Abstract
Endonuclease V (endo V) recognizes and cleaves deoxyinosine in deaminated DNA. These enzymatic activities are precursors of DNA repair and are fueled by metal ions such as Ca2+ and Mg2+, with the former being associated with protein binding and the latter with DNA cleavage. Using the technique of fluorescence resonance energy transfer (FRET) we determined the single-molecule kinetics of endo V in a catalytic cycle using a substrate of deoxyinosine-containing single-stranded DNA (ssDNA). The ssDNA was labeled with TAMRA, a fluorescence donor, while the endo V was labeled with Cy5, a fluorescence acceptor. The time lapses of FRET, resulting from the sequential association, recognition, and dissociation of the deoxyinosine by the endo V, were determined at 5.9 s, 14.5 s, and 9.1 s, respectively, in the presence of Mg2+. In contrast, the process of deoxyinosine recognition appeared little affected by the metal type. The prolonged association and dissociation events in the presence of the Ca2+-Mg2+ combination, as compared to that of Mg2+ alone, support the hypothesis that endo V has two metal binding sites to regulate its enzymatic activities.
Our genetic blueprint is under constant assault from adverse environmental and cellular influences (1, 2). These attacks may alter the physical and chemical properties of DNA by introducing pyrimidine dimers, base-pair mismatches, double-strand breakages, and miscellaneous forms of DNA base modifications such as deamination, in which cytosine, adenine, and guanine are converted to uracil, hypoxanthine, xanthine, and oxanine, respectively (3, 4). Fortunately these abnormalities of DNA are largely recognized and timely counteracted in vivo by a range of enzymes and proteins to prevent mutagenesis and cancer (5). Naturally, the study of DNA damage and repair marks one of the most fundamental research areas in molecular biology. However, the mechanisms of DNA damage and repair are not yet well understood and the corresponding biochemical studies at the single-molecule level are lacking (6-10).
Endo V is an enzyme involved in the repair of deaminated DNA by hydrolyzing its second phosphodiester bond 3′ from the base lesion (11-16). The ensemble kinetics of endo V indicates that it is a single turnover enzyme that retains tight binding to both deoxyinosine-containing double-stranded DNA (dsDNA) and their corresponding nicked products (13, 17). It has been proposed that this single turnover behavior may help endo V recruit downstream protein(s) to facilitate additional lesion removal (13, 18). Mutational analyses have defined amino acid residues in conserved motifs that play important roles in substrate binding, catalysis, and metal coordination (18, 19). D43, E89, D110 and H214 in Thermotoga maritima endo V have been identified as active site residues that are involved in metal coordination (20). A biochemical analysis of DNA cleavage activities in the presence of Mn2+ has resulted in the hypothesis that endo V contains two metal binding sites, a high affinity site required for catalysis and a relatively low affinity site involved in the regulation of non-specific activities (20). Biochemical analysis, using a different combination of metal ions, has substantiated the validity of this catalytic and regulatory two-metal model (20).
Single-molecule techniques have become increasingly important tools for the elucidation of the structural, functional, and mechanochemical properties of biomolecules and their complexes (21-29). These techniques can probe the highly localized nano-environments of biological systems and extract the dynamics of single molecules and their interactions (24). Utilizing the technique of FRET, the so-called “molecular ruler” owing to its nanometric sensitivity (24, 25), we have performed a first single-molecule study of lesion recognition and cleavage by endo V, as well as metal ion dependence in these processes. Our approach was compared with previous biochemistry studies to offer new information on the functional and kinetic features of endo V in DNA damage recognition and repair.
Materials and Methods
Wild type (wt) Thermotoga maritima endo V was purified as previously described (13, 18). To label endo V which contains two unconserved cysteine residues, 0.25 mg of Cy5-maleimide (Amersham) was dissolved in 120 μL of dimethyl sulfoxide and incubated with 250 μL of 50 μM endo V for 2 h at room temperature. The reaction mixture was then passed through a Microcon YM-3 spin column (Millipore) and washed with TE buffer (pH 7.5) to remove unreacted Cy5-maleimide. The labeled endo V was concentrated to about 80 μL in TE buffer, frozen in liquid nitrogen and stored at -80°C prior to use. The single-stranded deoxyoligonucleotide substrate (5′-Biotin-CC GCA ACA AGA CTA GAG GAT CAA ACT ATG ACA ACT IAC GC-TAMRA-3′; I, deoxyinosine) and its complementary strand (5′--TA CCC CAG CGT CTG CGG TGT TGC GTC AGT TGT CAT AGT TTG ATC CTC TAG TCT TGT TGC GGG TTC C-3′) were synthesized and HPLC purified by Integrated DNA Technologies. To obtain dsDNA the two complementary oligo strands of 1:1 ratio were hybridized at 85°C for 3 min and gradually cooled to room temperature. During the experiment, the biotin-oligonucleotide was first immobilized via the 5′-biotin bound to the streptavidin on a quartz slide. As endo V began to bind to the deoxyinosine site, FRET was induced between the TAMRA and Cy5, as illustrated in Figure 1. The cleavage of the ssDNA by endo V occurred at the site one nucleotide 3′ to the lesion.
