Abstract
In this study we characterized the extension, reciprocal arrangement, and orientation of syntenic chromosomal segments in the lar gibbon (Hylobates lar, HLA) by hybridization of a panel of ∼1000 human BAC clones. Each lar gibbon rearrangement was defined by a splitting BAC clone or by two overlapping clones flanking the breakpoint. A reconstruction of the synteny arrangement of the last common ancestor of all living lesser apes was made by combining these data with previous results in Nomascus leucogenys, Hoolock hoolock, and Symphalangus syndactylus. The definition of the ancestral synteny organization facilitated tracking the cascade of chromosomal changes from the Hominoidea ancestor to the present day karyotype of Hylobates and Nomascus. Each chromosomal rearrangement could be placed within an approximate phylogenetic and temporal framework. We identified 12 lar-specific rearrangements and five previously undescribed rearrangements that occurred in the Hylobatidae ancestor. The majority of the chromosomal differences between lar gibbons and humans are due to rearrangements that occurred in the Hylobatidae ancestor (38 events), consistent with the hypothesis that the genus Hylobates is the most recently evolved lesser ape genus. The rates of rearrangements in gibbons are 10 to 20 times higher than the mammalian default rate. Segmental duplication may be a driving force in gibbon chromosome evolution, because a consistent number of rearrangements involves pericentromeric regions (10 events) and centromere inactivation (seven events). Both phenomena can be reasonably supposed to have strongly contributed to the euchromatic dispersal of segmental duplications typical of pericentromeric regions. This hypothesis can be more fully tested when the sequence of this gibbon species becomes available. The detailed synteny map provided here will, in turn, substantially facilitate sequence assembly efforts.
In contrast to the relatively well-conserved karyotypes of the other hominoids (great apes and humans), lesser apes are characterized by extremely rapid chromosome evolution. Lesser apes (family Hylobatidae) include at least 12 species divided into four genera: Hoolock (formerly Bunopithecus), Hylobates, Symphalangus, and Nomascus (Brandon-Jones et al. 2004; Mootnick and Groves 2005). Each genus has a different diploid number: Hoolock, 2n = 38; Hylobates, 2n = 44; Symphalangus, 2n = 50; and Nomascus, 2n = 52. Chromosome translocations in each karyomorph have been documented using human and cross-species painting probes (Jauch et al. 1992; Koehler et al. 1995a, b; Arnold et al. 1996; Nie et al. 2001; Muller et al. 2002, 2003; Hirai et al. 2003; Mrasek et al. 2003; Ferguson-Smith et al. 2005). Recently, the chromosomes of the white-cheeked crested gibbon (Nomascus leucogenys, NLE) were defined in greater detail using high-resolution methods (Carbone et al. 2006; Roberto et al. 2007). In particular, synteny conservation and orientation were defined by a large panel of human BAC clones hybridized on NLE chromosomes, and by in silico mapping of the BAC end sequences of an NLE BAC library against the human genome reference sequence (Roberto et al. 2007).
In the present study we report detailed data on the synteny organization of the lar gibbon (white-handed gibbon, Hylobates lar, HLA), using a panel of almost 1000 human BAC clones. These results permit a clear picture of the flow of chromosomal changes from the recently proposed Hominoid ancestral karyotype (Stanyon et al. 2008) to the extant Hylobates lar. We tested recent hypotheses concerning the ancestral karyotype of Hylobatidae by comparing our results with data on the chromosome organization of Hylobates lar, Nomascus leucogenys, Hoolock hoolock (HHO), and Symphalangus syndactylus (SSY).
The precise phylogenetic and taxonomic relationships between lesser ape taxa are still controversial (Muller et al. 2003; Takacs et al. 2005; Whittaker et al. 2007). However, recent biomolecular studies either place Hoolock or Nomascus as basal (Roos and Geissmann 2001; Muller et al. 2003; Takacs et al. 2005; Baena et al. 2007), and most conclude that the genera Hylobates was the last and most recent genus to diverge (Takacs et al. 2005; Baena et al. 2007). In the present study, we have defined, using molecular cytogenetic tools, a high-resolution picture of synteny conservation and orientation in Hylobates lar. The results were then utilized to clarify the complex flow of chromosomal changes, which occurred in evolutionary lines leading to extant Hylobates lar and Nomascus leucogenys. Finally, our data allows us to test recent hypotheses about the ancestral Hylobatidae karyotype.
Results
Synteny segments definition
In order to define the synteny segments arrangements (SSA) of Hylobates lar, a total of about 980 human BAC clones were cohybridized in fluorescence in situ hybridization (FISH) experiments on metaphase chromosomes of this lesser ape. Examples of FISH experiments are reported in Figure 1. Comparisons were facilitated by the fact that most of these BACs were already used to characterize the SSA of another lesser ape species, NLE (Roberto et al. 2007). All of the mapping data of the latter study refer to the human genome assembly hg17 (May 2004 release). For this reason, the present study refers to the hg17 release. Chromosomes 5, 8, 11, and 13–21, however, remained unchanged in the hg18 assembly (March 2006), and only slight differences may occur for the other chromosomes. Further, BAC clones present in the hg17 release, but not present on the hg18, were avoided. Whenever possible, clones were selected in nonduplicated regions to avoid interpretation problems.
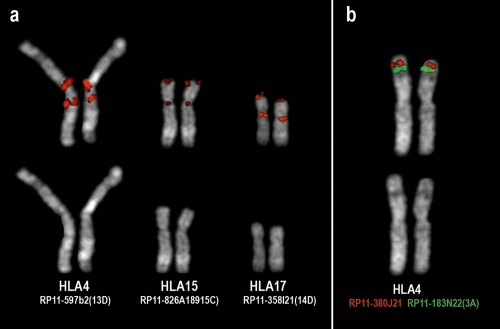
Figure 1.
Examples of FISH experiments on Hylobates lar metaphases using human BAC clones as probes. (a) Three examples of inversions, occuring in Hylobatidae ancestor, in which one of the two breakpoints fall in the centromeric/pericentromeric region. The figure shows the signals of the splitting BAC defining the euchromatic breakpoint. The code, in parenthesis on the right of each BAC name, refers to Supplemental Tables ST1 and ST2, where the BAC precise position on the human sequence is also reported. (b) The two BACs RP11-380J21 (red signal) and RP11-183N22 (green signal) are located, in humans, at chr3:64,053,486–64,212,751 and chr3:4,328,222–4,493,696, respectively, suggesting a rearrangement with respect to humans. On the contrary, the rearrangement occurred in the lineage leading to humans.
Additional human BAC clones were utilized in reiterative FISH experiments to define 12 HLA-specific rearrangements. The aim was to identify BACs yielding split signals or closely flanking each HLA-specific breakpoint. We also took advantage of sequenced Nomascus BAC-ends (CHORI-271 library), whose position on the human reference sequence was bioinformatically defined (see Roberto et al. 2007). A selection of these clones was used to confirm HLA-specific breakpoints. The overall SSAs, defined by specific BACs, are reported in detail in three Supplemental Tables (ST1, ST2, and ST3), assembled according to different perspectives. Table ST1 establishes the correspondence of human chromosomes with respect to HLA and Nomascus chromosomes. Tables ST2 and ST3 report SSA of Hylobates and Nomascus chromosomes, respectively, with reference to the human chromosomes and to the Hylobatidae Ancestral Karyotype (HyAK; see below) (Supplemental Figs. 1 and 2, respectively). The Nomascus synteny organization is essentially based on the data from Roberto et al. (2007). The data reported in Supplemental Table ST2 are also displayed, in a more figurative way, in Figure 2 and at the website www.biologia.uniba.it/lar.
