Abstract
Cytosolic and peroxisomal enzymes necessary for methanol assimilation are synthesized when Pichia pastoris is grown in methanol. Upon adaptation from methanol to a glucose environment, these enzymes are rapidly and selectively sequestered and degraded within the yeast vacuole. Sequestration begins when the vacuole changes shape and surrounds the peroxisomes. The opposing membranes then fuse, engulfing the peroxisome. In this study, we have characterized a mutant cell line (glucose-induced selective autophagy), gsa7, which is defective in glucose-induced selective autophagy of peroxisomes, and have identified the GSA7 gene. Upon glucose adaptation, gsa7 cells were unable to degrade peroxisomal alcohol oxidase. We observed that the peroxisomes were surrounded by the vacuole, but complete uptake into the vacuole did not occur. Therefore, we propose that GSA7 is not required for initiation of autophagy but is required for bringing the opposing vacuolar membranes together for homotypic fusion, thereby completing peroxisome sequestration. By sequencing the genomic DNA fragment that complemented the gsa7 phenotype, we have found that GSA7 encodes a protein of 71 kDa (Gsa7p) with limited sequence homology to a family of ubiquitin-activating enzymes, E1. The knockout mutant gsa7Δ had an identical phenotype to gsa7, and both mutants were rescued by an epitope-tagged Gsa7p (Gsa7-hemagglutinin [HA]). In addition, a GSA7 homolog, APG7, a protein required for autophagy in Saccharomyces cerevisiae, was capable of rescuing gsa7. We have sequenced the human homolog of GSA7 and have shown many regions of identity between the yeast and human proteins. Two of these regions align to the putative ATP-binding domain and catalytic site of the family of ubiquitin activating enzymes, E1 (UBA1, UBA2, and UBA3). When either of these sites was mutated, the resulting mutants [Gsa7(ΔATP)-HA and Gsa7(C518S)-HA] were unable to rescue gsa7 cells. We provide evidence to suggest that Gsa7-HA formed a thio-ester linkage with a 25–30 kDa protein. This conjugate was not observed in cells expressing Gsa7(ΔATP)-HA or in cells expressing Gsa7(C518S)-HA. Our results suggest that this unique E1-like enzyme is required for homotypic membrane fusion, a late event in the sequestration of peroxisomes by the vacuole.
INTRODUCTION
Autophagy is a tightly regulated nonselective and sometimes selective process for the degradation of cellular proteins and organelles in eukaryotic cells. Mammalian cells and yeast are capable of responding to environment changes and nutrient stress by activating the degradation of proteins via microautophagy and macroautophagy (Veenhuis et al., 1983; Lardeux and Mortimore, 1987; Mortimore et al., 1989; Kopitz et al., 1990; Tuttle et al., 1993; Baba et al., 1994; Dunn, 1994; Tuttle and Dunn, 1995; Chiang et al., 1996). Microautophagy is the process whereby cellular components are surrounded by invaginations of the lysosomal membrane or by finger-like protrusions of the lysosome or yeast vacuole. The resulting intralysosomal vesicles that contain the sequestered cellular components are eventually degraded by the hydrolytic enzymes (Ahlberg et al., 1985; Mortimore et al., 1988). Macroautophagy is the process whereby cellular components are first sequestered within an autophagosome that we have shown arises from an invagination of the rough endoplasmic reticulum (Dunn, 1990a). The autophagosome then fuses with a lysosome or vacuole, thereby delivering its contents to the acid hydrolases (Dunn, 1990b).
In Saccharomyces cerevisiae, autophagy is constitutive and regulated by nitrogen starvation and glucose adaptation (Takeshige et al., 1992; Egner et al., 1993; Baba et al., 1994, 1997; Chiang et al., 1996). Precursor aminopeptidase I (API)1 is constitutively sequestered into 140- to 160-nm double-membrane–bound vesicles that then fuse with the vacuole (Baba et al., 1997). Once inside the vacuole, pro-API is activated to API by a proteinase digestion (Harding et al., 1995; Scott et al., 1997). Nitrogen starvation stimulates the nonselective sequestration of cytosolic proteins into 300- to 900-nm double-membrane–bound autophagosomes (Baba et al., 1997; Lang et al., 1998). Upon fusion of the autophagosome with the vacuole, the contents of the autophagosome are completely degraded. Although the constitutive and regulated autophagic pathways are morphologically and functionally different, they are genetically similar. That is, some of the cytosol-to-vacuole targeting (cvt) (Harding et al., 1995), autophagy (apg) (Tsukada and Ohsumi, 1993), and autophagy (aut) (Thumm et al., 1994) mutations are allelic (Harding et al., 1996; Scott et al., 1996). This suggests that these pathways share common molecular machinery. Indeed, those proteins required for the formation of the autophagosome would likely be the same regardless of the environmental signal. An alternative pathway for the glucose-enhanced selective delivery of cytosolic fructose 1,6-bisphosphatase into the vacuole also exists in S. cerevisiae. Fructose 1,6-bisphosphatase is sequestered into 30- to 40-nm single-membrane–bound vesicles that appear to fuse with the vacuole (Huang and Chiang, 1997). The genetic overlap of this pathway with autophagy has yet to be examined.
We and others have shown that selective autophagy plays an important role in the adjustment of peroxisomal and cytosolic protein levels to meet environment changes in Hansenula polymorpha and Pichia pastoris. When these methylotrophic yeast are grown in methanol, their peroxisomes become very large because of the increased production of those enzymes necessary to assimilate methanol. One such enzyme is alcohol oxidase (AOX), the first enzyme in the methanol assimilation pathway. When the carbon source is switched from methanol to glucose or ethanol (glucose or ethanol adaptation), the yeast peroxisomes are degraded rapidly within the vacuole by a process analogous to autophagy (Veenhuis et al., 1983; Tuttle et al., 1993; Tuttle and Dunn, 1995). When H. polymorpha adapt from methanol medium to glucose or ethanol, the peroxisomes are engulfed within membranes of unknown origin. The resulting autophagosomes then fuse with the vacuole, and the peroxisomes are degraded (Veenhuis et al., 1983; Tuttle et al., 1993). During adaptation from methanol to glucose in P. pastoris, peroxisomes are selectively engulfed by the vacuole (microautophagy) (Tuttle et al., 1993; Tuttle and Dunn, 1995). The ability of these methylotrophic yeasts to modulate their peroxisome contents in response to nutritional adaptation makes them an excellent model by which to investigate the molecular events of peroxisome degradation. Titorenko et al. (1995) have identified two genes (PDD1 and PDD2) required for selective macroautophagy of peroxisomes in H. polymorpha. We have identified eight genes (GSA1–GSA8) required for glucose-induced selective autophagy of peroxisomes in P. pastoris (our unpublished results). We have shown previously that GSA1 encodes phosphofructokinase I, which is required for the initiation of glucose-induced autophagy (Yuan et al., 1997).
In this article, we have characterized GSA7 required for the vacuolar sequestration of peroxisomes during glucose adaptation. Gsa7p was identified as a unique protein with limited sequence homology to the family of ubiquitin-activating enzymes, E1. This protein is not required for initiation of autophagy but for the completion of the sequestration event. Furthermore, we have shown that a putative ATP-binding site and catalytic site are required for Gsa7p function.
MATERIALS AND METHODS
Yeast Strains and Media
Parental strains, GS115 (his4), GS190 (arg4), and PPF1 (his4, arg4), were a generous gift of Dr. J. M. Cregg (Oregon Graduate Institute, Beaverton, OR) and were routinely cultured at 30°C in YPD (1% Bacto yeast extract, 2% Bacto peptone, and 2% dextrose). For induction of peroxisomes the cells were grown in YNM (6.7 g/l yeast nitrogen base, 0.4 mg/l biotin, and 0.5% vol/vol methanol). For degradation of peroxisomes the cells were transferred to YND (6.7 g/l yeast nitrogen base, 0.4 mg/l biotin, and 2% glucose). The sporulation and mating medium was composed of 0.5% sodium acetate, 1% KCl, and 1% glucose. The electroporation medium was composed of 1 M Sorbitol, 2% glucose, 6.7 g/l yeast nitrogen base, and 0.4 mg/l biotin. All media contained 2% agar when made as plates. Histidine or arginine or both were added at 40 mg/l when needed. Nitrogen starvation medium contained 6.7 g/l yeast nitrogen base (without amino acids or NH4SO4) and 2% glucose. Both Escherichia coli DH5α and Epicurian coli XL2-blue (Stratagene, La Jolla, CA) were used to amplify plasmids. Cells were grown at 37°C in 0.5% Bacto yeast extract, 1% Bacto tryptone, and 1% NaCl and selected in 0.5% Bacto yeast extract, 1% Bacto tryptone, and 1% NaCl containing 100 μg/ml ampicillin.
