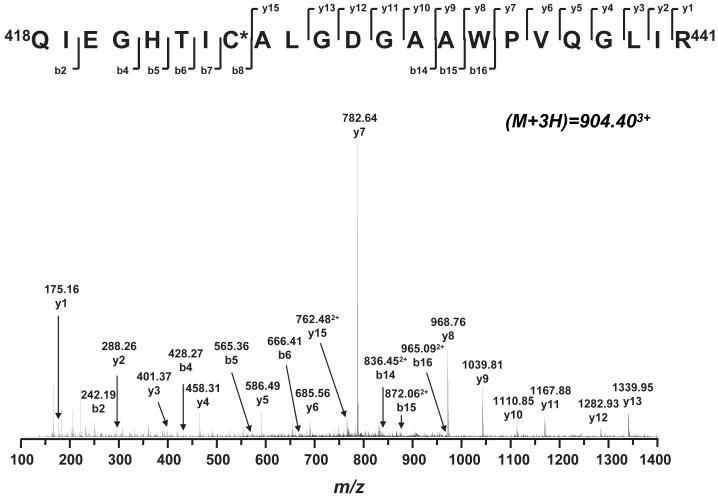
In addition to the tryptic peptides GSC206 and GSC187, the other tryptic peptide, GSC425, 418QIEGHTIC425ALGDGAAWPVQGLIR440 (aa 418-440), also exhibited a mass difference of 305 Da. The triply protonated molecular ion of GSC425 was observed at m/z 904.40, corresponding to a mass shift of 305 Da compared to that of the parent ion [m/z 2405.24 for (M+H)+]. These data suggest that one GSH is covalently bound to one of the residues of the GSC425 peptide in the NDH-51 kDa subunit.
In the MS/MS spectrum of the (GSC425)3+ ion (m/z 904.403+, Fig. 7), the sequence determined for this GS-binding peptide is mostly matched to the expected amino acid sequence 418QIEGHTIC425ALGDGAAWPVQGLIR440. The sequence informative ions, including y1-y15, b2, and b4-b8, were identified in the spectra (Fig. 7 and Table 1). Additionally, the structurally informative ions involving GS-binding C425, such as b8, b14, b15, and b16, were, detected as doubly protonated with weak intensity (Fig. 7 and Table 1).
Fig. 7. Tandem mass spectrum (MS/MS) of the triply protonated molecular ion of the GS-binding peptide (418QIETHTIC425ALGDGAAWPVQGLIR441) of the 51 kDa subunit from the GS-NDH.
The sequence-specific ions are labeled as y and b ions on the spectrum. The amino acid residues involved in GS-binding are identified by asterisks.
Effect of protein S-glutathiolation on the electron transfer activity and superoxide generation activity of NQR
Protein S-glutathiolation has been implicated as a mechanism to regulate the functions of the protein (29-31). Therefore, significant changes in the electron transfer and O2·- generaton activities of NQR are expected due to the site-specific S-glutathiolation involved in the 75 kDa and 51 kDa subunits.
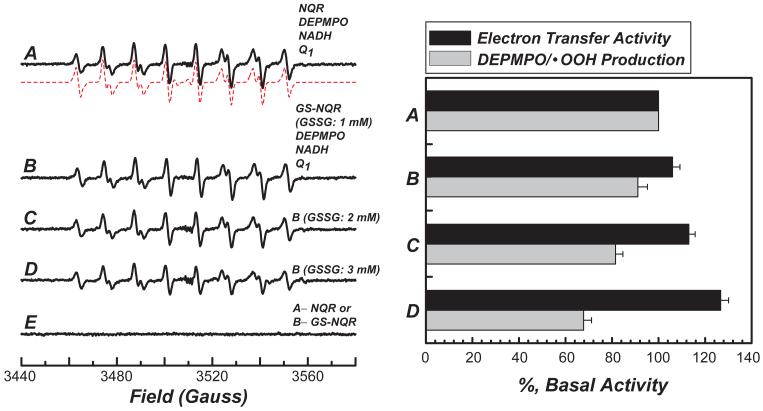
In order to learn the effect of these reactive cysteines on the mediation of NQR-derived electron transfer and O2·- generation activities, we titrated NQR with various amounts of GSSG. NQR (2.8 mg/ml) in PBS was incubated with GSSG (0-3 mM, NQR/GSSG ratio = 0-1,000) at room temperature for 1 h and determined the NQR-mediated O2·- production under enzyme turnover conditions using EPR spin-trapping with DEPMPO. Excess GSSG was removed by dialysis of the protein against 50 mM potassium phosphate buffer, pH 7.4. The S-glutathiolated NQR (GS-NQR) obtained was allowed to generate O2·- under conditions of enzyme turnover (in the presence of NADH and Q1) and the products analyzed by EPR spin trapping and subsequent spin quantitation based on the spectra obtained from computer simulation (Fig. 8A, dashed line). The titration curve indicated that NQR-mediated electron transfer was progressively increased and superoxide production was progressively decreased as the dosage of GSSG increased (supporting information Figure 5). As indicated in Fig. 8A, the NQR-mediated O2·- production detected by EPR revealed a multi-line spectrum of the DEPMPO/·OOH adduct. The detected DEPMPO/·OOH adduct was totally dependent on the presence of NQR or GS-NQR, thus confirming enzyme-mediated O2·- generation (Fig 8E). The signal from this adduct decreased progressively as the dosage of GSSG increased (Figs. 8B-8D). The GS-NQR was further subjected to analysis of the electron transfer activity in catalyzing NADH oxidation by Q1. Enhancement of the NQR-derived electron transfer activity was detected and gradually increased as the dosage of GSSG was raised (Figs. 8B-8D, right panel). This result implies that protein S-glutathiolation enhanced the electron transfer efficiency and subsequently decreased the electron leakage for O2·- generation mediated by NQR.
