Abstract
A stringent branch-site codon model was used to detect positive selection in vertebrate evolution. We show that the test is robust to the large evolutionary distances involved. Positive selection was detected in 77% of 884 genes studied. Most positive selection concerns a few sites on a single branch of the phylogenetic tree: Between 0.9% and 4.7% of sites are affected by positive selection depending on the branches. No functional category was overrepresented among genes under positive selection. Surprisingly, whole genome duplication had no effect on the prevalence of positive selection, whether the fish-specific genome duplication or the two rounds at the origin of vertebrates. Thus positive selection has not been limited to a few gene classes, or to specific evolutionary events such as duplication, but has been pervasive during vertebrate evolution.
How important has positive selection been in the evolution of vertebrate genes? While polymorphism data indicate high levels of positive selection in the Drosophila genus (Eyre-Walker 2006), much lower levels are found in mammals, which have smaller population sizes (Zhang and Li 2005; Gojobori et al. 2007). Moreover, the most recent use of likelihood tests of codon evolution has identified only two genes out of 13,888 under positive selections in the human lineage (Bakewell et al. 2007). Less stringent studies had found as many as 9% (Bustamante et al. 2005; Jorgensen et al. 2005; Nielsen et al. 2005; Arbiza et al. 2006). It is not obvious whether these results can be extended to deeper vertebrate evolution.
The study of vertebrate evolution includes two complicating factors relative to mammals or flies. First, the time scale is much larger, with the divergence between ray finned fishes and tetrapodes estimated at 416–422 million years ago (Mya) (Benton and Donoghue 2006). Second, whole genome duplications may have induced changes in selective regimes, either at the origin of vertebrates (2R) (Putnam et al. 2008) or at the origin of teleost fishes, the largest clade of vertebrates (Jaillon et al. 2004). It is expected that for both copies of a gene to be kept after duplication, there should be either fixation of a new function or complementary loss of subfunctions (Force et al. 1999; Lynch et al. 2001). The first implies increased positive selection; the second, relaxation of purifying selection. Although some studies have taken asymmetry of selective pressure as some degree of support for the former (Brunet et al. 2006; Byrne and Wolfe 2006), the evidence is not conclusive (He and Zhang 2005; Scannell and Wolfe 2007).
In this work, we use a rigorous branch-site specific likelihood test (Zhang et al. 2005) to quantify positive selection during several episodes of bony vertebrate evolution. To evaluate the role of duplication, we contrast the evidence for positive selection after duplication to the evidence after speciation. We find that although it affects a small proportion of sites, positive selection is pervasive in vertebrate evolution, and surprisingly, whole genome duplication has no measurable effect on its incidence.
Results
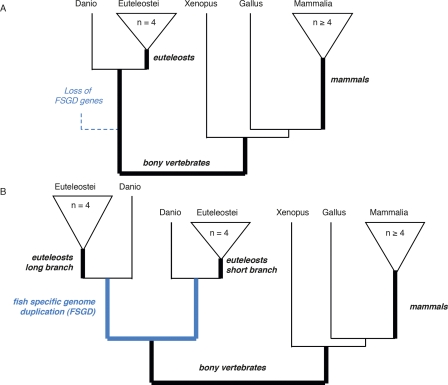
To investigate the impact of positive selection in vertebrate evolution, we have analyzed all gene families that include orthologs from chicken, Xenopus, five fish species, and at least four mammalian species (Fig. 1). Within this data set (884 gene families), we have distinguished strict one to one orthologs, with no duplication detected (“singletons”), and paralogs from the fish-specific whole genome duplication. We tested three to five internal branches, chosen because of their biological relevance and because they separate at least four sequences on each side, giving us sufficient power for statistical testing.
Figure 1.
Tree topologies studied. Schematic representation of the tree topologies selected. (Black) speciation branches; (blue) duplication branches. Branches in bold were used as “foreground branches” in branch-site tests for positive selection. Bold italics indicate branch names used in the text and the tables. (A) “Singleton” tree type, with no duplication allowed. The dotted line represents gene loss after whole genome duplication. (B) “Fish-specific whole genome duplication” tree type. Duplication on other branches may be excluded or allowed. Because differences in dN/dS ratio have been reported between paralogs (Brunet et al. 2006; Scannell and Wolfe 2007), with higher dN/dS in the fast evolving paralog, we distinguish the longer and shorter branch after duplication, based on the PhyML protein phylogeny.