FIGURE 1.
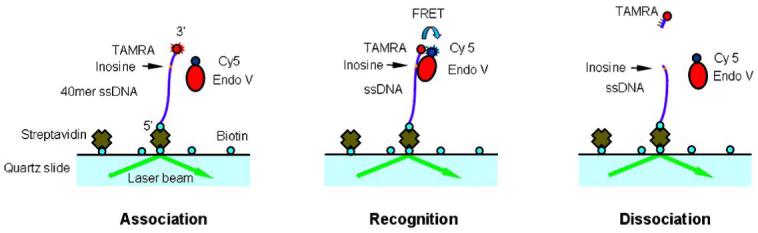
Schematic diagram of the FRET system configuration. A 40 mer ssDNA is anchored to the quartz via 5′ biotin-streptavidin bonding. A deoxyinosine site is represented in yellow and is 4 nt away from the 3′ of the ssDNA. FRET is induced when the deoxyinosine site is recognized by the endo V. The dissociation of the deoxyinosine quenches the fluorescence from both the donor TAMRA and the acceptor Cy5.
A quartz slide was immersed in a 2% Micro90 laboratory cleaning solution and incubated in a sonicator for 10 min. After a thorough wash with deionized water (Millipore), the slide was baked in a kiln at 420°C for 2 h. A double-sided tape was used to create a flow channel between the quartz slide and a coverslip. A 10 μL mixture of biotin-BSA (1 μg/μL in a buffer containing 50 mM NaCl and 10 mM Tris at pH 8.0) was added to the channel and incubated for 10 min. The mixture was washed with buffer STE (10 mM Tris, 50 mM NaCl, and 1 mM EDTA), and a 10 μL of streptavidin (0.2 μg/μL in buffer STE) was then added to the channel, left to incubate for 10 min, and again washed thoroughly with buffer STE.
Two μL of endo V (1 nM), 2 μL of oligonucleotides (ssDNA or dsDNA) (0.1 nM), 2 μL of dI buffer (10 mM HEPES, pH 7.4; 1 mM DTT; 2% glycerol), and 14 μL of H2O were combined in a vial and incubated at 65°C for 30 min. Propyl gallate (4% in glycerol) of 0.1 μL, an oxygen scavenger, was added to the vial to prevent photobleaching of the fluorophores. After cooling for 5 min, 10 μL of the reaction mixture was injected using a 10 μL syringe from the side of the prepared slide sandwich. The slide was left to incubate for 10 min to immobilize the DNA and again washed with buffer STE. Mg2+ (5 mM) was added to the sample channel immediately prior to imaging.
Our measurements were taken at room temperature with a prism-type total-internal-reflection microscope (Figure 1). The sample quartz slide was mounted on the prism via immersion oil. A Nd:YAG laser (532 nm) was used to excite the TAMRA attached to the immobilized oligonucleotides. The donor and acceptor emissions, collected with a 60× water objective (Olympus, NA = 1.2), were split into two optical channels and recombined onto a CCD camera (Roper, Cascade 512B). A lateral misalignment between the two channels was intentionally created to permit their viewing simultaneously. An LS2Z2 Uniblitz shutter controlled by a VMMT1 timer (Vincent Associates) was used to extend the single-molecule fluorescence to approximately 30 min, a time span much longer than the time lapses studied. The rate of imaging was set at 2.0 frames/s with an exposure of 0.23 s for each frame. In our experiment, we first located the fluorescence spots of immobilized TAMRA-labeled DNA and adjusted our microscope focus prior to the addition of metal ions. This method maximized our observation of endo V-DNA interaction. The recognition and cleavage kinetics of endo V were obtained by analyzing the intensity traces against the frame rate in the acceptor channel, while monitoring the anti-correlation signal in the acceptor channel. Data analysis was performed using programs written in MatLab.