Fig. 8. Effect of protein S-glutathiolation of NQR on NQR-mediated superoxide generation and its electron transfer activity.
The isolated NQR (2.8 mg/ml) in PBS was incubated with various concentrations of GSSG (0-3 mM) at room temperature for 1 h. The mixture was then subjected to dialysis against PBS for 8 h with one change of buffer at 4 °C. The protein concentration of dialysate containing NQR or GS-NQR was determined by the Lowry method (38). An aliquot of NQR or GS-NQR (127 μg/ml final concentration) was added to a mixture containing NADH (0.5 mM), Q1 (0.2 mM), DEPMPO (20 mM), and DTPA (1 mM) prior to EPR measurement. The DEPMPO/·OOH in each spectrum was quantitated by double integration of the simulation spectrum (dashed line)(7, 23). For measuring the electron transfer activity of NQR or GSN-QR, an aliquot of dialysate was withdrawn and assayed as described in the “Materials and Methods.” A, EPR spectrum obtained from a complete system containing NQR, NADH, Q1 and DEPMPO. B, the same as A except that NQR was replaced with GS-NQR. The GS-NQR was prepared using 1 mM GSSG. C, the same as B except that GS-NQR was prepared using 2 mM GSSG. D, the same as B except that GS-NQR was prepared using 3 mM GSSG. E, the same as A, but NQR was omitted from the system .
Effect of protein S-glutathiolation of the 51 kDa subunit on the electron transfer and superoxide generation activities of NADH Dehydrogenase
Previous results demonstrated that the specific S-glutathiolation of NADH dehydrogenase was involved in the residues of C187, C206, and C425 of the 51 kDa subunit. Therefore, significant changes in the enzymatic activities of NADH dehydrogenase were expected.
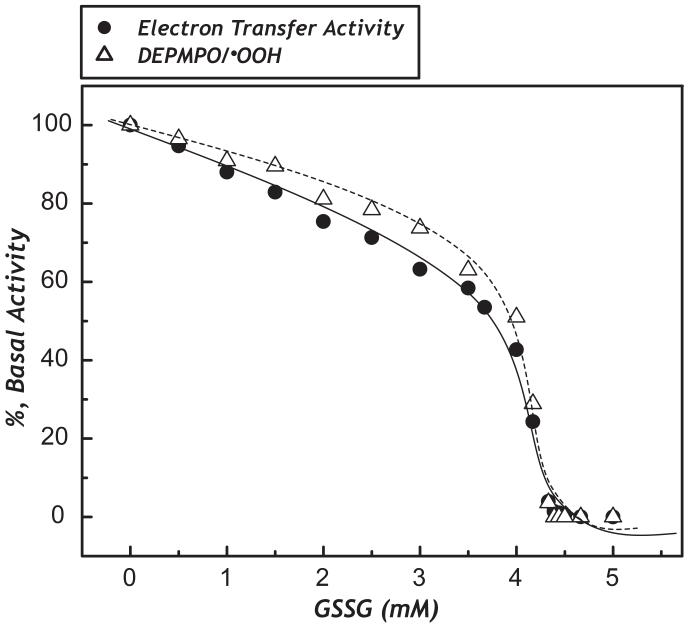
C206 is the reactive/regulatory thiol which has been shown to be involved in protein thiyl radical formation during oxidative attack (7). As C425 is one of the ligands involved in the [4Fe-4S] binding on the 51 kDa subunit (3), the S-glutathiolation of C425 would affect the NADH-derived electron transfer and O2·- generation activities. To address this issue, NDH (0.23 mg/ml) in PBS was incubated with various amounts (0-5 mM) of GSSG at room temperature for 1 h. Excess GSSG was removed by passing the protein through a MicroBioSpin-6 column (BioRad, Hercules, CA). The S-glutathiolated NDH (GS-NDH) was analyzed for NADH-induced O2·- generation in the absence of Q1 by EPR spin trapping with DEPMPO and subsequent spin quantitation (7). Fig. 9 is the titration curve, the enzyme-mediated O2·- production detected as DEPMPO/·OOH progressively declined as the dosage of GSSG increased (dashed line of Fig. 9). Complete inhibition was taken place at the concentration of GSSG of 4.25 mM (dashed line of Fig. 9). The extent of S-glutathiolation of the 51 kDa is correlated with the dosage of GSSG. Likewise, the GS-NDH was further subjected to analysis of the electron transfer activity in catalyzing NADH oxidation by Q1. Inhibition of the NDH-derived electron transfer activity was detected and the inhibitory effect was progressively enhanced as the dosage of GSSG increased (solid line of Fig. 9), implying that the impairment of NDH-derived electron transfer activity caused by S-glutathiolation at C425 contributed to the inhibition of O2·- generation mediated by NDH (7).
