Abstract
Abl interactor (Abi) 1 was first identified as the downstream target of Abl tyrosine kinases and was found to be dysregulated in leukemic cells expressing oncogenic Bcr-Abl and v-Abl. Although the accumulating evidence supports a role of Abi1 in actin cytoskeleton remodeling and growth factor/receptor signaling, it is not clear how it contributes to Bcr-Abl-induced leukemogenesis. We show here that Abi1 gene silencing by short hairpin RNA attenuated the Bcr-Abl-induced abnormal actin remodeling, membrane-type 1 metalloproteinase clustering and inhibited cell adhesion and migration on fibronectin-coated surfaces. Although the knock down of Abi1 expression did not affect growth factor-independent growth of Bcr-Abl-transformed Ba/F3 cells in vitro, it impeded competitive expansion of these cells in non obese diabetic (NOD)/ severe combined immuno-deficiency (SCID) mice. Remarkably, the knock down of Abi1 expression in Bcr-Abl-transformed Ba/F3 cells impaired the leukemogenic potential of these cells in NOD/SCID mice. Abi1 contributes to Bcr-Abl-induced leukemogenesis in part through Src family kinases, as the knock down of Abi1 expression attenuates Bcr-Abl-stimulated activation of Lyn. Together, these data provide for the first time the direct evidence that supports a critical role of Abi1 pathway in the pathogenesis of Bcr-Abl-induced leukemia.
Introduction
More than 95% of human chronic myelogenous leukemia and a subset of acute lymphocytic leukemia are caused by expression of Bcr-Abl, a fusion oncogene generated by reciprocal t(9;22)(q34;q11) chromosome translocation (1–3). Bcr-Abl-positive leukemias are characterized by premature release of myeloid and lymphoid lineage cells from bone marrow, followed by the expansion of these cells in peripheral blood and infiltration of organs such as spleen, liver and lung (1,3). The progression of these diseases involves not only accelerated cell proliferation and enhanced cell survival but also increased cell motility and active invasion of leukemic cells through blood vessel and matrix barriers (4,5). Although numerous studies have revealed that Bcr-Abl may activate multiple signaling pathways that are involved in cytoskeletal functions (4–14), precise mechanisms by which Bcr-Abl induces abnormal cytoskeletal functions in leukemic cells are not completely understood.
Bcr-Abl oncoproteins exert their oncogenic potential in cooperation with additional cytoplasmic and nuclear effectors such as those involved in the regulation of mitogenic and apoptotic pathways. They are also capable of binding to cytoskeleton proteins as well as the proteins involved in the regulation of cell adhesion and migration (4,5,10). Among these proteins is Abl interactor (Abi) (15,16), a key regulator of Rac-dependent actin polymerization (17,18). Mammalian Abi proteins consist of three members: Abi1 (also known as e3b1), Abi2 and Abi3 (also known as new molecule including SH3) (15,16,19). These proteins are present in cells as a complex with WASP-family verprolin-homologous (WAVE) proteins, Nck-associated protein, specifically Rac-associated protein, and hematopoietic stem progenitor cell 300 (17,20–22). The micromolecular complex (Abi–WAVE complex) regulates initiation of actin polymerization in response to Rac activation. In addition to the interactions with Abl, WAVE and Nck-associated protein, Abi proteins were also found to interact with a variety of other signaling molecules that are involved in the control of cell proliferation, apoptosis and cytoskeletal functions (23–37). Accumulating evidence suggests that Abi proteins may be involved in the signal transduction from membrane receptors to small guanosine triphosphate (GTP)-binding proteins and phosphoinositide 3-kinase (PI3 kinase) as well (26,27,36).
In hematopoietic cells transformed by Bcr-Abl, Abi1 is tyrosine phosphorylated and Abi2 is degraded (38,39). Recent studies have shown that Abi1 plays a critical role in Bcr-Abl-induced remodeling of actin cytoskeleton as well as clustering of adhesion molecules and membrane-type 1 metalloproteinase (MT1-MMP) (40,41). Blockade of the signal transduction from Bcr-Abl to Abi1 appeared to impair Bcr-Abl-stimulated cell adhesion and migration in vitro (40). Although these studies provide evidence in support of the role of Abi1 in Bcr-Abl transformation, it is not clear how and whether Abi1 contributes to Bcr-Abl-induced leukemogenesis in vivo. In present studies, we show that silencing the Abi1 gene by sequence-specific short hairpin RNA (shRNA) inhibited Bcr-Abl-stimulated cell adhesion and migration. Notably, the knock down of Abi1 expression impaired Bcr-Abl-induced leukemogenesis in vivo, possibly by impeding leukemic cell expansion and homing.
Materials and methods
Cell lines and reagents
Ba/F3 cells were grown in RPMI containing 10% fetal bovine serum (FBS) and 15% WEHI3-conditioned medium as a source of IL3. The Ba/F3 cell lines expressing wild-type p185Bcr-Abl (p185wt) or expressing both p185wt and Abi1 shRNA were cultured in RPMI containing 10% FBS. Retroviral packaging cell line GP2-293 (ClonTech, Mountain View, CA) was grown in Dulbecco’s modified Eagle’s medium containing 10% FBS. The preparation of rabbit polyclonal antibodies against Abi1 and Abi2 has been described previously (38,42). The antibodies against WAVE2, Lyn and phosphotyrosine-containing proteins were purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and the monoclonal antibodies for Abl were obtained from BD PharMingen (San Diego, CA). The antibody against phospho-Src family (Tyr 416) (including phospho-Lyn kinase) was purchased from Cell Signaling Technology (Danvers, MA). The monoclonal anti-β-actin antibody and the protease inhibitor cocktail were purchased from Sigma (St Louis, MO). The recombinant rat IL3 was purchased from PeproTech (Rocky Hill, NJ).
Plasmids and retroviral infection
Transfection of Ba/F3 cells with pSRαp185wt was described previously (38). Three murine stem cell virus-based pSM2 retroviral vectors containing complementary DNAs encoding for Abi1-specific shRNA, as well as a control vector encoding non-silencing shRNA, were purchased from Open Biosystems (Huntsville, AL). The sequences in Abi1 that are targeted by three shRNAs are as follows—shRNA A: ATGTCTACCTGTAAGCATA; shRNA B: CTCGAAGAGAGATTGGTAT and shRNA C: GTGCAATCATCTATGTTAT. The expression of shRNA was driven by U6 promoter. Amplification and purification of plasmid DNA were performed as specified by the manufacturer’s instruction. MSCV-green fluorescence protein (GFP) retroviral vector expressing enhanced green fluorescence protein was generated by subcloning an EcoR1/Not1 complementary DNA fragment encoding for enhanced green fluorescence protein into MSCV vector. Preparation of retroviruses and infection of Ba/F3 and Ba/F3p185wt cell lines were performed as described previously (39). Stable cell lines were selected by addition of puromycin into media (2 μg/ml).