Results
A FRET system for single-molecule study of endo V
We established a single-pair FRET system by labeling both endo V and the 3′-end of a 40-nt deoxyinosine-containing single-stranded oligonucleotide. Endo V was labeled with Cy5-maleiimide via its two unconserved cysteine residues. The fluorescently labeled endo V was still found to be catalytically active as demonstrated by cleavage of inosine-containing DNA by our conventional cleavage assay (data not shown). A TAMRA moiety, coupled at the 3′-end of the ssDNA, did not seem to interfere with interactions of endo V as suggested by the gel mobility shift assay (data not shown). A schematic diagram of our system is illustrated in Figure 1. Fluorescence excitation of the TAMRA was induced by a laser beam incident at a quartz/water interface under conditions of total-internal-reflection. Metal ions were injected to the sample cell containing endo V and ssDNA immediately before the measurement. It was discovered that as endo V approached the ssDNA, the acceptor Cy5 on endo V emitted fluorescence excited by the energy transferred from the donor TAMRA within its close proximity (Figure 1). Once endo V cleaved at the second phosphodiester bond 3′ to the deoxyinosine, the three-nt-TAMRA fragment was expected to dissociate from the ssDNA, causing the reduction and eventual disappearance of the fluorescence signal in the acceptor channel (Figure 1).
Single-molecule reaction profiles
In the following we describe the recognition and cleavage kinetics of endo V, in the presence of metal ions Ca2+, Mg2+ or Ca2+-Mg2+ combination. Upon the inclusion of Ca2+ with endo V and the ssDNA substrate (case 1), an initial rise followed by an extension was observed for the acceptor signal (Figure 2A). The rise of the acceptor signal represents the association of endo V with the substrate, while the extension indicates the binding of endo V to the deoxyinosine site. Here, association is defined as specific binding of endo V to its lesioncontaining DNA substrate. This observation was consistent with previous ensemble studies (13) indicating that Ca2+ alone induces only deoxyinosine recognition and not cleavage. In the presence of Mg2+ (case 2), the measured reaction profile is characterized by a rise, an extension, and a fall of the acceptor signal (Figure 2B). The rise can be similarly explained by the specific association of endo V with the deoxyinosine site over a time lapse of T1. The extension (T2) is the step when the enzyme completes the recognition processes prior to strand cleavage. The fall (T3) represents the dissociation of the small 3′ TAMRA fragment from the Cy5-labeled enzyme. While the cleavage event is expected to occur near the end of T2 and in the beginning of T3 this event is not clearly discernable from our data. A reaction profile similar to that of Fig. 2B was obtained with Ca2+-Mg2+ combination (case 3). Approximately 50 ssDNA-endo V pairs were analyzed for each case.
FIGURE 2.
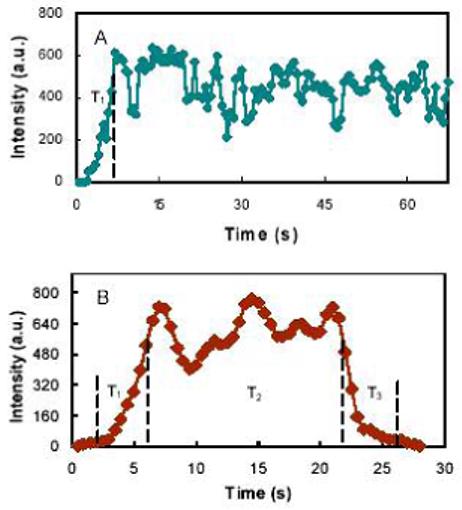
Exemplified time lapses of the association and recognition of deoxyinosine by endo V with various metal ions. The initial and ending points for each of the three time durations were taken from time-averaged signal and background noise. T1, T2, and T3 denoted association, recognition, and dissociation events. (A) with Ca2+. (B) with Mg2+.
Single-molecule reaction kinetics
Our case 1 assay was performed subsequent to the incubation of the sample of endo V and ssDNA with Ca2+ (5 mM), immediately prior to the measurement. As shown in Figure 3A (green curve), the rise of the acceptor signal, corresponding to the association of endo V with the deoxyinosine site, had a duration of approximately 4.9±1.8 s according to the Gaussian fitting curve. Here, the number after “±” represents the half-width at the half maximum of the fitted curve. The rise was followed by an extension of the acceptor signal for tens of minutes until it was subdued by photobleaching (Figure 2A). Like the previous biochemical studies, this result suggests that in the presence of Ca2+, endo V remained bound to the substrate once the deoxyinosine in a DNA sequence is recognized.
