5′-Methylthioadenosine/S-adenosylhomocysteine nucleosidase (MTAN) is a bacterial enzyme of the N-ribosyl transferase family. It catalyzes the biologically irreversible hydrolytic deadenylaton of 5′-methylthioadenosine (MTA) or S-adenosylhomocysteine to form adenine and methylthioribose (MTR) or S-ribosylhomocysteine, respectively (Figure 1). MTAN is involved in bacterial quorum sensing pathways and thus is a potential target for the design of antibiotics.1-4 There is no MTAN in humans, where 5′-methylthioadenosine phosphorylase (MTAP) catalyzes the reversible phosphorolysis of MTA to adenine and methylthioribose-1-phosphate. MTAP has been targeted in the design of anti-cancer drugs on the basis of its involvement in polyamine biosynthesis and purine salvage pathways.5-6 Probing transition state structures of these enzymatic reactions provides an approach for the design of transition state analogues as powerful inhibitors. This strategy has been successfully applied in the development of inhibitors for E. coli and S. pneumoniae MTANs with dissociation constants to the femtomolar range.7,8 Here we report that Neisseria meningitides MTAN forms a transition state unique from other MTANs. This surprising result adds another example of closely related enzymes with distinct transition state structures.9-12 Transition state analogues of bacterial quorum sensing pathways have potential for blocking virulence gene expression without affecting growth, thus minimizing the probability of generating bacterial strains resistant to quorum sensing antibiotics.
Figure 1.
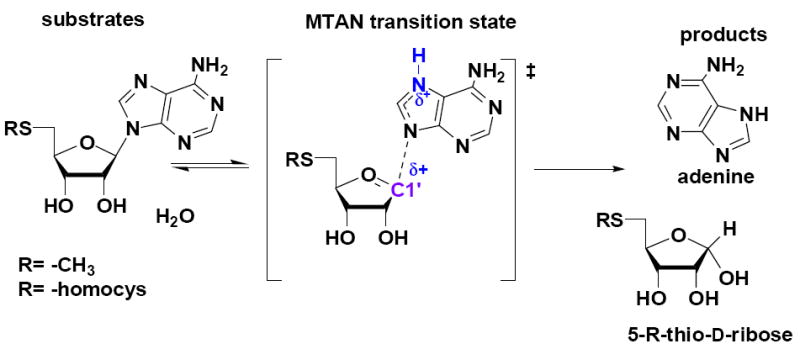
Hydrolysis of MTA by N. meningitides MTAN and the proposed transition state structure. Cationic features of the transition state are indicated in blue.
The transition states of E. coli and S. pneumoniae MTANs were recently solved using kinetic isotope effects (KIE).10,11 They were shown to have dissociative SN1 transition states with little or no bond order to the leaving adenine or to the attacking hydroxyl nucleophile. However, a key distinction between these two transition states is the protonation state of adenine N7 at the transition state. In E. coli MTAN, N7 is protonated to give a neutral leaving group. The N7 of adenine is not protonated at the transition state of S. pneumoniae MTAN, creating a relatively unfavorable anionic leaving group. This feature differentiates S. pneumoniae and E. coli MTAN transition states and explains why E. coli MTAN is catalytically ~103-fold more efficient than S. pneumoniae MTAN. The transition state of human MTAP also exhibits significant dissociative character in which the N-glycosidic bond is fully broken and the leaving group adenine is anionic.13 However, a unique feature of human MTAP in comparison with E. coli and S. pneumoniae MTANs is that human MTAP involves significant participation of the phosphate nucleophile.
The properties of ribose sugars at the transition states of E. coli and S. pneumoniae MTANs and human MTAP also make these enzymes different from other enzymes of the N-ribosyl transferase family. The transition states of human and Plasmodium falciparum purine nucleoside phosphorylases (PNPs) are fully dissociated ribocations while that for bovine PNP is an early SN1 transition state.9,14 In contrast, the transition states of MTANs from E. coli and S. pneumoniae and human MTAP appear to be zwitterionic, cationic at the anomeric carbon and anionic at the 3′-hydroxyl oxygen. The transition state of bovine PNP differs from other PNPs in that the nucleoside reactant retains substantial bond order to the hypoxanthine leaving group. This difference in transition state structure occurs despite the 87% sequence identity between bovine and human PNPs and their completely conserved catalytic sites.9,14,15 This surprising variety in the N-ribosyltransferase transition states led to the present analysis of the MTAN from N. meningitides.