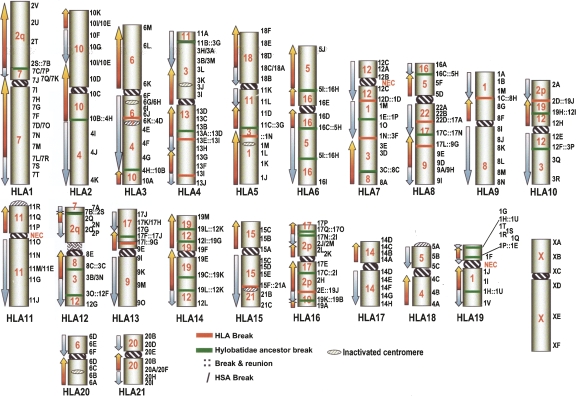
Figure 2.
HLA ideogram showing the synteny block correspondence with the human genome. Internal red numbers indicate the human corresponding chromosome. Blue and red arrows indicate concordant (blue) and inverse (red) sequence polarity (orientation) with respect to humans. Black numbers (chromosome) followed by a letter (BAC), on the right, indicate specific BACs as reported in the Supplemental Table S1. Breakpoints in HLA are indicated by red bars (as in Supplemental Table S2). Breaks that occurred in HyA are represented by green bars (see Supplemental Table S1). (∷) Breakage and reunion in HLA and/or HyA, and the most proximal clones are reported. Rearrangements in HSA generate apparent breaks in HLA. These apparent breaks are indicated as clones facing each other (3M/3D, for example).
A fuller understanding of HLA synteny arrangement presupposes the elucidation of the cascade of rearrangements that each chromosome underwent during evolution, from the hominoid ancestor to the present-day synteny organization in HLA. We reconstructed the synteny arrangement of HyAK based on the reconstruction by Muller et al. (2003) by comparing the HLA synteny data we established above with previous data on Nomascus leucogenys (Roberto et al. 2007), and considering additional painting data from Hoolock hoolock (Nie et al. 2001) and Symphalangus syndactylus (Muller et al. 2003; Ferguson-Smith et al. 2005). Details of the reconstruction are summarized in the Supplemental Table ST4, and graphically illustrated in Figure 3. The numbers reported to the right of each chromosome represent arbitrarily chosen landmarks useful in defining the chromosome composition and facilitate the description of rearrangements. The reference to the HyAK was also inserted in Supplemental Tables ST2 and ST3. We made several improvements to the Hylobatidae ancestral karyotype proposed by Muller et al. (2003). Differences were essentially due to the higher resolution power of our BAC–FISH analysis. The newly discovered rearrangements all occurred in the common ancestor of Nomascus and Hylobates and consisted of three inversions (involving Hominoidea ancestral chromosomes ANC6, ANC9, and ANC13), and two translocations that involved the ancestral Hominoidea chromosomes ANC2q/ANC7 and ANC3/ANC11 (see Supplemental file SF1). We propose that these common changes can be incorporated into the Hylobatidae ancestral karyotype. Further minor changes may be eventually made to the HyAK once high-resolution studies of Symphalangus and especially Hoolock are accomplished.
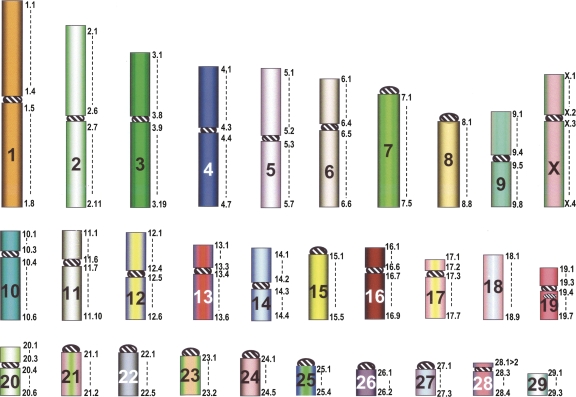
Figure 3.
Hylobatidae Ancestral karyotype (HyAK). The numbers reported to the right of each chromosome represent arbitrarily chosen landmarks useful in defining the chromosome composition and in facilitating the description of rearrangements. They indicate the extension of each chromosomal arm, according to the data reported in Supplemental Table ST4, first column.
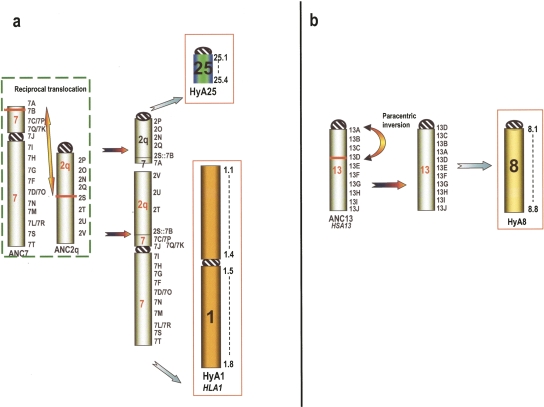
The hypothesized HyAK was then compared with the hominoid ancestral karyotype we recently presented (Stanyon et al. 2008) and graphically displayed in Figure 4. Supplemental Table ST5 describes the organization of Hominoidea chromosomes compared with Hylobates, Nomascus, and HyAK. Comparison with the chromosomal organization of Hominoidea ancestor was crucial (1) to discriminate rearrangements that occurred in the Hylobatidae ancestor before branching and (2) to pinpoint changes that occurred, after Hylobatidae divergence, in the lineage leading to human. Because our objective was to test the already published Hylobatidae ancestral karyotype (Muller et al. 2003), no computer-assisted reanalysis was necessary or done. Instead, new synteny segments found due to our higher-resolution BAC–FISH approach were incorporated into the previous results. In each case we applied the maximum parsimony criteria and manually selected the rearrangements and breakpoints. Using these principles we were able to decide which chromosome forms were probably present in the Hylobatidae ancestral karyotype departing from extant chromosome forms and compared them with the chromosome hypothesized to be present in the ancestral hominoid karyotype (Stanyon et al. 2008). Two examples are provided in Figure 5. Figure 5a shows the origin of HyA1 and HyA25, while Figure 5b shows the inversion present in ANC13 necessary to derive HyA8. Note in Figure 5a that Muller et al. (2003), due to the limitations of chromosome painting, were not able to detect the small segment of ANC7 in NLE20, whose short arm corresponds to Hy25 (see Supplemental File SF3). Nie et al. (2001) used the NLE20 whole-chromosome painting as a probe on HHO metaphases and found the hybridization signal associated with HSA2 on HHO4. These investigators, however, do not report a 2/7 association for this chromosome, even if we now know that a small segment of HSA 7 is present in NLE20. BAC–FISH results directly indicate that this segment is present in NLE and indirectly show that it is present in HHO; therefore, the Hylobatidae ancestral karyotype must be modified accordingly. Figure 5b shows the inversion detected in both HLA and NLE. The only open question is the position of the centromere in HyA8; in other words, whether the inversion is paracentric or pericentric. The inversion appears pericentric in NLE (Roberto et al. 2007), while paracentric in HLA (this study) and, very likely, also in Hoolock: in HHO, the short arm of chromosome 2 is hybridized by NLE9 and NLE5, which derive from HyA8. The position of the two paint hybridizations suggests that the ancestral form, HyA8, was acrocentric. The present interpretation was favored because of parsimony reasons. The hypothesized evolutionary changes necessary to derive the Hylobatidae-ancestor forms from the Hominoidea-ancestor chromosomal forms are graphically reported in the figures contained in the Supplemental File SF1. Similarly, the flow of changes from Hylobatidae ancestor to Hylobates lar are illustrated in Supplemental File SF2, while the flow from HyA to NLE is summarized in Supplemental File SF3. Rearrangements were introduced following the maximum parsimony criteria. The summary of SSA arrangement of HLA and NLE with respect to the Hylobatidae ancestor is depicted in Supplemental Figures SF1 and SF2, respectively. The consistency of the reciprocal orientation of synteny blocks, which resulted from the hypothesized chromosomal rearrangements, especially inversions, was carefully tested against data derived from the cohybridization FISH experiments.