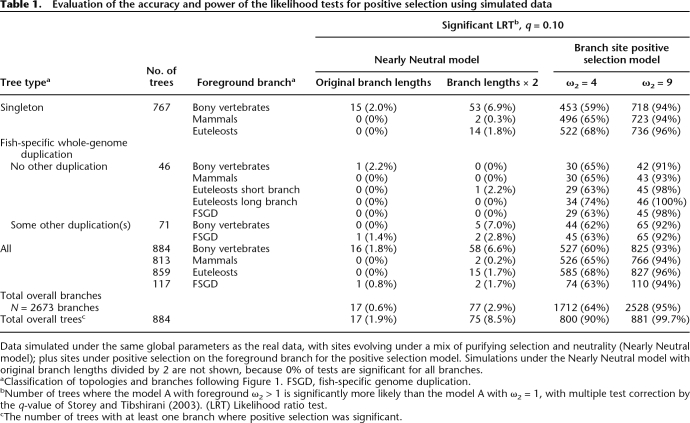
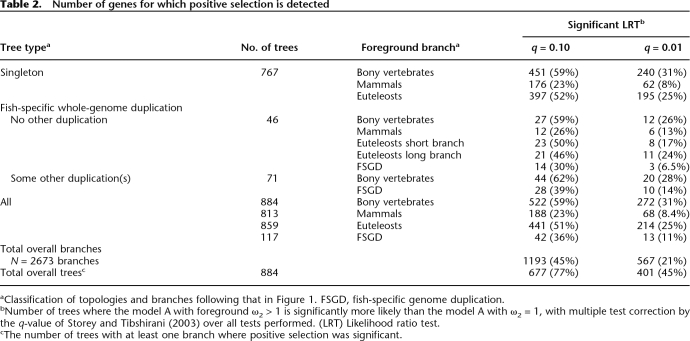
We use a branch-site model of positive selection for which the branch to be tested needs to be specified a priori (Yang 1998; Yang and Nielsen 2002). It is also possible in the absence of a specific biological hypothesis to use this test to scan for positive selection over several branches, on condition of correcting for multiple testing (Anisimova and Yang 2007). We use the q-value method to control for false discovery rates (Storey and Tibshirani 2003), as it is well adapted to large numbers of tests and has been shown to be powerful when applied to the branch-site test of positive selection (Anisimova and Yang 2007). To evaluate the specificity and power of this methodology, we performed simulations that reproduce the original data as closely as possible (Table 1). We obtain 59%–99.7% of true positives when positive selection is simulated and 0%–8.5% of false positives on simulations with no positive selection. Using this test on the real data set, we find that positive selection has affected most genes during bony vertebrate evolution: 77% with the commonly used q = 10% threshold of false positives, and still 45% with a stringent q = 1% (Table 2). In the following analyses, we use q = 10% (P ≤ 0.078), as it provides more data, and trends in all parameters considered are consistent if we use the more restrictive cut-off. The complete data, including all P- and q-values, are available as Supplemental material (Supplemental Table 1).
Table 1.
Evaluation of the accuracy and power of the likelihood tests for positive selection using simulated data
Data simulated under the same global parameters as the real data, with sites evolving under a mix of purifying selection and neutrality (Nearly Neutral model); plus sites under positive selection on the foreground branch for the positive selection model. Simulations under the Nearly Neutral model with original branch lengths divided by 2 are not shown, because 0% of tests are significant for all branches.
aClassification of topologies and branches following Figure 1. FSGD, fish-specific genome duplication.
bNumber of trees where the model A with foreground ω2 > 1 is significantly more likely than the model A with ω2 = 1, with multiple test correction by the q-value of Storey and Tibshirani (2003). (LRT) Likelihood ratio test.
cThe number of trees with at least one branch where positive selection was significant.
Table 2.
Number of genes for which positive selection is detected
aClassification of topologies and branches following that in Figure 1. FSGD, fish-specific genome duplication.
bNumber of trees where the model A with foreground ω2 > 1 is significantly more likely than the model A with ω2 = 1, with multiple test correction by the q-value of Storey and Tibshirani (2003) over all tests performed. (LRT) Likelihood ratio test.
cThe number of trees with at least one branch where positive selection was significant.
Detection of positive selection on 77% of genes corresponds to only 45% of phylogenetic branches tested. Most genes appear to have experienced positive selection during some periods of their evolution, but not during others. Evidence for positive selection is not evenly distributed across the branches we tested but is higher on the longer branches: The more evolutionary change accumulates, the greater the chance of detecting an episode of positive selection. Thus the most positive selection in our data set is detected for the divergence of tetrapodes and teleost fishes (∼352 million years [Myr] of cumulated evolution); the less, for the base of the mammals (∼150 Myr) (Fig. 1). Over all branches tested, there is a significant correlation between branch length (in amino acid substitutions/site) and the result of the likelihood test for positive selection (Spearman’s ρ = −0.37, P < 2.2 × 10−16).