All previously-reported MTAN transition states are characterized by fully dissociated leaving groups with no participation of the water nucleophile. However, associative transition states are found in other enzymes of the N-ribosyl transferase family and we explored this possibility for a family of MTANs. We screened several MTANs with MT-Immucillin-A, a transition state analogue of early SN1 transition states for MTANs, in comparison to inhibition by MT-DADMe-Immucillin-A, an analogue of fully dissociated transition states.7-11 N. meningitides MTAN gave an inhibition profile suggesting an early transition state. Catalytic turnover of N. meningitides MTAN (kcat = 8 s-1) is similar to that of E. coli MTAN (kcat = 3 s-1).16
Kinetic isotope effect analysis and computational modeling were used to solve the transition state structure of N. meningitides MTAN. The study establishes that the N. meningitides MTAN has an early SN1 transition state with residual N-glycosidic bond order at the transition state. The early transition state of N. meningitides MTAN is consistent with its inhibition by transition state analogues of MTANs.16 The N. meningitides MTAN is the first reported MTA hydrolase to have an early transition state with significant bond order to the adenine leaving group.
The transition state of N. meningitides MTAN was probed with pairs of isotopically MTAs, synthesized enzymatically.10 Experimental KIEs were measured for MTAs labeled at [1′-3H], [1′-14C], [2′-3H], [4′-3H], [5′-3H2], [9-15N] and [Me-3H3]. Experimental KIEs were measured for MTAs under competitive conditions known to give rise to kcat/Km (V /K) kinetic parameters.17 KIEs were measured by comparing the 3H/14C ratio in products formed from each pair of isotopically labeled MTAs (supporting material).10 [5’-14C]MTA was used as the isotopically silent remote label to examine the 3H isotope effects and both [4’-3H]- and [5′-3H2]MTAs were used as the remote labels for measuring [1′-14C] and [9-15N, 5′-14C] KIEs. The [1′-14C] and [9-15N, 5′-14C] KIEs were subsequently corrected for the remote [3H] KIE. All KIEs were corrected to represent the isotope effects extrapolated to 0% hydrolysis according to the equation:
where f is the fraction of reaction progress, Rf and Ro are ratios of heavy to light isotope at partial and total reaction completion, respectively. The V/K KIEs obtained above were further corrected for forward commitment using Northorp’s equation modified for irreversible reaction: , where H (V / K) is the observed heavy atom isotope effect measured experimentally, Cf is the forward commitment to catalysis and Hk is the intrinsic isotope effect.17 The forward commitment to catalysis was measured using the isotope partition method as described previously for E. coli MTAN. 10,18 MTAN from N. meningitides exhibits a relatively large forward commitment factor of 0.66 ± 0.027 (supporting information), consistent with its small Km of 1.4 μM.16 The V/K KIEs were corrected for forward commitment to obtain the intrinsic KIEs for [1′-3H]-, [1′-14C]-, [2′-3H]-, [4′-3H]-, [9-15N]-, [5′-3H2]- and [Me-3H3] MTAs (Figure 2).
Figure 2.
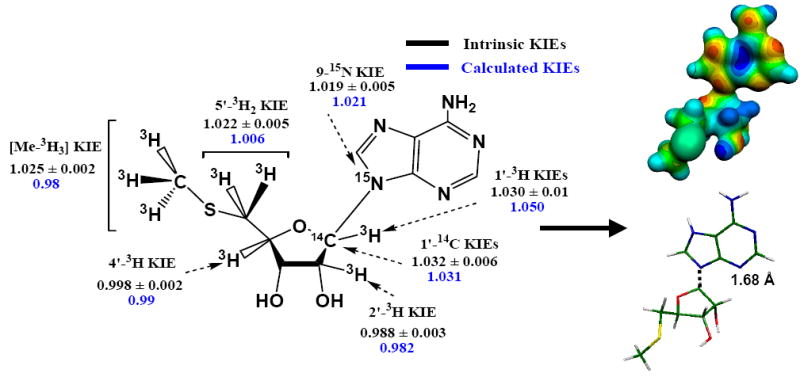
Intrinsic and calculated KIEs (left) and the modeled transition state (right) of N. meningitides MTAN. The MEP model shown top on the right was calculated at HF/STO3G (Gaussian 98/cube) for the geometry optimized at the B3LYP/6-31G(d,p) level of theory and visualized with Molekel 4.020 at density of 0.15 electrons/Å3. The stick model shown below on the right has the same geometry as the MEP surface.