Biochemical assays
Quantitative real-time reverse transcription–polymerase chain reaction (PCR) was performed on MyiQ single-color real-time PCR detection system (Bio-Rad, Hercules, CA) using mouse Abi1 primers (forward: 5′-AATCGCACCCGCAAATATG-3′ and reverse: 5′-GTTCTCGACAATGTGCCAGTTC-3′) combined with SYBR Green Master Mix (Applied Biosystems, Foster City, CA). Briefly, total RNA was extracted from cells using RNeasy mini kit (Qiagen, Valencia, CA) and complementary DNA was subsequently generated using SuperScript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA). The PCRs began with 10 min at 95°C for AmpliTaq Gold activation followed by 40 cycles at 95°C for 15 s for denaturation then 60°C for 1 min for annealing/extension. Relative quantification was done using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (forward primer: 5′-AACGACCCCTTCATTGAC-3′ and reverse primer: 5′-TCCACGACATACTCAGCAC-3′) as an endogenous control.
Immunoprecipitation and western blot analyses were performed as described previously (40). Briefly, control Ba/F3 cells and the Ba/F3 cells expressing p185wt and p185wt plus Abi1 shRNA were lysed in lysis buffer (20 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.2, 150 mM NaCl, 1% Triton X-100 and 10% glycerol) and incubated with appropriate antibodies bound to Sepharose beads. The immunoprecipitates were separated on sodium dodecyl sulfate–polyacrylamide gel electrophoresis, transferred to nitrocellulose and immunoblotted with appropriate antibodies.
In vivo leukemogenesis studies
A suspension of 1 × 106 Ba/F3 cells or Ba/F3 cells expressing p185wt alone or p185wt plus Abi1 shRNA was injected into 6- to 8-week-old female NOD/SCID mice through tail vein. The mice were followed for disease development as judged by symptoms such as abnormal gait and labored breathing. Moribund animals were killed by CO2 asphyxiation and were examined for tumors or other visible abnormalities. Collection of spleens, livers and bone marrow cells was performed immediately after killing. All protocols used were approved by Institutional Animal Review Committee at the Texas Tech University Health Sciences Center.
To examine the capacity of in vivo competitive expansion, the Ba/F3p185wt cells transduced with MSCV-based retroviruses expressing either GFP or Abi1 shRNA were mixed in vitro at 1:1 ratio and then injected into mice through tail vein. The Ba/F3-derived leukemic cells were rescued from peripheral blood, spleens and bone marrow of the diseased mice by culturing them in RPMI containing 10% FBS for 2–7 days under selection with puromycin.
Adhesion and migration assays
For adhesion assay, Ba/F3 cells and Ba/F3 cells expressing either p185wt alone or p185wt plus Abi1 shRNA were plated in six-well plates (2.5 ml per well) coated with fibronectin (BD Biosciences, Bedford, MA) and incubated at 37°C/5% CO2 for 16 h. Non-adherent cells were removed and adherent cells were washed three times with 1 ml prewarmed RPMI medium. The adherent cells then were trypsinized and collected. Both non-adherent and adherent cells were counted to determine the percentage of adherent cells.
The cell migration assay was performed as described previously (39). The inserts of Transwell plates (8 μm pores, Corning Costar Corp., Cambridge, MA) were coated with human fibronectin (Sigma). The control Ba/F3 cells and the Ba/F3 cells expressing p185wt alone or pl85wt plus Abi1 shRNA were resuspended in RPMI containing 0.1% bovine serum albumin at a concentration of 1 × 106 cells/ml. A suspension of 0.1 ml cells was added into fibronectin-coated insert and cells were allowed to migrate at 37°C in 5% CO2 incubator for 6–8 h.
Fluorescence microscopy and flow cytometry analysis
Cultured Ba/F3 cell lines were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min, permeabilized in 0.2% Triton X-100/PBS for 5 min and stained with 50 μg/ml tetramethyl rhodamine iso-thiocyanate (TRITC)-conjugated phalloidin (Sigma) in PBS. After washing extensively with PBS and a brief staining with 4′,6-diamidino-2-phenylindole (Sigma) to visualize nuclei, 5–10 × 103 cells were loaded per slide by cytospin and mounted with Vectashield mounting medium (Vector, Burlingame, CA). Images were captured and analyzed using Nikon TE-2000 microscope with Image software associated. For fluorescence-activated cell sorting of GFP-positive cells, Ba/F3 cells transfected with p185wt and MSCV-GFP were resuspended in PBS and subjected to flow cytometry (FACS Calibur Flow Cytometer, BD Biosciences) analysis using the software associated. GFP-positive cells were collected with >95% purity. These cells were grown in vitro and used for in vivo competitive expansion assay. To determine the percentage of GFP-positive cells in those cells rescued from diseased mice, rescued cells were grown in puromycin selection media for 2–7 days to morphologically homogenous. The cells were collected, washed and resuspended in PBS for flow cytometry analysis.
Statistical analysis
Descriptive statistics were generated for all quantitative data with the presentation of means ± SDs. Significance of comparisons between experimental groups was tested using the Student’s t-test.
Results
ShRNA-induced Abi1 gene silencing in Bcr-Abl-transformed Ba/F3 cells
We have shown previously that Abi1 is tyrosine phosphorylated in hematopoietic cells transformed by Bcr-Abl (39). To test the role of Abi1 in Bcr-Abl-induced cellular transformation and leukemogenesis, shRNAs with either scrambled sequences or the sequences that specifically target different regions of Abi1 transcript were introduced into p185Bcr-Abl-transformed Ba/F3 cells (p185wt cells) by retroviral transduction. Stable cell populations were selected by puromycin and tested for the effect of shRNA on Abi1 expression. Remarkably, the expression of Abi1 in p185wt cells transduced with Abi1 shRNA was reduced to a level undetectable by western blot analysis, as compared with p185wt cells transduced with scrambled shRNA (Figure 1A, compare lanes 3–5 with lane 2). To compare the efficacy of the Abi1 knockdown by different shRNA, quantitative real-time PCR analysis was performed. As shown in Figure 1B (lower panel), expression of Abi1 shRNAs B and C in p185wt cells resulted in a decrease of Abi1 messenger RNA level by 95 and 90%, respectively, as compared with the p185wt cells expressing scrambled shRNA (p185wt control cells). Consistently, immunoprecipitation followed by western blot analysis using Abi1-specific antibody revealed that among three shRNA constructs tested, shRNA A induces ∼70% reduction of Abi1 protein level, whereas the shRNAs B and C caused >90% reduction of Abi1. The p185wt Abi1 shRNA B and C cells (p185wt Abi1 shRNA cells), therefore, were chosen for further studies.
