Abstract
This special issue of AGE showcases powerful alternative or unconventional approaches to basic aging research, including the use of exceptionally long-lived animal model species and comparative methods from evolutionary biology. In this opening paper, we introduce several of these alternative aging research themes, including the comparative phylogenetic approach. This approach applies modern inferential methods for dissecting basic physiological and biochemical mechanisms correlated with phenotypic traits including longevity, slow aging, sustained somatic maintenance, and repair of molecular damage. Comparative methods can be used to assess the general relevance of specific aging mechanisms—including oxidative processes—to diverse animal species, as well as to assess their potential clinical relevance to humans and other mammals. We also introduce several other novel, underexploited approaches with particular relevance to biogerontology, including the use of model animal species or strains that retain natural genetic heterogeneity, studies of effects of infectious disease and parasites on aging and responses to caloric restriction, studies of reproductive aging, and naturally occurring sex differences in aging. We emphasize the importance of drawing inferences from aging phenomena in laboratory studies that can be applied to clinically relevant aging syndromes in long-lived, outbred animals, including humans.
Keywords: Aging, Animal models, Biogerontology, Comparative method, Evolution, Free radical theory, Longevity, Oxidative damage hypothesis, Phylogenetic comparative method
Introduction
This issue of AGE showcases a group of papers representing comparative or otherwise somewhat unconventional or alternative approaches and animal models for investigating basic aging processes. The inspiration for this special issue arose during the June 2007 meetings of the American Aging Association in San Antonio, Texas. Along with presentations by most of the contributors featured here, these meetings featured a debate on the evolution of aging between two respected authorities on very different aspects of aging: Daniel Promislow (comparative and evolutionary biology of aging) and Valter Longo (molecular biology of aging in the yeast Saccharomyces cerevisiae). Issues raised in this debate, as well as during a number of the other talks at this meeting, reflect the variety of ways in which biogerontologists representing different primary biological specialties frame hypotheses and develop research and experimental strategies for understanding organismal senescence.
Some of the key questions raised by this debate can be expressed as follows:
▪ How does an evolutionary or comparative approach contribute to our understanding of basic aging processes?
▪ How can we frame and test rigorous evolutionary hypotheses about organismal aging?
▪ Is aging “programmed” by natural selection?
▪ How can research on extremely long- or short-lived animal species contribute to our understanding of basic aging processes?
▪ Does aging occur differently in free-living, outbred organisms vs inbred laboratory animals maintained under controlled, disease-free conditions?
Historically, the biology of aging has benefited from a broad, interdisciplinary perspective. At its best, the aging research community encourages communication among scientists from a variety of intellectual backgrounds, and integrates state-of-the art approaches to understanding “ultimate” (evolutionary via comparative, and population-level studies), as well as “proximate” (molecular, biochemical, cellular or physiological) mechanisms responsible for organismal senescence, aging-related dysfunction and disease. Biogerontology currently represents a dynamic synthesis of diverse intellectual perspectives on aging patterns and mechanisms; these include evolutionary theory, population biology and demography, comparative biology and bioenergetics, as well as developmental biology, molecular genetics and clinical biomedical sciences (see, for example, Finch 1990, 2007; Rose 1991; Kenyon et al. 1993; Kirkwood and Austad 2000; Tatar et al. 2003; Partridge et al. 2005; Speakman 2005a; Austad and Masoro 2006).
At the same time, as a rule, biogerontologists currently employ a few indispensable, short-lived laboratory models: baker’s yeast (Saccharomyces cerevisiae), free-living nematodes (Caenorhabditis elegans), fruit flies (Drosophila melanogaster), and inbred laboratory mice (Mus musculus). For in vitro cellular studies, fibroblasts are the most commonly studied animal cell type, with most fibroblast studies using cells derived only from skin rather than a range of organs, and originating from humans or mice. There are now well developed molecular toolkits and transgenic technologies for these traditional models (Austad and Masoro 2006; Conn 2006). These standard model systems are characterized by conveniently short lifespans and highly genetically inbred (“isogenic”) lines of organisms maintained in immaculate, virtually disease- and parasite-free facilities. In the interest of rigorous experimental controls, animals in aging studies typically are maintained in a non-reproductive state, and are physically inactive, socially isolated, and exposed to as few natural stressors as possible (except, in the case of calorie restriction [CR] studies, energy stress).
In this special issue, we emphasize some powerful alternative aging research themes. The most prominent of these, the comparative approach, consists of a collection of inferential and statistical methods that now constitute mainstream practice for zoologists and evolutionary biologists, but remain much less familiar to many biogerontologists, despite a long-standing interest in comparing aging of diverse animal taxa (Harvey and Pagel 1991; Pagel 1992; Garland and Adolph 1994; Garland et al. 2005) (see also Furness and Speakman; Hulbert; Buffenstein et al.; Gavrilova et al., this issue). In the broadest sense, this approach involves systematic comparison of aging-related phenomena among different strains, populations, species or other biologically meaningful groups. It is indispensable not only for testing hypotheses about the evolutionary processes giving rise to particular aging, developmental, and life history patterns, but is also very useful for identifying pertinent aging biomarkers, as well as putative cellular or molecular mechanisms for aging or its prevention. Just as experimental manipulations (e.g., CR) can provide a reliable way of investigating and intervening in the aging process, extremely short- or long-lived populations, mutants, or species can provide an invaluable window on the range of adaptations organisms have for promoting longevity, adaptive somatic maintenance and repair of molecular damage.