Fig. 9. Effect of protein S-glutathiolation of NDH on NDH-mediated electron transfer activity its superoxide generation activity (dashed line).
The isolated NDH (0.23 mg/ml) in PBS was incubated with various concentrations of GSSG (0-5 mM) at room temperature for 1 h. Excess GSSG was removed by passing the sample through a MicroBioSpin-6 column. An appropriate amount of NDH or GS-NDH enzyme solution was withdrawn and subjected to ETA measurement as described in “Materials and Methods.” For measuring the superoxide generation by NDH or GS-NDH, the experimental approach of EPR was the same as that described in the legend of Fig. 8, except that Q1 was omitted from the reaction mixture. The absolute basal activity of NDH-mediated superoxide generation activity is 28.1 nmol O O2·- production min-1 mg-1 using 4-Hydroxy-2,2,6,6-tetramethylpiperidinyloxy (TEMPOL) as a standard for spin quantitation. Each data point represents the average of enzymatic assay from two batches of NDH preparation.
DISCUSSION
In the current investigation, we have identified and characterized protein S-glutathiolation at both the 51 kDa and 75 kDa subunits of NQR with a combination of immunoblotting and mass spectrometry. Furthermore, we have clarified the relevance of this event in the mediation of electron transfer and O2·- generation activities catalyzed by NQR and NDH. The immunoblotting study with anti-GSH monoclonal antibody has shown that NQR-derived PrSSG induced by GSSG is specifically located on the subunits of 51 kDa and 75 kDa (Fig. 1A). This result is basically consistent with those reported in the literature (13, 14). The 51 kDa subunit was specifically involved in the above redox modification when the Fp subcomplex was exposed to GSSG (Fig. 1B). In agreement with our previous study, the 51 kDa subunit of NDH was also involved in the specific protein thiyl radical formation by oxidative attack as probed by immunospin trapping (7).
Protein S-glutathiolation at the 51 kDa subunit of NQR
As probed by LC/MS/MS, C206 and C187 are the specific cysteinyl residues identified to be the GS-binding sites of the 51 kDa subunit in both NQR and its Fp subcomplex. In the previous study, C206 was verified to be a target susceptible to oxidative attack of oxygen free radicals, forming a protein-derived thiyl radical (7). From this result, together with the results of the current study, we conclude that the C206 on the 51 kDa subunit of NQR plays the unique role of the reactive/regulatory thiol of this subunit of mitochondrial Complex I. The bovine protein has 12 cysteine residues, but only 5 of them are conserved. The first conserved cysteine is C206, which is separated from the others by 172 residues. The four remaining conserved cysteine residues are involved in the ligands of the [4Fe-4S] cluster. C206 is conserved among the proteins from E. coli (C180 of NUO F), bovine heart (C206 of 51 kDa), Thermus thermophilus (C182 of NQO1) and Neurospora crassa (C218 of 51 kDa) (1, 3).
Residue C187 is not conserved between mammalian and bacterial proteins. In the previous study, C187 was not found to be involved in the protein radical formation (7). However, C187 is clearly involved in the S-glutathiolation as evidenced by LC/MS/MS (Fig. 5B). Currently, the hydrophilic domain of Complex I from Thermus thermophilius is the only high resolution X-ray structure available (32). Using the ExPASy Sim alignment tool, it was found that the Nqo1 subunit of T. thermophilius is 46.4% identical to the bovine 51 kDa subunit. In the region containing residues of 180-210 of the bovine subunit, there is 70% identity of the subunits from the two species. Thus, it is highly likely that the overall folding and structure of the enzymes in this region are similar. Examination of the T. thermophilius X-ray structure suggests that C187 and C206 are probably on the surface of protein. It should be noted that the T. thermophilius Nqo1 does not contain a cysteine residue at position 187 (equivalent to Phenylalanine 163 of T. thermophilius), but rather has a phenylalanine residue at this position. The region including C187 could be located based on the T. thermophilius structure appears to be on the surface of protein, suggesting that C187 of the bovine enzyme may be surface exposed and thus susceptible to attack by GSSG. Based on the X-ray crystal structure of T. thermophilius, C206 of 51 kDa subunit from bovine protein is very near the FMN binding site (∼ 6 Å). This may explain the reactivity of this specific thiol to GSSG.