Fig. 1.
The shRNA-mediated Abi1 gene silencing. (A) Abi1 expression in Ba/F3 cells (BaF3) and p185wt cells transduced with either non-silencing shRNA (p185wt) or Abi1 shRNAs (shRNAs A, B and C). Total lysates from 1 × 106 cells were separated on sodium dodecyl sulfate–polyacrylamide gel electrophoresis and were transferred to nitrocellulose membrane. The membrane was probed with the antibodies as indicated. The p185 Bcr-Abl and c-Abl were indicated by the arrow and arrowhead, respectively. (B) Efficacy of Abi1 knockdown by different Abi1 shRNAs. The lysates (2 × 107 cells) from the indicated cell lines were immunoprecipitated (IP) by Abi1 antibody and the immunoprecipitates were analyzed by western blotting (WB) using Abi1 antibody as probe (upper panel). Total RNAs from the indicated cell lines were isolated and the complementary DNAs were synthesized. Real-time quantitative PCR (lower panel) was performed to determine Abi1 messenger RNA (mRNA) levels, which are expressed as the levels relative to that of GAPDH. Data represent mean ± SD of triplicate experiments; *P < 0.001. (C) Expression of Abi2 and WAVE2 in Ba/F3 cells and p185wt cells transduced with either non-silencing shRNA or Abi1 shRNAs. Total lysates from 1 × 106 cells were separated on sodium dodecyl sulfate–polyacrylamide gel electrophoresis and were transferred to nitrocellulose membrane. The membrane was probed with the antibodies as indicated.
The expression of Bcr-Abl in hematopoietic cells induces downregulation of Abi2 (38). To determine if Abi1 knockdown affects the Bcr-Abl-induced Abi2 downregulation, we examined the protein level of Abi2 in p185wt Abi1 shRNA cells. Consistent with the previous report (38), the expression of Abi2 is lost in p185wt control cells as compared with Ba/F3 cells (Figure 1C). Similarly, no Abi2 was detected in p185wt Abi1 shRNA cells (Figure 1C), suggesting that Bcr-Abl-induced downregulation of Abi2 is not affected by Abi1 knockdown. We also examined the protein level of WAVE2 in p185wt Abi1 shRNA cells. In line with the results reported by other investigators (18,20,43), the knock down of Abi1 in p185wt cells resulted in a reduction of WAVE2 protein level (Figure 1C).
Abi1 knockdown does not affect Bcr-Abl-induced protein tyrosine phosphorylation and IL3-independent growth of Ba/F3 cells
Abi proteins have been shown capable of regulating the tyrosine kinase activity of cAbl (44,45). To determine if the Abi1 knockdown affects the tyrosine kinase activity of Bcr-Abl, we examined the tyrosine phosphorylation profile of the p185wt Abi1 shRNA cells. Expression of p185wt in Ba/F3 cells resulted in elevated protein tyrosine phosphorylation, as compared with parental Ba/F3 cells (Figure 2A). It appeared that Abi1 knockdown had little, if any, effect on Bcr-Abl-induced protein tyrosine phosphorylation (Figure 2A, compare lane 3 with lanes 1 and 2).
Fig. 2.
Abi1 knockdown did not affect Bcr-Abl-induced protein tyrosine phosphorylation and IL3-independent growth. (A) Profiles of protein tyrosine phosphorylation in Ba/F3 cells and p185wt cells transduced with either non-silencing shRNA (p185wt) or Abi1 shRNAs (Abi1R). Indicated cell lines were starved in RPMI 1640 plus 0.1% bovine serum albumin for 6 h prior to harvest. Total lysates from 1 × 106 cells were separated on 8% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and subjected to western blot analysis with anti-phosphotyrosine antibody. (B) IL3-independent growth of Ba/F3 cells as well as the p185wt cells transduced with either non-silencing shRNA (p185wt) or Abi1 shRNA (Abi1R).
Next, we tested if the Abi1 knockdown affects Bcr-Abl-induced IL3-independent growth of Ba/F3 cells. The Ba/F3, p185wt control and p185wt Abi1 shRNA cells were grown in RPMI 1640 media containing 10% FBS with no addition of IL3. Unlike parental Ba/F3 cells, which failed to grow and died within 72 h in IL3-free medium (Figure 2B), the p185wt control cells and the p185wt Abi1 shRNA cells grew equally well in this medium. Thus, the knock down of Abi1 expression had no effect on Bcr-Abl-induced IL3-independent growth of Ba/F3 cells.
Abi1 knockdown inhibits Bcr-Abl-stimulated abnormal cytoskeleton remodeling, MT1-MMP clustering, as well as cell adhesion and migration
Given that Abi1 is a key regulator of actin polymerization, we tested if the Abi1 knockdown affects Bcr-Abl-induced abnormal cytoskeletal functions, such as abnormal actin cytoskeleton remodeling, cell adhesion and migration. Expression of p185wt in Ba/F3 cells induced a profound actin cytoskeleton remodeling. Specifically, a spot intensively stained by phalloidin, indicative of filament actin (F-actin) aggregates, was observed in p185wt cells, but not in Ba/F3 cells and the Ba/F3 cells transfected by a construct expressing a kinase-deficient Bcr-Abl (p185K671R, Figure 3A). Expression of Abi1 shRNA in p185wt cells resulted in a 3-fold reduction in the formation of abnormal F-actin-enriched structures (Figure 3A).
Fig. 3.
Abi1 knockdown inhibited Bcr-Abl-stimulated actin cytoskeleton remodeling, MT1-MMP clustering, as well as cell adhesion and migration on fibronectin-coated surfaces. (A) Inhibition of Bcr-Abl-induced abnormal actin remodeling by Abi1 knockdown. Ba/F3 cells and the Ba/F3 cells expressing p185wt, p185K671R and p185wt plus Abi1 shRNA were fixed and stained with TRITC-conjugated phalloidin. The cells with F-actin-rich structures (invadopodia-like structures) were visualized by fluorescence microscopy as shown by arrowheads (upper panel) and were counted (lower panel, represented as mean ± SD percentage of three randomly picked areas). (B) The p185wt cells expressing GFP–MT1-MMP were transduced with either control retrovirus (control) or the retrovirus expressing Abi1 shRNA. The knock down of Abi1 expression was confirmed by western blotting (data not shown). The distribution of GFP–MT1-MMP was visualized by fluorescence microscopy. A similar result has been described previously (41). (C) Effects of Abi1 knockdown on Bcr-Abl-stimulated cell adhesion (lower panel) and migration (upper panel) on fibronectin-coated surfaces. Ba/F3 cells and the p185wt cells transduced with either non-silencing shRNA or Abi1 shRNAs were grown in fibronectin-coated six-well plate (2.5 × 105 per well) for 16 h. The total cells and the cells that were adherent to fibronectin-coated surfaces were counted and the percentage of adherent cells calculated. The vertical axis shows the percentage of the adherent cells and is expressed as the mean ± SD of triplicate wells. For cell migration on fibronectin-coated membrane, 1 × 105 cells were tested in Transwell migration assay. The vertical axis shows the percentage of the migrated cells and is expressed as the mean ± SD of triplicate wells; *P < 0.01.