We also feature contributions from researchers examining aging processes in unconventional aging models—animal species or strains that are wild, exotic, outbred, or exceptionally long-lived for their taxa, body sizes or metabolic rates. These include wild mice (Ungvari et al.), naked mole-rats (Buffenstein et al.), monkeys (Sitzmann et al.), snakes (Bronikowksi), birds (Furness and Speakman) and bees (Remolina and Hughes). We also include several contributions from researchers that use non-traditional approaches—outbred laboratory mouse strains, along with wild-derived Mus musculus (Harper) and Peromyscus (Ungvari et al.), and studies of infectious disease CR (Kristan). As in the biology of aging as a whole, a common thread running through these contributions is a discussion of the free radical theory and oxidative damage hypotheses of aging (Harman 1956, 1972; Beckman and Ames 1998; Muller et al. 2007), which posit that aging processes are linked to ways in which animals produce, prevent, or repair oxidative damage to tissues, cells and molecules. Some researchers are now questioning whether this theory can adequately explain natural variation in organismal aging (see for example Buffenstein and Hulbert, this issue).
Much of what contributors to this issue have to say about aging-related phenomena in free-living, reproducing wild animals is clinically relevant to humans, and serves to emphasize how a comparative and evolutionary perspective can have important medical implications (Finch 2007; Stearns and Koella 2008). In this opening paper, we provide additional background and intellectual perspective on comparative and other alternative approaches and animal models for aging studies. In general, our aim in this special issue is to promote a wide range of “phenotypic variation” in the scientists who study aging, as well as in the research models they employ.
Comparative methods and the biology of aging
Historical and general perspective
A comparative theme has been woven into the basic biology of aging since the late 1800s, when August Weismann employed a comparative perspective in his thinking about the diversity of animal lifespans and aging patterns (Weismann 1989). Medawar, Comfort and Sacher were the most visible proponents of the comparative perspective the biology of aging from the 1950s through the 1970s (Medawar 1952; Comfort 1964; Sacher 1978; reviewed in Rose 1991). In addition to discussing plausible aging mechanisms, both proximate and ultimate, these three great biological polymaths also developed conventions for the analysis of survivorship and mortality statistics and compared developmental and longevity data from a wide range of animal species, from mollusks to deer mice, and guppies to Mexican cave fish. Comparative analyses of the allometric relationships between lifespans and the sizes of organisms’ bodies, organs and various other structures during this period complemented those of Calder and others investigating relationships between bioenergetics (i.e., metabolism, locomotion) and lifespan (Schmidt-Nielsen 1970; Lindstedt and Calder 1976; Calder 1984a; reviewed in Finch 1990). More or less concurrently, evolutionary biologists—most notably George Williams—used examples from natural history and comparative zoology (e.g., the extremely long lifespans of many birds and bats) in developing the theoretical framework still widely accepted by evolutionary biologists for explaining natural variation in animal lifespans and aging patterns, incorporating mathematical models and theoretical tools from evolutionary biology, demography and population genetics (Williams 1957; Hamilton 1966; Charlesworth 1980; Rose 1991; Kirkwood and Austad 2000; Reznick et al. 2004).
Since the 1950s, biologists of various stripes have amassed a taxonomically diverse body of data on animal lifespans, metabolism and aging rates (Calder 1984b; Finch 1990; Schmidt-Nielsen 1997; Speakman 2005a, b). In gerontology, comparative analyses have been included in the wide variety of approaches used to address mechanistic ideas about aging, including the “rate of living” theory (Rubner 1908; Pearl 1928) and its more recent incarnations, the free radical and oxidative stress theories (Harman 1956, 1972; Beckman and Ames 1998; Muller et al. 2007). Moreover, comparative data have been invaluable for testing hypotheses about evolutionary life history trade-offs relevant to aging processes (Stearns 1992; Garland et al. 2005), as well as for identifying animal populations or species that are exceptionally long- or short-lived for their sizes, metabolic rates, or taxonomic groups, and that hence represent potentially useful models for aging studies (see, for example, Austad and Fischer 1991; Austad 1993, 2001; Holmes and Austad 1995; Miller et al. 2002; Brunet-Rossinni and Austad 2004; Buffenstein 2005).
In general, however, gerontologists remain relatively unschooled in the potential inferential pitfalls associated with comparative studies of aging (Pagel 1992; Garland and Adolph 1994; Austad and Holmes 1999; Garland et al. 2005; Ives et al. 2007). Over the past 20 years, standards for the application of comparative methods in evolutionary biology have undergone aggressive revision, becoming much more analytically sophisticated (for a recent review, see Garland et al. 2005). Among evolutionary biologists, the term “comparative analysis” now implies the application of rigorous and specialized inferential techniques designed to incorporate information about phylogeny (i.e., evolutionary relationships) into statistical tests of hypotheses about physiological or other functional differences between groups of organisms. These techniques include, but are not limited to, the use of “phylogenetic contrasts” (also sometimes referred to as “phylogenetically independent contrasts”) (Felsenstein 1985; Harvey and Pagel 1991); they can be referred to collectively as the “comparative phylogenetic method (Sanford et al. 2002; Garland et al. 2005). These analytical approaches are not for the statistically faint of heart; considerable expertise is required to employ the analytical approaches that now constitute “best practice” in evolutionary physiology, even as these methods continue to undergo discussion and refinement (for an introduction to these issues, see Garland et al. 2005; Ives et al. 2007).