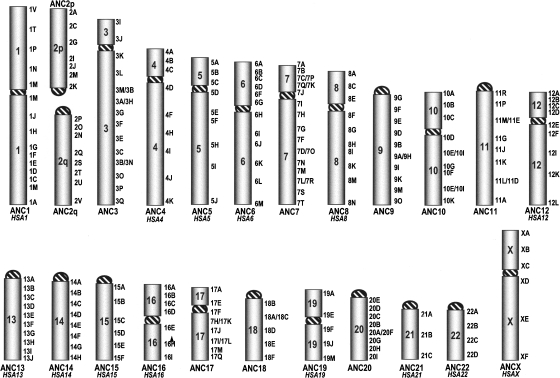
Figure 4.
Hominoidea ancestral karyotype. The number plus letter on the right indicate specific human BACs reported in the Supplemental Table S5. Synteny arrangement differences with respect to humans are only due to peri/paracentric inversions (with the exception of the 2p–2q fusion). These breaks are indicated as clones facing each other (3M/3D, for instance). When a chromosome remained unchanged in humans, its human nomenclature is reported in italics.
Figure 5.
(a) Example of the evolutionary chromosomal changes leading, from Hominoidea ancestor (ANC7 and ANC2q), to chromosomes HyA1 and HyA25 in Hylobatidae ancestor. (b) Shows the inversion in ANC13 necessary to derive HyA8 (see text for details). Numbers followed by a letter on the right of chromosomes indicate a specific human BAC as reported in both ST1 and ST5 Supplemental Tables. The numbers on the right of the final Hylobatidae ancestor chromosome (HyA) represent arbitrarily chosen points useful in indicating its composition, as reported in the ST4 Supplemental Table. Usually, they correspond to a BAC, but when found at the borders of the synteny blocks, they identify two BACs facing each other following a rearrangement. The Hylobatidae Ancestral Kayotype (HyAK) is depicted in Fig. 3.
The rearrangements that occurred in Hylobatidae are grouped in Table 1 according to their phylogenetic position: from the Hominoidea ancestor to the Hylobatidae ancestor, from the Hylobatidae ancestor to HLA, and from the Hylobatidae ancestor to NLE. They are also grouped according to the rearrangement type: reciprocal translocations, Robertsonian (centric) fissions, Robertsonian fusions (centric translocations), fissions, fusions, inversions, and evolutionary new centromeres. In several instances the rearrangement triggered centromere inactivation. Table 2 summarizes these events, which are also displayed in the figures.
Table 1.
Type and rate of rearrangements occurred in gibbons
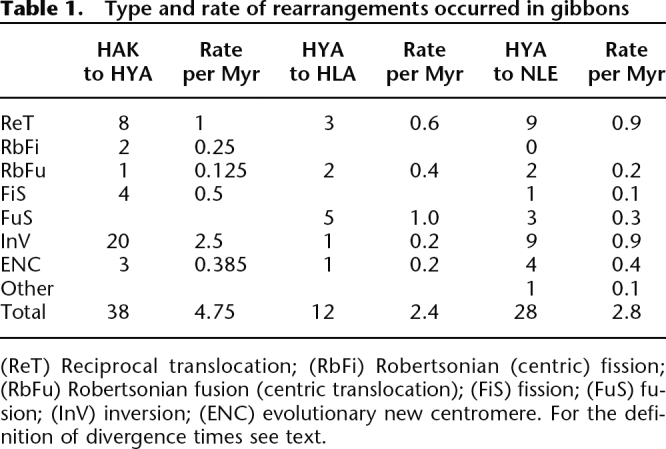
(ReT) Reciprocal translocation; (RbFi) Robertsonian (centric) fission; (RbFu) Robertsonian fusion (centric translocation); (FiS) fission; (FuS) fusion; (InV) inversion; (ENC) evolutionary new centromere. For the definition of divergence times see text.
Table 2.
HLA and NLE chromosomes harboring inactivated centromeres
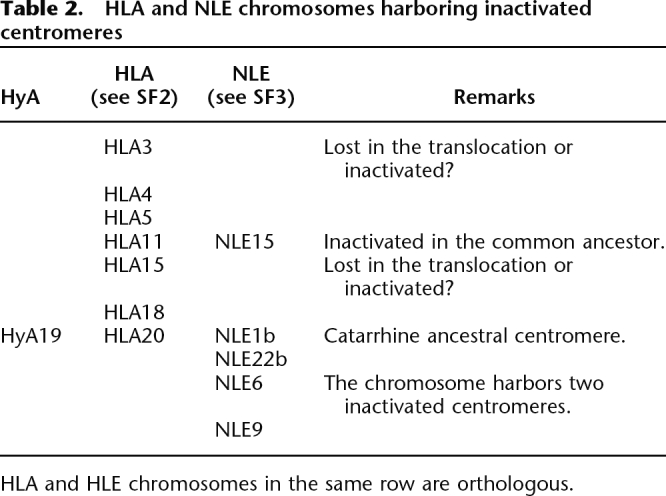
HLA and HLE chromosomes in the same row are orthologous.
Centromere repositioning is the movement of the centromere along a chromosome without marker order variation. This phenomena produces novel centromeres, also termed as new evolutionary centromere (NEC), and is quite widespread in primates and other mammalian orders (see Cardone et al. 2006, and references therein). We detected three clear examples of NECs in HLA (see Table 1).
Some break regions appear to be independently utilized as breakpoints in rearrangements, which occurred in other mammalian species. Supplemental Table 6 summarizes these data.
Discussion
We have cohybridized in situ a large panel of human BAC clones to HLA metaphases to gain detailed information on the extension, reciprocal arrangement, and orientation of syntenic chromosomal segments in the lar gibbon with respect to humans. The borders of each segment were defined by a splitting BAC or by two overlapping BACs spanning each breakpoint. These results can be considered as the major achievement of the present work because they provide a much higher level of resolution than previous cytogenetic comparisons between the genomes of humans and the lar gibbon.
Our approach discriminated between breakpoint-spanning probes and multiple signals due to segmental duplications, because all breakpoints were determined following reiterative FISH experiments approaching the break from both sides. Our detailed and high-resolution analysis unveiled 12 previously undetected, mostly subtelomeric rearrangements; the high plasticity of subtelomeric regions and their proneness to reciprocal translocations are well known (Flint and Knight 2003; Linardopoulou et al. 2005).