Yeast Transformation
Cells grown overnight in YPD to a density of A600 = 1.0 were harvested and treated with 10 mM DTT in YPD containing 25 mM HEPES, pH 8, for 15 min at 30°C. The cells were washed twice in ice-cold water and once in 1 M sorbitol resuspended in a small volume of 1 M sorbitol. Cells (40 μl) were mixed with 0.2–1 μg DNA and transferred to a 0.25-mM gap Bio-Rad (Bio-Rad, Hercules, CA) cuvette, and the DNA was introduced by electroporation at 1.5 kV, 25 μF, 400Ω (Gene Pulser, Bio-Rad). The cells were placed on electroporation plates for 3–5 d before colonies appeared.
Isolation and Rescue of gsa7
Mutant strain WDY7 was generated by mutagenizing GS115 cells with nitrosoguanidine (Tuttle and Dunn, 1995). Mutagenized colonies grown on YPD plates were replica-plated to YNMH (YNM containing 40 μg/ml histidine) plates, and the colonies were allowed to grow for 4–5 d. The colonies were replica-plated onto nitrocellulose and placed on YNDH (YND containing 40 μg/ml histidine) plates for 12–16 h. The putative mutant colonies maintained AOX, as identified by the purple reaction product of the direct colony assay (Tuttle and Dunn, 1995). Those mutant strains were isolated, and their inability to degrade AOX during glucose adaptation was verified by liquid AOX assay (Tuttle and Dunn, 1995). Mutant strains were backcrossed to GS190 (arg4) and GS115 (his4) twice each, and complementation analysis placed WDY7 in complementation group gsa7. WDY7 (his4, gsa7) was then rescued by transformation with the episomal shuttle vector pYM8 containing a genomic library of P. pastoris and the HIS4 gene of S. cerevisiae (a generous gift of Dr. J. M. Cregg). Putative rescued colonies were first identified as white colonies in the AOX direct colony assay and then verified by AOX liquid assay.
Plasmid Isolation and DNA Sequencing
Rescued colonies were grown overnight in YPD before the cells were pelleted and resuspended in 2% Triton X-100, 1% SDS, 1 mM EDTA, and 100 mM NaCl in 10 mM Tris/HCl, pH 8.0. An equal volume of phenol:chloroform:isoamyl alcohol (25:24:1) was added, and the cells were disrupted by vortexing in the presence of acid-washed glass beads. After centrifugation, a small aliquot of the upper phase containing the plasmid was used to transform E. coli DH5α cells (Ausubel et al., 1988). The plasmid was then isolated from the the cultured E. coli by Promega Wizard Plus miniprep (Promega, Madison, WI), and the genomic DNA insert was sequenced at the DNA Sequencing Facility at the University of Florida (Gainesville, FL). The DNA sequences were assembled, and all six reading frames were compared with protein sequence databases using the blastx program of the National Center for Biotechnology Information.
Isolation of GSA7 Knockout
A 200 bp HindIII/BglII fragment of GSA7 gene was replaced with a 3-kb genomic fragment of S. cerevisiae containing ARG4. A 5.1-kb ApaI/ScaI fragment containing the ARG4 gene flanked by 400-bp 5′ and 1.2-kbp 3′ GSA7 was excised and introduced into PPF1 (his4, arg4). Colonies growing on electroporation plates with histidine that were defective in degradation of AOX were detected by direct colony assay and verified by liquid medium assay (see below). Genomic DNA was isolated, and the site of ARG4 insertion was determined by the size of the DNA fragment generated by PCR.
Measurements of Glucose-induced AOX Degradation
Direct Colony Assay.
Cells were plated on YNM plates and allowed to grow for 4–5 d before replicas on nitrocellulose were placed on YND plates for 12–16 h. The nitrocellulose blots were then placed on two sheets of Whatman paper circles saturated with 10 mM DTT, 10 mM EDTA, and 67 mM potassium phosphate buffer, pH 7.5, for 5 min, before transfer to paper saturated with 67 mM potassium phosphate buffer, pH 7.5, containing 5 U/ml Zymolase 20T (Seikagaku Corp., Tokyo, Japan), and incubated at 37°C for 1 h. The blots were rinsed in 33 mM potassium phosphate buffer, pH 7.5, and color was developed by incubation on paper saturated with 33 mM potassium phosphate buffer, pH 7.5, containing 3400 U/ml HRP, 5.6 mg/ml 2,2′-azino-bis(3-ethylbenz-thazoline-6-sulfonic acid), and 0.13% methanol at room temperature.
Liquid Medium Assay.
Cells were grown for 40 h on methanol as the sole carbon source (YNM) before addition of 2% glucose. Cells (8.0 OD600 U) at 0- and 6-h glucose adaptation were resuspended in 1 ml 20 mM Tris, pH 7.5, containing 50 mM NaCl, 1 mM EDTA, 1 mM PMSF, and 500 μl glass beads and vortexed three times for 1 min each (Tuttle et al., 1993). Alcohol oxidase was measured by adding 50 μl of extract to 3 ml reaction mixture containing 10 U HRP and 1.6 mg 2,2′-azino-bis(3-ethylbenz-thazoline-6-sulfonic acid) in 33 mM potassium phosphate buffer, pH 7.5 (Tuttle et al., 1993). The reaction was started by adding 10 μl methanol, stopped by adding 200 μl 4 M HCl, and read at 410 nm. Formate dehydrogenase (FDH) was measured by adding 50 μl of extract to 750 μl of a reaction mixture containing 3 mM NAD and 240 mM sodium formate in 50 mM potassium phosphate buffer, pH 7.5, and the formation of NADH was read at 10-s intervals at 340 nm (Kato, 1990).
Measurements of Protein Degradation
The degradation of cellular proteins during nitrogen starvation was performed as described previously (Tuttle and Dunn, 1995). Cellular proteins were radiolabeled with 1 μCi/ml 14C-valine for 16 h in 6.7 g/l YND, 2% glucose, 40 μg/l biotin, and 40 mg/l histidine (if needed). The cells were then washed and switched to a YND medium supplemented with 2% glucose and 10 mM valine that either contained histidine and NH4SO4 or lacked histidine and NH4SO4. The production of trichloroacetic (TCA)-soluble radioactivity was measured during 2–24 h of chase by the addition of 20% TCA. Soluble and insoluble radioactivity was separated by centrifugation and measured by scintillation counting.
Immunoblotting (Western Blotting)
Cells (5–10 OD600 U) were pelleted and resuspended into 150 μl of sample buffer with proteinase inhibitors (67 mM Tris/HCl pH 6.8, 2% SDS, 10% glycerol, 0.01% bromphenol blue, ±1.5% DTT, 0.5 mM PMSF, 1 μg/ml pepstatin A, and 0.5 μg/ml leupeptin). The cells were broken by vortexing in the presence of 200 μl glass beads and heated at 100°C for 3–5 min, and the proteins were separated by SDS-PAGE. The proteins were transferred to 0.45 μ Nitropure nitrocellulose membranes (Micron Separations, West Borough, MA). The blots were blocked in 5% nonfat dried milk in PBS for 1 h at room temperature, incubated at room temperature for 1–2 h with rabbit anti-HA (Babco, Richmond, CA) or mouse anti-HA (Babco) antibodies in PBS containing 0.1% Tween 20 (PBS-T), and for 1 h with HRP-conjugated secondary antibody in PBS-T with 2% dried milk. After each step the blots were washed four times in PBS-T. The antibodies were detected by enhanced chemiluminescence (Amersham, Arlington Heights, IL) and Kodak X-Omat LS film (Kodak, New Haven, CT).