Why bother with phylogeny in aging research?
Evolutionary biology provides both a theoretical and an experimental framework for explaining the existence of aging, as well as natural variation in aging patterns. In the simplest of evolutionary terms, organismal senescence can be viewed as the result of declining natural selection with advancing age after puberty (Charlesworth 1980; Rose 1991; Partridge and Barton 1993; Martin et al. 1996; Kirkwood and Austad 2000). Evolutionary biology also provides a particular set of intellectual and experimental tools for probing the physiological, biochemical and molecular mechanisms responsible for organismal aging. Comparative methods rank among the most powerful that evolutionary biology includes in its toolkit.
To modern evolutionary biologists, effective use of the comparative approach in the biology of aging reflects the idea that comparing differences and similarities among groups of organisms can provide insight into the evolutionary processes producing them—including the process of natural selection resulting in specific forms of adaptation (Futuyma 2005). A comparative approach can be useful in the biology of aging in several ways (reviewed in Austad 1997; Austad and Holmes 1999). First, surveying physiological or other characteristics in a variety of species with a range of aging rates can help in identifying putative aging mechanisms (e.g., oxidative or glycoxidative processes; failure of DNA repair, etc.) (Ku and Sohal 1993; Shay 1995; Barja 1998; Monnier et al. 1999). It should be noted, however, that such surveys can identify correlation, but not actual causative mechanisms. Second, comparative methods can be used to assess the general relevance of particular aging mechanisms—that is, to elucidate whether mechanisms seen in one or a few strains or species are idiosyncratic or can be generalized to others (Martin et al. 1996). For example, cell signaling pathways involving insulin and insulin-like growth factor I (IGF-1) and the genes controlling them have been shown to be important correlates of aging in representative animal models of three evolutionarily distant phyla: roundworms, fruit flies, and house mice (Tatar et al. 2003). The fact that these animals are from widely divergent taxa suggests these mechanisms are evolutionarily conserved and hence important in a wide range of animals, including humans. Finally, evolutionary comparative methods can help pinpoint the most important putative mechanisms of aging from an array of plausible mechanisms. One way to do this is to select or create experimental strains, mutants or populations of animals differing in their aging patterns (e.g., through comparative surveys, CR experiments, gene knockouts, or artificial selection experiments) or to use different species found in nature, and then to identify concordant differences in particular physiological, biochemical or molecular processes as candidate aging mechanisms. When using such an approach, however, multiple differences associated with aging patterns may occur between experimental groups, making it difficult to infer specific aging or anti-aging mechanisms with certainty.
It is in this third case, when dealing with the phenomenon of “multiple differences,” where several traits of interests appear to be closely correlated but causation cannot clearly be inferred through direct experiment, that modern evolutionary or phylogenetic comparative methods become particularly valuable. As several of the papers in this special issue demonstrate, the field of aging is interested in exploring the biological basis of exceptionally long lifespans observed in some homeothermic vertebrates (e.g., birds, bats, naked mole-rats, and outbred, wild-derived mouse strains). Animal species or strains with exceptionally long lifespans, especially coupled with small body size and high metabolic rates or lifetime energy expenditures, like bats and birds—are ostensibly the superheroes of the aging world. Hence the field of aging is—quite rightly—interested in how understanding extreme variability in species lifespans among vertebrates could elucidate the cellular or molecular mechanisms promoting slow aging and long-term health in these unusual animals.
In the past, traditional comparative studies in gerontology have explored associations between lifespan, body size, metabolic rates or other physiological, biochemical or molecular parameters potentially relevant to basic aging processes, often applying conventional statistical approaches (e.g., standard linear regression models) to compare these traits in species representing different groups of vertebrates. However, a number of potential inferential confounds are associated with a conventional statistical approach that treats data points (species averages, for example) as statistically independent. In reality, values used to represent mean “physiological phenotypes” (such as differences in aging patterns) for different species or other taxa are neither statistically nor taxonomically independent, since some are likely to be more similar simply by virtue of shared evolutionary ancestry, or phylogeny. Evolutionary biology now provides statistical tools to incorporate the best available information about phylogeny from biosystematics into comparative studies of multiple species. In these phylogenetic comparative approaches, relationships in the form of branching, phylogenetic “trees” are actually built into appropriate statistical models (Felsenstein 1985; Garland et al. 2005), providing information about the effects of phylogenetic inertia on our focal dependent variable. Thus, building information about patterns of evolutionary relationships into comparative analyses helps to sort out effects of taxonomy and shared ancestry from the actual physiological or biochemical aging phenotypes or correlates of interest.
Expressed succinctly in a recent review, “Empirically, more closely related species do indeed tend to resemble one another...hummingbirds look like hummingbirds, and turtles look like turtles, and the same is true for [these animals’] physiological traits (Blomberg et al. 2003; Garland et al. 2005). Hence the long lifespans seen in many small songbirds or bats, vs those of similar-sized mammals, may not be attributed in a straightforward way to the peculiarities of avian mitochondrial oxidative metabolism (or, alternatively, to the metabolic demands of flight) without taking into account the hierarchical phylogenetic relationships and evolutionary history of these groups of animals. For this reason, proponents of rigorous evolutionary comparative methods strongly discourage tests of hypotheses employing comparisons of only two species or taxa along a given phenotypic continuum of interest (Rose 1991; Garland and Adolph 1994; Austad and Holmes 1999). While two-species comparisons can be heuristic for generating hypotheses about possible aging mechanisms (e.g., Ku and Sohal 1993; Barja 1998; Pamplona et al. 2005), these comparisons have limited value for making strong inferences about physiological adaptation vs phylogenetic effects.