Extra evidence of surface-exposed nature of the residues C206 and C187 at the 51 kDa from bovine NQR was provided by the studies of soluble thiol modification agent, iodoacetamide (ICH2CONH2). Incubation of NQR with iodoacetamide (1 mM) at room temperature for 1-h resulted in carbamoylmethylation of NQR. LC/MS/MS analysis of trypsinolytic digests of 51 kDa indicated two doublet and two triplet ions involved in the carbamoylmethylated peptides containing C206 and C187. They are (M+2H)2+ = 1034.84 and (M+3H)3+ = 690.48 for the peptide 200GAGAYICGEETALIESIEGK219 and (M+2H)2+ = 853.80 and (M+3H)3+ = 569.60 for 185NACGSGYDFDVFVVR199. Likewise, LC/MS/MS analysis of trypsinolytic digest of the 75 kDa from the carbarmoylmethylated NQR revealed one triplet ion (M+3H)3+ = 785.51 involved in the carbamoylmethylation at the C367 of the peptide 361VDSDTLCTEEVFPTAGAGTDLR382, thus confirming the surface-exposed nature of the residue C367 at the 75 kDa subunit of bovine NQR .
C206 is near FMN-binding site where the major catalysis of electron transfer and O2·- production occur. As demonstrated in the previous study, the electron transfer coupled with O2·- generation as induced by NADH was tightly controlled by the FMN cofactor and the FMN-binding site at the 51 kDa subunit (7). Therefore, specific S-glutathiolation at C206 seems likely to induce a small conformational change near the FMN/NADH binding site which might marginally increase the efficiency of electron transfer from the FMN to the center of 4Fe-4S clusters (N3 center) and subsequently reduce the electron leakage under enzyme turnover conditions (Fig. 8).
Physiologically, it is likely that S-glutathiolation of C206 and C187 is an early consequence of mitochondrial oxidative stress. The purpose of this event may be to help maintain the ratio of GSH/GSSG during oxidative stress by scavenging the GSSG formed (Equation 1) (10). The S-glutathiolation of protein can be reversed by the enzymes glutaredoxin and thioredoxin to restore the GSH pool (10, 12, 17). Alternatively, this event may play an antioxidant role and make an adaptive response to combat oxidative injury (29).
In the enzymatic system of the glutathiolated Fp subcomplex, GS-NDH, LC/MS/MS revealed an additional cysteinyl residue, C425, to be S-glutathiolated apart from C206 and C187. C425 is involved in the 4Fe-4S binding motif (3), CXXCXXCX39C (C, cysteine residue; X, any aa residue) corresponding to aa 379-425 at the 51 kDa subunit (Fig. 6). C425 is one of the ligands for the 4Fe-4S cluster (N3 center), which was also verified by X-ray crystal structure (32). In the previous study, we demonstrated that destruction of iron-sulfur centers of NDH by p-chloromercuribenzoate inhibited the electron transfer activity and enzyme-mediated O2·- generation (7). Likewise, S-glutathiolation of C425 resulted in a decrease in NDH-derived electron transfer and superoxide generation activities (Fig. 9).
C425 glutathiolation was not detected in GS-NQR, presumably because this structural thiol involved in the 4Fe-4S binding is buried inside the NQR and structurally protected by the 75 kDa subunit (see discussion below), a location that GSSG cannot access. Most of iron-sulfur clusters are usually protected by not exposed to the protein surface. When the Fp subcomplex was isolated from NQR, it becomes artificially exposed in the isolated NDH and was susceptible to redox modification.
Protein S -glutathiolation in the 75 kDa subunit of NQR
The bovine 75 kDa polypeptide is encoded by nuclear DNA. The DNA sequence encodes an N-terminal signal peptide containing 23 amino acid residues (aa 1-23 of Fig. 2). This N-terminal extension acts as a mitochondrial import sequence, which has been removed in the mature protein (33). The mature N-terminal sequence is TATAASNLIE, which was verified by LC/MS/MS in this study (Fig. 2).
The 75 kDa subunit contains 17 cysteines, of which 11 are conserved throughout the proteins from N. crassa, T. thermophilus, R. capsulatus, and E. coli. These cysteines, found in the N-terminal domain of the protein, have been assigned to the ligands of Fe-S clusters including N4, N1b, and N5 respectively (Figure 2) (1).
C367 represents the only reactive/regulatory thiol in the 75 kDa subunit to be involved in GS-binding probed by LC/MS/MS (Figs. 2 and 3). This specific cysteine residue C367 is not observed or conserved in T. thermophilius (Nqo3 subunit) and fungal enzymes (NuoG subunit), but rather has an Ala at this position (A351 in T. thermophilius enzyme). In the region of identified GS-binding (residues 361-382, Fig. 3) of bovine protein, there is only 27.2% identity from two species. However, C367 and the corresponding GS-binding domain (Fig. 3) are highly conserved (>90%) in mammalian enzymes. C367 of bovine protein should be surface exposed based on the A367 location in the X-ray structure of T. thermophilius enzyme. Therefore, C367-derived S-glutathiolation induced by GSSG was via thiol-disulfide exchange (Equation 1).