Electron Microscopy
Ultrastructural analysis was performed using potassium permanganate fixation (Veenhuis et al., 1983). Cells were harvested by centrifugation, washed in water, and fixed in 1.5% KMnO4 in veronal-acetate buffer (0.3 mM sodium acetate, 0.3 mM sodium barbital, pH 7.6) for 20 min at room temperature. The specimens were dehydrated by washing with increasing concentrations of ethanol followed by two washes with 100% propylene oxide. The cells were then infiltrated with a 50:50 mix of propylene oxide and the POLY/BED 812 (Polysciences, Inc., Warrington, PA) for 2 d. The preparations were dried in vacuum overnight, infiltrated with 100% POLY/BED with accelerator 2,4,6-Tri(dimethylaminomethyl) phenol (DMP-30, Polysciences, Inc.) for another 2 d, and then incubated in an oven overnight at 60°C. The resulting samples were mounted on blocks, sectioned, and prepared for examination on a JEOL 100CX II transmission electron microscope.
Preparation of HA-Epitope–tagged GSA7/APG7 and Mutagenesis
We constructed a GSA7 gene containing its endogenous promoter, a peptide epitope (YPYDVPDYA) derived from influenza hemagglutinin (HA) at its C terminus, and a unique 3′ restriction site (ClaI). This was performed by PCR using the plasmid pYWP7-4 containing the genomic GSA7 and primers synthesized by the DNA Synthesis Laboratory at the University of Florida. The resulting DNA was digested with ClaI and MstI and inserted into the ClaI and EcoRV sites of pYM4 (pGSA7HA). The C-terminal sequence of the recombinant Gsa7-HA was DEDSEWINYPYDVPDYA, with the original C terminus being DEDSEWIN. Gsa7(ΔATP), which lacked amino acid residues 317 through 345, was constructed by inverse PCR using pGSA7HA as a template and two primers (with SmaI sites) that flanked the deletion. The resulting linear DNA was cut with SmaI to ensure the correct ORF. The vector was then ligated and amplified in E. coli DH5α cells. Site-directed mutagenesis was performed using the Stratagene QuikChange Site-Directed Mutagenesis Kit (Stratagene). Mutated Gsa7-HA was prepared by PCR using pGSA7HA as a template and two complementary oligonucleotide primers that contained the desired mutations (C518S and C562S). The template was eliminated by DpnI endonuclease digestion, and the “nicked” plasmid containing the mutated gsa7 was amplified in Epicurian Coli XL1-Blue cells. The mutations were verified by sequencing performed by the DNA Sequencing Facility at the University of Florida.
We constructed an APG7 gene containing its endogenous promoter, the HA peptide at its C terminus, and unique SmaI (5′) and ClaI (3′) restriction sites. This was performed by PCR using genomic DNA from S. cerevisiae as a template and primers synthesized by the DNA Synthesis Laboratory at the University of Florida. The resulting DNA was digested with SmaI and ClaI and inserted into the ClaI and EcoRV sites of pYM4 (pAPG7HA).
RESULTS
Microautophagy of Peroxisomes Is Impaired in the gsa7 Mutant
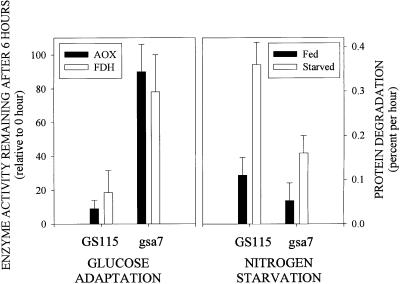
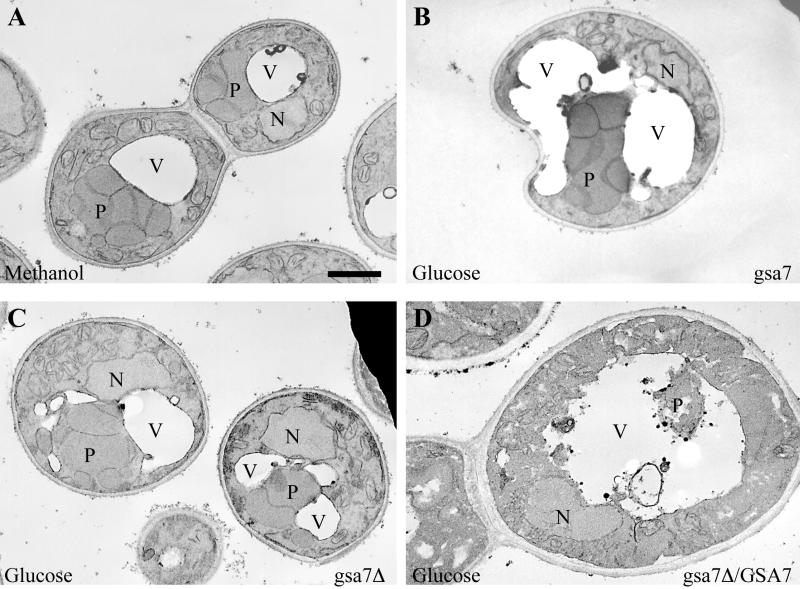
The P. pastoris parental strain GS115 and the mutant strain WDY7 (gsa7) were grown on methanol as the sole carbon source until they reached stationary growth, and then they were switched to glucose medium. During the following adaptation, >90% of the peroxisomal AOX and 80% of the cytosolic FDH were degraded by parental cells, whereas the level of AOX and FDH in gsa7 remained relatively unchanged (Figure 1). The impaired glucose-induced autophagy in gsa7 cells was verified next by electron microscopy. Cells grown on methanol as the sole carbon source contain numerous large peroxisomes clustered together (Figure 2A). At 3-h glucose adaptation, the vacuoles in virtually all of the gsa7 cells examined were extended around the peroxisomes (Figure 2B); however, complete engulfment and hence degradation of the peroxisomes could not be seen. It therefore seems that gsa7 is not defective in the initiation of autophagy, but rather at a later step involving the completion of the sequestration. This is unlike gsa1/pfk1 cells, which we showed previously to be defective in the initiation of sequestration leading to a vacuolar morphology at 3-h glucose adaptation undistinguishable from that seen at 0 h (Yuan et al., 1997).
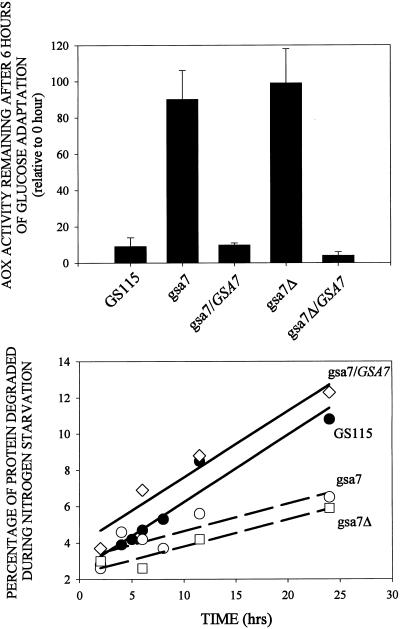
Figure 1.
Peroxisome degradation during nutrient adaptation and protein degradation during nitrogen starvation in parental GS115 and gsa7–1 mutant. Cells were grown for 24–36 h in methanol induction medium. At 0 and 6 h of glucose (2%) adaptation, cell-free extracts were prepared, and alcohol oxidase (AOX) and formate dehydrogenase (FDH) activities were assayed as described in MATERIALS AND METHODS. The values represent the mean ± SD of three or more determinations and are presented as a percentage of the activity measured at 0 h. The degradation of cellular proteins during nitrogen starvation was measured as described in MATERIALS AND METHODS. Cellular proteins were radiolabeled and then chased in medium containing histidine and nitrogen (Fed) or lacking amino acids and nitrogen (Starved). The production of TCA-soluble radioactivity was measured during 2–24 h of chase, and the rates of degradation were determined by linear regression. The values are presented as the percentage degraded per hour ± SE of the linear regression.
Figure 2.