FIGURE 3.
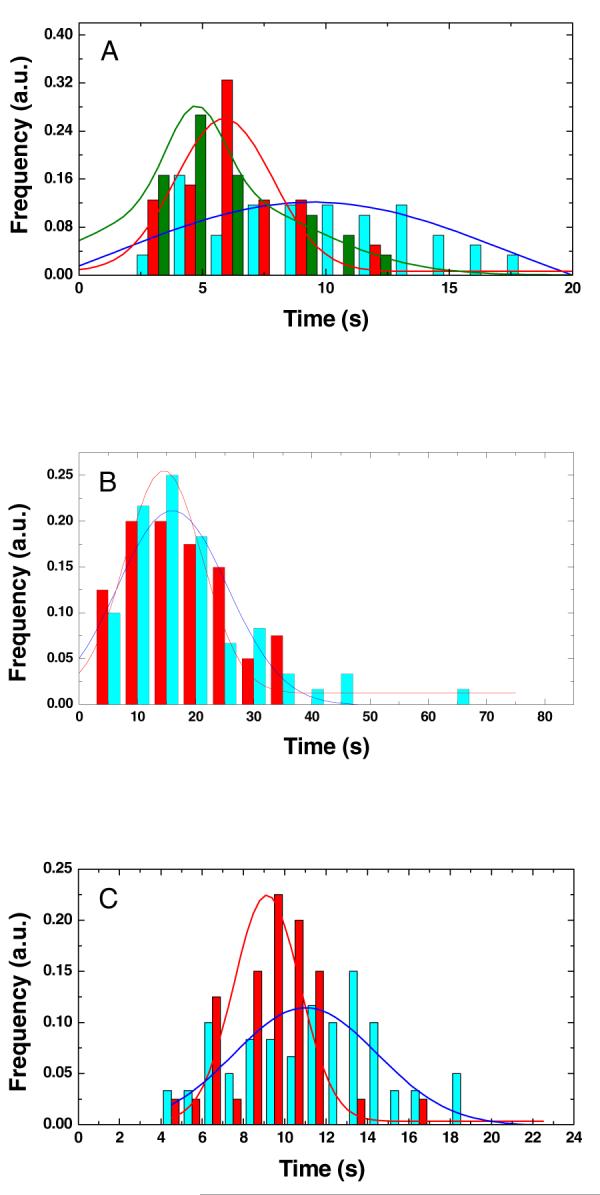
Time lapses with their Gaussian fitting curves for (A) association, (B) recognition, and (C) dissociation by endo V, activated by Ca2+ (case 1, green), Mg2+ (case 2, red), and Ca2+-Mg2+ (case 3, blue), respectively. The vertical axes are normalized in each case. The fitted time lapses T1 for the association events in (A) are 4.9±1.8 s, 5.9±2.0 s, and 9.5±7.9 s for cases 1, 2, and 3. The fitted time lapses T2 for the recognition events in (B) are 14.5±6.6 s and 16.0±9.5 s for cases 2 and 3. The fitted time lapses T3 for the dissociation events in (C) are 9.1±1.6 s and 11.0±3.4 s for cases 2 and 3.
The experimental procedures in case 2 are identical to those in case 1, except that Ca2+ was replaced with Mg2+ (5 mM). The time lapses of the acceptor signal in case 2 were fitted at approximately 5.9±2.0 s, 14.5±6.6 s, and 9.1±1.6 s (red curves in Figure 3), corresponding to the events of association, recognition, and dissociation of endo V, respectively. The entire catalytic cycle from endo V association with the deoxyinosine site to the dissociation of the small 3′ TAMRA fragment is approximately 29.5 s.
Case 3 (blue curves in Figure 3) denotes when the sample of endo V and deoxyinosine was first incubated with Ca2+ (5 mM) for 30 min and then refilled with Mg2+ (10 mM) immediately prior to the measurement. The final concentrations of Ca2+ and Mg2+ in the sample cell are estimated at 2.5 mM and 5 mM, respectively. The association of the deoxyinosine site with endo V in case 3 lasted approximately 9.5±7.9 s. Interestingly, the dissociation of deoxyinosine by endo V, when comparing case 3 and case 2, was somehow hindered by preincubation of the sample with Ca2+, by 2 s.