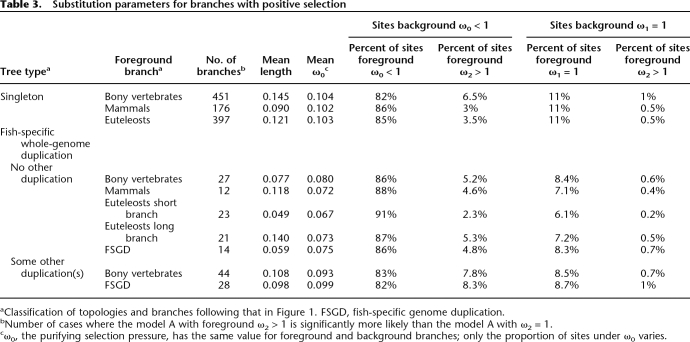
For branches on which positive selection is detected, it only concerns a minority of sites (Table 3). The mean over all significant branches gives 5.6% of sites under positive selection. Assuming that zero sites are positively selected when the test is not significant and weighting by gene length, we obtain a mean of 2.7% sites under positive selection (30,210 sites under positive selection/1,129,328 sites analyzed). This number corresponds to an “average branch,” but the number of sites predicted to be under positive selection varies among branches (Table 3). Computing the mean for each branch separately, we obtain between 0.9% for the mammalian branch and 4.7% for the bony vertebrate branch. It is difficult to compute the proportion of sites that have been under positive selection over all of vertebrate evolution, since (1) we do not test all possible branches; (2) our data provide insufficient power to identify all the specific sites under positive selection, so we cannot determine whether the same sites are repeatedly under selection or not; and (3) the bony vertebrate branch is unrooted, thus combining selection that occurred in the ancestor of tetrapodes and in the ancestor of teleost fishes. But while we cannot from these data give a specific number of sites affected by positive selection overall, it is clear that it is a small proportion during each evolutionary period tested.
Table 3.
Substitution parameters for branches with positive selection
aClassification of topologies and branches following that in Figure 1. FSGD, fish-specific genome duplication.
bNumber of cases where the model A with foreground ω2 > 1 is significantly more likely than the model A with ω2 = 1.
cω0, the purifying selection pressure, has the same value for foreground and background branches; only the proportion of sites under ω0 varies.
We tested for the influence of saturation of synonymous sites in two ways: by removing extreme data points and by simulation. First, we removed all genes with at least one branch of dS ≥ 2 synonymous substitutions/codon. This leads to removal of 237 genes (25% of the total), but the proportion of genes with positive selection detected is almost unchanged, at 79%; results per branch are also not affected (data not shown). Repeating this with a limit of dS ≥ 1 leads to removal of 88% of genes, but the proportions are again hardly changed, with positive selection detected on 77% of genes left (data not shown).
Second, we performed simulations that reproduce the distribution of gene families from the real data set (see Methods). Reassuringly, we recover the simulated total tree length with good accuracy, both in dN (mean absolute value of error 14%) and in dS (mean absolute value of error 13%). This stands in contrast to saturation problems if we were to use pairwise comparisons instead of computation over the whole tree. For example, on the simulated stickleback–rat data corresponding to ENSGACG00000006060 and ENSRNOG00000014240 (family HBG000007 of Supplemental Table 1), the simulated “true” dS of 4.158 is estimated at 16.2 by pairwise comparison. But a whole tree analysis yields a value of 4.155. The longest single branch in this case has a true value of 1.52 and is estimated at 1.51. Thus the use of more sequences does help to break long branches with saturation issues. Moreover, if the results were due in large part to saturation of dS, we expect spurious detection of positive selection on long branches. This is not the case, with only 0.6% of the 2673 branches tested significant for positive selection (q = 10%) (Table 1), when simulated without positive selection. To investigate further the effect of potential saturation, we conducted additional simulations with modified branch lengths. Dividing all branch lengths by 2 led to zero branches significant for positive selection (data not shown), unsurprisingly. Multiplying all branch lengths by 1.5 (data not shown) or 2 (Table 1) led to 2.0% and 2.9%, respectively, of branches significant for positive selection, largely under the accepted q = 10% of false positives. To also verify the power of the branch site test on such divergent sequences, we added 6% of sites under positive selection on one branch at a time. With ω = 4 (as in Anisimova and Yang 2007), 64% of the branches tested were found significant. With ω = 9, which is our observed median value, 95% of the branches tested were found significant (Table 1). Thus it appears that the test used is robust to large sequence divergence, and our results are not due to dS saturation.