Physiologically, this event may play a role in buffering the GSH pool during oxidative stress since the GSH is regenerated from GSSG [Equation 1 and reference (10)]. Furthermore, it is likely that S-glutathiolation of C367 may also play a role in protecting the 4Fe-4S cluster of the 51 kDa subunit from oxidative damage via C425 S-glutathiolation when NQR is exposed to a redox change in the GSH pool. Walker et. al. has suggested that the 75 kDa subunit is associated structurally with the 51- and 24 kDa subunits of the Fp subcomplex based on the structural homology related to the α (51 and 24 kDa) and γ (75 kDa) subunits of the NAD-reducing hydrogenases and that the NuoE, F and G subunits of E. coli Complex I can be co-expressed to form a catalytically active recombinant subcomplex (1, 34, 35). The information of X-ray structure from T. thermophilius Complex I supports this prediction (32).
Superoxide Generation by NQR in the presence of GSSG
(a) Analyzed by Cytochrome c Reduction Assay
Taylor et al. have reported that reversible glutathiolation of Complex I at a very high ratio of GSSG/NQR (20 mM of GSSG used) increases O2·- formation as measured by the cytochrome c reduction assay (14). This result seems contradictory our results obtained from EPR measurement at low concentration of GSSG (1-3 mM). It is necessary to clarify and discuss the reasons why different assays lead to different results. We have re-examined the assay of NQR-derived O2·- production by the cytochrome c reduction assay (Supporting Information Figure 3). Purified NQR (10 μg/ml and myxothiazol pretreatment) in 50 mM phosphate buffer, pH 7.5 containing 1 mM EGTA and 0.1 mM DTPA (PED buffer) was incubated with native cytochrome c (50 μM). The O2·- production was initiated with NADH (0.2 mM) and inhibited by pre-addition of Zn,Cu-SOD (300 U), which indicates that 12.3±4.9 % (n=5) of cytochrome c reduction was sensitive to Zn,Cu-SOD. Therefore, 80 -90% cytochrome c reduction was derived from the NADH cytochrome c reductase (NCR) activity of Complex I. To test the effect of S-glutathiolation, GSSG (3 mM) was pre-incubated with the assay mixture containing NQR for 10 min prior to NADH initiation. It was observed that pre-incubation of NQR with GSSG significantly enhanced [22.1 ± 5.3%, n=5] the activity of NADH cytochrome c reducatse, but only 10.4±5.5% of cytochrome c reduction is sensitive to pre-addition of Zn, Cu-SOD. However, it is not easy to draw a conclusion of how S-glutathiolation affects the NQR-mediated O2·- generation by this assay since the electron transfer activity from NADH to cytochrome c is so dominant in this assay and controls most of the cytochrome c reduction.
If the cytochrome c was replaced with acetylated cytochrome c in the assay mixture, it was observed that 65-75% of acetylated cytochrome c reduction was sensitive to Zn, Cu-SOD. In the presence of GSSG (1-3 mM), the acetylated cytochrome c reduction caused by O2·- generation is marginally decreased (5-25%). Basically, the results obtained by this assay were in line with those from EPR assay under the conditions of GSSG dosage of 1-3 mM.
(b) Analyzed by EPR Spin Trapping with DEPMPO
DEPMPO assay provides direct measurement of O2·-, and this measurement is not affected by the activity of NADH cytochrome c reductase of Complex I. Furthermore, this assay does not require pre-incubation of Zn,Cu-SOD. The advantages include: (i) the DEPMPO assay is 40-fold more sensitive than the cytochrome c reduction assay for the detection of O2·- (36) (ii) DEPMPO traps O2·- with an efficiency of 60-70% (36) (iii) The detected spectrum of DEPMPO/·OOH can be well simulated for spin quantitation.
The O2·- generation mediated by NQR under the same conditions was further analyzed by EPR spin trapping. It was observed that pre-incubation of GSSG (1-3 mM) with NQR (10 μg/ml and myxothiazol pretreatment) marginally decreased (from 11.52±3.05% to 28.15±4.05% inhibition, n=5, Supporting Information Figure 4) the generated DEPMPO/·OOH adduct when the concentration of GSSG was gradually increased (1-3 mM). Pre-incubation of 20 mM GSSG with the same amount of NQR completely abolished the O2·- production detected as DEPMPO/·OOH.
In summary, EPR spin-trapping with DEPMPO provided direct and reliable measurements of O2·- generation mediated by NQR. Whereas, the assay of O2·- generation with cytochrome c reduction is an indirect measurement; and is greatly affected by the electron transfer activity of NADH cytochrome c reductase.