Intrinsic KIEs were used as constraints to match a transition state structure of N. meningitides MTAN calculated from B3LYP functional density theory and the 6-31G (d, p) basis set implemented in Gaussian98 or Gaussian03.19 Transition state optimization for N. meningitides was initiated by making small perturbations to the C1′-N9 bond of MTA. The KIEs were calculated for each perturbation and compared to the intrinsic KIEs. The transition state optimization process was completed when the calculated isotope effects were closely matched to the intrinsic KIEs (Figure 2).
The calculated transition state for hydrolysis of MTA by N. meningitides MTAN shows good agreement with the intrinsic KIEs. At the transition state the C1′-N9 glycosidic bond is weakened with an increase in bond length from 1.47 Å for MTA to 1.68 Å. This bond length corresponds to a Pauling bond order of 0.50, the earliest transition state known for any dissociative N-ribosyltransferase, since others have C1’-N9 bond orders of 0.38 to <0.01.7,9,11,13 This early transition state differs from the fully dissociated transition states of E. coli and S. pneumoniae MTANs where their C1′-N9 bond lengths are 3.0 Å or more, corresponding to a Pauling bond order of 0.01 or less.10,11 The significant bond order at the TS of N. meningitides MTAN is shown in the molecular electrostatic map, with continuous electron density between methylthioribose and the leaving group adenine (Figure 2). The early N. meningitides MTAN transition state generates a significant primary [1′-14C] KIE of 1.032 and a small [1′-3H] KIE of 1.030. The relatively large [1′-14C] KIE and small [1′-3H] KIE are expected for an early transition state because reaction coordinate motion contributes to the [1′-14C] KIE. The small 1′-3H of 1.030 is also consistent with the early transition state because the partial C1′-N9 bond sterically restricts enhanced out-of-plane bending modes and prevents the expression of large [1′-3H] KIE values. N-ribosyltransferases or acid-catalyzed solvolysis of adenine nucleotides with fully developed ribooxacarbenium ion transition states give rise to [1′-3H] KIE of 1.10 to 1.35 as a consequence of enhanced out-of-plane bending modes following dissociation of the C1′-N9 bond.10,11,13,21
The [9-15N] KIE is sensitive to both dissociation of the C1′-N9 bond and N7 protonation.11 A fully dissociated (at the C1′-N9 bond) adenine intermediate of MTA is known to give an equilibrium [9-15N] KIE of 1.036 which decreases to 1.027 upon N7 protonation.11 Correlating the calculated [1′-14C] KIE to the intrinsic KIE indicated a C1′-N9 bond length of 1.68 Å and a bond order that corresponds to a [9-15N] KIE of 1.012 for unprotonated adenine. Protonation at N7 of adenine in this partly dissociated transition state increases the calculated [9-15N] KIE to 1.021, consistent with the experimental intrinsic KIE (Figure 2). At this transition state, a fully N7-protonated adenine with residual N9-C1’ bond order is partially cationic. The electrostatic repulsion between the partially cationic methylthioribosyl and adenyl groups drives the reaction along the reaction coordinate, causing the dissociation of the N-glycosidic bond of MTA. After the transition state barrier is passed, dissociation of the C1′-N9 bond decreases the cationic character of the adenine leaving group and nucleophilic attack by a hydroxide ion neutralizes the positive charge on methylthioribosyl to generate adenine and methylthioribose products. The N7 of adenine is also protonated at the TS of E. coli MTAN, however unlike N. meningitides MTAN, the N7 protonated adenine is neutral in E. coli MTAN due to the well-advanced dissociation of the C1′-N9 bond.