An issue that is not covered by the simulations is the impact of alignment uncertainty (Landan and Graur 2007; Wong et al. 2008). We expect this impact to be limited in our study because (1) we selected very conserved gene families, (2) we realigned selected sequences using one of the best available multiple alignment algorithms, and (3) the detection of positive selection is done using only columns without gaps. To verify this, we performed two controls. For each control, results were recomputed on the “bony vertebrate” branch, the most divergent of our study. First, we realigned selected sequences with MAFFT (Katoh and Toh 2008). Results are strongly correlated between the MAFFT and MUSCLE alignments (correlation of ΔLnL values: r = 0.87, P < 10−16). The significance of the test changes for only 6% of genes. Second, we filtered both MUSCLE and MAFFT alignments with Gblocks (Castresana 2000) to exclude poorly aligned sites from the computations. On average, only 4.4% of sites are excluded. The correlation between results with and without Gblocks is high (MUSCLE: r = 0.88; MAFFT: r = 0.94), with changes in significance for 7% of genes based on either MUSCLE or MAFFT alignments. Changes in significance occur in both directions, so that the proportion of genes with significant positive selection on the “bony vertebrate” branch is approximately the same (59%–64%) with MUSCLE or MAFFT, with or without Gblocks. In summary, alignment errors do not appear to have a major impact on our results.
What can these results teach us about the occurrence of positive selection? First, we contrasted the Gene Ontology and Panther ontology categories of genes that show positive selection on a given branch, to those that do not. Although the usual suspects of positive selection are slightly overrepresented (e.g., GO terms “response to external stimulus,” “defense response,” “immune system process,” Panther terms “signal transduction,” “immunity and defense”), no term varies significantly according to positive selection, on any branch (for GO, all adjusted P-values = 1; for Panther, all q-values > 0.53; data not shown). Second, we contrasted the categories of our data set to the complete set of genes from the human genome. This tests whether our procedure of data selection may have introduced a functional bias leading to excess detection of positive selection. On the contrary, our data set appears enriched in genes implicated in core cellular and physiological processes (Supplemental Tables 2, 3), and depleted in categories most often reported as targets of positive selection (Vallender and Lahn 2004; Yang 2006). This is consistent with our relatively conservative selection procedure and indicates that positive selection in vertebrates is not restricted to a small subset of fast evolving genes.
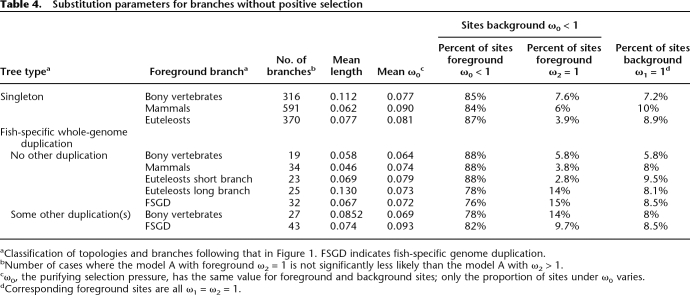
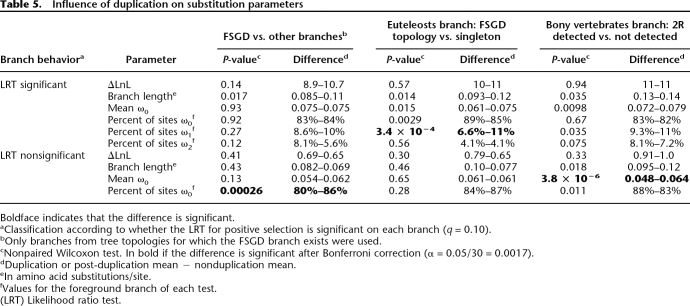
Another surprising observation is that the branches following the fish-specific whole genome duplication do not stand out in our results. For each parameter computed, they are within the range observed for the other branches tested (Tables 2–4). We searched for weaker but potentially significant effects, first by contrasting parameter values for the duplication branches (“FSGD”) to those for all other branches, on the same trees (Table 5; column FSGD vs. Other Branches). The only significant effect is weaker purifying selection in some cases. Next, we contrasted speciation branches that follow a duplication event to the same speciation branches without a prior duplication (Table 5) to quantify longer term effects. In addition to the fish-specific genome duplication, we verified for such effects due to the whole genome duplications at the origin of vertebrates (2R in Table 5). Consistent with the previous results, most comparisons are not significant. It should be noted that the “singleton” branches include duplication followed by loss, as well as cases where one paralog was not detected by the methods used. The only significant differences indicate more purifying selection for genes kept in double after the duplication, consistent with known biased retention (Davis and Petrov 2004; Brunet et al. 2006). If there is more positive selection after duplication, as expected for neofunctionalization of sequences, we expect a better fit of the positive selection model. This should translate into higher ΔLnL values, the difference in log likelihood between the models with and without positive selection. But these values never differ significantly according to the presence or absence of duplication, on or before the branches tested, thus providing no evidence for protein sequence neofunctionalization.
Table 4.