In previous studies, we have shown that MT1-MMP, a member of transmembrane metalloproteinases that is responsible for the degradation of a variety of extracellular matrix, is clustered in Ba/F3 cells upon Bcr-Abl transformation (41). We therefore tested if the Abi1 is required for Bcr-Abl-induced clustering of MT1-MMP. We introduced retroviral vectors expressing either control shRNA or Abi1 shRNA into p185wt cells that express GFP-tagged MT1-MMP (41). Consistent with our previous results (41), GFP-tagged MT1-MMP displayed a clustered distribution in control p185wt cells (Figure 3B). In contrast, clustering of GFP–MT1-MMP was reduced in p185wt Abi1 shRNA cells (Figure 3B), suggesting an involvement of Abi1 in Bcr-Abl-induced clustering of MT1-MMP.
We then tested the effect of Abi1 knockdown on Bcr-Abl-stimulated cell adhesion and migration. As shown in Figure 3C, expression of p185wt in Ba/F3 cells stimulated cell adhesion and spontaneous cell migration on fibronectin-coated surfaces. However, expression of Abi1 shRNA in p185wt cells inhibited the Bcr-Abl-stimulated cell adhesion and migration on fibronectin-coated surfaces (Figure 3C). Together, these results strongly suggest that Abi1 pathway plays an important role in Bcr-Abl-induced abnormalities of cytoskeletal functions of leukemic cells.
Bcr-Abl-induced leukemogenesis is impaired by Abi1 knockdown
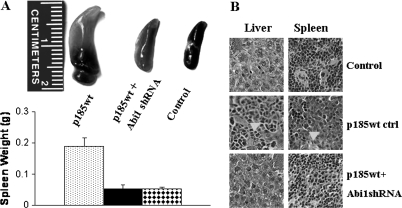
The shRNA-mediated Abi1 knockdown in p185wt Abi1 shRNA cells is stable. After 5-week culture in vitro, expression of Abi1 remained undetectable in total lysate of p185wt cells transduced with Abi1 shRNA (Figure 4A). This allowed us to test if Abi1 knockdown affects Bcr-Abl-induced leukemogenesis in vivo. Ba/F3, p185wt control and p185wt Abi1 shRNA cells were injected intravenously into NOD/SCID mice through tail vein. The recipient mice were followed for the development of leukemia. The mice injected with Ba/F3 cells were healthy with no signs of leukemia for up to 3 months (Table I and Figure 4B). In contrast, the mice injected with p185wt control cells developed leukemia in 2–3 weeks. These mice either died or became moribund (Figure 4B) with a mean latency of 17.5 days (Table I). The mice injected with p185wt Abi1 shRNA cells survived longer with a mean latency of 35.8 days (Figure 4B). Gross pathology analysis revealed that all the mice injected with p185wt control cells developed splenomegaly and hepatomegaly (Table I and Figure 5A and B), whereas no apparent splenomegaly or hepatomegaly was observed in mice injected with p185wt Abi1 shRNA cells (Table I and Figure 5A). Histopathology analysis showed that the destruction of normal cytoarchitecture in the spleen and liver, due to the massive accumulation of tumor cells, was observed in the mice injected with p185wt control cells (Figure 5B). In contrast, no apparent abnormality of the splenic and hepatic cytoarchitecture was observed in the mice injected with p185wt Abi1 shRNA cells (Figure 5B).
Fig. 4.
Bcr-Abl-induced leukemogenesis was impaired by Abi1 knockdown. (A) Stable knock down of Abi1 expression in p185wt cells transduced with Abi1 shRNA. The Ba/F3 cells and the p185wt cells transduced with a non-silencing shRNA or Abi1 shRNA were grown in vitro for indicated time. Total lysates from 1 × 106 cells were separated on sodium dodecyl sulfate–polyacrylamide gel electrophoresis, transferred to nitrocellulose membrane and subjected to western blot analysis using indicated antibodies. (B) Survival of the mice injected with Ba/F3 cells, control p185wt cells and the p185wt cells expressing Abi1 shRNA.
Table I.
Summary of the disease development in mice injected with p185wt/BaF3 cells and the p185wt/BaF3 cells expressing Abi1 shRNA
| Mouse | Latencya (days) | Spleen weight (g) | Liver weight (g) |
| Ba/F3 injected | |||
| A1 | >90b | 0.04 | 1.18 |
| A2 | >90b | 0.05 | 1.35 |
| A3 | >90b | 0.05 | 1.51 |
| A4 | >90b | 0.05 | 1.36 |
| A5 | >90b | 0.06 | 1.45 |
| A6 | >90b | 0.05 | 1.73 |
| p185wt injected | |||
| A1 | 18 | —c | —c |
| A2 | 20 | 0.16 | 3.00 |
| A3 | 20 | 0.18 | 2.98 |
| A4 | 20 | 0.16 | 3.39 |
| B1 | 13 | —c | —c |
| B2 | 14 | —c | —c |
| B3 | 16 | 0.21 | 3.24 |
| B4 | 16 | 0.19 | 2.88 |
| B5 | 16 | 0.18 | 3.29 |
| B6 | 19 | —c | —c |
| Abi1 shRNA injected | |||
| A1 | 32 | —c | —c |
| A2 | 34 | 0.03 | 1.89 |
| A3 | 37 | 0.04 | 1.23 |
| A4 | 37 | 0.06 | 1.15 |
| A5 | 37 | 0.05 | 1.70 |
| A6 | 37 | 0.06 | 1.76 |
| A7 | 37 | 0.07 | 1.63 |
Latency is defined as the time after injection that mice died or become moribund.
The mice injected with Ba/F3 cells survived >90 days without any disease observed.
The mice dead prior to the pathology analysis.
Fig. 5.
Pathology analysis of the mice injected with Ba/F3 cells, control p185wt cells and p185wt cells transduced with Abi1 shRNA. (A) Spleen weight of mice injected with Ba/F3 cells (control) and the p185wt cells expressing with (p185wt + Abi1 shRNA) or without (p185wt) Abi1 shRNA. (B) Histology of spleens and livers from the mice receiving Ba/F3 cells (control) and the p185wt cells expressing with (p185wt + Abi1 shRNA) or without (p185wt ctrl) Abi1 shRNA. Arrowheads indicate the massively expanded p185wt cells.