A full understanding of the synteny arrangement and chromosome evolution in HLA can only be obtained by reference to the organization of both the ancestral Hominoidea and ancestral Hylobatidae karyotype. The ancestral hominoid karyotype, including marker order, was recently published by Stanyon et al. (2008). A reconstruction of the ancestral Hylobatidae karyotype based on reciprocal chromosome painting was also recently presented (Muller et al. 2003). This hypothetical ancestral Hylobatidae karyotype can be tested by comparing the high-resolution, BAC–FISH synteny data on HLA presented here with that of Nomascus leucogenys (Roberto et al. 2007). Our data mostly support previous reconstructions of the ancestral Hylobatidae karyotype, but also allows us to propose a number of improvements due to the higher resolution of our approach (see Fig. 4; Supplemental Table ST4; Supplemental File SF1; www.biologia.uniba.it/lar).
Once we have a proposal for the ancestral hominoid karyotype and the ancestral Hylobatidae karyotype, it is possible to reconstruct the flow of chromosomal changes from the Hominoidea ancestor to HLA, through the Hylobatidae ancestor. Likewise, previous high-resolution data published by our group on NLE permitted the same analysis of the rearrangements necessary to derive this species’ chromosomes. It should be noted that only chromosome painting data are available for Hoolock and Symphalangus. If Nomascus and Symphalangus are sister species, as suggested by various investigators (Muller et al. 2003; Whittaker et al. 2007), then the lack of BAC–FISH data on Symphalangus is not critical. However, it would be desirable to have a higher resolution study of both the Symphalangus and especially Hoolock genomes in order to provide full confidence in the reconstruction of the Hylobatidae ancestral karyotype. Certainly, our reconstruction is valid, as a few additional changes will be necessary once high-resolution studies of the remaining two karyomorphs are completed.
All of the rearrangements we detected could be grouped into four categories: (1) those that occurred after the divergence of the lesser ape lineage from the other hominoids (great apes and humans) and were present in the hypothesized Hylobatidae ancestral karyotype; (2) those that occurred apomorphically in HLA; (3) chromosome rearrangements that occurred apomorphically in the lineage leading to NL; (4) rearrangements that occurred in the line leading to humans, but could be incorrectly assigned as gibbon breaks because they were detected using human probes.
These distinctions were possible only in the framework of a detailed evolutionary history of lineages leading to human and to HLA gibbon after Hylobatidae/Hominidae branching and were crucial to avoid misinterpretations when the human chromosome is derived. For example, the portion of human chromosome 3 present on HLA4 might appear highly rearranged when compared with humans but, on the contrary, the ancestral order is perfectly conserved in HLA (see the example in Fig. 1b). The difference is due to two successive inversions that specifically occurred in the lineage leading to humans after orangutan divergence (Ventura et al. 2004). Figures depicting the reconstructed evolutionary history of chromosomes can be found in Supplemental Files SF1, SF2, and SF3. Some points, concerning chromosomes of special interest or discrepancies with the literature, will be discussed in detail below. More specific points are reported in the legends to the Supplemental Files.
The majority of the chromosomal differences between HLA and humans are due to rearrangements that occurred in the Hylobatidae ancestor (see Table 1). The HLA genome showed only 12 HLA-specific chromosomal events, while changes it inherited from the ancestor were 38. This result seems congruent with the conclusion from molecular work that Hylobates is the most recently evolved lesser ape genus (Roos and Geissmann 2001; Takacs et al. 2005; Baena et al. 2007; Whittaker et al. 2007). Molecular research converges on a date for lesser apes origins around 17 to 18 million years ago (Mya) (Caccone and Powell 1989; Bailey et al. 1991; Stauffer et al. 2001; Hasegawa et al. 2003; Raaum et al. 2005; Baena et al. 2007). According to these molecular analyses, crown species originated much later and Hoolock, Symphalangus, and Nomascus, whatever the exact order, diverged between 10 and 12 Mya. Finally, extant species forming the genus Hylobates began to diverge around 5 Mya. This time frame allows us to estimate rearrangement rates in different lineages and at different periods in the evolution of lesser apes, summarized in Table 1. Because all living species of the genus Hylobates have essentially the same karyotype, it is clear that these rearrangements occurred in the last common ancestor of the 44 chromosome gibbons before they began to diverge 5 Mya, which provides 5 million years (Myr) to accumulate the rearrangements in Hylobates. The line leading to Nomascus diverged around 10 Mya, leaving 8 Myr (given an origin of the lesser ape line at 18 Mya) for the accumulation of rearrangements found in the common ancestor of these two species.
Rates and types of chromosome evolution differ in the diverse lineages. Rearrangements that occurred in their common ancestor were essentially translocations and inversions (29 out of 38 or 76%). In NLE, translocations and inversions were also predominant: 21 out of 28 (75%). In contrast, the HLA-specific rearrangements are qualitatively different: 10 of the 12 (83%) HLA-specific rearrangements were chromosomal fusions. Indeed, the lar gibbon has 44 chromosomes, while the hypothesized Hylobatidae ancestor had a diploid number of 2n = 64 (Muller et al. 2003).
The overall rate of evolution was more rapid after the divergence of lesser apes from hominids. In about 8 Myr, 38 rearrangements accumulated in the common ancestor of living lesser apes (4.75 events per million years). Rates slowed to about half in the lineage leading to the genera Hylobates (2.4) and Nomascus (2.8). All of these evolutionary rates are quite impressive, because the default rate of mammalian chromosome evolution is about one rearrangement every 4 Myr (0.25). The gibbon rates are at least a magnitude higher (10–20 times higher) and up to twice those found in the mouse (1.71–2.76) and the canine (2.1) lineages (Murphy et al. 2005). Higher rates of evolution are only found in some gerbils (Dobigny et al. 2005) and in the karyotypic evolution of onager, donkey, and zebras (Trifonov et al. 2008).
Hylobates/Hominoidea chromosomal differences affected all HLA autosomes. Even though Hominoidea chromosomes 13, 14, 15, 18, 20, 21, and 22 constitute a single, uninterrupted chromosomal block in the lar gibbon, most of them are part of larger chromosomes and/or show internal rearrangements. These differences appear even more impressive if compared with the relatively high chromosomal stability of the other Hominoidea lineages. Human and orangutan, for instance, share with the Hominoidea ancestor 14 and 11 unchanged chromosomes, respectively. In HLA, the only conserved chromosome is the X, which appeared to be colinear to the human X, and probably, to the mammal ancestral form (Murphy et al. 1999). The X chromosome conservation, however, is not unexpected. The exclusive inactivation mechanism that compensates gene expression in mammalian females make this chromosome refractory to rearrangement with autosomes (Homolka et al. 2007). Incident-balanced X/autosome rearrangements, indeed, easily become functionally unbalanced when passed to the progeny, drastically precluding any evolutionary perspective.
In humans, some clusters of segmental duplications present in nonpericentromeric regions are remains of inactivated centromeres (Ventura et al. 2003, 2004). In the present work, we unveiled, in Hylobatidae, a significant number of inactivated centromeres triggered by the high number of rearrangements (Table 2). Additionally, a substantial number of breakpoints involved centromeric/pericentromeric regions (see the evolutionary history of HLA chromosomes, Supplemental File SF2). As a consequence, some ancestral pericentromeric regions became euchromatic, thus contributing to the dispersal of segmental duplications in euchromatic regions. In turn, euchromatic regions, ancestrally located far from centromeres, became contiguous with centromeres, where they were then exposed to shuffling processes typical of these regions (Ventura et al. 2007). We counted 10 such events (centromeres of HLA4, HLA5, HLA9, HLA10, HLA11, HLA16, HLA17, HLA19, HLA20, and HLA21). Taking into account these data, it would not be a surprise if the complete genome assembly of HLA or NLE eventually shows a high presence of interspersed segmental duplications.