Morphology of gsa7 and gsa7Δ cells during glucose adaptation. gsa7Δ cells were grown in methanol (A) and gsa7, gsa7Δ, and gsa7Δ cells transformed with the normal GSA7 gene (gsa7Δ/GSA7) were grown in methanol induction medium then adapted to glucose for 3 h (B–D). Cells were harvested, fixed in potassium permanganate, and prepared for electron microscopy as described in MATERIALS AND METHODS. N, nucleus; V, vacuole; P, peroxisome. Bar, 0.5 μm.
We next examined the ability of gsa7 cells to degrade proteins during nitrogen starvation. Nitrogen starvation has been shown to stimulate macroautophagy in S. cerevisiae (Baba et al., 1994). In P. pastoris, protein degradation was enhanced threefold when cells were starved for nitrogen (Figure 1). As was reported for S. cerevisiae, autophagosomes were visualized in GS115 cells starved for nitrogen (Dunn, unpublished data). This starvation-induced degradation of proteins was suppressed in gsa7 cells (Figure 1). Our data suggest that gsa7 cells were defective in both glucose-induced microautophagy of peroxisomes and starvation-induced macroautophagy of endogenous proteins
GSA7 Encodes a Unique 71-kDa Protein
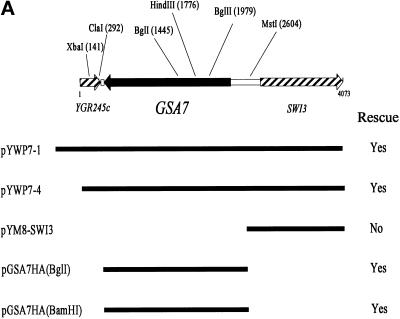
gsa7 cells were transformed with the episomal shuttle vector pYM8 containing a genomic library from P. pastoris (a generous gift of Dr. J. M. Cregg). The transformed clones were harvested and replated on YNM plates, and 30,000 colonies were screened for their ability to degrade AOX by the direct colony assay. Thirty clones were selected and rescreened by the liquid medium assay, and out of these, three had the ability to degrade AOX as efficiently as GS115 cells. The rescuing plasmids (pYWP7-1, pYWP7-4, and pYWP7-5) were isolated and shown by restriction map analysis to contain overlapping DNA sequences. pYWP7-1 had a genomic DNA insert of 4.5 kb, pYWP7-4 of 4 kb, and pYWP7-5 of 10 kb. The 4-kb insert from pYWP7-4 was sequenced, and all six reading frames were analyzed and compared with the S. cerevisiae genome database (Figure 3A). The insert encoded a protein homologous to YHR171w in S. cerevisiae, a 104 amino acid C-terminal fragment homologous to YGR245c and a 340 amino acid N-terminal fragment homologous to the transcription factor SWI3. We have shown that the SWI3 fragment did not rescue gsa7 cells. We next constructed the putative GSA7 (YHR171w) with an HA epitope tag and accompanying 300 bp of upstream promoter into the pYM4 vector. The insertion of the GSA7HA gene into the GSA7 locus was expedited by cutting pGSA7HA with BglI (within the GSA7 gene) or into the HIS4 locus by cutting with BamHI (within the HIS4 gene). Those gsa7 cells transformed with pGSA7HA(BglI) (see Figure 7) or pGSAHA(BamHI) (Figure 4 and see Figure 7) were able to efficiently degrade AOX during glucose adaptation. In addition, the rate of protein degradation under nitrogen starvation conditions in those gsa7 cells transformed with pGSA7HA(BamHI) was comparable to that observed in GS115 cells (Figure 4). GSA7 encoded a unique protein of 654 amino acids (Figure 3B). The amino acid sequence did not reveal any transmembrane domains but did contain a limited sequence homologue to the family of ubiquitin-activating enzymes (see below).
Figure 3.
Nucleotide and predicted amino acid sequence of GSA7. gsa7 cells were stably transformed with pYWP7-1 (containing a 4.5-kb genomic DNA insert), pYWP7-4 (containing a 4.1-kb genomic DNA insert), pGSA7HA (containing a 2.3-kb ORF of GSA7 with an HA epitope at its C terminus), and pWP-SWI3 (containing a 1.5-kb 3′ fragment of the original 4.1-kb genomic DNA). pGSA7HA(BglI) was linearized within the GSA7gene with BglI to promote insertion into the gsa7 locus. pGSA7HA (BamHI) was linearized within the HIS4 gene with BamHI to promote insertion into the his4 locus. The resulting transformants were analyzed for their ability to rescue the mutant phenotype as described previously (Yuan et al., 1997). The GSA7 ORF encodes a protein of 989 amino acids with a molecular mass of 71 kDa. The GSA7 sequence has a GenBank accession number of AF098976.
Figure 7.
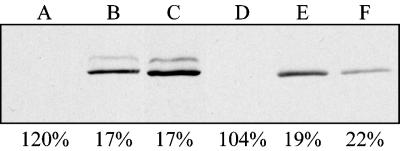
Complementation of gsa7 by APG7. gsa7 cells were stably transformed with pGSA7HA(BamHI) (lanes A and B), pGSA7HA(BglI) (lane C), or pAPG7HA(BamHI) (lanes D–F) behind their respective endogenous promoters. Expression of Gsa7-HA and Apg7-HA was evaluated by Western blotting with a monoclonal antibody to the HA epitope. Clones expressing and nonexpressing Gsa7-HA and Apg7-HA were grown in methanol induction medium and adapted to glucose for 6 h. Cells were lysed, and the extracts were assayed for AOX activity (see MATERIALS AND METHODS). The numbers represent the percentage of AOX activity present at 6 h relative to that measured at 0 h.
Figure 4.
Rescue of gsa7 and gsa7Δ by Gsa7p. gsa7 and gsa7Δ cells were stably transformed with pGSA7HA(BamHI). Those colonies that grew on His− medium were screened for expression of Gsa7-HA by Western blotting with an antibody to the HA epitope. The nontransformed (gsa7 and gsa7Δ) and transformed (gsa7/GSA7 and gsa7Δ/GSA7) cells were then grown in methanol induction medium and adapted to glucose for 6 h. The AOX activities are presented as a percentage of the activities measured at 0 h and represent the mean ± SD of four or more determinations. The degradation of cellular proteins during nitrogen starvation was performed as described previously in MATERIALS AND METHODS. Cellular proteins were radiolabeled and then chased in starvation medium lacking histidine and nitrogen. The production of TCA-soluble radioactivity was measured at 2–24 h of chase.
Degradation of Peroxisomes by Microautophagy Requires GSA7
We next isolated a null mutant of Gsa7p (Figure 5). This was performed by replacing a 200-bp fragment of GSA7 with a 3-kb genomic DNA fragment containing the S. cerevisiae ARG4 with its promoter (Figure 5). The ARG4 insert flanked by 0.4-kb 5′ and a 1.2-kb 3′ fragments of GSA7 was excised and introduced into PPF1 (his4, arg4). Histidine auxotrophs defective in AOX degradation were identified by direct colony assay and verified by liquid medium assay. A single clone was isolated, and the insertion of ARG4 into the GSA7 gene was verified by PCR (Figure 5). PCR analysis of the genomic DNA was performed using primers located inside and outside the gsa7::ARG4 insert, and the sizes of the resulting PCR products were compared by agarose gel electrophoresis (Figure 5). A PCR fragment of 2.5 kb corresponding to the normal GSA7 with promoter was observed in GS115 when using primers a and b; however, the 5-kb PCR fragment obtained from gsa7Δ was consistent with the ARG4 gene residing within the GSA7 locus, thereby disrupting its coding frame. Further PCR analysis of the genomic DNA of gsa7Δ revealed the expected fragments of 3.3 kb (primers a and d) and 6.1 kb (primers a and c) for the ARG4 gene being inserted into the GSA7 locus. gsa7Δ cells were unable to degrade alcohol oxidase during glucose adaptation (Figure 4). gsa7Δ was then transformed with pGSA7HA(BamHI). The resulting transformant was shown to efficiently degrade AOX during glucose adaptation (Figure 4).
Figure 5.