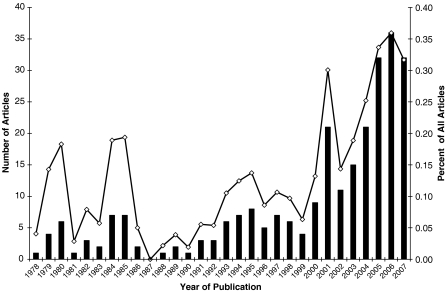
A growing number of investigators in biogerontology are now exploiting relevant phylogenetic relationships in studies of basic aging mechanisms, as exemplified by several papers in this issue (e.g., Speakman, Hulbert, Buffenstein). When we conducted a recent search of PubMed using the keywords “phylogeny” and “aging” by year to uncover papers published including these keywords over the last 30 years, we found that both the absolute number of articles and the proportion of total articles (of those found using the keyword “aging” only) has increased noticeably since 2000 (Fig. 1). These publications, while not limited to aging journals, do seem to reflect a trend for scientists focusing on aging to at least consider phylogeny in the context of their studies. Further use of actual comparative statistical approaches may expand our interpretation of theories of biological aging, as noted in the following example of a recent study of putative associations between long life span, key aspects of oxidative metabolism and the potential for oxidative damage.
Fig. 1.
Number of articles (bars) retrieved in a PubMed search using both “phylogeny” and “aging” as search terms. Percentages of articles (open circles) that incorporated “phylogeny” were calculated as the proportion of all articles retrieved using only “aging” as the keyword
The free radical hypothesis meets the comparative phylogenetic method
The free radical and oxidative stress hypotheses posit that aging-related disease and deterioration arise as a result of the action of reactive oxygen species (ROS) or other reactive molecules arising as a normal by-product of oxidative metabolism (Harman 1956, 1972; Beckman and Ames 1998; Muller et al. 2007). Consider again our long-lived “superhero” species: birds, bats, and naked mole-rats. According to these hypotheses, we would predict that these animals have some means of preventing, repairing, or otherwise circumventing the cumulative damage to cells and macromolecules expected to arise from sustained oxidative metabolism. Hence well-conceived comparative studies of these species are warranted, with a focus on such phenotypic variables as mitochondrial ROS production, resistance of cell membranes to damage, accumulation of oxidative adducts or other aging phenotypes or biomarkers.
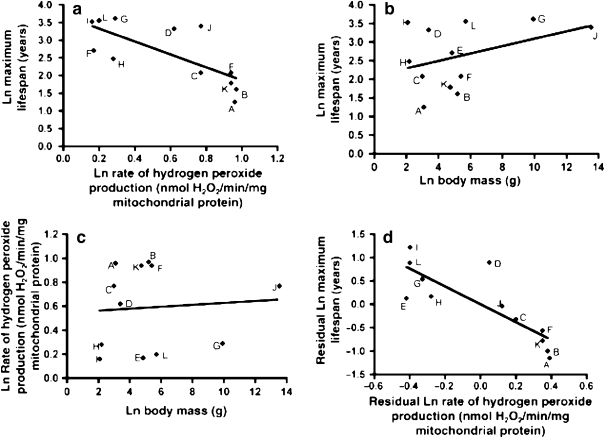
A recent study by Lambert et al. (2007) shows how phylogenetic comparative methods can be applied to refine tests of the free radical hypothesis using data gathered through in vitro mitochondrial biochemistry. These authors investigated the oft-reported inverse correlation between mitochondrial ROS production and maximum species lifespans in homeotherms, comparing rates of in vitro hydrogen peroxide (H2O2) production by mitochondria from heart tissue in an assortment of ten mammalian (mouse, rat, white-footed mouse, naked mole-rat, Damara mole-rat, guinea pig, baboon, little brown bat, Brazilian free-tailed bat and domestic ox) and two avian (pigeon, quail) species (for scientific names, see Fig. 2). In keeping with standards for evolutionary comparative studies, the species in their analyses included a wide range of maximum lifespans, adult body sizes and metabolic rates.
Fig. 2.