Supplemental Table 6 summarizes the reuse of some chromosomal regions as breakpoints in different species. The reuse of some regions as places of “chromosomal events” like rearrangements, segmental duplications, fragile sites, and centromere repositioning has been pointed out in a number of studies (Armengol et al. 2003; Pevzner and Tesler 2003; Bailey et al. 2004a; Ventura et al. 2004; Ruiz-Herrera et al. 2005).
Centromere movements
Many examples of centromere repositioning events have been reported in primates (Ventura et al. 2004, and references therein). The evolutionary history of each HLA centromere was therefore accurately tracked for consistency with the flanking markers. The centromeres of HLA chromosomes 7, 11, and 19 were found clearly displaced with respect to their position in the Hominoidea ancestor, and can be considered as evolutionarily novel.
Concluding remarks
Pioneering studies by Jauch et al. (1992) revealed a high plasticity of the lar gibbon genome at the chromosomal level. The present study provides, at a much higher resolution, an accurate definition of synteny block extension, arrangement, and orientation that could prove very helpful for a sequencing effort for this species. As stated, the rearrangement rate of gibbons is at least an order of magnitude higher with respect to the average rate in mammals, and only comparable, as far as the sequenced genomes are concerned, to rates found in mouse and dog. The sequencing of the lar gibbon could therefore prove useful toward a deeper understanding of the mechanisms of genome instability. This topic is particularly intriguing because several factors at distinct levels may have contributed to this high rate: at the sequence level (segmental duplications for instance), at meiotic level (meiotic drive), and at social organization level (low Ne promoted by monogamy). The present data point to a high level of segmental duplications in gibbons, H. lar in particular, contrary to what we found in mouse (Bailey et al. 2004b) and dog (Lindblad-Toh et al. 2005). This consideration further points to the sequencing of the lar gibbon genome as an appealing task that will facilitate hypothesis tests of lesser ape and genome evolution in general.
Methods
Fluorescence in situ hybridization (FISH) experiments
Metaphase preparations were obtained from a male lymphoblastoid cell lines from Hylobates lar. Occasionally, FISH experiments were performed on metaphases from a lymphoblastoid cell line of Macaca mulatta. BAC clones were from (1) the RPCI11 and CDD human library and (2) Nomascus leucogenys BAC library (CHORI-271) (for both libraries, see http://www.chori.org/bacpac/). Extraction of total DNA from BACs was performed according to standard methods. Chromosome preparations were hybridized in situ with probes directly labeled with Cy3-dCTP, FluorX-dCTP, DEAC, Cy5-dCTP by nick-translation, essentially as described by Lichter et al. (1990), with minor modifications.
Briefly: 300 ng of labeled probe (total BAC DNA) was used for the FISH experiments; hybridization was performed at 37°C in 2 SSC, 50% (v/v) formamide, 10% (w/v) dextran sulfate, 5 mg of COT1 DNA (Roche), and 3 mg of sonicated salmon sperm DNA, in a volume of 10 mL. Post-hybridization washes of FISH experiments were performed at lower stringency: 37°C in 2 SSC-50% formamide (three times), followed by three washes at 42°C in 2 SSC (three times). Chromosome identification was obtained by simultaneous DAPI staining, producing a Q-banding pattern. Some lar gibbon chromosomes, however, are difficult to distinguish on the basis of DAPI banding. The short and long arms of some lar gibbon metacentric chromosomes are also hard to distinguish (see a DAPI-banded karyotype at www.biologia.uniba.it/lar). In these cases, an appropriate BAC clone was always cohybridized, as a reference, to unambiguously identify the chromosome and/or the chromosome arm under study.
Digital images were obtained using a Leica DMRXA2 epifluorescence microscope equipped with a cooled CCD camera (Princeton Instruments). Cy3 (red), FluorX (green), and DAPI (blue) fluorescence signals, detected with specific filters, were recorded separately as grayscale images. Pseudocoloring and merging of images were performed using Adobe Photoshop software.
Acknowledgments
This work was supported by PRIN (Programmi di Ricerca di Interesse Nazionale) to M.R. and N.A.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.078295.108.
References
- Armengol L., Pujana M.A., Cheung J., Scherer S.W., Estivill X., Pujana M.A., Cheung J., Scherer S.W., Estivill X., Cheung J., Scherer S.W., Estivill X., Scherer S.W., Estivill X., Estivill X. Enrichment of segmental duplications in regions of breaks of synteny between the human and mouse genomes suggest their involvement in evolutionary rearrangements. Hum. Mol. Genet. 2003;12:2201–2208. doi: 10.1093/hmg/ddg223. [DOI] [PubMed] [Google Scholar]
- Arnold N., Stanyon R., Jauch A., O'Brien P., Wienberg J., Stanyon R., Jauch A., O'Brien P., Wienberg J., Jauch A., O'Brien P., Wienberg J., O'Brien P., Wienberg J., Wienberg J. Identification of complex chromosome rearrangements in the gibbon by fluorescence in situ hybridization (FISH) of a human chromosome 2q specific microlibrary, yeast artificial chromosomes, and reciprocal chromosome painting. Cytogenet. Cell Genet. 1996;74:80–85. doi: 10.1159/000134387. [DOI] [PubMed] [Google Scholar]
- Baena A., Mootnick A.R., Falvo J.V., Tsytskova A.V., Ligeiro F., Diop O.M., Brieva C., Gagneux P., O'Brien S.J., Ryder O.A., Mootnick A.R., Falvo J.V., Tsytskova A.V., Ligeiro F., Diop O.M., Brieva C., Gagneux P., O'Brien S.J., Ryder O.A., Falvo J.V., Tsytskova A.V., Ligeiro F., Diop O.M., Brieva C., Gagneux P., O'Brien S.J., Ryder O.A., Tsytskova A.V., Ligeiro F., Diop O.M., Brieva C., Gagneux P., O'Brien S.J., Ryder O.A., Ligeiro F., Diop O.M., Brieva C., Gagneux P., O'Brien S.J., Ryder O.A., Diop O.M., Brieva C., Gagneux P., O'Brien S.J., Ryder O.A., Brieva C., Gagneux P., O'Brien S.J., Ryder O.A., Gagneux P., O'Brien S.J., Ryder O.A., O'Brien S.J., Ryder O.A., Ryder O.A., et al. Primate TNF promoters reveal markers of phylogeny and evolution of innate immunity. PLoS ONE. 2007;2:e621. doi: 10.1371/journal.pone.0000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey W.J., Fitch D.H.A., Tagle D.A., Czelusniak J., Slightom J.L., Goodman M., Fitch D.H.A., Tagle D.A., Czelusniak J., Slightom J.L., Goodman M., Tagle D.A., Czelusniak J., Slightom J.L., Goodman M., Czelusniak J., Slightom J.L., Goodman M., Slightom J.L., Goodman M., Goodman M. Molecular evolution of the psi eta-globin gene locus: Gibbon phylogeny and the hominoid slowdown. Mol. Biol. Evol. 1991;8:155–184. doi: 10.1093/oxfordjournals.molbev.a040641. [DOI] [PubMed] [Google Scholar]