Preparation and characterization of the GSA7 knockout. The S. cerevisiae ARG4 gene was inserted into HindIII and BglII sites within the ORF of GSA7. The ARG4 insert included the ORF of ARG4 (arrow) and 1300 bp of 5′ and 500 bp of 3′ noncoding regions. The 5′ noncoding region includes the ARG4 promoter, whereas the 3′ noncoding region presumably lacks a promoter sequence, because the next gene downstream of ARG4 is 2200 bp away. In addition, the 3′ noncoding region likely contains a transcription terminator. The knockout fragment (5.1 kb) was cut out of the shuttle vector by ApaI and ScaI digestion and used to transform PPF1 (his4, arg4). Stable Arg+ transformants were isolated, gsa mutants were identified by direct colony assay, and the recombination of this construct with the genomic DNA was confirmed by PCR. Genomic DNA was isolated from GS115 and gsa7Δ, and the site of insertion of the ARG4 gene was verified by PCR analysis using the primers inside (b and d) and outside (a and c) the insert. The expected fragments of 5.0 kb (a → b), 6.1 kb (a → c), and 3.3 kb (a → d) were consistent with the ARG4 gene being inserted into the GSA7 locus of gsa7Δ cells.
At 3 h of glucose adaptation, gsa7Δ cells and the rescued clone gsa7Δ/GSA7 were processed for electron microscopy. gsa7Δ (Figure 2C) cells displayed a morphology indistinguishable from the gsa7 mutants (Figure 2B). The vacuole was observed surrounding the peroxisomes as if autophagy were proceeding normally, but complete engulfment and degradation of the peroxisomes could not be detected. In the rescued gsa7Δ/GSA7 clone (Figure 2D), only remnants of peroxisomes inside the vacuole could be seen. The vacuole being enlarged and with a ruffling border displayed a morphology identical to parental GS115 strains undergoing glucose adaptation (Yuan et al., 1997).
We next examined the ability of gsa7Δ to degrade proteins under nitrogen starvation conditions (Figure 4). In nitrogen-starved GS115 cells, endogenous protein was degraded at a rate of 0.37% of the total cellular protein per hour; however, the rates of protein degradation in starved gsa7 and gsa7Δ cells were much slower (0.15%/h). The data are consistent with Gsa7p being required for both glucose-induced and starvation-induced autophagy in P. pastoris.
GSA7 Homologues in S. cerevisiae and H. sapiens
We originally indicated that Gsa7p had homology to S. cerevisiae YHR171w. YHR171w has recently been characterized and identified as APG7, a gene required for nitrogen-starvation–induced autophagy in S. cerevisiae (Tanida, et al., 1999), as well as CVT2, a gene necessary for constitutive delivery of pro-API from the cytosol to the vacuole by autophagy in S. cerevisiae (Kim, et al., 1999). The amino acid sequences of Gsa7p and Apg7p/Cvt2p were compared in Figure 6. Although the overall homology showed 44% identity and 66% similarity, the C-terminal half of the protein (residues 269 to 654) was 58% identical and 74% similar. A protein of unknown function with homology to Gsa7p/Apg7p was also found in Schizosaccharomyces pombe (GenBank accession number: AL021838)
Figure 6.
Comparison of GSA7 genes. The amino acid sequence of GSA7 from P. pastoris (see Figure 3B) was aligned to APG7 from S. cerevisiae, and GSA7 was aligned from H. sapiens (HsGSA7). The sequence of HsGSA7 has a GenBank accession number of AF094516. Amino acid identity represented by dashed lines was evident within many regions of the sequence. Gaps represented by dots were inserted to optimize amino acid alignments.
We next examined the ability of AGP7 to complement gsa7 to verify that these proteins have similar functional roles. Genomic DNA from S. cerevisiae was used as a template for PCR using a 3′ primer adding the HA tag to the protein and an 5′ primer 300-bp upstream of the start codon including the promoter region. The PCR fragment was put into the nonepisomal vector pYM4, and the plasmid linearized in the P. pastoris HIS4 gene was used to transform the gsa7 cells. gsa7 cells were transformed with pGSA7HA(BamHI), pGSA7HA(BglI), or pAPG7HA(BamHI), the expression of Gsa7-HA and Apg7-HA was verified by Western blotting, and the ability of the transformed cells to degrade AOX was measured by the liquid medium assay (Figure 7). Both Gsa7-HA and Apg7-HA migrated as 72-kDa proteins. A gsa7 clone that had been transformed, but was not expressing Gsa7-HA, was unable to degrade AOX during glucose adaptation (Figure 7, lane A). Alternatively, those gsa7 cells transformed with pGSA7HA(BamHI) (Figure 7, lane B) or pGSA7HA(BglI) (Figure 7, lane C) and expressing Gsa7-HA efficiently degraded AOX. The expression of Apg7-HA in P. pastoris behind its S. cerevisiae promoter was only slightly less than that seen with Gsa7-HA behind its endogenous promoter. In addition, Apg7-HA rescued the gsa7 mutant phenotype. That is, in gsa7 cells expressing Apg7p-HA (Figure 7, lanes E and F), only 19–22% of the AOX activity remained after 6 h of glucose adaptation as compared with 104% in cells not expressing Apg7-HA (Figure 7, lane D) and 17% in gsa7 cells expressing Gsa7-HA (Figure 7, lanes B and C). The results indicated that Apg7p is a functional homologue of Gsa7p. In addition, Gsa7p was able to partially rescue the cvt2 phenotype (Kim et al., 1999).
We next searched the expressed sequence tagged (EST) databases. Mouse ESTs from eight-cell embryo (AU020314), blastocyst (AA399856), and liver (AI099344) were found to encode proteins homologous to Gsa7p/Apg7p. Additional homologous proteins were identified in human ESTs from tissues as diverse as brain (F10693), heart (AA630317), testis (AA393419), lung (N77821), melanocyte (H99680), olfactory epithelium (H65101), and placenta (D78810) as well as from kidney (AA975772) and colon (AA569145) tumors and the HeLa carcinoma cell line (AA630317). The high homology between the amino acid sequences encoded by these ESTs and Gsa7p suggested that a protein with a similar function likely exists in mammals. Therefore, we sequenced the cDNA (IMAGE Clone ID:23293; ATCC #327890) isolated from a human infant brain cDNA library and compared its amino acid sequence with Gsa7p and Apg7p (Figure 6). The human Gsa7p sequence was 38% identical and 55% similar to Gsa7p and Apg7p. Again, the homology is very strong in the C-terminal half of the protein, having almost 50% identity to both Gsa7p and Apg7p. Other regions of high homology near the N terminus can be seen in Figure 6.
Gsa7p Displays E1-like Enzyme Properties
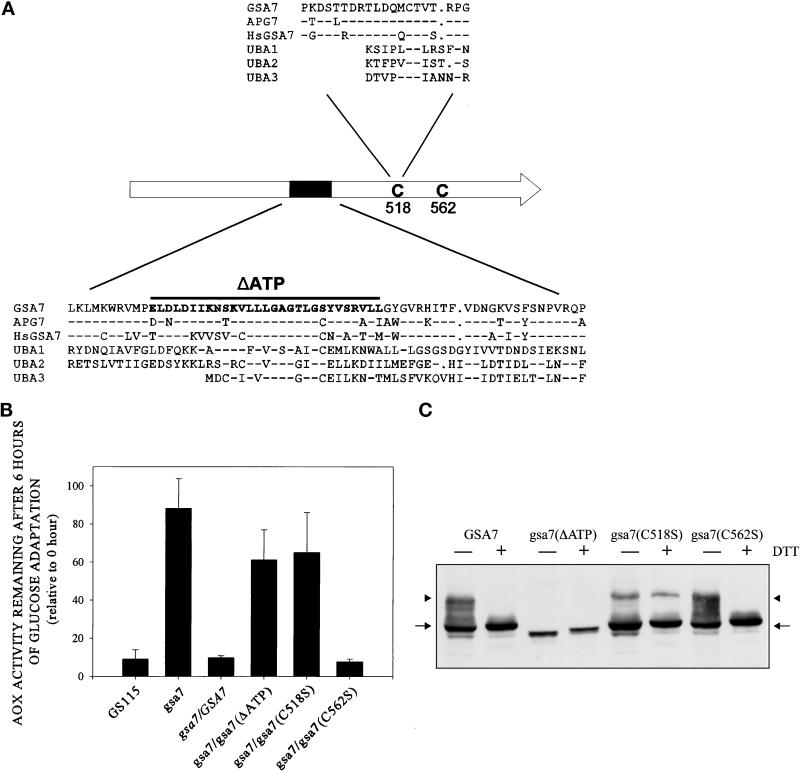
The high conservation in the C-terminal region of Gsa7p between yeast and mammals suggests that this portion of the protein may have some kind of enzymatic activity. A comparison of these protein sequences against the nonredundant protein database at National Center for Biotechnology Information revealed similarity to the family of ubiquitin-activating enzymes E1, particularly Uba2p and Uba3p. Uba2p and Uba3p have recently been shown to activate the ubiquitin-like proteins Smt3p/SUMO-1 and Rub1p/NEDD-8, respectively, in an E1-like manner (Lammer et al., 1998; Liakopoulos et al., 1998; Mahajan et al., 1998; Matunis et al., 1998; Schwarz et al., 1998). A highly conserved region of Gsa7p (residues 332 to 337) aligned with the putative E1 ATP-binding site GxGxxG (Figure 8A). The GxGxxG motif is found in many ATP-binding proteins (Koonin, 1993) and in all E1 enzymes (Haas and Siepmann, 1997). In addition, Gsa7p and the E1 enzymes have conserved positively charged amino acids (K or R) flanking the GxGxxG motif. We then constructed a Gsa7-HA mutant protein lacking 29 amino acids (residues 317 to 345) around and including the GxGxxG motif Gsa7(ΔATP)-HA (Figure 8A). AOX degradation remained impaired in gsa7 cells expressing this mutant protein (Figure 8B), suggesting that this domain is important for Gsa7p function, possibly in binding ATP.