a Mitochondrial H2O2 production during succinate respiration, including analysis of residual values after correcting for body mass. b ln maximum lifespan vs ln body mass; c ln H2O2 production; d residual values for ln maximum lifespan vs residuals for ln rate of H2O2 production after correcting for body mass. The correlation in a is significant (r2 = 0.54, F1,10 = 11.7, P = 0.007); the correlation in d is significant (r2 = 0.69, F1,10 = 22.5, P = 0.0008). Upper case letters denote data for the following species or strains: A mouse; Mus musculus (C57BL/6, BALB/c, and wild Idaho strains); B rat, Rattus norvegicus; C white-footed mouse, Peromyscus leucopus; D naked mole-rat, Hetereocephalus glaber; E Damara mole-rat, Cryptomys dearness; F guinea pig, Cavia porcellus; G baboon, Papio cynocephalus; H Brazilian free-tailed bat, Tadarida brasiliensis; I little brown bat, Myotis lucifugus; J domestic ox, Bos taurus; K Japanese quail, Coturnix japonica; L domestic pigeon Columba livia. Reprinted with permission from Lambert et al. 2007
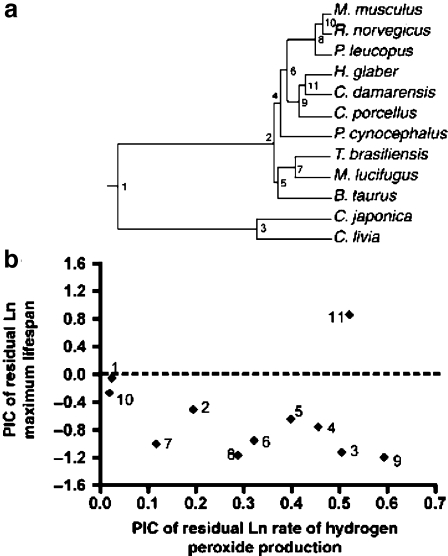
In this study, as well as in previous studies by other laboratories, H2O2 production at mitochondrial complex I during reverse electron transport (at high succinate concentrations) was shown to be negatively correlated with maximum life span (Fig. 2a). After correcting for body mass, this correlation was even more pronounced (Fig. 2d). Additionally, Lambert et al. (2007) included the first analysis of data of this type employing phylogenetic independent contrasts (Felsenstein 1985), in order to incorporate the effects of phylogenetic similarities between species in their analysis statistically (Fig. 3). Using this approach, the inverse association between maximum lifespan and hydrogen peroxide production, while still present, failed to achieve statistical significance in a regression analysis, although a sign test showed this trend to be significant (Fig. 3b). Particularly important, the failure in the regression analysis to achieve significance with phylogenetic contrasts clearly could be attributed to the inclusion of the two mole-rat species, which have exceptionally long lifespans for their sizes and rates of hydrogen peroxide production. Therefore, while the results of this study in general supported the free radical hypothesis, and show that lifespan and mitochondrial ROS production are inversely correlated, they also suggest that mole-rats may represent important exceptions to the free radical theory, and that the biochemical bases of longevity in these rodents may not be related in a simple or direct way to the production of fewer ROS (see also Buffenstein and Hulbert, this issue). Analyses like this can play a pivotal role in directing researchers toward groups of animals with unusual anti-aging adaptations, or have particular utility for understanding both the mechanistic and evolutionary bases of aging and somatic maintenance.
Fig. 3.
Comparative analysis of ln maximum lifespan vs residual ln rate of mitochondrial H2O2 production during succinate respiration using phylogenetic independent contrasts (PIC). a Construction of a phylogenetic “tree” for 12 mammalian and avian species results in 11 contrasts (nodes numbered 1–11) based on phylogenetic distance. The plot in b represents the PIC of residual ln maximum lifespan (corrected for body mass) vs PIC of residual ln H2O2 production rate (corrected for body mass) with the x-axis ‘positivized’ (Garland et al. 1992). For 10 of the 11 points in b, H2O2 production is negatively correlated with maximum lifespan, emphasized by the fact that 10 of the 11 points fall below the dashed line at y = 0. This relationship achieves significance in a sign test (P = 0.011; sign test), but not in a regression analysis due to the outlier at node 11 representing the contrast between two mole-rat species. Reprinted with permission from Lambert et al. 2007
Additional alternative animal models and experimental approaches
Traditional models have promoted in-depth characterization of molecular changes during aging that have allowed biogerontologists to refine theoretical predictions and empirical understanding of the aging process. However, the relatively small number of model species and cell types typically used may limit the inferences that can be made about basic aging processes, particularly in longer-lived species. Also, the use of domesticated animals and established cell lines long removed from their natural environments further limits the ability to extrapolate aging phenomena in these systems to those in long-lived, outbred animals, including humans (see Harper, this issue). It is quite possible that the use of additional animal models and cell types will reveal entirely new responses to standard biogerontological experimental paradigms, such as effects of CR, and help test and revise well-supported evolutionary and proximate theories of aging.
For example, comparative studies using traditional aging model taxa have revealed the importance of insulin-like signaling pathways to biological aging, responses to CR, and tradeoffs between reproduction and somatic maintenance (Weindruch and Walford 1988; Tatar et al. 2003; Partridge et al. 2005; Tu et al. 2005; Finch 2007). New animal models can be used to test whether observations obtained using traditional models can be generalized to new taxa. Testing for common mechanisms of biological aging among animals will require strategic selection of new animal models, including both invertebrates and vertebrates. It also should include outbred animals that retain their natural genetic heterogeneity (see Harper, this issue) and studies of wild animal populations in the field, as well as in controlled laboratory settings (Bronikowski et al., this issue), to assess the effects of environmental variation on physiological responses that contribute to biological aging (Bronikowski and Promislow 2005; Promislow et al. 2006; Austad and Finch 2008). Additionally, there are still many aspects of biological aging that remain poorly investigated, even after decades of research using traditional models. For example, we still have only a cursory understanding of naturally occurring sex differences in aging (vomSaal et al. 1994; Austad 2006; Clutton-Brock and Isvaran 2007) (Sitzmann et al., this issue). Comparative studies that employ closely related species differing in their sex-specific characteristics and differences may yield productive insights here. Given the wealth of information known about C. elegans, which is hermaphroditic (or, alternatively, male), it may be useful to investigate related dioecious nematode species to examine differences in aging among closely related species that differ with respect to sex-determination systems.