- Bailey J.A., Baertsch R., Kent W.J., Haussler D., Eichler E.E., Baertsch R., Kent W.J., Haussler D., Eichler E.E., Kent W.J., Haussler D., Eichler E.E., Haussler D., Eichler E.E., Eichler E.E. Hotspots of mammalian chromosomal evolution. Genome Biol. 2004a;5:R23. doi: 10.1186/gb-2004-5-4-r23. http://genomebiology.com/2004/5/4/R23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J.A., Church D.M., Ventura M., Rocchi M., Eichler E.E., Church D.M., Ventura M., Rocchi M., Eichler E.E., Ventura M., Rocchi M., Eichler E.E., Rocchi M., Eichler E.E., Eichler E.E. Analysis of segmental duplications and genome assembly in the mouse. Genome Res. 2004b;14:789–801. doi: 10.1101/gr.2238404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon-Jones D., Eudey A.A., Geissmann T., Groves C.P., Melnick D.J., Morales J.C., Shekelle M., Stewart C.B., Eudey A.A., Geissmann T., Groves C.P., Melnick D.J., Morales J.C., Shekelle M., Stewart C.B., Geissmann T., Groves C.P., Melnick D.J., Morales J.C., Shekelle M., Stewart C.B., Groves C.P., Melnick D.J., Morales J.C., Shekelle M., Stewart C.B., Melnick D.J., Morales J.C., Shekelle M., Stewart C.B., Morales J.C., Shekelle M., Stewart C.B., Shekelle M., Stewart C.B., Stewart C.B. Asian primate classification. Int. J. Primatol. 2004;25:97–164. [Google Scholar]
- Caccone A., Powell J.R., Powell J.R. DNA divergence among hominoids. Evolution Int. J. Org. Evolution. 1989;43:925–942. doi: 10.1111/j.1558-5646.1989.tb02540.x. [DOI] [PubMed] [Google Scholar]
- Carbone L., Vessere G.M., ten Hallers B.F., Zhu B., Osoegawa K., Mootnick A., Kofler A., Wienberg J., Rogers J., Humphray S., Vessere G.M., ten Hallers B.F., Zhu B., Osoegawa K., Mootnick A., Kofler A., Wienberg J., Rogers J., Humphray S., ten Hallers B.F., Zhu B., Osoegawa K., Mootnick A., Kofler A., Wienberg J., Rogers J., Humphray S., Zhu B., Osoegawa K., Mootnick A., Kofler A., Wienberg J., Rogers J., Humphray S., Osoegawa K., Mootnick A., Kofler A., Wienberg J., Rogers J., Humphray S., Mootnick A., Kofler A., Wienberg J., Rogers J., Humphray S., Kofler A., Wienberg J., Rogers J., Humphray S., Wienberg J., Rogers J., Humphray S., Rogers J., Humphray S., Humphray S., et al. A high-resolution map of synteny disruptions in gibbon and human genomes. PLoS Genet. 2006;2:e223. doi: 10.1371/journal.pgen.0020223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone M.F., Alonso A., Pazienza M., Ventura M., Montemurro G., Carbone L., de Jong P.J., Stanyon R., D'Addabbo P., Archidiacono N., Alonso A., Pazienza M., Ventura M., Montemurro G., Carbone L., de Jong P.J., Stanyon R., D'Addabbo P., Archidiacono N., Pazienza M., Ventura M., Montemurro G., Carbone L., de Jong P.J., Stanyon R., D'Addabbo P., Archidiacono N., Ventura M., Montemurro G., Carbone L., de Jong P.J., Stanyon R., D'Addabbo P., Archidiacono N., Montemurro G., Carbone L., de Jong P.J., Stanyon R., D'Addabbo P., Archidiacono N., Carbone L., de Jong P.J., Stanyon R., D'Addabbo P., Archidiacono N., de Jong P.J., Stanyon R., D'Addabbo P., Archidiacono N., Stanyon R., D'Addabbo P., Archidiacono N., D'Addabbo P., Archidiacono N., Archidiacono N., et al. Independent centromere formation in a capricious, gene-free domain of chromosome 13q21 in Old World monkeys and pigs. Genome Biol. 2006;7:R91. doi: 10.1186/gb-2006-7-10-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobigny G., Yang F., O'Brien P.C., Volobouev V., Kovacs A., Pieczarka J.C., Ferguson-Smith M.A., Robinson T.J., Yang F., O'Brien P.C., Volobouev V., Kovacs A., Pieczarka J.C., Ferguson-Smith M.A., Robinson T.J., O'Brien P.C., Volobouev V., Kovacs A., Pieczarka J.C., Ferguson-Smith M.A., Robinson T.J., Volobouev V., Kovacs A., Pieczarka J.C., Ferguson-Smith M.A., Robinson T.J., Kovacs A., Pieczarka J.C., Ferguson-Smith M.A., Robinson T.J., Pieczarka J.C., Ferguson-Smith M.A., Robinson T.J., Ferguson-Smith M.A., Robinson T.J., Robinson T.J. Low rate of genomic repatterning in Xenarthra inferred from chromosome painting data. Chromosome Res. 2005;13:651–663. doi: 10.1007/s10577-005-1002-9. [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith M.A., Yang F., Rens W., O’Brien ,, Yang F., Rens W., O’Brien ,, Rens W., O’Brien ,, O’Brien , The impact of chromosome sorting and painting on the comparative analysis of primate genomes. Cytogenet. Genome Res. 2005;108:112–121. doi: 10.1159/000080809. [DOI] [PubMed] [Google Scholar]
- Flint J., Knight S., Knight S. The use of telomere probes to investigate submicroscopic rearrangements associated with mental retardation. Curr. Opin. Genet. Dev. 2003;13:310–316. doi: 10.1016/s0959-437x(03)00049-2. [DOI] [PubMed] [Google Scholar]
- Hasegawa M., Thorne J.L., Kishino H., Thorne J.L., Kishino H., Kishino H. Time scale of eutherian evolution estimated without assuming a constant rate of molecular evolution. Genes Genet. Syst. 2003;78:267–283. doi: 10.1266/ggs.78.267. [DOI] [PubMed] [Google Scholar]
- Hirai H., Mootnick A.R., Takenaka O., Suryobroto B., Mouri T., Kamanaka Y., Katoh A., Kimura N., Maeda N., Mootnick A.R., Takenaka O., Suryobroto B., Mouri T., Kamanaka Y., Katoh A., Kimura N., Maeda N., Takenaka O., Suryobroto B., Mouri T., Kamanaka Y., Katoh A., Kimura N., Maeda N., Suryobroto B., Mouri T., Kamanaka Y., Katoh A., Kimura N., Maeda N., Mouri T., Kamanaka Y., Katoh A., Kimura N., Maeda N., Kamanaka Y., Katoh A., Kimura N., Maeda N., Katoh A., Kimura N., Maeda N., Kimura N., Maeda N., Maeda N. Genetic mechanism and property of a whole-arm translocation (WAT) between chromosomes 8 and 9 of agile gibbons (Hylobates agilis) Chromosome Res. 2003;11:37–50. doi: 10.1023/a:1022006015379. [DOI] [PubMed] [Google Scholar]