It is important to note that 20 mM of GSSG in the reaction mixture is not likely to occur under physiological or patho-physiological conditions. The physiologically or patho-physiological relevant concentration of GSSG should be in the range of 0.1-3 mM. Normally, the GSH/GSSG ratio in mitochondria is kept high; typically 95-99% is reduced (10, 12), which results in GSSG concentrations of 0.1-0.5 mM assuming that mitochondria contain 10 mM GSH. The ratio of GSH/GSSG was reportedly increased to 2.06±0.93 during oxidative stress such as in ischemia-reperfusion injury (37), which showed GSSG concentrations of 2-3 mM. Site-specific S-glutathiolation can only occur under physiologically relevant concentrations of GSSG.
In current studies, we did not detect S-glutathiolation involved in the ubiquinone-binding region which is another important site in controlling superoxide generation by NQR. Nevertheless, it should not eliminate the possibility of S-glutahiolation of ubiquinone-binding region in vivo. This topic is left for the future study. Furthermore, development of amphiphilic spin trap which can target to mitochondria may facilitate this study. This work is under the progress in our laboratory.
Physiological Implications and Conclusions
The present studies provide the molecular mechanism of NQR-derived S-glutathiolation and the way this event modulates its electron transfer and O2·- generation activities. The mechanism addressed here provides a useful concept for understanding the fundamental question of how mitochondrial Complex I utilizes its redox thiols to address situations of oxidative stress and to regulate its enzymatic functions. Clearly, the major role of this event is to regulate the GSH pool of mitochondria, which is perhaps involved in the regulation of the redox signal caused by oxidative stress. The secondary role of this event is perhaps to combat oxidative injury by increasing electron transfer efficiency and decreasing electron leakage to molecular oxygen. In agreement to the in vitro results of this study, our recent research progress using the model of in vivo post-ischemic rat heart has indicated that 51 kDa subunit-derived S-glutathiolation was enhanced during ischemia-reperfusion injury2. Recognition of the molecular mechanism addressed in this work is important in understanding the fundamental basis by which oxidants modulate apoptosis, cell senescence, or proliferation through communicating the internal redox state of mitochondria with redox-sensitive signaling pathways in the cytoplasm.
ACKNOWLEDGMENT
The authors thank Ms. Nan Kleinholz and Ms. Rhonda Pitsch (CCIC, The Ohio State University) for assistance in mass spectrometry and Drs Chang-An Yu and Linda Yu (Oklahoma State University, Stillwater, OK) for valuable advice in preparation of NDH.
This work was supported by RO1 grants from National Institutes of Health [(HL083237, to Y-R C) and (HL63744, to JLZ)].
Abbreviations
- NQR
NADH ubiquinone reductase, or mitochondrial Complex I
- GS-NQR
glutathiolated NQR
- NDH
NADH dehydrogenase or flavin protein subcomplex of Complex I
- GS-NDH
glutathiolated NDH
- O2·-
superoxide anion radical
- ETC
electron transport chain
- SMP
submitochondrial particles
- DEPMPO
5-diethoxylphosphoryl-5-methyl-1-pyrroline N-oxide
- FMN
flavin mononucleotide
- GSH
glutathione
- GSSG
oxidized glutathione
- SOD
superoxide dismutase
- SDS-PAGE
SDS polyacrylamide gel electrophoresis
- EPR
electron paramagnetic resonance
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- PBS
phosphate buffered saline
- β-ME
β-mercaptoethanol
Footnotes
C.L. Chen, J.L. Zweier, and Y.R. Chen, unpublished results.
REFERENCES
- (1).Hirst J, Carroll J, Fearnley IM, Shannon RJ, Walker JE. The nuclear encoded subunits of complex I from bovine heart mitochondria. Biochim Biophys Acta. 2003;1604:135–50. doi: 10.1016/s0005-2728(03)00059-8. [DOI] [PubMed] [Google Scholar]
- (2).Carroll J, Fearnley IM, Shannon RJ, Hirst J, Walker JE. Analysis of the subunit composition of complex I from bovine heart mitochondria. Mol Cell Proteomics. 2003;2:117–26. doi: 10.1074/mcp.M300014-MCP200. [DOI] [PubMed] [Google Scholar]
- (3).Walker JE. The NADH:ubiquinone oxidoreductase (complex I) of respiratory chains. Q Rev Biophys. 1992;25:253–324. doi: 10.1017/s003358350000425x. [DOI] [PubMed] [Google Scholar]