Single-stranded DNA versus double-stranded DNA
A comparison of the dsDNA and ssDNA binding patterns may facilitate our understanding of the structural roles of endo V in DNA damage and repair. Our previous studies (13, 19) found that the recognition of deoxyinosine by endo V was equally efficient for both dsDNA and ssDNA. A consistent result was obtained from our single-molecule study in the presence of Ca2+ alone, as shown in Figure 4 (green bars). However, we noticed a ∼27% increase of endo V efficiency in cleaving dsDNA than ssDNA, in the presence of Mg2+ (Figure 4, red bars).
FIGURE 4.
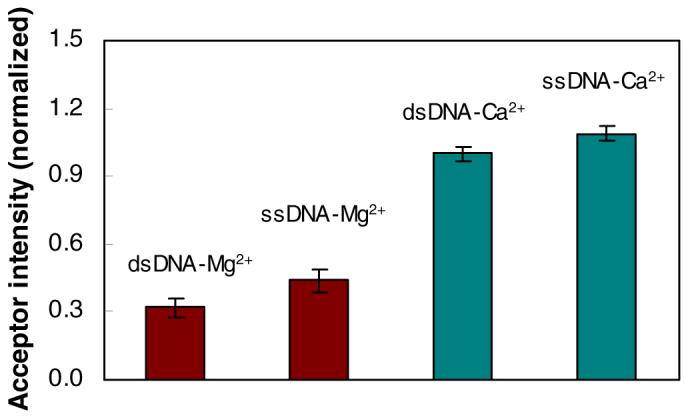
A comparison of the acceptor intensity for ssDNA and dsDNA, in the presence of Mg2+ and Ca2+. The intensities were averaged over the whole acceptor channel and normalized by their corresponding donor intensities. The higher acceptor intensities for Ca2+ (green bars) were induced by the recognition of dexyoinsine by endo V, while the lower acceptor intensities for Mg2+ (red bars) were caused by the dissociation of deoxyinosine.
Discussion
Association kinetics
The association time lapse T1 was measured at 4.9±1.8 s and 5.9±2.0 s for Ca2+ and Mg2+, respectively (green and red curves in Figure 3A). This suggests that both metal ions are equally efficient in mediating endo V-DNA interactions. The recognition of deoxyinosine by the enzyme could be a consequence of two scenarios: 1) endo V identifies the deoxyinosine site through random diffusion, or hopping; 2) endo V first locates the ssDNA substrate through diffusion and identifies the damaged site by sliding along the substrate. Compared with diffusion-driven deoxyinosine recognition, the second scenario is more likely as the largely monotonic time lapse T1 in Figure 2 would be otherwise replaced by random fluctuations.
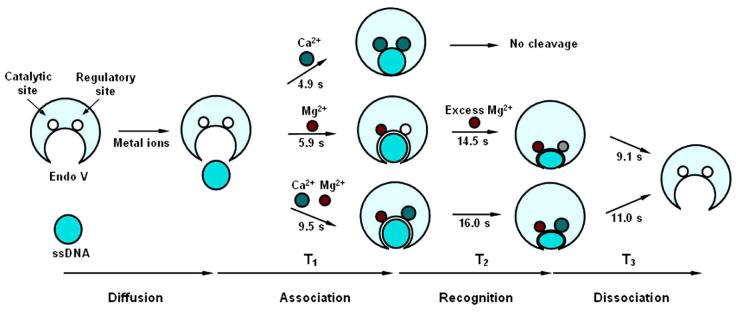
Since the sample of endo V and ssDNA in case 3 was pre-incubated with Ca2+ for 30 min prior to the injection of Mg2+, the registered association events in case 3 must be derived from either the binding of endo V and ssDNA following their previous dissociation events, or from association events anew. The former is possible with the release of Ca2+ from endo V, followed by the binding of Ca2+ and/or Mg2+ driven by diffusion and enzyme-substrate interaction. The latter is possible when the Ca2+ concentration was too low to induce frequent encountering of endo V with deoxyinosine, or when the initially hidden deoxyinosine site on ss DNA was exposed to endo V resulting from conformational changes. However, the mM metal ion concentrations in our experiment and the non-stochastic histogram of case 1 in Figure 3A regard the latter case as trivial. Previously, we have proposed a catalytic and regulatory two-metal model to account for the unique catalytic mechanism of endo V (20). The prolonged association of endo V in the presence of Ca2+ and Mg2+, as compared to that in case 1 or 2, may be explained by assigning two-metal binding sites to endo V (Figure 5). The association of endo V with deoxyinosine is most efficient when both metal binding sites are occupied by Ca2+ and least efficient when both sites are bound with Ca2+ and Mg2+. Our previous Mg2+-Ca2+ assay (13) showed that Ca2+ has little influence on Mg2+-mediated DNA cleavage, indicating that the catalytic site was occupied by Mg2+. Thus, the observed association events can perhaps be activated by three metal binding combinations: Ca2+ (catalytic site)-Ca2+ (regulatory site) (case 1), Mg2+-Mg2+ (case 2), and Mg2+-Ca2+ (Figure 5). The distribution of time lapse T1 in case 3 could be a superposition of the three possible kinetics and is found to be broader than that in either case 1 or case 2 (Figure 3A, blue vs. green or red).