- Homolka D., Ivanek R., Capkova J., Jansa P., Forejt J., Ivanek R., Capkova J., Jansa P., Forejt J., Capkova J., Jansa P., Forejt J., Jansa P., Forejt J., Forejt J. Chromosomal rearrangement interferes with meiotic X chromosome inactivation. Genome Res. 2007;17:1431–1437. doi: 10.1101/gr.6520107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauch A., Wienberg J., Stanyon R., Arnold N., Tofanelli S., Ishida T., Cremer T., Wienberg J., Stanyon R., Arnold N., Tofanelli S., Ishida T., Cremer T., Stanyon R., Arnold N., Tofanelli S., Ishida T., Cremer T., Arnold N., Tofanelli S., Ishida T., Cremer T., Tofanelli S., Ishida T., Cremer T., Ishida T., Cremer T., Cremer T. Reconstruction of genomic rearrangements in great apes and gibbons by chromosome painting. Proc. Natl. Acad. Sci. 1992;89:8611–8615. doi: 10.1073/pnas.89.18.8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler U., Arnold N., Wienberg J., Tofanelli S., Stanyon R., Arnold N., Wienberg J., Tofanelli S., Stanyon R., Wienberg J., Tofanelli S., Stanyon R., Tofanelli S., Stanyon R., Stanyon R. Genomic reorganization and disrupted chromosomal synteny in the siamang (Hylobates syndactylus) revealed by fluorescence in situ hybridization. Am. J. Phys. Anthropol. 1995a;97:37–47. doi: 10.1002/ajpa.1330970104. [DOI] [PubMed] [Google Scholar]
- Koehler U., Bigoni F., Wienberg J., Stanyon R., Bigoni F., Wienberg J., Stanyon R., Wienberg J., Stanyon R., Stanyon R. Genomic reorganization in the concolor gibbon (Hylobates concolor) revealed by chromosome painting. Genomics. 1995b;30:287–292. doi: 10.1006/geno.1995.9875. [DOI] [PubMed] [Google Scholar]
- Lichter P., Tang Chang C.-J., Call K., Hermanson G., Evans G.A., Housman D., Ward D.C., Tang Chang C.-J., Call K., Hermanson G., Evans G.A., Housman D., Ward D.C., Call K., Hermanson G., Evans G.A., Housman D., Ward D.C., Hermanson G., Evans G.A., Housman D., Ward D.C., Evans G.A., Housman D., Ward D.C., Housman D., Ward D.C., Ward D.C. High resolution mapping of human chromosomes 11 by in situ hybridization with cosmid clones. Science. 1990;247:64–69. doi: 10.1126/science.2294592. [DOI] [PubMed] [Google Scholar]
- Linardopoulou E.V., Williams E.M., Fan Y., Friedman C., Young J.M., Trask B.J., Williams E.M., Fan Y., Friedman C., Young J.M., Trask B.J., Fan Y., Friedman C., Young J.M., Trask B.J., Friedman C., Young J.M., Trask B.J., Young J.M., Trask B.J., Trask B.J. Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature. 2005;437:94–100. doi: 10.1038/nature04029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Chang J.L., Kulbokas E.J., Zody M.C., Kulbokas E.J., Zody M.C., Zody M.C., et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Mootnick A., Groves C., Groves C. A new generic name for the hoolock gibbon (Hylobatidae) Int. J. Primatol. 2005;26:971–975. [Google Scholar]
- Mrasek K., Heller A., Rubtsov N., Trifonov V., Starke H., Claussen U., Liehr T., Heller A., Rubtsov N., Trifonov V., Starke H., Claussen U., Liehr T., Rubtsov N., Trifonov V., Starke H., Claussen U., Liehr T., Trifonov V., Starke H., Claussen U., Liehr T., Starke H., Claussen U., Liehr T., Claussen U., Liehr T., Liehr T. Detailed Hylobates lar karyotype defined by 25-color FISH and multicolor banding. Int. J. Mol. Med. 2003;12:139–146. [PubMed] [Google Scholar]
- Muller S., Neusser M., Wienberg J., Neusser M., Wienberg J., Wienberg J. Towards unlimited colors for fluorescence in-situ hybridization (FISH) Chromosome Res. 2002;10:223–232. doi: 10.1023/a:1015296122470. [DOI] [PubMed] [Google Scholar]
- Muller S., Hollatz M., Wienberg J., Hollatz M., Wienberg J., Wienberg J. Chromosomal phylogeny and evolution of gibbons (Hylobatidae) Hum. Genet. 2003;113:493–501. doi: 10.1007/s00439-003-0997-2. [DOI] [PubMed] [Google Scholar]
- Murphy W.J., Sun S., Chen Z.Q., Pecon-Slattery J., O'Brien S.J., Sun S., Chen Z.Q., Pecon-Slattery J., O'Brien S.J., Chen Z.Q., Pecon-Slattery J., O'Brien S.J., Pecon-Slattery J., O'Brien S.J., O'Brien S.J. Extensive conservation of sex chromosome organization between cat and human revealed by parallel radiation hybrid mapping. Genome Res. 1999;9:1223–1230. doi: 10.1101/gr.9.12.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy W.J., Larkin D.M., Everts-van der Wind A., Bourque G., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Larkin D.M., Everts-van der Wind A., Bourque G., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Everts-van der Wind A., Bourque G., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Bourque G., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Chowdhary B.P., Galibert F., Gatzke L., Galibert F., Gatzke L., Gatzke L., et al. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 2005;309:613–617. doi: 10.1126/science.1111387. [DOI] [PubMed] [Google Scholar]
- Nie W., Rens W., Wang J., Yang F., Rens W., Wang J., Yang F., Wang J., Yang F., Yang F. Conserved chromosome segments in Hylobates hoolock revealed by human and H. leucogenys paint probes. Cytogenet. Cell Genet. 2001;92:248–253. doi: 10.1159/000056912. [DOI] [PubMed] [Google Scholar]
- Pevzner P., Tesler G., Tesler G. Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution. Proc. Natl. Acad. Sci. 2003;100:7672–7677. doi: 10.1073/pnas.1330369100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaum R.L., Sterner K.N., Noviello C.M., Stewart C.B., Disotell T.R., Sterner K.N., Noviello C.M., Stewart C.B., Disotell T.R., Noviello C.M., Stewart C.B., Disotell T.R., Stewart C.B., Disotell T.R., Disotell T.R. Catarrhine primate divergence dates estimated from complete mitochondrial genomes: Concordance with fossil and nuclear DNA evidence. J. Hum. Evol. 2005;48:237–257. doi: 10.1016/j.jhevol.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Roberto R., Capozzi O., Wilson R.K., Mardis E.R., Lomiento M., Tuzun E., Cheng Z., Mootnick A.R., Archidiacono N., Rocchi M., Capozzi O., Wilson R.K., Mardis E.R., Lomiento M., Tuzun E., Cheng Z., Mootnick A.R., Archidiacono N., Rocchi M., Wilson R.K., Mardis E.R., Lomiento M., Tuzun E., Cheng Z., Mootnick A.R., Archidiacono N., Rocchi M., Mardis E.R., Lomiento M., Tuzun E., Cheng Z., Mootnick A.R., Archidiacono N., Rocchi M., Lomiento M., Tuzun E., Cheng Z., Mootnick A.R., Archidiacono N., Rocchi M., Tuzun E., Cheng Z., Mootnick A.R., Archidiacono N., Rocchi M., Cheng Z., Mootnick A.R., Archidiacono N., Rocchi M., Mootnick A.R., Archidiacono N., Rocchi M., Archidiacono N., Rocchi M., Rocchi M., et al. Molecular refinement of gibbon genome rearrangement. Genome Res. 2007;17:249–257. doi: 10.1101/gr.6052507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos C., Geissmann T., Geissmann T. Molecular phylogeny of the major hylobatid divisions. Mol. Phylogenet. Evol. 2001;19:486–494. doi: 10.1006/mpev.2001.0939. [DOI] [PubMed] [Google Scholar]