The [2′-3H] KIE is caused by hyperconjugation of the C2′-H2′ bonding (σ) electrons to the p-orbital on the anomeric carbon (in the C1′-N9 bond). The magnitude of the [2′-3H] KIE is proportional to the overlap between the σ (C2′-H2′) orbital and the partially empty p-orbital as well as to the relative emptiness of the p-orbital.11 Occupancy of the p-orbital is directly affected by the degree of the C1′-N9 bond dissociation, as is the change in hybridization of the anomeric carbon. The small inverse [2′-3H] KIE of 0.988 for N. meningitides MTAN is consistent with both a limited dissociation of the C1′-N9 bond and disrupted orbital overlap by an unfavorable dihedral angle between the C1’-N9 and C2’-H2’ bonds. Partial dissociation of the C1′-N9 bond at the transitions state of N. meningitides MTAN results in incomplete rehybridization of the anomeric carbon. Gaussian calculations indicate that the anomeric carbon is sp2.62 hybridized in the substrate compared to sp2.28 at the transition state and the p-orbital remains significantly occupied at the transition state. Decreased electron occupancy of the p-orbital at the transition state of N. meningitides MTAN would cause a significant change in the σ C2′-H2′ to p-orbital hyperconjugation at the transition state if the bonds were in conjugation. The small inverse [2′-3H] KIE is therefore due to geometrically unfavorable hyperconjugation with the C1′-N9 bond and may also be influenced by the geometry of the 2′-hydroxyl oxygen and σ* C2′-H2′ at the transition state. 11
The [4′-3H] KIE reports on both the dissociation of the C1′-N9 bond and the polarization of the 3′-hydroxyl group.11 Dissociation of the C1′-N9 bond influences the [4′-3H] KIE by affecting the hyperconjugative interaction between the lone pair (np) of the ring oxygen (O4′) and σ* (C4′-H4′) antibonding orbital. 11 The [4′-3H] KIE is close to unity and is consistent with incomplete dissociation of the C1′-N9 bond where hyperconjugative interaction between np of O4′ and σ* (C4′-H4′) is relatively unchanged at the transition state compared to the substrate state. A small inverse [4′-3H] KIE arising from C1′-N9s bonding loss is easily reversed by 3′-hydroxyl group geometry. However, an inverse [4′-3H] KIE establishes that the 3′-hydroxyl is not ionized and is in contrast to the normal [4′-3H] KIE found for S. pneumoniae MTAN and human MTAP.10,11 Polarization of the 3′-hydroxyl causes a large and normal [4′-3H] KIE by increasing the accumulation of the negatives charge on the ring oxygen.11
The [5′-3H2] and [Me-3H3] KIEs arise from hyperconjugative interactions between the np of sulphur and σ* antibonding orbitals of each individual C-H bond.10 This hyperconjugative interaction and therefore the [5′-3H2] and [Me-3H3] KIEs are influenced by the rotation of the C4′-C5′-S-CMe torsional angle. Hence, 5′-3H2 and [Me-3H3] KIEs determine the conformation of the 5′-methylthio group at the transition state. This approach often leads to inconclusive geometries for the 5′-methylthio residue because multiple conformations can cause similar KIE values. Consequently, the accurate determination of the 5′-methylthio conformation relies upon the combined information of [5′-3H2], [Me-3H3] KIEs and crystal structures of MTANs bound to a transition state analogue.
The transition state of N. meningitides MTAN from KIEs and computational chemistry is early in a SN1 reaction path, unlike the later transition states of S. pneumoniae and E. coli MTANs. An early transition state for N. meningitides MTAN is also consistent with its inhibition by MTAN transition state analogue inhibitors.16 N. meningitides MTAN is the first MTA hydrolase and only the second enzyme of the N-ribosyltransferase family to exhibit an early transition state. Late dissociative SN1 transition states dominate the N-ribosyltransferases. Knowledge of this early transition state structure is highly relevant for developing specific transition state analogues for the quorum sensing pathways of this pathogenic bacterium.
Supplementary Material
Complete author list for reference 19, experimental details for KIEs and commitment factor, molecular geometries, coordinates and normal modes of the transition state. This material is available free of charge via the internet at http://pubs.acs.org
Acknowledgments
This work was supported by NIH research grant GM41916.
Contributor Information
Vipender Singh, Department of Biochemistry, Albert Einstein College of Medicine, 1300 Morris Park Avenue, Bronx, New York 10461.
Minkui Luo, Department of Biochemistry, Albert Einstein College of Medicine, 1300 Morris Park Avenue, Bronx, New York 10461.