Figure 8.
Complementation of gsa7 by normal and mutant forms of recombinant Gsa7-HA. (A) GSA7, APG7, and HsGSA7 proteins were aligned to three ubiquitin-activating enzymes (UBA1, UBA2, and UBA3). The amino acid alignment around the putative catalytic domain (C518) and ATP-binding region (K327–R342) are indicated. Amino acid identity represented by dashed lines was evident within these regions. Gaps represented by dots were inserted to optimize amino acid alignments. (B) gsa7 cells were stably transformed with GSA7-HA, gsa7(ΔATP), gsa7(C518S), and gsa7(C562S). The cells were then grown in methanol induction medium then adapted to glucose for 6 h. Cell extracts were prepared, and AOX assays were performed. The resulting values are presented as a percentage of the activity measured at 0 h and represent the mean ± SD of four or more determinations. (C) Aliquots of cell extracts prepared at 3 h of glucose adaptation were solubilized in 2% SDS and boiled for 3 min (−DTT) or solubilized in 2% SDS with 1.5% DTT and boiled for 5 min (+DTT), and the proteins were separated by SDS-PAGE. After transfer to nitrocellulose, the epitope-tagged Gsa7 proteins were identified using polyclonal antibodies that recognized the HA epitope. Gsa7-HA migrated as a 72-kDa protein (arrow). A thio-ester conjugate of Gsa7-HA was observed at ∼100 kDa (arrowhead).
A second conserved region of Gsa7p aligns with the putative active site of E1 (Figure 8A). The catalytic cysteine is required for a thio-ester bond between the sulfhydryl moiety of cysteine and the carboxyl moiety of the C-terminal glycine of ubiquitin or a ubiquitin-like protein. Cysteine 518 in Gsa7p and 509 in Apg7p aligned with the catalytic sites in Uba1p (C600), Uba2p (C177), and Uba3p (C168). This site is flanked by prolines and contained a conserved CTx region with “x” being a valine, leucine, or isoleucine (Figure 8A). We examined the functional importance of this cysteine by mutating it to a serine, Gsa7(C518S). gsa7 cells expressing Gsa7(C518S)-HA were unable to efficiently degrade AOX during glucose adaptation (Figure 8B). As a control we also mutated cysteine 562, which was found in an area of high homology between Gsa7p, Apg7p, and HsGsa7p, but which itself was not conserved. gsa7 cells expressing Gsa7(C562S)-HA efficiently degraded AOX during glucose adaptation (Figure 8B).
The inability of Gsa7(ΔATP)-HA and Gsa7(C518S)-HA to complement gsa7 cells was not due to a lack of protein expression. Gsa7-HA, Gsa7(ΔATP)-HA, Gsa7(C518S)-HA, and Gsa7(C562S)-HA were all expressed at comparable levels (Figure 8C). The ubiquitin-activating enzyme forms a DTT-sensitive thio-ester linkage with ubiquitin (Jonnalagadda et al., 1988; Hochstrasser, 1996). This conjugation requires ATP (Jonnalagadda et al., 1988; Hochstrasser, 1996). Cells expressing Gsa7-HA contained two predominant HA-tagged proteins at ∼72 and 100 kDa when samples were not treated with DTT. Under reduced conditions, the 100-kDa protein was not observed. Similar findings were observed in cells expressing Gsa7(C562S)-HA; however, the 100-kDa protein was not detected in cells expressing Gsa7(ΔATP)-HA. The presence of the 100-kDa protein was coincident with the ability of these proteins to rescue the gsa7 mutants. The data suggest that the upper band might represent the intermediate conjugate of Gsa7p to the protein it is activating. It has recently been shown that in S. cerevisiae this protein is the 21-kDa gene product of APG12 (Mizushima et al., 1998). Therefore, we suggest that the 100-kDa protein is a thio-ester conjugate between Gsa7p and a putative homologue of Apg12p. Cells expressing Gsa7(C518S)-HA contained both 72- and 100-kDa proteins, regardless of DTT treatment. The mutation of the presumed active site cysteine 518 to a serine appears to yield a stable ester instead of a thio-ester intermediate. We propose that the stable ester linkage prevents the transfer of Apg12p from Gsa7p to the appropriate E2 (i.e., Apg10p). This would ultimately result in an inactivation of the Gsa7p/Apg7p conjugation pathway consistent with the inability of Gsa7(C518S)-HA to rescue gsa7 cells.
In addition to the loss of the 100-kDa protein, DTT reduction also caused a small but significant decrease in the migration of normal and mutant forms of Gsa7-HA (Figure 8C). This change in migration may reflect a change in the tertiary structure of Gsa7p brought about by sulfhydryl reduction.
DISCUSSION
When P. pastoris adapt from methanol to glucose, the peroxisomes are sequestered within “finger-like” extensions of the vacuole by a process analogous to microautophagy (Tuttle and Dunn, 1995). We have isolated eight glucose-induced selective autophagy (gsa) mutants (our unpublished data). We have shown that Gsa1p is a phosphofructokinase and required for the onset of microautophagy (Yuan et al., 1997). We report here the identification and characterization of Gsa7p. GSA7 encodes a unique 71-kDa protein with homologues in S. cerevisiae and H. sapiens (Figure 6). gsa7 and gsa7Δ cells were unable to degrade peroxisomal AOX and cytosolic FDH during glucose adaptation or cellular proteins during amino acid and nitrogen starvation (see Figures 1 and 4). During glucose adaptation, the peroxisomes of both mutants were surrounded by “finger-like” extensions of the vacuole but not sequestered into the vacuole (Figure 2). In addition, the ability of the cells to degrade peroxisomes was coincident with the presence of a thio-protein conjugate of Gsa7-HA (Figure 8). The data suggest that Gsa7p is required for a late event in the microautophagic sequestration of peroxisomes.