FIGURE 5.
A proposed two-metal model at the single-molecule level. The regulatory site (top right circle on endo V) may be activated by either Ca2+ or Mg2+, the catalysis site or the catalytic site (top left circle on endo V) is only activated by Mg2, but may be occupied by Ca2+ or Mg2+. The time lapses T1, T2, and T3 corresponding to the single-molecule association, recognition, and dissociation events are illustrated for the three cases (Ca2+, Mg2+, and Ca2+-Mg2+), respectively. The regulatory site in case 2 could be occupied by Mg2+ or left empty, as indicated by the gray circle.
Recognition kinetics
It is known that DNA repair enzymes usually experience considerable conformation changes in lesion recognition, including the event of flipping the damaged base out of the helix (30, 31). Endo V is likely to have adopted a similar base flipping recognition mechanism. The consistent time durations in cases 2 and 3 suggest that Ca2+ or Mg2+ facilitates conformational changes with the same efficiency.
Dissociation kinetics
Studies performed by Huang et al. (13) showed no enhancement of deoxyinosine cleavage in Ca2+ and Mg2+ competition. In contrast, our results in Figure 3C indicate an improved efficiency of deoxyinosine dissociation in case 2 over case 3. This increased activity with Mg2+ alone reinforces the possibility that endo V possesses two metal binding sites (20), with one for the regulation of nonspecific activities during time lapses T1 and T2 and the other mainly for the catalysis of deaminated DNA. As shown in Figure 2B, the dissociation signal is noticeably noisier than the association process. This could be due to i) dissociation of the short 3′ TAMRA fragment from the enzyme-product complex or ii) dissociation of endo V from the enzyme-substrate or enzyme-product complex. The catalytic site of endo V may be occupied by either Mg2+ or Ca2+, but the catalysis of lesion cleavage was shown to only occur when this site is bound with Mg2+ (Figure 5), which agrees with our biochemical assays (13, 20).
Others factors concerning the kinetics
Our further tests on the endo V-ssDNA system, in the absence of metal ions, showed neither recognition nor cleavage activity. This observation is consistent with that of biochemical assays reinforcing the necessity of metal ions for endo V-DNA interactions (13). In our experiment, the endonuclease events were found to mostly occur during the initial 5-10 min subsequent to the introduction of metal ions into the sample cell. This is in agreement with the kinetics analysis of gel mobility shift assay, though it was conducted for a substrate of dsDNA and the ratio of enzyme to substrate was at 1:10 to 2:10 (19). In comparison, our ratio of endo V to ssDNA was approximately 10:1, a seemingly large value intending to promote frequent recognition and cleavage events. However, since endo V mostly diffused into the sample volume of ∼1 fL while ssDNA were largely immobilized to a quartz slide, the endo V molecules involved in both the endonuclease events and FRET must be within the evanescent wave field for fluorescence excitation. The depth of the evanescent wave was ∼100 nm as compared to the thickness of the sample cell of ∼100 μm. Given these results, the actual ratio of endo V to ssDNA probed in our assay is expected to be significantly below 10:1 and resemble that in the biochemical assays.
This preference of dsDNA to ssDNA by endo V is possible due to the fact that DNA in nature mostly adopts the double-stranded form (Figure 4). Since the persistence length P, a parameter to evaluate the rigidity of biopolymers, is ∼50 nm for dsDNA as opposed to that of 1 nm for ssDNA at the physiological salt strength (10), ssDNA curves more than dsDNA and therefore could make its lesion site(s) less accessible to endo V for binding and cleavage.