In addition to novel animal species, it may be valuable to consider new experimental approaches that would employ either traditional or new animal models. One general area waiting to be explored relates to genetic manipulations. Modern molecular tools allow the conversion of traditional animal models into new aging models via direct manipulation of the genome. For example, Flurkey et al. created a new mouse model (Pohn strain) for extended female lifespan (Flurkey et al. 2007), and Van Remmen et al. (2004) have created mouse strains with knockouts of specific antioxidant enzymes to test the oxidative stress theory of aging. Non-traditional models may well further the development of these types of genomic approaches. For example, the lipid membrane profile of naked mole-rats may contribute to their extraordinary longevity (Buffenstein and Hulbert, this issue). Conceivably, a laboratory mouse could be developed with lipid membrane composition similar to that of naked mole-rats, enabling us to test the effects of this one physiological feature on lifespan in the mouse, an otherwise extremely well-characterized animal model.
Another new approach would be to more fully consider that during their evolution animals lived in fluctuating, often unpredictable, environments very different from the laboratory settings for most scientific research. Pertinent environmental manipulations could include allowing for natural reproduction, housing animals in temperature and light environments to mimic seasonal variation in nature, and allowing social interactions with conspecifics, all of which could be applied systematically to assess effects on biological aging. Variation in reproductive effort is clearly related to variation in mortality patterns (and, probably, aging rates) in natural populations of animals (including humans) (Partridge et al. 2005; Finch 2007), yet normal reproductive activity is very rarely a focus of aging studies. Environmental forces that actively change an animal’s requirements for physiological homeostasis, while part of normal evolutionary pressures and very relevant to human health, have rarely been incorporated into studies of longevity. For example, even though most animals in nature must cope with parasite infection (Cox 2001), the effects of intact pathogens and the associated immune response on biological aging are rarely considered.
In the interest of experimental control, the gold standard for studies of aging in laboratory mice is the rearing and maintenance of animals in a specific-pathogen-free facility. However, we know that infectious diseases and parasites (and even the internal microbiota) constitute an integral part of the environment of free-living humans and wild, outbred animals, making the aging picture far more complex in the real world. Effects of infection on organismal health can begin during prenatal life (reviewed in Finch 2007) and extend through adult life, affecting clinical health and reproduction in humans (Van Bodegom et al. 2007), as well as in terms of evolutionary fitness measures (Koella et al. 1998). In this issue, Kristan discusses studies in which long-term CR increases susceptibility of laboratory mice to experimental infections with a range of pathogen taxa, despite some measures of enhanced immune response in CR mice. These findings underline the importance of considering the beneficial effects of undernutrition without malnutrition (as in experimental CR) against a backdrop of infectious disease in nature.
Emerging syntheses of aging, health and clinical disease propose that infectious disease and resulting inflammatory and immune responses rank among the key causes of aging-related disease and dysfunction, including diabetes, cardiovascular disease and Alzheimer’s disease (Finch 2007). Given this perspective, broadening our biogerontological focus to include animal models exposed to environmental challenges from pathogens and parasites is essential. Furthermore, tradeoffs between development, reproduction and somatic maintenance (including immune defenses against infectious disease) are one of the cornerstones of evolutionary aging theory (Partridge and Barton 1993; Kirkwood and Austad 2000) and should be investigated in environments that are as relevant to evolutionary forces in nature as possible.
Closing remarks
In our view, more communication and collaboration are needed between evolutionary biologists and biogerontologists, as well as an increased understanding of evolutionary approaches—including well-developed comparative tools—to advance the field of biology of aging. Certainly, as the diversity of animal models and approaches expands and the use of current comparative tools becomes more commonplace, our understanding of how novel animal models fit into established understanding of biological aging also should increase. We hope that this special issue promotes the growth of a broader, better integrated research perspective in the biology of aging.