- (4).Galkin A, Brandt U. Superoxide radical formation by pure complex I (NADH:ubiquinone oxidoreductase) from Yarrowia lipolytica. J Biol Chem. 2005;280:30129–35. doi: 10.1074/jbc.M504709200. [DOI] [PubMed] [Google Scholar]
- (5).Kudin AP, Bimpong-Buta NY, Vielhaber S, Elger CE, Kunz WS. Characterization of superoxide-producing sites in isolated brain mitochondria. J Biol Chem. 2004;279:4127–35. doi: 10.1074/jbc.M310341200. [DOI] [PubMed] [Google Scholar]
- (6).Turrens JF, Boveris A. Generation of superoxide anion by the NADH dehydrogenase of bovine heart mitochondria. Biochem J. 1980;191:421–7. doi: 10.1042/bj1910421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Chen YR, Chen CL, Zhang L, Green-Church KB, Zweier JL. Superoxide generation from mitochondrial NADH dehydrogenase induces self-inactivation with specific protein radical formation. J Biol Chem. 2005;280:37339–48. doi: 10.1074/jbc.M503936200. [DOI] [PubMed] [Google Scholar]
- (8).Lambert AJ, Brand MD. Inhibitors of the quinone-binding site allow rapid superoxide production from mitochondrial NADH:ubiquinone oxidoreductase (complex I) J Biol Chem. 2004;279:39414–20. doi: 10.1074/jbc.M406576200. [DOI] [PubMed] [Google Scholar]
- (9).Ohnishi ST, Ohnishi T, Muranaka S, Fujita H, Kimura H, Uemura K, Yoshida K, Utsumi K. A possible site of superoxide generation in the complex I segment of rat heart mitochondria. J Bioenerg Biomembr. 2005;37:1–15. doi: 10.1007/s10863-005-4117-y. [DOI] [PubMed] [Google Scholar]
- (10).Costa NJ, Dahm CC, Hurrell F, Taylor ER, Murphy MP. Interactions of mitochondrial thiols with nitric oxide. Antioxid Redox Signal. 2003;5:291–305. doi: 10.1089/152308603322110878. [DOI] [PubMed] [Google Scholar]
- (11).Hurd TR, Filipovska A, Costa NJ, Dahm CC, Murphy MP, Hurd TR, Costa NJ, Dahm CC, Beer SM, Brown SE, Filipovska A, Murphy MP. Disulphide formation on mitochondrial protein thiols Glutathionylation of mitochondrial proteins. Biochem Soc Trans. 2005;33:1390–3. doi: 10.1042/BST0331390. [DOI] [PubMed] [Google Scholar]
- (12).Hurd TR, Costa NJ, Dahm CC, Beer SM, Brown SE, Filipovska A, Murphy MP. Glutathionylation of mitochondrial proteins. Antioxid Redox Signal. 2005;7:999–1010. doi: 10.1089/ars.2005.7.999. [DOI] [PubMed] [Google Scholar]
- (13).Beer SM, Taylor ER, Brown SE, Dahm CC, Costa NJ, Runswick MJ, Murphy MP. Glutaredoxin 2 catalyzes the reversible oxidation and glutathionylation of mitochondrial membrane thiol proteins: implications for mitochondrial redox regulation and antioxidant DEFENSE. J Biol Chem. 2004;279:47939–51. doi: 10.1074/jbc.M408011200. [DOI] [PubMed] [Google Scholar]
- (14).Taylor ER, Hurrell F, Shannon RJ, Lin TK, Hirst J, Murphy MP. Reversible glutathionylation of complex I increases mitochondrial superoxide formation. J Biol Chem. 2003;278:19603–10. doi: 10.1074/jbc.M209359200. [DOI] [PubMed] [Google Scholar]
- (15).Beltran B, Orsi A, Clementi E, Moncada S. Oxidative stress and S-nitrosylation of proteins in cells. Br J Pharmacol. 2000;129:953–60. doi: 10.1038/sj.bjp.0703147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Clementi E, Brown GC, Feelisch M, Moncada S. Persistent inhibition of cell respiration by nitric oxide: crucial role of S-nitrosylation of mitochondrial complex I and protective action of glutathione. Proc Natl Acad Sci U S A. 1998;95:7631–6. doi: 10.1073/pnas.95.13.7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Galante YM, Hatefi Y. Purification and molecular and enzymic properties of mitochondrial NADH dehydrogenase. Arch Biochem Biophys. 1979;192:559–68. doi: 10.1016/0003-9861(79)90126-7. [DOI] [PubMed] [Google Scholar]
- (18).Vinogradov AD, King TE. The Keilin-Hartree heart muscle preparation. Methods Enzymol. 1979;55:118–27. doi: 10.1016/0076-6879(79)55017-4. [DOI] [PubMed] [Google Scholar]
- (19).Redfearn ER, Whittaker PA. The determination of the oxidation-reduction states of ubiquinone (coenzyme Q) in rat-liver mitochondria. Biochim Biophys Acta. 1966;118:413–8. doi: 10.1016/s0926-6593(66)80050-4. [DOI] [PubMed] [Google Scholar]
- (20).Hatefi Y. Preparation and properties of NADH: ubiquinone oxidoreductase (complexI), EC 1.6.5.3. Methods Enzymol. 1978;53:11–4. doi: 10.1016/s0076-6879(78)53006-1. [DOI] [PubMed] [Google Scholar]