Substitution parameters for branches without positive selection
aClassification of topologies and branches following that in Figure 1. FSGD indicates fish-specific genome duplication.
bNumber of cases where the model A with foreground ω2 = 1 is not significantly less likely than the model A with ω2 > 1.
cω0, the purifying selection pressure, has the same value for foreground and background sites; only the proportion of sites under ω0 varies.
dCorresponding foreground sites are all ω1 = ω2 = 1.
Table 5.
Influence of duplication on substitution parameters
Boldface indicates that the difference is significant.
aClassification according to whether the LRT for positive selection is significant on each branch (q = 0.10).
bOnly branches from tree topologies for which the FSGD branch exists were used.
cNonpaired Wilcoxon test. In bold if the difference is significant after Bonferroni correction (α = 0.05/30 = 0.0017).
dDuplication or post-duplication mean − nonduplication mean.
eIn amino acid substitutions/site.
fValues for the foreground branch of each test.
(LRT) Likelihood ratio test.
Discussion
Detection of positive selection in vertebrate evolution
This is to our knowledge the first such scan for ancient positive selection using the rigorous branch-site model as improved by Zhang et al. (2005). We find that while only 0.9%–4.7% of codons have experienced positive selection on different branches of vertebrate evolution, such episodes of positive selection have affected 77% of genes investigated. This high number is found, although we use a conservative test (e.g., Bakewell et al. 2007) and do not test all branches of the vertebrate tree. It should be noted that the gene data set analyzed here is not representative of the whole genome. Indeed, to avoid misalignments artifacts, we restricted our analyses to gene families for which orthologs could be aligned over at least 80% of their length. Moreover, to limit problems of saturation, we selected only gene families for which orthologs could be identified in at least 11 species (five fishes, Xenopus, chicken, and at least four mammals). Hence gene families with frequent gene duplications and losses or with a high rate of amino acid substitution are underrepresented in our data set. Because of this bias toward highly conserved genes, our results are probably an underestimate of the true frequency of genes that are subject to positive selection.
Our results stand in some contrast to reports of rare positive selection in mammals (Endo et al. 1996; Clark et al. 2003; Jorgensen et al. 2005; Zhang and Li 2005; Arbiza et al. 2006; Bakewell et al. 2007; Gibbs et al. 2007) and are more reminiscent of results in lineages with larger population sizes (Nielsen and Yang 2003; Eyre-Walker 2006; Drosophila 12 Genomes Consortium 2007; Sawyer et al. 2007). Interestingly, the most recent Drosophila study (Drosophila 12 Genomes Consortium 2007) found 2% of codons under positive selection, a very similar proportion to our observations. Estimates of the proportion of changes driven by positive selection in Drosophila vary considerably according to methodology (Sawyer et al. 2007; Shapiro et al. 2007).
Most codon model studies in mammals have used relatively few sequences per gene, testing for selection either by pairwise comparison or on a tree with few sequences. Simulations indicate that likelihood tests tend to be overly conservative when few sequences are used (Anisimova et al. 2002; Anisimova and Yang 2007). The design of our tree patterns allowed us to test exclusively internal branches with at least four sequences on each side (Fig. 1). Moreover, we tested longer branches than intramammalian studies. We observe a positive correlation between detection of selection and branch length, which is consistent with previous reports that more positive selection can be detected when longer branches are tested (Anisimova and Yang 2007; Gibbs et al. 2007), as long as saturation is not reached. A few previous reports have illustrated the power of likelihood tests to detect positive selection in ancient vertebrate evolution (e.g., Bielawski and Yang 2004). Saturation of dS would be problematic in pairwise comparisons of sequences as divergent as human and zebrafish. But our simulations, as well as those of Anisimova and colleagues (Anisimova et al. 2002; Anisimova and Yang 2007), show that the maximum likelihood estimate as we use it is robust to dS saturation. This appears to be due to the use of more sequences, which break the long branches of the gene tree.
Our results are consistent with a model of recurrent adaptive amino acid substitutions, driven by weak positive selection, as modeled recently in fly (Andolfatto 2007). This model notably predicts more selective substitutions in rapidly evolving genes, which is consistent with the correlation with dN that we observe. It also predicts that such selection will be difficult to detect in genome scans, as is the case. This model, and simulation results (Anisimova et al. 2002; Anisimova and Yang 2007), indicate that our results are not contradictory to the reports of rare positive selection in mammals. Rather, testing over relatively short time intervals provides evidence for the strongest signals of positive selection only, on a small subset of genes, whereas testing over longer time enables us to detect events which are rarer, but eventually affect most genes. It remains to be seen whether the same sites are repeatedly under positive selection in different lineages or whether different sites are affected. Our data provide insufficient power to test this, as specific sites under weak positive selection are difficult to identify.