Abi1 knockdown impedes in vivo competitive expansion of Bcr-Abl-transformed cells
Although the mice received p185wt Abi1 shRNA cells showed longer survival as compared with those received p185wt control cells, they eventually developed disease and became moribund ∼5 weeks after injection. Because the MSCV vector used for expression of control or Abi1 shRNA also contains the gene that confers puromycin resistance, we were able to recover the transduced cells from the diseased mice. The cells capable of growing in IL3-free medium containing puromycin were readily recovered from the peripheral blood, bone marrow and spleen of the diseased mice received p185wt control cells. These cells became predominant in 3–5 days in culture under puromycin selection and displayed the morphology similar to that of the p185wt control cells grown in vitro. Similarly, the puromycin-resistant cells were readily recovered from the peripheral blood and bone marrow of the diseased mice injected with p185wt Abi1 shRNA cells. With a prolonged culture period (7–10 days), the puromycin-resistant cells were also recovered from the spleen of some diseased mice injected with p185wt Abi1 shRNA cells. In contrast, no puromycin-resistant cells were recovered from the mice injected with control BaF3 cells.
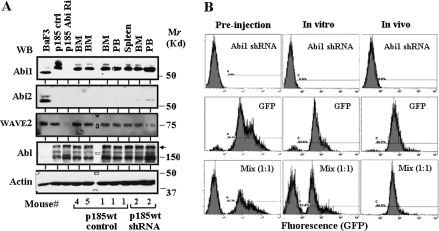
Western blot analysis was performed to compare the expression of Abi1, Abi2 and Bcr-Abl among cultured Ba/F3, p185wt control and p185wt Abi1 shRNA cells, as well as the cells recovered from the diseased mice. As expected, the puromycin-resistant cells recovered from the diseased mice express Bcr-Abl (Figure 6A). Expression of Abi2 was greatly reduced in these cells as compared with cultured Ba/F3 cells (Figure 6A), possibly due to the Bcr-Abl-induced Abi2 degradation (38). Like p185wt control cells cultured in vitro, the cells recovered from the diseased p185wt mice expressed Abi1 (Figure 6A). Interestingly, Abi1 was also detected in the cells recovered from the diseased mice injected with p185wt Abi1 shRNA (Figure 6A). This is different from the p185wt Abi1 shRNA cells cultured in vitro, in which no Abi1 expression was detected even after the cells grew in vitro for >5 weeks (Figures 4A and 6A). This result may be explained by a selective expansion occurred in vivo for those cells that regained the expression of Abi1. Thus, the data suggest that the Abi1 knockdown may impede in vivo competitive expansion of the p185wt Abi1 shRNA cells. To test this, we transduced p185wt cells with a retroviral vector expressing GFP. The GFP-positive p185wt cells (p185wt GFP cells) were sorted by fluorescence-activated cell sorting to a purity of >95% (Figure 6B, middle panel in column preinjection). These cells remain GFP positive after they grew in vitro for >30 days (Figure 6B, middle panel in column in vitro). We mixed p185wt GFP cells with p185wt Abi1 shRNA cells at 1:1 ratio (Figure 6B, lower panel in column preinjection) and injected them into recipient mice. We also injected either the p185wt Abi1 shRNA or p185wt GFP cells alone into the recipient mice. The puromycin-resistant cells were recovered from the diseased mice and analyzed by flow cytometry. As shown in Figure 6B, all cells recovered from the mice injected with p185wt Abi1 shRNA cells alone were GFP negative (upper panel in column in vivo), whereas >95% of the cells recovered from the mice injected with p185wt GFP cells alone were GFP positive (middle panel in column in vivo). Remarkably, the cells recovered from the mice injected with the mixed cells were mostly, if not all, GFP positive (Figure 6B, lower panel in column in vivo). However, when grown in vitro for the same time period, the mixed cells remain at a ratio of 1:1 (Figure 6B, lower panel in column in vitro). These results suggest that, although there is no growth advantage of p185wt GFP cells over p185wt Abi1 shRNA cells in vitro under the culture condition we used, p185wt GFP cells exhibited growth advantage over p185wt Abi1 shRNA cells in vivo. Thus, the data support the notion that the knock down of Abi1 expression impedes in vivo competitive expansion of the p185wt Abi1 shRNA cells.
Fig. 6.
Abi1 knockdown impeded in vivo competitive expansion of p185wt cells. (A) Expression of Abi1 in Ba/F3 cells, p185wt cells transduced with non-silencing shRNA (p185wt control) or Abi1 shRNA (p185wt Abi1 Ri), as well as the cells rescued from bone marrow (BM), peripheral blood (PB) and spleen of the diseased mice injected with p185wt cells transduced with either non-silencing shRNA or Abi1 shRNA. The 100 μg total proteins from these cells were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, transferred to nitrocellulose membrane and western blotted (WB) with indicated antibodies. Molecular markers are indicated. The arrow indicates p185Bcr-Abl. (B) In vivo competitive expansion of p185wt GFP cells and p185wt Abi1 shRNA cells. The p185wt cells were transduced with MSCV-based retroviruses expressing either GFP or Abi1 shRNA. The GFP-positive p185wt cells were sorted by fluorescence-activated cell sorting to a purity of >95%. The GFP-positive cells then were mixed with p185wt cells expressing Abi1 shRNA at a ratio of 1:1. The mixed cells were either cultured in vitro for 5 weeks or injected into Balb/c mice (1 × 106 cells per mouse) through tail vein. The cells derived from p185wt cells were rescued from bone marrow and peripheral blood (data not shown) of the diseased mice by selection with puromycin. The rescued cells and the cells cultured in vitro were subjected to flow cytometry analysis.
Abi1 is required for Bcr-Abl-induced activation of Lyn kinases
Abi1 has been shown to play a role in signal transduction trigged by growth factors (26,27,32,36). It has been widely accepted that Src family kinases are critical players that act downstream of growth factor/cytokine receptors to stimulate cell growth, survival and homing. To determine the mechanism by which Abi1 contributes to Bcr-Abl-induced leukemogenesis, we examined the effect of Abi1 knockdown on activation of Lyn, a member of Src family kinases that is expressed in hematopoietic cells and required for Bcr-Abl-induced leukemogenesis (46). Lyn is mostly inactive in starved Ba/F3 cells but its activity is increased upon IL3 stimulation, as revealed by increased level of phosphorylated tyrosine 416 (Figure 7, compare lane 1 with lane 4). In p185wt-transformed Ba/F3 cells, Lyn is constitutively activated (Figure 7, lanes 2 and 5). Knock down of the Abi1 expression in p185wt cells resulted in a reduction of Lyn activation (Figure 7, compare lane 3 with lane 2). Moreover, while the IL3 is a potent stimulator of Lyn activity in Ba/F3 cells, it failed to stimulate Lyn activation in p185wt Abi1 shRNA cells (Figure 7A, compare lane 6 with lane 3).