Abbreviations
- PIC
Phylogenetic independent contrasts
- ROS
Reactive oxygen species
- CR
Calorie restriction
References
- Austad SN (1993) The comparative perspective and choice of animal models in aging research. Aging (Milano) 5:259–267 [DOI] [PubMed]
- Austad SN (2001) An experimental paradigm for the study of slowly aging organisms. Exp Gerontol 36:599–605 doi:10.1016/S0531-5565(00)00229-1 [DOI] [PubMed]
- Austad SN (2006) Why women live longer than men. Gend Med 3:79–92 doi:10.1016/S1550-8579(06)80198-1 [DOI] [PubMed]
- Austad SN (1997) Why we age. Wiley, New York
- Austad SN, Finch CE (2008) The evolutionary context of human aging and degenerative disease. In: Stearns SC (ed) Evolution in health and disease. Oxford University Press, New York
- Austad SN, Fischer KE (1991) Mammalian aging, metabolism, and ecology: evidence from the bats and marsupials. J Gerontol Biol Sci 46:B47–B53 [DOI] [PubMed]
- Austad SN, Holmes DJ (1999) Evolutionary approaches to probing aging mechanisms. In: Yu BP (ed) Methods in aging research. CRC, Boca Raton, pp 437–452
- Austad SN, Masoro EJ (2006) Handbook of the biology of aging
- Barja G (1998) Mitochondrial free radical production and aging in mammals and birds. Ann N Y Acad Sci 854:224–238 doi:10.1111/j.1749-6632.1998.tb09905.x [DOI] [PubMed]
- Beckman KB, Ames BN (1998) The free radical theory of aging matures. Physiol Rev 78:547–581 [DOI] [PubMed]
- Blomberg SP, Garland T Jr, Ives AR (2003) Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57:717–745 [DOI] [PubMed]
- Bronikowski AM, Promislow DEL (2005) Testing evolutionary theories of aging in wild populations. Trends Ecol Evol 20:271–273. doi:10.1016/j.tree.2005.03.011 [DOI] [PubMed]
- Brunet-Rossinni AK, Austad SN (2004) Ageing studies on bats: a review. Biogerontology 5:211–222 doi:10.1023/B:BGEN.0000038022.65024.d8 [DOI] [PubMed]
- Buffenstein R (2005) The naked mole-rat: a new long-living model for human aging research. J Gerontol A Biol Sci Med Sci 60:1369–1377 [DOI] [PubMed]
- Calder WA (1984a) Body size and life histories. Dover, New York
- Calder WA (1984b) Size, function and life history. Harvard University Press, Cambridge, MA
- Charlesworth B (1980) Evolution in age-structured populations. Cambridge University Press, Cambridge, UK
- Clutton-Brock TH, Isvaran K (2007) Sex differences in ageing in natural populations of vertebrates. Proc Biol Sci 274:3097–3104 doi:10.1098/rspb.2007.1138 [DOI] [PMC free article] [PubMed]
- Comfort A (1964) Ageing: the biology of senescence. Routledge and Kegan Paul, London
- Conn PM (2006) Handbook of models for human aging. Academic, New York
- Cox FEG (2001) Concomitant infections, parasites and immune responses. Parasitology 122:S23–S38 [DOI] [PubMed]
- Felsenstein J (1985) Phylogenies and the comparative method. Am Nat 126:1–25
- Finch CE (1990) Longevity, senescence, and the genome. University of Chicago Press, Chicago
- Finch CE (2007) The biology of human longevity: inflammation, nutrition, and aging in the evolution of lifespans. Academic (Elsevier), Amsterdam
- Flurkey K, Brandvain Y, Klebanov S, Austad SN, Miller RA, Yuan R et al (2007) PohnB6F1: a cross of wild and domestic mice that is a new model of extended female reproductive life span. J Gerontol A Biol Sci Med Sci 62:1187–1198 [DOI] [PubMed]
- Futuyma D (2005) Evolution. Sinauer, Sunderland
- Garland T, Adolph SC (1994) Why not to do two-species comparative studies: limitations on inferring adaptation. Physiol Zool 67:797–828
- Garland T Jr, Harvey PH, Ives AR (1992) Procedures for the analysis of comparative data using phylogenetically independent contrasts. Systematic Biol 41:18–32 [DOI]
- Garland T Jr, Bennett AF, Rezende EL (2005) Phylogenetic approaches in comparative physiology. J Exp Biol 208:3015–3035 doi:10.1242/jeb.01745 [DOI] [PubMed]
- Hamilton WD (1966) The moulding of senescence by natural selection. J Theor Biol 12:12–45 doi:10.1016/0022-5193(66)90184-6 [DOI] [PubMed]
- Harman D (1956) Aging: a theory based on free radical and radiation chemistry. J Gerontol 11:298–300 [DOI] [PubMed]
- Harman D (1972) A biologic clock: the mitochondria? J Am Geriatr Soc 20:145–147 [DOI] [PubMed]
- Harvey PH, Pagel MD (1991) The comparative method in evolutionary biology. Oxford University Press, Oxford
- Holmes DJ, Austad SN (1995) Birds as animal models for the comparative biology of aging: a prospectus. J Gerontol Biol Sci 50A:59–66 [DOI] [PubMed]
- Ives AR, Midford PE, Garland T Jr (2007) Within-species variation and measurement error in phylogenetic comparative methods. Syst Biol 56:252–270 doi:10.1080/10635150701313830 [DOI] [PubMed]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R (1993) A C. elegans mutant that lives twice as long as wild type. Nature 366:461–464 doi:10.1038/366461a0 [DOI] [PubMed]
- Kirkwood TBL, Austad SN (2000) Why do we age? Nature 408:233–238 [DOI] [PubMed]
- Koella JC, Agnew P, Michalakis Y (1998) Coevolutionary interactions between host life histories and parasite life cycles. Parasitology 116(Suppl):S47–S55 [DOI] [PubMed]