Several investigators have noted that a high dN/dS ratio can be caused by low dS, due to local constraints on synonymous substitutions (Pond and Muse 2005; Chamary et al. 2006; Schattner and Diekhans 2006; Mayrose et al. 2007; Parmley and Hurst 2007), while Friedman and Hughes (2007) have also argued for an impact of GC content. To evaluate more in detail the influence of such potential confounding factors on our results, we adjusted a linear model explaining the test results (ΔLnL) for each branch by global characteristics of the tree (Supplemental Table 4). An ANOVA on this model shows that (1) for all branches, the most significant contributors to our results are the number of sites and the dN value. This confirms that with more amino acid substitutions, more positive selection can be detected. (2) In some cases GC content and dS contribute significantly to test results, but always explain little variance. (3) More than 60% of variance in test results is explained by none of the global parameters included in the model. Of note, two measures of alignment quality, the proportion of gaps and the number of sites excluded by Gblocks, have little to no effect on test results. The variation in GC content along each branch, which could indicate changes in codon usage or in recombination rates, also has no effect. In contrast to a report of bias of dN/dS tests (Wyckoff et al. 2005), we find the expected weak negative correlation of global ω with dS length of the tree (r = −0.19, P = 1.1 × 10−8), and strong positive correlation with dN (r = 0.82, P < 2.2 × 10−16). Excluding genes with at least one very high dS branch did not change results. Thus, as far as we can tell, we seem to be effectively detecting branch-site specific positive selection, not some bias of the alignment or the tree.
Two functional categories of genes tend to be overrepresented in reports of positive selection (Vallender and Lahn 2004; Yang 2006): genes involved in host defense and immunity or in evading these defenses, and genes involved in sexual reproduction. Such trends have been confirmed in several genomic scans for positive selection, notably in primates, where they typically also include neuronal function and perception (Bustamante et al. 2005; Nielsen et al. 2005; Biswas and Akey 2006; Voight et al. 2006; Wang et al. 2006; Gibbs et al. 2007). This seems contradictory with our results: Positively selected genes do not differ in functional categories from other genes, while our total sample is in fact biased toward basic cellular processes (Supplemental Tables 2, 3). First, we note that in some previous studies, functional categories such as metabolism genes were reported as under positive selection (Roth and Liberles 2006; Voight et al. 2006; Petersen et al. 2007). Second, different studies may measure different selection modes. Polymorphism studies in primates typically report genes that are under recent selection, whereas we have scanned for selection in more ancient vertebrate evolution. The branch-site test is not intended to detect continuous selective pressure, which would likely characterize arms race genes. Moreover, most interspecific studies have used less stringent model testing and may report as being under positive selection genes that are under weak purifying selection (Zhang et al. 2005; Bakewell et al. 2007). Indeed, the most stringent primate study so far found positive selection mostly in genes involved in basic cellular functions (Bakewell et al. 2007). Finally, in a recent study of Drosophila genomes (Drosophila 12 Genomes Consortium 2007), positive selection was found to affect all functional categories, albeit more strongly “defense response.” Similarly, our results do not exclude that positive selection be strongest on fast evolving (e.g., immunity) genes, absent from our data set. But they do indicate that episodes of positive selection affect all categories of genes, in vertebrates as in flies.
The impact of genome duplication on patterns of selection
In addition to the functional categories already discussed, duplicate genes are also overrepresented in reports of positive selection (Yang 2006). There have been several reports of higher dN/dS ratios on branches following duplications (Jordan et al. 2004; He and Zhang 2005; Brunet et al. 2006; Byrne and Wolfe 2006; Johnston et al. 2007) and even on branches preceding duplications (Johnston et al. 2007). But these studies used global measures of dN/dS, which do not distinguish between relaxed negative selection, and positive selection on a few sites. In the analysis of the macaque genome, an excess of duplicate genes among those with positive selection was noted, but on quite a small sample (Gibbs et al. 2007). Thus this study is to our knowledge the first large-scale quantification of positive selection after duplication. By using only duplicates from whole genome duplication, we constrain the duplication branches to represent the same divergence time. And by contrasting branches of the same gene tree, we control for biased retention of duplicates (Davis and Petrov 2004; Brunet et al. 2006).