Fig. 7.
The knock down of Abi1 expression attenuated Bcr-Abl-induced activation of Lyn kinases. Ba/F3 cells (lanes 1 and 4), control p185wt cells (lanes 2 and 5) and p185wt Abi1 shRNA cells (lanes 3 and 6) were starved in RPMI 1640 plus 0.1% bovine serum albumin for 6 h. The cells were then treated without (lanes 1–3) or with (lanes 4–6) recombinant rat IL3 (20 ng/ml) for 10 min. The lysates (2 × 107 cells) from indicated cell lines were immunoprecipitated (IP) with anti-Lyn antibodies, separated on 8% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and subjected to western blot (WB) analysis using anti-Src p416 antibody, which recognizes activated Lyn (upper panel). The membrane then was stripped and reprobed with anti-Lyn antibodies (lower panel). Two Lyn isoforms, p53Lyn and p56Lyn, were indicated.
Discussion
To examine the role of Abi1 in Bcr-Abl-induced leukemogenesis, we knocked down its expression in p185wt Ba/F3 cells by shRNA-mediated gene silencing. The messenger RNA and protein levels of Abi1 were decreased by at least 90% in p185wt cells transduced with Abi1 shRNA. In these cells, the expression of Abi2 is also dramatically downregulated due to Bcr-Abl-induced activation of proteolytic pathways (38). Furthermore, the expression of Abi3 (also known as new molecule including SH3) in these cells is relatively low as determined by DNA microarray analysis (Weidong Yu and Zonghan Dai, unpublished data). Thus, the p185wt cells transduced with Abi1 shRNA may provide a simplified system for analysis of the role of Abi pathway in Bcr-Abl-induced leukemogenesis. Knock down of Abi1 expression did not affect tyrosine kinase activity of Bcr-Abl, nor did it have any effect on Bcr-Abl-induced IL3-independent growth in vitro. However, Abi1 knockdown inhibited Bcr-Abl-stimulated actin cytoskeleton remodeling, MT1-MMP clustering as well as cell adhesion and migration in vitro. More importantly, knock down of the expression of Abi1 impaired Bcr-Abl-induced leukemogenesis in vivo. Remarkably, the mice receiving p185wt Abi1 shRNA cells did not develop splenomegaly, which is a common pathologic characteristic of Bcr-Abl-induced leukemogenesis. These results are consistent with our previous report shown that p185ΔSH3ΔC, a mutant Bcr-Abl defective in signaling to Abi pathway, failed to stimulate cell migration in vitro and to induce chronic myelogenous leukemia-like disease in vivo (39). Together, these studies highlight the importance of Abi1 in Bcr-Abl-induced leukemogenesis.
Abi1 plays a key role in regulation of actin polymerization, a fundamental cellular process that controls cell adhesion and motility. In cultured fibroblast cells and melanoma cells, blockade of Abi1 pathway abrogated growth factor-stimulated membrane ruffling and the formation of lamellipodia (36,47,48). In p185wt-transformed Ba/F3 cells, Abi1 is tyrosine phosphorylated and translocated to a site adjacent to membrane where an F-actin-enriched structure was assembled (40). More recent studies have shown that this F-actin-enriched structure shares some similarities with invadopodium, a specialized adhesive/invasive structure found in metastatic breast cancer and melanoma cells (49,50). Like invadopodium, the F-actin-enriched structures found in p185wt cells are associated with the membrane and enriched with multiple structural/regulatory proteins involved in the regulation of actin cytoskeleton assembly, cell adhesion and extracellular matrix degradation (40,41). It is widely believed that invadopodia play a crucial role in tumor cell invasion and migration (50,51). Knock down of Abi1 in p185wt cells significantly inhibited the formation of invadopodia-like structure and clustering of MT1-MMP (41). Interestingly, the decreased formation of the invadopodia-like structures as a consequence of Abi1 knockdown correlated with a decrease of cell adhesion and migration on fibronectin-coated surfaces. Given the importance of the cytoskeletal functions and MT1-MMP activities in control of hematopoietic cell invasion and homing, it is conceivable that defects in Abi1-mediated cytoskeletal functions and MT1-MMP clustering in p185wt Abi1 shRNA cells may account in part for the failure of these cells to develop splenomegaly in the mice.
Although the knock down of Abi1 expression did not affect the IL3-independent growth of p185wt cells in vitro, it impeded the in vivo competitive expansion of these cells. In addition to the regulation of actin polymerization, Abi1 is also involved in multiple signaling pathways important for cell growth. It has been shown that Abi1 is capable of binding to a variety of signaling molecules involved in control of cell proliferation, apoptosis and cytoskeletal functions. These include c-Abl (16), Abl-related gene product (24), epidermal growth factor receptor pathway substrate 8 (23), cytoskeleton protein spectrin (25), guanine nucleotide exchange factors Sos (26,27) and Pix (33), adaptor protein Grb4 (28), membrane metalloproteinase ADAM19 (35), PI3 kinase p85 subunit (36), reduced nicotinamide dinucleotide phosphate (NADPH) oxidase adapter protein p47 (30), cyclin-dependent kinase cdc2 (29), p21-activated kinase Pak2 (31,34) and E3 ubiquitin ligase cbl (32). The ability to interact with multiple signaling molecules places Abi1 at a position that may link signals from membrane receptors to intracellular networks. It is likely that the knock down of Abi1 may impair these signaling pathways and therefore confer p185wt Abi1 shRNA cell disadvantage in an in vivo environment where the cells are exposed to numerous factors including cytokines, growth factors and extracellular matrix. Indeed, it has been reported that in addition to the regulation of cytoskeleton remodeling, the Abi–WAVE pathway is also involved in T cell receptor-mediated cell proliferation (52). Consistently, we found that at the presence of IL3, the p185wt Abi1 shRNA cells grew slower in vitro as compared with control p185wt cells (data not shown). In an effort to determine the molecular mechanism by which Abi1 contributes to Bcr-Abl-induced leukemogenesis, we found that the knock down of Abi1 expression attenuates Bcr-Abl-stimulated activation of Lyn kinases. Given the importance of Lyn kinases in regulation of hematopoietic cell growth, survival and homing, our findings may provide an explanation why the Abi1 knockdown impeded in vivo expansion of p185wt Abi1 shRNA cells. More importantly, because recent studies suggest that Lyn kinases may play a key role in leukemia development (46,53), our observation that Abi1 is involved in abnormal activation of Lyn kinases in Bcr-Abl-positive leukemic cells may provide insight into the development of novel therapeutic strategies for treatment of human leukemia.
Funding
National Institutes of Health/National Cancer Institute (R01 CA094921) to Z.D.; National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases (K01 DK067191) to Y.T.