In this study we have provided evidence to suggest that Gsa7p is an enzyme homologous to ubiquitin-activating enzyme E1 and capable of protein–protein conjugation, which is vital for microautophagy of peroxisomes in P. pastoris. Protein conjugation as a regulatory mechanism for degradation of proteins has been known for a long time. Ubiquitination as a signal for recognition and degradation by the proteasome starts with the activation of ubiquitin by ubiquitin-activating enzyme E1 (Hochstrasser, 1996). The activation of ubiquitin by Uba1p proceeds by coupling the C-terminal glycine of ubiquitin to adenosine monophosphate (Jonnalagadda et al., 1988; Hochstrasser, 1996; Haas and Siepmann, 1997). The “activated” ubiquitin is then transferred to cysteine 600 and forms a thio-ester complex. The ubiquitin is subsequently transferred to a substrate through an E2 enzyme intermediate, with or without the aid of an E3 ligase (Hochstrasser, 1996). Similar scenarios have been proposed for the conjugation of Smt3p/SUMO-1 to Ran GTPase-activating protein (RanGAP1), which requires the E1, UBA2, and E2, UBC9, and Rub1p/NEDD-8 to CDC53/cullin, which requires the E1, UBA3, and E2, UBC12 (Matunis et al., 1996; Liakopoulos et al., 1998; Matunis et al., 1998). Gsa7p bears important homologies to the putative ATP-binding motif as well as the catalytic domain of this family of protein-activating E1 enzymes. Interestingly, Uba1p has two ATP-binding domains, whereas Uba2p, Uba3p, and Gsa7p/Apg7p have only one. We have shown here that the ATP-binding domain and the functional C518, which is analogous to C507 in Apg7p, were necessary for Gsa7p function in peroxisome autophagy. Mizushima et al. (1998) have shown that the Gsa7p homologue Apg7p catalyzes the conjugation of the C-terminal glycine of Apg12p to lysine 149 of Apg5p with the assistance of Apg10p, which is presumed to be an E2-like protein. Mutations within the ATP-binding site (G333A) or cysteine active site (C507A) of Apg7p inhibited its ability to interact with Apg12p (Tanida et al., 1999). In addition, they have shown that G333 and C507 were required for autophagy of cytosolic alkaline phosphatase and API (Tanida et al., 1999). We have shown that Gsa7p is required for glucose-induced autophagy of peroxisomes and starvation-induced autophagy of cellular proteins. In addition, our data suggest that Gsa7p probably forms a thio-ester linkage with a putative homologue of Apg12p; however, the requirements for APG12 and APG5 in peroxisome autophagy have yet to be explored.
It has been known that protein conjugation may serve purposes other than the signaling of protein degradation. For example, the ubiquitination of Ste2p is required for its internalization from the cell surface during endocytosis (Hicke and Riezman, 1996). In addition, two ubiquitin-like proteins have been found in both S. cerevisiae and mammals. Smt3p/SUMO-1 is activated by UBA2, which until recently was thought to be a nuclear activator of ubiquitin, whereas Rub1p/NEDD-8 is activated by UBA3. Smt3p/SUMO-1 conjugation to RanGAP1 is necessary to target RanGAP1 to the nuclear pore complex (Matunis et al., 1996, 1998). It has been suggested that the conjugation of CDC53/cullin by Rub1p/NEDD-8 may modify the interaction between the SCF ubiquitin ligase and its substrates (i.e., Sic1p and Cln2p) (Liakopoulos et al., 1998). Gsa7p/Apg7p has been shown to be responsible for the conjugation of Apg12p to Apg5p (Mizushima et al., 1998). This protein complex is associated with membranes. The function of the conjugation activity of Gsa7p in glucose-induced autophagy remains to be fully elucidated. Gsa7p does not seem to be involved in regulation of initiation of autophagy or in the recognition and partial sequestration of the peroxisomes. It is therefore unlike gsa1/pfk1, which we previously have shown to be involved in initiation of microautophagy (Yuan et al., 1997). The morphology of the gsa7 mutant during adaptation from methanol to glucose suggests instead that Gsa7p is necessary either to bring the opposing vacuolar membranes together or to assist in the fusion of the opposing vacuolar membranes. Gsa7p function in glucose-induced peroxisome autophagy is likely mediated through the formation of the membrane-associated Apg12p-Apg5p complex; however, the functional role of GSA7-mediated protein conjugation in the fusion and/or prefusion events has yet to be defined. The implications of the conjugation of Agp12p to Apg5p are still unknown, but conjugation of these two membrane-associated proteins could of course bring two membranes close together, maybe as a step leading up to fusion.
Mechanistically, microautophagy and macroautophagy are quite different. Thus, it was surprising to find a protein that is required for both pathways. GSA7 is required for sequestering peroxisomes by the vacuole, whereas APG7 appears to be required for the formation of cvt vesicles and apg vacuoles (e.g., autophagosomes) (Mizushima et al., 1998; Kim et al., 1999; Tanida, et al., 1999). We propose that the homotypic fusion between opposing membranes of the sequestering organelles (e.g., vacuole membrane and autophagosome membrane) require at least some of the same molecular machinery. Without this fusion event the autophagosome will not be completely formed, and thus it would be difficult to distinguish morphologically a fusion mutant from those impaired in the initiation of autophagy. The same would apply if GSA7/APG7 functions to bring the membranes together for fusion. Therefore, we propose that gsa7/apg7 mutants would have partially formed autophagosomes. Indeed, Kim et al. (1999) have shown pro-API associated with subcellular membranes of cvt2 mutants; however, the pro-API was accessible to proteinase K digestion. The data are consistent with pro-API in cvt vesicles that had failed to close (Kim, et al., 1999). We propose that the homotypic fusion events required for sequestration of peroxisomes into the vacuole and cytosolic proteins into cvt vesicles or autophagosomes require the Gsa7p/Apg7p conjugation pathway.
Several of the proteins described in this protein conjugation pathway appear to have mammalian counterparts. We have identified and sequenced a human homologue of GSA7 from an infant brain cDNA library, and human homologues of APG5 and APG12 have also been identified and sequenced (Hammond et al., 1998; Mizushima et al., 1998). These proteins appear to be present in a number of different tissues, which underlines their importance in cellular events. Whether these proteins are required for autophagy in mammalian cells remains to be seen. It is particularly interesting that several mouse GSA7 ESTs come from embryogenic cDNA libraries. This suggests a role for the APG7/GSA7 conjugation pathway and possibly autophagy in the cellular remodeling that occurs during development. APG5 was identified as a protein being associated with the onset of apoptosis at a step downstream of caspase activation (Hammond et al., 1998). This finding implies that the APG7/GSA7 protein conjugation pathway may have a role in apoptosis or that apoptosis is accompanied by or dependent on autophagy. Indeed, the cell death induced by antiestrogens in mammary carcinoma cells and by toxins in Madin–Darby canine kidney cells can be suppressed by drugs (e.g., cycloheximide and 3-methyladenine) that inhibit autophagy (Sandvig and van-Deurs, 1992; Bursch et al., 1996). Further experiments are necessary to define the role of APG7/GSA7 and autophagy in apoptosis.
In summary, we have identified a unique E1-like protein, which we call Gsa7p, that is required for glucose-induced and starvation-induced autophagy in P. pastoris. The S. cerevisiae homologue (Apg7p) has been identified and shown to be required for constitutive (cvt pathway) and starvation-induced (apg pathway) autophagy (Scott et al., 1996; Mizushima et al., 1998). In addition, we have found the mammalian homologue in mouse embryonic tissues and various human organs. We propose that this enzyme is responsible for a protein–protein conjugation that serves a dual role in the fusion of the vacuolar membranes during sequestration of the peroxisomes in P. pastoris and in the fusion of the autophagosome membranes during sequestration of cytosolic components in S. cerevisiae. Our studies combined with those reported by Mizushima et al. (1998), Kim et al. (1999), and Tanida et al. (1999) suggest that the APG7/GSA7 conjugation pathway is necessary for the sequestration of cytosolic proteins and organelles into the vacuole/lysosome for degradation.
ACKNOWLEDGMENTS
This work was supported by grants from National Institutes of Health to W.A.D. (AM-33326) and the Norwegian Cancer Society to P.E.S. We are greatly indebted to Dr. James M. Cregg for the generous gifts of yeast strains, genomic DNA library, and the helpful conversations.