The result of this careful testing is striking: The substitution parameters after duplication differ very little, if at all, from those after speciation (Table 5). The few significant differences are consistent with previous reports of biased retention of genes under stronger purifying selection, after whole genome duplication (Davis and Petrov 2004; Brunet et al. 2006), and of some relaxation of purifying selection, rather than with any increase in positive selection. Of note, the faster evolving paralog does not show evidence of more positive selection (Table 2), as would be expected if the asymmetry were due to neofunctionalization at the protein level (Brunet et al. 2006; Byrne and Wolfe 2006). This result shows the importance of controlling with speciation branches before attributing an effect to duplication. A study solely conducted on the duplication branch might have concluded erroneously that 36% of genes were subject to positive selection because of the duplication, whereas this proportion is not higher than in comparable branches without duplication. We note that previous studies that contrasted duplication and speciation branches, while not explicitly identifying positive selection, also found that differences are more slight than expected (Seoighe et al. 2003; Conant et al. 2007; Hellsten et al. 2007; Johnston et al. 2007; Scannell and Wolfe 2007). The weak difference between protein coding gene evolution after speciation and after duplication may reflect the importance of other levels of function, such as expression, on the divergence of duplicates (Hellsten et al. 2007; Hughes 2007). Indeed, a recent study of whole genome duplication in yeast (Wapinski et al. 2007) found that duplicates diverged mostly in regulation, and much less in biochemical function, which is what we can detect at the level of amino acid sequences.
Conclusion
An important conclusion of this work is that the most stringent test does detect positive selection in significant amounts in vertebrate evolution. Thus, adaptive evolution at the molecular level does appear significant (Hoekstra and Coyne 2007), although we note that our results are in no way exclusive of functional evolution by regulatory mutations (Hughes 2007; Nei 2007; Prud’homme et al. 2007). Our results are supportive of a model of widespread but transient positive selection. Finally, we do not find a large difference of evolutionary modes of protein coding sequences after duplication, relative to speciation. It remains to be tested whether this is due to divergence at other levels or to a lesser impact of duplication on gene evolution than expected.
Methods
Data
Gene families were obtained from the database HOMOLENS version 3 (http://pbil.univ-lyon1.fr/databases/homolens.html), which is based on Ensembl release 41 (October 2006) (Hubbard et al. 2007). HOMOLENS is built on the same model as HOVERGEN (Duret et al. 1994) or HOBACGEN (Perriere et al. 2000), with genes organized in families, which include precalculated alignments and phylogenies. In HomolEns version 3, alignments are computed with MUSCLE (Edgar 2004) (with default parameters), and phylogenetic trees with PHYML (substitution model = JTT, estimated proportion of invariable sites, four categories, estimated gamma, initial tree with BIONJ) (Guindon and Gascuel 2003). Phylogenies are computed on conserved blocks of the alignments selected with Gblocks (Castresana 2000).
Using the TreePattern functionality of the FamFetch client for HOMOLENS, which allows scanning for gene tree topologies (Dufayard et al. 2005), we selected three sets (Fig. 1): (1) a set of “singleton” genes, where duplication is strictly forbidden along the tree; (2) a strict set of “fish-specific genome duplication” genes, where paralogs are retained in all fishes after the whole genome duplication but other duplication is forbidden; and (3) a relaxed set of “fish-specific genome duplication” genes, where paralogs are retained in all fishes after the whole genome duplication and other duplication is allowed. In all cases, we imposed that all five fishes, the Xenopus, the chicken, and at least four mammals be represented in the tree. In addition, to clarify an eventual effect of older whole genome duplications at the origin of vertebrates (known as 2R for two rounds of duplication), we selected all genes with duplications specific to vertebrates, predating the teleost fish–tetrapode split. This allowed us to define the subset of genes in our data that were kept in duplicate after these older genome duplications.
For the families thus recovered, we restricted alignments and trees to the selected phylogenetic pattern, notably excluding more distant paralogs (e.g., from duplications basic to vertebrates). We removed species with low genome coverage. The restricted alignments were refined with MUSCLE (Edgar 2004). Computations were then done on the new alignment, after removing all columns with at least one gap. DNA alignments are calculated from the protein alignments, with RevTrans (Wernersson and Pedersen 2003). To evaluate the impact of alignment uncertainty, the restricted alignments were also refined with MAFFT (Katoh and Toh 2008), and high-quality alignments were selected from both MUSCLE and MAFFT alignments using Gblocks (type = codons) (Castresana 2000). For the manipulations of sequences and trees, we combined scripts in Python, BioPython, Jalview (Clamp et al. 2004), and the R library APE (Paradis et al. 2004).
Our data set includes 10 species of tetrapodes: the frog Xenopus tropicalis (DoE Joint Genome Institute, unpubl.); the chicken Gallus gallus (International Chicken Genome Sequencing Consortium 2004); the seven mammals Monodelphis domestica (Mikkelsen et al. 2007), Bos taurus (HGSC at Baylor College of Medicine, unpubl.), Canis familiaris (Lindblad-Toh et al. 2005), Mus musculus (Waterston et al. 2002), Rattus norvegicus (Rat Genome Sequencing Project Consortium 2004), Macaca mulatta (Gibbs et al. 2007), Pan troglodytes (The Chimpanzee Sequencing and Analysis Consortium 2005), and Homo sapiens (International Human Genome Sequencing Consortium 2001, 2004); and five species of teleost fishes: the zebrafish Danio rerio (Zebrafish Sequencing Group at the Sanger Institute, unpubl.); and the four euteleosts Gasterosteus aculeatus (The Broad Institute, unpubl.), Oryzias latipes (Kasahara et al. 2007), Tetraodon nigroviridis (Jaillon et al. 2004), and Takifugu rubripes (Aparicio et al. 2002).