- Ruiz-Herrera A., Garcia F., Giulotto E., Attolini C., Egozcue J., Ponsa M., Garcia M., Garcia F., Giulotto E., Attolini C., Egozcue J., Ponsa M., Garcia M., Giulotto E., Attolini C., Egozcue J., Ponsa M., Garcia M., Attolini C., Egozcue J., Ponsa M., Garcia M., Egozcue J., Ponsa M., Garcia M., Ponsa M., Garcia M., Garcia M. Evolutionary breakpoints are co-localized with fragile sites and intrachromosomal telomeric sequences in primates. Cytogenet. Genome Res. 2005;108:234–247. doi: 10.1159/000080822. [DOI] [PubMed] [Google Scholar]
- Stanyon R., Rocchi R., Capozzi O., Roberto R., Misceo D., Ventura M., Cardone M., Bigoni F., Archidiacono N., Rocchi R., Capozzi O., Roberto R., Misceo D., Ventura M., Cardone M., Bigoni F., Archidiacono N., Capozzi O., Roberto R., Misceo D., Ventura M., Cardone M., Bigoni F., Archidiacono N., Roberto R., Misceo D., Ventura M., Cardone M., Bigoni F., Archidiacono N., Misceo D., Ventura M., Cardone M., Bigoni F., Archidiacono N., Ventura M., Cardone M., Bigoni F., Archidiacono N., Cardone M., Bigoni F., Archidiacono N., Bigoni F., Archidiacono N., Archidiacono N. Primate chromosome evolution: Ancestral karyotypes, marker order and neocentromeres. Chromosome Res. 2008;16:17–39. doi: 10.1007/s10577-007-1209-z. [DOI] [PubMed] [Google Scholar]
- Stauffer R.L., Walker A., Ryder O.A., Lyons-Weiler M., Hedges S.B., Walker A., Ryder O.A., Lyons-Weiler M., Hedges S.B., Ryder O.A., Lyons-Weiler M., Hedges S.B., Lyons-Weiler M., Hedges S.B., Hedges S.B. Human and ape molecular clocks and constraints on paleontological hypotheses. J. Hered. 2001;92:469–474. doi: 10.1093/jhered/92.6.469. [DOI] [PubMed] [Google Scholar]
- Takacs Z., Morales J.C., Geissmann T., Melnick D.J., Morales J.C., Geissmann T., Melnick D.J., Geissmann T., Melnick D.J., Melnick D.J. A complete species-level phylogeny of the Hylobatidae based on mitochondrial ND3-ND4 gene sequences. Mol. Phylogenet. Evol. 2005;36:456–467. doi: 10.1016/j.ympev.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Trifonov V.A., Stanyon R., Nesterenko A.I., Beiyuan F.B., Perelman P.L., O’Brien P.C., Stone G., Rubtsova N.V., Houck M.L., Robinson T.J., Stanyon R., Nesterenko A.I., Beiyuan F.B., Perelman P.L., O’Brien P.C., Stone G., Rubtsova N.V., Houck M.L., Robinson T.J., Nesterenko A.I., Beiyuan F.B., Perelman P.L., O’Brien P.C., Stone G., Rubtsova N.V., Houck M.L., Robinson T.J., Beiyuan F.B., Perelman P.L., O’Brien P.C., Stone G., Rubtsova N.V., Houck M.L., Robinson T.J., Perelman P.L., O’Brien P.C., Stone G., Rubtsova N.V., Houck M.L., Robinson T.J., O’Brien P.C., Stone G., Rubtsova N.V., Houck M.L., Robinson T.J., Stone G., Rubtsova N.V., Houck M.L., Robinson T.J., Rubtsova N.V., Houck M.L., Robinson T.J., Houck M.L., Robinson T.J., Robinson T.J., et al. Multidirectional cross-species painting illuminates the history of karyotypic evolution in Perissodactyla. Chromosome Res. 2008;16:89–107. doi: 10.1007/s10577-007-1201-7. [DOI] [PubMed] [Google Scholar]
- Ventura M., Mudge J.M., Palumbo V., Burn S., Blennow E., Pierluigi M., Giorda R., Zuffardi O., Archidiacono N., Jackson M.S., Mudge J.M., Palumbo V., Burn S., Blennow E., Pierluigi M., Giorda R., Zuffardi O., Archidiacono N., Jackson M.S., Palumbo V., Burn S., Blennow E., Pierluigi M., Giorda R., Zuffardi O., Archidiacono N., Jackson M.S., Burn S., Blennow E., Pierluigi M., Giorda R., Zuffardi O., Archidiacono N., Jackson M.S., Blennow E., Pierluigi M., Giorda R., Zuffardi O., Archidiacono N., Jackson M.S., Pierluigi M., Giorda R., Zuffardi O., Archidiacono N., Jackson M.S., Giorda R., Zuffardi O., Archidiacono N., Jackson M.S., Zuffardi O., Archidiacono N., Jackson M.S., Archidiacono N., Jackson M.S., Jackson M.S., et al. Neocentromeres in 15q24-26 map to duplicons which flanked an ancestral centromere in 15q25. Genome Res. 2003;13:2059–2068. doi: 10.1101/gr.1155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M., Weigl S., Carbone L., Cardone M.F., Misceo D., Teti M., D'Addabbo P., Wandall A., Björck E., de Jong P., Weigl S., Carbone L., Cardone M.F., Misceo D., Teti M., D'Addabbo P., Wandall A., Björck E., de Jong P., Carbone L., Cardone M.F., Misceo D., Teti M., D'Addabbo P., Wandall A., Björck E., de Jong P., Cardone M.F., Misceo D., Teti M., D'Addabbo P., Wandall A., Björck E., de Jong P., Misceo D., Teti M., D'Addabbo P., Wandall A., Björck E., de Jong P., Teti M., D'Addabbo P., Wandall A., Björck E., de Jong P., D'Addabbo P., Wandall A., Björck E., de Jong P., Wandall A., Björck E., de Jong P., Björck E., de Jong P., de Jong P., et al. Recurrent sites for new centromere seeding. Genome Res. 2004;14:1696–1703. doi: 10.1101/gr.2608804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M., Antonacci F., Cardone M.F., Stanyon R., D'Addabbo P., Cellamare A., Sprague L.J., Eichler E.E., Archidiacono N., Rocchi M., Antonacci F., Cardone M.F., Stanyon R., D'Addabbo P., Cellamare A., Sprague L.J., Eichler E.E., Archidiacono N., Rocchi M., Cardone M.F., Stanyon R., D'Addabbo P., Cellamare A., Sprague L.J., Eichler E.E., Archidiacono N., Rocchi M., Stanyon R., D'Addabbo P., Cellamare A., Sprague L.J., Eichler E.E., Archidiacono N., Rocchi M., D'Addabbo P., Cellamare A., Sprague L.J., Eichler E.E., Archidiacono N., Rocchi M., Cellamare A., Sprague L.J., Eichler E.E., Archidiacono N., Rocchi M., Sprague L.J., Eichler E.E., Archidiacono N., Rocchi M., Eichler E.E., Archidiacono N., Rocchi M., Archidiacono N., Rocchi M., Rocchi M. Evolutionary formation of new centromeres in macaque. Science. 2007;316:243–246. doi: 10.1126/science.1140615. [DOI] [PubMed] [Google Scholar]
- Whittaker D.J., Morales J.C., Melnick D.J., Morales J.C., Melnick D.J., Melnick D.J. Resolution of the Hylobates phylogeny: Congruence of mitochondrial D-loop sequences with molecular, behavioral, and morphological data sets. Mol. Phylogenet. Evol. 2007;45:620–628. doi: 10.1016/j.ympev.2007.08.009. [DOI] [PubMed] [Google Scholar]