- (21).Duling DR. Simulation of multiple isotropic spin-trap EPR spectra. J Magn Reson B. 1994;104:105–10. doi: 10.1006/jmrb.1994.1062. [DOI] [PubMed] [Google Scholar]
- (22).Frejaville C, Karoui H, Tuccio B, Le Moigne F, Culcasi M, Pietri S, Lauricella R, Tordo P. 5-(Diethoxyphosphoryl)-5-methyl-1-pyrroline N-oxide: a new efficient phosphorylated nitrone for the in vitro and in vivo spin trapping of oxygen-centered radicals. J Med Chem. 1995;38:258–65. doi: 10.1021/jm00002a007. [DOI] [PubMed] [Google Scholar]
- (23).Vasquez-Vivar J, Kalyanaraman B, Martasek P, Hogg N, Masters BS, Karoui H, Tordo P, Pritchard KA., Jr. Superoxide generation by endothelial nitric oxide synthase: the influence of cofactors. Proc Natl Acad Sci U S A. 1998;95:9220–5. doi: 10.1073/pnas.95.16.9220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Jocelyn PC. The standard redox potential of cysteine-cystine from the thiol-disulphide exchange reaction with glutathione and lipoic acid. Eur J Biochem. 1967;2:327–31. doi: 10.1111/j.1432-1033.1967.tb00142.x. [DOI] [PubMed] [Google Scholar]
- (25).Scheraga HA, Konishi Y, Rothwarf DM, Mui PW. Toward an understanding of the folding of ribonuclease A. Proc Natl Acad Sci U S A. 1987;84:5740–4. doi: 10.1073/pnas.84.16.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Rothwarf DM, Scheraga HA. Equilibrium and kinetic constants for the thiol-disulfide interchange reaction between glutathione and dithiothreitol. Proc Natl Acad Sci U S A. 1992;89:7944–8. doi: 10.1073/pnas.89.17.7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Roepstorff P, Fohlman J. Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed Mass Spectrom. 1984;11:601. doi: 10.1002/bms.1200111109. [DOI] [PubMed] [Google Scholar]
- (28).Biemann K. Contributions of mass spectrometry to peptide and protein structure. Biomed Environ Mass Spectrom. 1988;16:99–111. doi: 10.1002/bms.1200160119. [DOI] [PubMed] [Google Scholar]
- (29).Giustarini D, Rossi R, Milzani A, Colombo R, Dalle-Donne I. S-glutathionylation: from redox regulation of protein functions to human diseases. J Cell Mol Med. 2004;8:201–12. doi: 10.1111/j.1582-4934.2004.tb00275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Cotgreave IA, Gerdes RG. Recent trends in glutathione biochemistry--glutathione-protein interactions: a molecular link between oxidative stress and cell proliferation? Biochem Biophys Res Commun. 1998;242:1–9. doi: 10.1006/bbrc.1997.7812. [DOI] [PubMed] [Google Scholar]
- (31).Thomas JA, Poland B, Honzatko R. Protein sulfhydryls and their role in the antioxidant function of protein S-thiolation. Arch Biochem Biophys. 1995;319:1–9. doi: 10.1006/abbi.1995.1261. [DOI] [PubMed] [Google Scholar]
- (32).Sazanov LA, Hinchliffe P. Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science. 2006;311:1430–6. doi: 10.1126/science.1123809. [DOI] [PubMed] [Google Scholar]
- (33).Runswick MJ, Gennis RB, Fearnley IM, Walker JE. Mitochondrial NADH:ubiquinone reductase: complementary DNA sequence of the import precursor of the bovine 75-kDa subunit. Biochemistry. 1989;28:9452–9. doi: 10.1021/bi00450a031. [DOI] [PubMed] [Google Scholar]
- (34).Braun M, Bungert S, Friedrich T. Characterization of the overproduced NADH dehydrogenase fragment of the NADH:ubiquinone oxidoreductase (complex I) from Escherichia coli. Biochemistry. 1998;37:1861–7. doi: 10.1021/bi971176p. [DOI] [PubMed] [Google Scholar]
- (35).Pilkington SJ, Skehel JM, Gennis RB, Walker JE. Relationship between mitochondrial NADH-ubiquinone reductase and a bacterial NAD-reducing hydrogenase. Biochemistry. 1991;30:2166–75. doi: 10.1021/bi00222a021. [DOI] [PubMed] [Google Scholar]
- (36).Roubaud V, Sankarapandi S, Kuppusamy P, Tordo P, Zweier JL. Quantitative measurement of superoxide generation using the spin trap 5-(diethoxyphosphoryl)-5-methyl-1-pyrroline-N-oxide. Anal Biochem. 1997;247:404–11. doi: 10.1006/abio.1997.2067. [DOI] [PubMed] [Google Scholar]
- (37).Ucar G, Topaloglu E, Kandilci HB, Gumusel B. Effect of ischemic preconditioning on reactive oxygen species-mediated ischemia--reperfusion injury in the isolated perfused rat lung. Clin Biochem. 2005;38:681–4. doi: 10.1016/j.clinbiochem.2005.04.002. [DOI] [PubMed] [Google Scholar]
- (38).Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–75. [PubMed] [Google Scholar]