Rosemary L. Brown, Institute of Molecular Biosciences, Massey University, Private Bag 11222, Palmerston North, New Zealand
Gillian E. Norris, Institute of Molecular Biosciences, Massey University, Private Bag 11222, Palmerston North, New Zealand
Vern L. Schramm, Department of Biochemistry, Albert Einstein College of Medicine, 1300 Morris Park Avenue, Bronx, New York 10461, vern@aecom.yu.edu
References
- 1.Schauder S, Shokat K, Surette MG, Bassler BL. Mol Microbiol. 2001;41:463–476. doi: 10.1046/j.1365-2958.2001.02532.x. [DOI] [PubMed] [Google Scholar]
- 2.Ragione D, Porcelli FM, Carteni-Farina M, Zappia V, Pegg AE. Biochem J. 1985;232:335–341. doi: 10.1042/bj2320335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parsek MR, Val DL, Hanzelka BL, Cronan JE, Jr, Greenberg EP. Proc Natl Acad Sci. 1999;96:4360–4365. doi: 10.1073/pnas.96.8.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Withers H, Swift HS, Williams P. Curr Opin Microbiol. 2001;4:186–193. doi: 10.1016/s1369-5274(00)00187-9. [DOI] [PubMed] [Google Scholar]
- 5.Harasawa H, Yamada Y, Kudoh M, Sugahara K, Soda H, Hirakata Y, Sasaki H, Ikeda S, Matsuo T, Tomonaga M, Nobori T, Kamihira S. Leukemia. 2002;16:1799. doi: 10.1038/sj.leu.2402570. [DOI] [PubMed] [Google Scholar]
- 6.Tabor CW, Tabor H. Methods Enzymol. 1983;94:294–297. [Google Scholar]; Basu I, Cordovano G, Das I, Belbin TJ, Guha C, Schramm VL. J Biol Chem. 2007;282:21477–21486. doi: 10.1074/jbc.M702287200. [DOI] [PubMed] [Google Scholar]
- 7.Singh V, Evans GB, Lenz DH, Painter GF, Tyler PC, Furneaux RH, Lee JE, Howell PL, Schramm VL. J Biol Chem. 2005;280:18265–18273. doi: 10.1074/jbc.M414472200. [DOI] [PubMed] [Google Scholar]
- 8.Singh V, Shi W, Almo SC, Evans GB, Furneaux RH, Tyler PC, Painter GF, Lenz DH, Mee S, Zheng R, Schramm VL. Biochemistry. 2006;45:12929–12941. doi: 10.1021/bi061184i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewandowicz A, Schramm VL. Biochemistry. 2004;43:1458–1468. doi: 10.1021/bi0359123. [DOI] [PubMed] [Google Scholar]
- 10.Singh V, Lee JE, Nunez S, Howell LP, Schramm VL. Biochemistry. 2005;44:11647–11659. doi: 10.1021/bi050863a. [DOI] [PubMed] [Google Scholar]
- 11.Singh V, Schramm VL. J Am Chem Soc. 2007;129:2783–2795. doi: 10.1021/ja065082r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo M, Singh V, Taylor EA, Schramm VL. J Am Chem Soc. 2007;129:8008–8017. doi: 10.1021/ja072122y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh V, Schramm VL. J Am Chem Soc. 2006;128:14691–14696. doi: 10.1021/ja065419p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kline PC, Schramm VL. Biochemistry. 1993;32:13212–13219. doi: 10.1021/bi00211a033. [DOI] [PubMed] [Google Scholar]
- 15.Taylor Ringia EA, Tyler PC, Evans GB, Furneaux RH, Murkin AS, Schramm VL. J Am Chem Soc. 2006;128:7126–7127. doi: 10.1021/ja061403n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutierrez J, Luo M, Singh V, Schramm VL. Unpublished results. [Google Scholar]
- 17.Northrop DB. Methods. 2001;24:117–124. doi: 10.1006/meth.2001.1173. [DOI] [PubMed] [Google Scholar]; Northrop DB. Annu Rev Biochem. 1981;50:103–131. doi: 10.1146/annurev.bi.50.070181.000535. [DOI] [PubMed] [Google Scholar]
- 18.Rose IA. Methods Enzymol. 1980;64:47–59. doi: 10.1016/s0076-6879(80)64004-x. [DOI] [PubMed] [Google Scholar]
- 19.Frisch MJ, et al. Gaussian 03, Revision B.04. Gaussian, Inc.; Pittsburgh, PA: 2003. [Google Scholar]
- 20.Flükiger P, Lüthi HP, Portmann S, Weber J. MOLEKEL 4.0. Swiss Center for Scientific Computing; Manno, Switzerland: 2000. [Google Scholar]
- 21.McCann JAB, Berti PJ. J Am Chem Soc. 2007;129:7055–7064. doi: 10.1021/ja067371l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complete author list for reference 19, experimental details for KIEs and commitment factor, molecular geometries, coordinates and normal modes of the transition state. This material is available free of charge via the internet at http://pubs.acs.org