In conclusion, we have demonstrated the use of FRET technique in unraveling the recognition and cleavage events of deoxyinosine by endo V. The recognition of a deoxyinosine site in an ssDNA molecule is comparably effective with Ca2+ or Mg2+. The recognition process is not discernible by biochemical methods and has been found in our study to be largely unaffected when switching Ca2+ to Mg2+. The cleavage of the deoxyinosine site by endo V is catalyzed by Mg2+ and the complete catalytic cycle ranging from association to dissociation from nicked DNA takes approximately half a minute. We have further shown that the recognition of deoxyinosine in either dsDNA or ssDNA is equally effective with Ca2+, and the dissociation of deoxyinosine in dsDNA is 27% more efficient than for ssDNA with Mg2+. In addition to providing reaction kinetics, our single-molecule approach supports the hypothesis (20) of two metal binding sites for the regulation of enzymatic activities.
Acknowledgements
The authors thank Rose Loughlin and Godfrey Kimball for critical reading of the manuscript and Qi Lu for assistance with data analysis.
Abbreviations
- endo V
endonuclease V
- FRET
Fluorescence Resonance Energy Transfer
- ssDNA
single-stranded DNA
- dsDNA
double-stranded DNA
Footnotes
This work was supported by CSREES/USDA (SC-1700274, technical contribution No. 5222), the NIH (GM 067744), the Concern Foundation, and DOD-Army Research Office (W911NF-05-1-0335).†
References
- 1.David SS. Structural biology: DNA search and rescue. Nature. 2005;434:569–570. doi: 10.1038/434569a. [DOI] [PubMed] [Google Scholar]
- 2.Rouse J, Jackson SP. Interfaces between the detection, signaling, and repair of DNA damage. Science. 2002;297:547–551. doi: 10.1126/science.1074740. [DOI] [PubMed] [Google Scholar]
- 3.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki T, Yamaoka R, Nishi M, Ide H, Makino K. Isolation and characterization of a novel product, 2′-deoxyoxanosine, from 2′-deoxyguanosine, oligodeoxynucleotide, and calf thymus DNA treated by nitrous acid and nitric oxide. J. Am. Chem. Soc. 1996;118:2515–2516. [Google Scholar]
- 5.Kow YW. Repair of deaminated bases in DNA. Free Radic. Biol. Med. 2002;33:886–893. doi: 10.1016/s0891-5849(02)00902-4. [DOI] [PubMed] [Google Scholar]
- 6.Yang Y, Sass LE, Du C, Hsieh P, Erie DR. Determination of protein-DNA binding constants and specificities from statistical analyses of single molecules: MutS-DNA interactions. Nucleic Acids Res. 2005;33:4322–4334. doi: 10.1093/nar/gki708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schallhorn KA, Freedman KO, Moore JM, Lin J, Ke PC. Single-molecule DNA flexibility in the presence of base-pair mismatch. Appl. Phys. Lett. 2005;87:033901–1-3. [Google Scholar]
- 8.Cloutier TE, Widom J. Spontaneous sharp bending of double stranded DNA. Molecular Cell. 2004;14:355–362. doi: 10.1016/s1097-2765(04)00210-2. [DOI] [PubMed] [Google Scholar]
- 9.Yan J, Marco JF. Localized single-stranded bubble mechanism for cyclization of short double helix DNA. Phys. Rev. Lett. 2004;93:108108–1-4. doi: 10.1103/PhysRevLett.93.108108. [DOI] [PubMed] [Google Scholar]
- 10.Rivetti C, Walker C, Bustamante C. Polymer chain statistics and conformational analysis of DNA molecules with bends or sections of different flexibility. J. Mol. Biol. 1998;280:41–59. doi: 10.1006/jmbi.1998.1830. [DOI] [PubMed] [Google Scholar]
- 11.Yao M, Hatahet Z, Melamede RJ, Kow YW. Purification and characterization of a novel deoxyinosine-specific enzyme, deoxyinosine 3′ endonuclease, from Escherichia coli. J. Biol. Chem. 1994;269:16260–16268. [PubMed] [Google Scholar]
- 12.Guo G, Weiss B. Endonuclease V (nfi) mutant of Escherichia coli K-12. J. Bacteriol. 1998;180:46–51. doi: 10.1128/jb.180.1.46-51.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang J, Lu J, Barany F, Cao W. Multiple cleavage activities of endonuclease V from Thermotoga maritima: recognition and strand nicking mechanism. Biochemistry. 2001;40:8738–8748. doi: 10.1021/bi010183h. [DOI] [PubMed] [Google Scholar]