Abbreviations used:
- AOX
alcohol oxidase
- API
aminopeptidase I
- EST
expressed sequence tag
- FDH
formate dehydrogenase
- HA
hemagglutinin
REFERENCES
- Ahlberg L, Berkenstam A, Henell F, Glaumann H. Degradation of short and long lived proteins in isolated rat liver lysosomes: effects of pH, temperature, and proteolytic inhibitors. J Biol Chem. 1985;260:5847–5854. [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Smith JA, Seidman JG, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1988. [Google Scholar]
- Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Takeshige K, Baba H, Ohsumi Y. Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J Cell Biol. 1994;124:903–913. doi: 10.1083/jcb.124.6.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursch W, Ellinger A, Kienzl H, Torok L, Pandey S, Sikorska M, Walker R, Hermann RS. Active cell death induced by the antiestrogens tamoxifen and ICI 164 384 in human mammary carcinoma cells (MCF-7) in culture: the role of autophagy. Carcinogenesis. 1996;17:1595–1607. doi: 10.1093/carcin/17.8.1595. [DOI] [PubMed] [Google Scholar]
- Chiang HL, Schekman R, Hamamoto S. Selective uptake of cytosolic, peroxisomal, and plasma membrane proteins into the yeast lysosome for degradation. J Biol Chem. 1996;271:9934–9941. doi: 10.1074/jbc.271.17.9934. [DOI] [PubMed] [Google Scholar]
- Dunn WA., Jr Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J Cell Biol. 1990a;110:1923–1933. doi: 10.1083/jcb.110.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn WA., Jr Studies on the mechanisms of autophagy: maturation of the autophagic vacuole. J Cell Biol. 1990b;110:1935–1945. doi: 10.1083/jcb.110.6.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn WA., Jr Autophagy and related mechanisms of lysosome-mediated protein degradation. Trends Cell Biol. 1994;4:139–143. doi: 10.1016/0962-8924(94)90069-8. [DOI] [PubMed] [Google Scholar]
- Egner R, Thumm M, Straub M, Simeon A, Schuller H-J, Wolf DH. Tracing intracellular proteolytic pathways: proteolysis of fatty acid synthase and other cytoplasmic proteins in the yeast Saccharomyces cerevisiae. J Biol Chem. 1993;268:27269–27276. [PubMed] [Google Scholar]
- Haas AL, Siepmann TJ. Pathways of ubiquitin conjugation. FASEB J. 1997;11:1257–1268. doi: 10.1096/fasebj.11.14.9409544. [DOI] [PubMed] [Google Scholar]
- Hammond EM, Brunet CL, Johnson GD, Parkhill J, Milner AE, Brady G, Gregory CD, Grand RJ. Homology between a human apoptosis specific protein and the product of APG5, a gene involved in autophagy in yeast. FEBS Lett. 1998;425:391–395. doi: 10.1016/s0014-5793(98)00266-x. [DOI] [PubMed] [Google Scholar]
- Harding TM, Hefner Gravink A, Thumm M, Klionsky DJ. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein targeting pathway. J Biol Chem. 1996;271:17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- Huang P-H, Chiang H-L. Identification of novel vesicles in the cytosol to vacuole protein degradation pathway. J Cell Biol. 1997;136:803–810. doi: 10.1083/jcb.136.4.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonnalagadda S, Ecker DJ, Sternberg EJ, Butt TR, Crooke ST. Ubiquitin carboxyl-terminal peptides. Substrates for ubiquitin activating enzyme. J Biol Chem. 1988;263:5016–5019. [PubMed] [Google Scholar]
- Kato N. Formaldehyde dehydrogenase from methylotrophic yeasts. Methods Enzymol. 1990;188:459–462. doi: 10.1016/0076-6879(90)88073-j. [DOI] [PubMed] [Google Scholar]
- Kim J, Dalton VM, Eggerton KP, Scott SV, Klionsky DJ. Apg7p/Cvt2p is required for the cytoplasm-to-vacuole targeting, macroautophagy, and peroxisomal degradation pathways. Mol Biol Cell. 1999;10:1337–1351. doi: 10.1091/mbc.10.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J Mol Biol. 1993;229:1165–1174. doi: 10.1006/jmbi.1993.1115. . (Errata: J. Mol. Biol. 232, 1013.) [DOI] [PubMed] [Google Scholar]
- Kopitz J, Kisen GO, Gordon PB, Bohley P, Seglen PO. Nonselective autophagy of cytosolic enzymes by isolated rat hepatocytes. J Cell Biol. 1990;111:941–953. doi: 10.1083/jcb.111.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammer D, Mathias N, Laplaza JM, Jiang W, Liu Y, Callis J, Goebl M, Estelle M. Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes Dev. 1998;12:914–926. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang T, Schaeffeler E, Bernreuther D, Bredschneider M, Wolf DH, Thumm M. Aut2p and Aut7p, two novel microtubule associated proteins are essential for delivery of autophagic vesicles to the vacuole. EMBO J. 1998;17:3597–3607. doi: 10.1093/emboj/17.13.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardeux BR, Mortimore GE. Amino acid and hormonal control of macromolecular turnover in perfused rat liver. Evidence for selective autophagy. J Biol Chem. 1987;262:14514–14519. [PubMed] [Google Scholar]
- Liakopoulos D, Doenges G, Matuschewski K, Jentsch S. A novel protein modification pathway related to the ubiquitin system. EMBO J. 1998;17:2208–2214. doi: 10.1093/emboj/17.8.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan R, Gerace L, Melchior F. Molecular characterization of the SUMO-1 modification of RanGAP1 and its role in nuclear envelope association. J Cell Biol. 1998;140:259–270. doi: 10.1083/jcb.140.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis MJ, Wu J, Blobel G. SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J Cell Biol. 1998;140:499–509. doi: 10.1083/jcb.140.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishil T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- Mortimore GE, Lardeux BR, Adams CE. Regulation of microautophagy and basal protein turnover in rat liver. Effects of short-term starvation. J Biol Chem. 1988;263:2506–2512. [PubMed] [Google Scholar]
- Mortimore GE, Poso AR, Lardeux BR. Mechanism and regulation of protein degradation in liver. Diabetes Metab Rev. 1989;5:49–70. doi: 10.1002/dmr.5610050105. [DOI] [PubMed] [Google Scholar]
- Sandvig K, van-Deurs B. Toxin-induced cell lysis: protection by 3-methyladenine and cycloheximide. Exp Cell Res. 1992;200:253–262. doi: 10.1016/0014-4827(92)90171-4. [DOI] [PubMed] [Google Scholar]
- Schwarz SE, Matuschewski K, Liakopoulos D, Scheffner M, Jentsch S. The ubiquitin-like proteins SMT3 and SUMO-1 are conjugated by the UBC9 E2 enzyme. Proc Natl Acad Sci USA. 1998;95:560–564. doi: 10.1073/pnas.95.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Baba M, Ohsumi Y, Klionsky DJ. Aminopeptidase I is targeted to the vacuole by a nonclassical vesicular mechanism. J Cell Biol. 1997;138:37–44. doi: 10.1083/jcb.138.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Hefner Gravink A, Morano KA, Noda T, Ohsumi Y, Klionsky DJ. Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc Natl Acad Sci USA. 1996;93:12304–12308. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119:301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Mizushima N, Kiyooka M, Ohsumi M, Veno T, Ohsumi Y, Kominami E. Apg7p/Cvt2p: a novel protein-activating enzyme essential for autophagy. Mol Biol Cell. 1999;10:1367–1379. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, Wolf DH. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- Titorenko VI, Keizer I, Harder W, Veenhuis M. Isolation and characterization of mutants impaired in the selective degradation of peroxisomes in the yeast Hansenula polymorpha. J Bacteriol. 1995;177:357–363. doi: 10.1128/jb.177.2.357-363.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- Tuttle DL, Dunn WA., Jr Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris. J Cell Sci. 1995;108:25–35. doi: 10.1242/jcs.108.1.25. [DOI] [PubMed] [Google Scholar]
- Tuttle DL, Lewin AS, Dunn WA., Jr Selective autophagy of peroxisomes in methylotrophic yeasts. Eur J Cell Biol. 1993;60:283–290. [PubMed] [Google Scholar]
- Veenhuis M, Douma A, Harder W, Osumi M. Degradation and turnover of peroxisomes in the yeast Hansenula polymorpha induced by selective inactivation of peroxisomal enzymes. Arch Microbiol. 1983;134:193–203. doi: 10.1007/BF00407757. [DOI] [PubMed] [Google Scholar]
- Yuan W, Tuttle DL, Shi YJ, Ralph GS, Dunn WA., Jr Glucose-induced microautophagy in Pichia pastoris requires the alpha-subunit of phosphofructokinase. J Cell Sci. 1997;110:1935–1945. doi: 10.1242/jcs.110.16.1935. [DOI] [PubMed] [Google Scholar]