Acknowledgments
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- Abi
Abl interactor
- F-actin
filament actin
- FBS
fetal bovine serum
- MT1-MMP
membrane-type 1 metalloproteinase
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- shRNA
short hairpin RNA
- WAVE
WASP-family verprolin-homologous
References
- 1.Druker BJ, et al. Chronic myelogenous leukemia. Hematology Am Soc Hematol Educ Program. 2001:87–112. doi: 10.1182/asheducation-2001.1.87. [DOI] [PubMed] [Google Scholar]
- 2.Melo JV, et al. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat. Rev. Cancer. 2007;7:441–453. doi: 10.1038/nrc2147. [DOI] [PubMed] [Google Scholar]
- 3.Hehlmann R, et al. Chronic myeloid leukaemia. Lancet. 2007;370:342–350. doi: 10.1016/S0140-6736(07)61165-9. [DOI] [PubMed] [Google Scholar]
- 4.Ren R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat. Rev. Cancer. 2005;5:172–183. doi: 10.1038/nrc1567. [DOI] [PubMed] [Google Scholar]
- 5.Van Etten RA. Oncogenic signaling: new insights and controversies from chronic myeloid leukemia. J. Exp. Med. 2007;204:461–465. doi: 10.1084/jem.20062335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verfaillie CM, et al. Mechanisms underlying abnormal trafficking of malignant progenitors in chronic myelogenous leukemia. Decreased adhesion to stroma and fibronectin but increased adhesion to the basement membrane components laminin and collagen type IV. J. Clin. Invest. 1992;90:1232–1241. doi: 10.1172/JCI115985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skorski T, et al. The SH3 domain contributes to BCR/ABL-dependent leukemogenesis in vivo: role in adhesion, invasion, and homing. Blood. 1998;91:406–418. [PubMed] [Google Scholar]
- 8.McWhirter JR, et al. Effect of Bcr sequences on the cellular function of the Bcr-Abl oncoprotein. Oncogene. 1997;15:1625–1634. doi: 10.1038/sj.onc.1201342. [DOI] [PubMed] [Google Scholar]
- 9.Salgia R, et al. BCR/ABL induces multiple abnormalities of cytoskeletal function. J. Clin. Invest. 1997;100:46–57. doi: 10.1172/JCI119520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salesse S, et al. Mechanisms underlying abnormal trafficking and expansion of malignant progenitors in CML: BCR/ABL-induced defects in integrin function in CML. Oncogene. 2002;21:8605–8611. doi: 10.1038/sj.onc.1206088. [DOI] [PubMed] [Google Scholar]
- 11.Salgia R, et al. The BCR/ABL oncogene alters the chemotactic response to stromal-derived factor-1alpha. Blood. 1999;94:4233–4246. [PubMed] [Google Scholar]
- 12.Salgia R, et al. CRKL links p210BCR/ABL with paxillin in chronic myelogenous leukemia cells. J. Biol. Chem. 1995;270:29145–29150. doi: 10.1074/jbc.270.49.29145. [DOI] [PubMed] [Google Scholar]
- 13.Sattler M, et al. Differential signaling after beta1 integrin ligation is mediated through binding of CRKL to p120(CBL) and p110(HEF1) J. Biol. Chem. 1997;272:14320–14326. doi: 10.1074/jbc.272.22.14320. [DOI] [PubMed] [Google Scholar]
- 14.Skourides PA, et al. Polarized distribution of Bcr-Abl in migrating myeloid cells and co-localization of Bcr-Abl and its target proteins. Oncogene. 1999;18:1165–1176. doi: 10.1038/sj.onc.1202407. [DOI] [PubMed] [Google Scholar]
- 15.Dai Z, et al. Abi-2, a novel SH3-containing protein interacts with the c-Abl tyrosine kinase and modulates c-Abl transforming activity. Genes Dev. 1995;9:2569–2582. doi: 10.1101/gad.9.21.2569. [DOI] [PubMed] [Google Scholar]
- 16.Shi Y, et al. Abl-interactor-1, a novel SH3 protein binding to the carboxy-terminal portion of the Abl protein, suppresses v-abl transforming activity. Genes Dev. 1995;9:2583–2597. doi: 10.1101/gad.9.21.2583. [DOI] [PubMed] [Google Scholar]
- 17.Eden S, et al. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature. 2002;418:790–793. doi: 10.1038/nature00859. [DOI] [PubMed] [Google Scholar]
- 18.Innocenti M, et al. Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nat. Cell Biol. 2004;6:319–327. doi: 10.1038/ncb1105. [DOI] [PubMed] [Google Scholar]
- 19.Miyazaki K, et al. Isolation and characterization of a novel human gene (NESH) which encodes a putative signaling molecule similar to e3B1 protein. Biochim. Biophys. Acta. 2000;1493:237–241. doi: 10.1016/s0167-4781(00)00158-5. [DOI] [PubMed] [Google Scholar]
- 20.Kunda P, et al. Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr. Biol. 2003;13:1867–1875. doi: 10.1016/j.cub.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 21.Steffen A, et al. Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. EMBO J. 2004;23:749–759. doi: 10.1038/sj.emboj.7600084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gautreau A, et al. Purification and architecture of the ubiquitous Wave complex. Proc. Natl Acad. Sci. USA. 2004;101:4379–4383. doi: 10.1073/pnas.0400628101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biesova Z, et al. Isolation and characterization of e3B1, an eps8 binding protein that regulates cell growth. Oncogene. 1997;14:233–241. doi: 10.1038/sj.onc.1200822. [DOI] [PubMed] [Google Scholar]
- 24.Wang B, et al. ArgBP2, a multiple Src homology 3 domain-containing, Arg/Abl-interacting protein, is phosphorylated in v-Abl-transformed cells and localized in stress fibers and cardiocyte Z-disks. J. Biol. Chem. 1997;272:17542–17550. doi: 10.1074/jbc.272.28.17542. [DOI] [PubMed] [Google Scholar]
- 25.Ziemnicka-Kotula D, et al. Identification of a candidate human spectrin Src homology 3 domain-binding protein suggests a general mechanism of association of tyrosine kinases with the spectrin-based membrane skeleton. J. Biol. Chem. 1998;273:13681–13692. doi: 10.1074/jbc.273.22.13681. [DOI] [PubMed] [Google Scholar]
- 26.Scita G, et al. EPS8 and E3B1 transduce signals from Ras to Rac. Nature. 1999;401:290–293. doi: 10.1038/45822. [DOI] [PubMed] [Google Scholar]