- Ku H, Sohal RS (1993) Comparison of mitochondrial pro-oxidant generation and anti-oxidant defenses between rat and pigeon: possible basis of variation in longevity and metabolic potential. Mech Ageing Dev 72:67–76 doi:10.1016/0047-6374(93)90132-B [DOI] [PubMed]
- Lambert AJ, Boysen HM, Buckingham JA, Yang T, Podlutsky A, Austad SN et al (2007) Low rates of hydrogen peroxide production by isolated heart mitochondria associate with long maximum lifespan in vertebrate homeotherms. Aging Cell 6:607–618 doi:10.1111/j.1474-9726.2007.00312.x [DOI] [PubMed]
- Lindstedt SL, Calder WA (1976) Body size and longevity in birds. Condor 78:91–94
- Martin GM, Austad SN, Johson TE (1996) Genetic analysis of ageing: role of oxidative damage and environmental stresses. Nat Genet 13:25–34 doi:10.1038/ng0596-25 [DOI] [PubMed]
- Medawar PB (1952) An unsolved problem of biology. Lewis, London
- Miller R, Harper J, Dysko R, Durkee S, Austad S (2002) Longer life spans and delayed maturation in wild-derived mice. Exp Biol Med 227:500–508 [DOI] [PubMed]
- Monnier VM, Fogarty JF, Monnier CS, Sell DR (1999) Glycation, glycoxidation, and other Maillard reaction products. In: Yu BP (ed) Methods in aging research. CRC, Boca Raton, pp 657–681
- Muller FL, Lustgarten MS, Jang Y, Richardson A, Van Remmen H (2007) Trends in oxidative aging theories. Free Radic Biol Med 43:477–503 doi:10.1016/j.freeradbiomed.2007.03.034 [DOI] [PubMed]
- Pagel MD (1992) A method for the analysis of comparative data. J Theor Biol 156:431–442 doi:10.1016/S0022-5193(05)80637-X [DOI]
- Pamplona R, Portero-Otin M, Sanz A, Ayala V, Vasileva E (2005) Protein and lipid oxidative damage and complex I are lower in the brain of budgerigar and canaries than in mice. AGE 27:267–280 [DOI] [PMC free article] [PubMed]
- Partridge L, Barton NH (1993) Optimality, mutation and the evolution of ageing. Nature 362:305–311 doi:10.1038/362305a0 [DOI] [PubMed]
- Partridge L, Gems D, Withers DJ (2005) Sex and death: what is the connection? Cell 120:461–472 doi:10.1016/j.cell.2005.01.026 [DOI] [PubMed]
- Pearl R (1928) The rate of living. Knopf, New York
- Promislow DEL, Fedorka KM, Burger JMS (2006) Evolutionary biology of aging: future directions. In: Masoro EJ, Austad SN (eds) Handbook of the biology of aging. Academic, New York, pp 217–242
- Reznick DN, Bryant MJ, Roff D, Ghalambor CK, Ghalambor DE (2004) Effect of extrinsic mortality on the evolution of senescence in guppies. Nature 431:1095–1099 doi:10.1038/nature02936 [DOI] [PubMed]
- Rose MR (1991) Evolutionary Biology of Aging. Oxford University Press, New York
- Rubner M (1908) Das Problem der Lebensdauer und seine beziehungen zum Wachstum und Erhahrung. Oldenburg, Munich
- Sacher GA (1978) Longevity and aging in vertebrate evolution. Bioscience 28:497–501 doi:10.2307/1307295 [DOI]
- Sanford GM, Lutterschmidt WI, Hutchison VH (2002) The comparative method revisited. Bioscience 52:830–836 doi:10.1641/0006-3568(2002)052[0830:TCMR]2.0.CO;2 [DOI]
- Schmidt-Nielsen K (1970) Energy metabolism, body size, and problems of scaling. Fed Proc 29:1524–1532 [PubMed]
- Schmidt-Nielsen K (1997) Animal physiology. Adaptation and environment. Cambridge University Press, Cambridge, UK
- Shay JW (1995) Aging and cancer: are telomeres and telomerase the connection? Mol Med Today 1:378–384 doi:10.1016/S1357-4310(95)93872-9 [DOI] [PubMed]
- Speakman JR (2005a) Body size, metabolism and lifespan. J Exp Biol 208:1717–1730 doi:10.1242/jeb.01556 [DOI] [PubMed]
- Speakman JR (2005b) Correlations between physiology and lifespan—two widely ignored problems with comparative studies. Aging Cell 4:167–175 doi:10.1111/j.1474-9726.2005.00162.x [DOI] [PubMed]
- Stearns SC (1992) The evolution of life histories. Oxford University Press, New York
- Stearns SC, Koella JC (2008) Evolution in health and disease. Oxford University Press, New York, p 368
- Tatar M, Bartke A, Antebi A (2003) The endocrine regulation of aging by insulin-like signals. Science 299:1346–1351 doi:10.1126/science.1081447 [DOI] [PubMed]
- Tu MP, Yin CM, Tatar M (2005) Mutations in insulin signaling pathway alter juvenile hormone synthesis in Drosophila melanogaster. Gen Comp Endocrinol 142:347–356 doi:10.1016/j.ygcen.2005.02.009 [DOI] [PubMed]
- Van Bodegom D, May L, Meij HJ, Westendorp RG (2007) Regulation of human life histories: the role of the inflammatory host response. Ann N Y Acad Sci 1100:84–97 doi:10.1196/annals.1395.007 [DOI] [PubMed]
- Van Remmen H, Qi W, Sabia M, Freeman G, Estlack L, Yang H et al (2004) Multiple deficiencies in antioxidant enzymes in mice result in a compound increase in sensitivity to oxidative stress. Free Radic Biol Med 36:1625–1634 doi:10.1016/j.freeradbiomed.2004.03.016 [DOI] [PubMed]
- vomSaal FS, Finch CE, Nelson JF (1994) Natural history and mechanisms of reproductive aging in humans, laboratory rodents, and other selected vertebrates. In: Knobil E, Neill JD (eds) The physiology of reproduction. Raven, New York, pp 1213–1314
- Weindruch R, Walford RL (1988) The retardation of aging and disease by dietary restriction. Thomas, Springfield
- Weismann A (1989) Essays upon heredity and kindred biological problems. Clarendon, Oxford
- Williams GC (1957) Pleiotrophy, natural selection and the evolution of senescence. Evolution 11:398–411 [DOI]