- 14.He B, Qing H, Kow YW. Deoxyxanthosine in DNA is repaired by Escherichia coli endonuclease V. Mutat. Res. 2000;459:109–114. doi: 10.1016/s0921-8777(99)00063-4. [DOI] [PubMed] [Google Scholar]
- 15.Feng H, Klutz AM, Cao W. Active site plasticity of endonuclease V from Salmonella typhimurium. Biochemistry. 2005;44:675–683. doi: 10.1021/bi048752j. [DOI] [PubMed] [Google Scholar]
- 16.Hitchcock TM, Gao H, Cao W. Cleavage of deoxyoxanosine-containing oligodeoxyribonucleotides by bacterial endonuclease V. Nucleic Acids Res. 2004;32:4071–4080. doi: 10.1093/nar/gkh747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao M, Kow YW. Interaction of deoxyinosine 3′-endonuclease from Escherichia coli with DNA containing deoxyinosine. J. Biol. Chem. 1995;270:28609–28616. doi: 10.1074/jbc.270.48.28609. [DOI] [PubMed] [Google Scholar]
- 18.Feng H, Dong L, Klutz AM, Aghaebrahim N, Cao W. Defining Amino Acid Residues Involved in DNA-protein Interactions and Revelation of 3′-Exonuclease Activity in Endonuclease V. Biochemistry. 2005;44:11486–11495. doi: 10.1021/bi050837c. [DOI] [PubMed] [Google Scholar]
- 19.Huang J, Lu J, Barany F, Cao W. Mutational analysis of endonuclease V from Thermotoga maritima. Biochemistry. 2002;41:8342–8350. doi: 10.1021/bi015960s. [DOI] [PubMed] [Google Scholar]
- 20.Feng H, Dong L, Cao W. Catalytic mechanism of endonuclease V: a catalytic and regulatory two-metal model. Biochemistry. 2006;45:10251–10259. doi: 10.1021/bi060512b. [DOI] [PubMed] [Google Scholar]
- 21.Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5 nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 22.Asbury CL, Fehr AN, Block SM. Kinesin moves by an asymmetric hand-over-hand mechanism. Science. 2003;302:2130–2134. doi: 10.1126/science.1092985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strick TR, Croquette V, Bensimon D. Single-molecule analysis of Topoisomerase II-DNA interactions. Nature. 2000;404:901–904. doi: 10.1038/35009144. [DOI] [PubMed] [Google Scholar]
- 24.Ha T, Enderle T, Ogletree DF, Chemla DS, Selvin PR, Weiss S. Probing the interaction between two single molecules: Fluorescence resonance energy transfer between a single donor and a single acceptor. Proc. Natl. Acad. Sci. USA. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhuang X, Bartley LE, Babcock HP, Russel R, Ha T, Herschlag D, Chu S. A single molecule study of RNA catalysis and folding. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 26.Morgan MA, Okamoto K, Kahn JD, English DS. Single-molecule spectroscopic determination of Lac repressor-DNA loop conformation. Biophys. J. 2005;89:2588–2596. doi: 10.1529/biophysj.105.067728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gemmen GJ, Sim R, Haushalter KA, Ke PC, Kadonaga JT, Smith DE. Forced unraveling of nucleosomes assembled on heterogeneous DNA using core histones, NAP-1, and ACF. J. Mol. Biol. 2005;351:89–99. doi: 10.1016/j.jmb.2005.05.058. [DOI] [PubMed] [Google Scholar]
- 28.Harada Y, Funatsu T, Murakami K, Nonoyama Y, Ishihama A, Yanagida T. Single molecule imaging of RNA polymerase-DNA interactions in real time. Biophys. J. 1999;76:709–715. doi: 10.1016/S0006-3495(99)77237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Noort SJT, van der Werf KO, Eker APM, Wyman C, de Grooth BG, van Hulst NF, Greve J. Direct visualization of dynamic protein-DNA interactions with a dedicated atomic force microscope. Biophys. J. 1998;74:2840–2849. doi: 10.1016/S0006-3495(98)77991-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huffman JL, Sundheim O, Tainer JA. DNA base damage recognition and removal: new twists and grooves. Mutat. Res. 2005;577:55–76. doi: 10.1016/j.mrfmmm.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 31.Roberts RJ, Cheng X. Base flipping. Annu. Rev. Biochem. 1998;67:181–198. doi: 10.1146/annurev.biochem.67.1.181. [DOI] [PubMed] [Google Scholar]