- 27.Fan PD, et al. Abl interactor 1 binds to sos and inhibits epidermal growth factor- and v-Abl-induced activation of extracellular signal-regulated kinases. Mol. Cell. Biol. 2000;20:7591–7601. doi: 10.1128/mcb.20.20.7591-7601.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cowan CA, et al. The SH2/SH3 adaptor Grb4 transduces B-ephrin reverse signals. Nature. 2001;413:174–179. doi: 10.1038/35093123. [DOI] [PubMed] [Google Scholar]
- 29.Lin TY, et al. Abi enhances Abl-mediated CDC2 phosphorylation and inactivation. J. Biomed. Sci. 2004;11:902–910. doi: 10.1007/BF02254375. [DOI] [PubMed] [Google Scholar]
- 30.Gu Y, et al. Induction of colonic epithelial cell apoptosis by p47-dependent oxidants. FEBS Lett. 2003;540:195–200. doi: 10.1016/s0014-5793(03)00262-x. [DOI] [PubMed] [Google Scholar]
- 31.Machuy N, et al. c-Abl-binding protein interacts with p21-activated kinase 2 (PAK-2) to regulate PDGF-induced membrane ruffles. J. Mol. Biol. 2007;370:620–632. doi: 10.1016/j.jmb.2007.04.080. [DOI] [PubMed] [Google Scholar]
- 32.Tanos BE, et al. Abi-1 forms an epidermal growth factor-inducible complex with Cbl: role in receptor endocytosis. Cell. Signal. 2007;19:1602–1609. doi: 10.1016/j.cellsig.2007.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Campa F, et al. A new interaction between Abi-1 and betaPIX involved in PDGF-activated actin cytoskeleton reorganisation. Cell Res. 2006;16:759–770. doi: 10.1038/sj.cr.7310091. [DOI] [PubMed] [Google Scholar]
- 34.Chi S, et al. Pak regulates calpain-dependent degradation of E3b1. Biochem. Biophys. Res. Commun. 2004;319:683–689. doi: 10.1016/j.bbrc.2004.05.042. [DOI] [PubMed] [Google Scholar]
- 35.Huang L, et al. Screen and identification of proteins interacting with ADAM19 cytoplasmic tail. Mol. Biol. Rep. 2002;29:317–323. doi: 10.1023/a:1020409217215. [DOI] [PubMed] [Google Scholar]
- 36.Innocenti M, et al. Phosphoinositide 3-kinase activates Rac by entering in a complex with Eps8, Abi1, and Sos-1. J. Cell Biol. 2003;160:17–23. doi: 10.1083/jcb.200206079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Innocenti M, et al. Abi1 regulates the activity of N-WASP and WAVE in distinct actin-based processes. Nat. Cell Biol. 2005;7:969–976. doi: 10.1038/ncb1304. [DOI] [PubMed] [Google Scholar]
- 38.Dai Z, et al. Oncogenic Abl and Src tyrosine kinases elicit the ubiquitin-dependent degradation of target proteins through a Ras-independent pathway. Genes Dev. 1998;12:1415–1424. doi: 10.1101/gad.12.10.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dai Z, et al. Deletion of the Src homology 3 domain and C-terminal proline-rich sequences in Bcr-Abl prevents Abl interactor 2 degradation and spontaneous cell migration and impairs leukemogenesis. J. Biol. Chem. 2001;276:28954–28960. doi: 10.1074/jbc.M101170200. [DOI] [PubMed] [Google Scholar]
- 40.Li Y, et al. Bcr-Abl induces abnormal cytoskeleton remodeling, beta1 integrin clustering and increased cell adhesion to fibronectin through the Abl interactor 1 pathway. J. Cell Sci. 2007;120:1436–1446. doi: 10.1242/jcs.03430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun X, et al. MT1-MMP as a downstream target of BCR-ABL/ABL interactor 1 signaling: polarized distribution and involvement in BCR-ABL-stimulated leukemic cell migration. Leukemia. 2007 doi: 10.1038/sj.leu.2404990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Courtney KD, et al. Localization and phosphorylation of Abl-interactor proteins, Abi-1 and Abi-2, in the developing nervous system. Mol. Cell. Neurosci. 2000;16:244–257. doi: 10.1006/mcne.2000.0865. [DOI] [PubMed] [Google Scholar]
- 43.Echarri A, et al. Abl interactor 1 (Abi-1) wave-binding and SNARE domains regulate its nucleocytoplasmic shuttling, lamellipodium localization, and wave-1 levels. Mol. Cell. Biol. 2004;24:4979–4993. doi: 10.1128/MCB.24.11.4979-4993.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Juang JL, et al. Drosophila abelson interacting protein (dAbi) is a positive regulator of abelson tyrosine kinase activity. Oncogene. 1999;18:5138–5147. doi: 10.1038/sj.onc.1202911. [DOI] [PubMed] [Google Scholar]
- 45.Tani K, et al. Abl interactor 1 promotes tyrosine 296 phosphorylation of mammalian enabled (Mena) by c-Abl kinase. J. Biol. Chem. 2003;278:21685–21692. doi: 10.1074/jbc.M301447200. [DOI] [PubMed] [Google Scholar]
- 46.Hu Y, et al. Requirement of Src kinases Lyn, Hck and Fgr for BCR-ABL1-induced B-lymphoblastic leukemia but not chronic myeloid leukemia. Nat. Genet. 2004;36:453–461. doi: 10.1038/ng1343. [DOI] [PubMed] [Google Scholar]
- 47.Innocenti M, et al. Mechanisms through which Sos-1 coordinates the activation of Ras and Rac. J. Cell Biol. 2002;156:125–136. doi: 10.1083/jcb.200108035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leng Y, et al. Abelson-interactor-1 promotes WAVE2 membrane translocation and Abelson-mediated tyrosine phosphorylation required for WAVE2 activation. Proc. Natl Acad. Sci. USA. 2005;102:1098–1103. doi: 10.1073/pnas.0409120102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen WT, et al. Membrane proteases as potential diagnostic and therapeutic targets for breast malignancy. Breast Cancer Res. Treat. 1994;31:217–226. doi: 10.1007/BF00666155. [DOI] [PubMed] [Google Scholar]
- 50.Courtneidge SA, et al. The SRC substrate Tks5, podosomes (invadopodia), and cancer cell invasion. Cold Spring Harb. Symp. Quant. Biol. 2005;70:167–171. doi: 10.1101/sqb.2005.70.014. [DOI] [PubMed] [Google Scholar]
- 51.Linder S. The matrix corroded: podosomes and invadopodia in extracellular matrix degradation. Trends Cell Biol. 2007;17:107–117. doi: 10.1016/j.tcb.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 52.Zipfel PA, et al. Role for the Abi/wave protein complex in T cell receptor-mediated proliferation and cytoskeletal remodeling. Curr. Biol. 2006;16:35–46. doi: 10.1016/j.cub.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 53.Dos Santos C, et al. A critical role for Lyn in acute myeloid leukemia. Blood. 2008;111:2269–2279. doi: 10.1182/blood-2007-04-082099. [DOI] [PubMed] [Google Scholar]