All alignments and trees can be viewed and downloaded at http://bioinfo.unil.ch/supdata/.
Detection of positive selection
We used the branch-site model A (Yang and Nielsen 2002; Zhang et al. 2005), which allows to detect positive selection that acts on a subset of sites in a specific lineage. Positive selection is detected by a dN/dS ratio ω > 1. This model has been reported to be more sensitive for the detection of positive selection than previous models (Yang 2006), such as branch models (Yang 1998) or site models (Yang et al. 2000).
The application of this model necessitates providing a phylogenetic tree and defining a priori the branch we want to test for positive selection. This branch is called the foreground branch (Fig. 1, bold), where positive selection may be allowed. All other branches in the tree represent the background branches, where sites are only allowed to evolve under purifying or neutral selection. The original formulation (Yang and Nielsen 2002) compared this branch-site model A (alternative hypothesis) to the Neutral site model M1 (null hypothesis). The problem in this test is that it does not discriminate between positive selection and relaxation of purifying selection (Zhang 2004). To avoid this problem of false positives, Zhang et al. (2005) proposed a stricter test, which contrasts the branch-site model A with ω2 ≥ 1 (alternative hypothesis) to the model A with ω2 = 1 fixed (null hypothesis). The test is done by comparing the difference of likelihood values 2 × ΔLnL to a χ2 distribution of 1 degree of freedom. This test has been reported to be very conservative (e.g., Bakewell et al. 2007), and it is the only one we used in our analysis. Of note, the model distinguishes two components of the positively selected set of sites: sites that are under purifying selection in the rest of the tree, and sites that are neutral in the rest of the tree. All computations are done using CODEML from the PAML package (v3.15) (Yang 1997).
We use the value 2 × ΔLnL as the best measure of outcome of the test for positive selection in all subsequent statistical analyses.
Statistical analysis
We used the web server FatiGO+ to perform statistical analysis on Gene Ontology terms (Al-Shahrour et al. 2007), with FDR correction for multiple testing (Benjamini and Hochberg 1995). We also used the PANTHER Classification System (Thomas et al. 2003), followed by QVALUE correction for multiple testing (A. Dabney and J.D. Storey, unpubl.).
To correct for CODEML testing on multiple branches in multiple phylogenetic trees, we control for false discovery by using the q-value (Storey and Tibshirani 2003). All P-values from our likelihood ratio tests were treated as one series of repetitions (m branches × n trees). Our P-values follow a bimodal distribution, because it is not rare that the alternative brings no improvement over the null hypothesis (thus P = 1), while in many other cases the difference of likelihoods is large (thus P ≈ 0). As recommended for a bimodal distribution (documentation of the QVALUE library), we used the bootstrap method for estimating π0 in the R package QVALUE (A. Dabney and J.D. Storey, unpubl.).
We used nhPhyml (topology fixed, transition/transversion [Ts/Tv] ratio estimated, alpha parameter estimated with four categories, GC equilibrium frequency optimized for each branch) (Boussau and Gouy 2006) to estimate the GC rate at third codon positions at each node of the phylogenetic trees. We then computed the ΔGC for each branch of interest, as the difference between GC at the nodes bracketing that branch.
All other statistical analyses were performed using R (R Development Core Team 2007).
Simulations
Simulated nucleotide alignments were generated using Evolver, from the PAML package (v3.15) (Yang 1997).
To test accuracy, we generated alignments under the null hypothesis of no positive selection. We used the Nearly Neutral model (M1a), allowing sites to be under purifying selection or neutral evolution. For each real data set, a simulated data set was generated using the same global parameters as the real data: number of sequences, sequence length, tree topology, branch lengths (defined as the number of nucleotide substitutions per codon), dN/dS ratio ω, Ts/Tv ratio κ, and codon usage. This procedure guaranties that the simulated data set has the same distribution of parameters as the real data set, including potential confounding factors such as codon usage or long branches. In addition, to control for the effect of potential underestimation of dS (saturation), we conducted simulations modifying the total length of the tree. Dividing all branch lengths by 2 provides an estimate of the behavior of the test with less divergent sequences, while multiplying by 1.5 or 2 provides estimates of the behavior of the test with even more divergent sequences. Alternatively, the latter simulations could correct for underestimation of branch lengths in the original analysis.
To test power, we generated alignments using the same procedure, plus specifying branch-site–specific positive selection on one branch. Thus we performed as many simulations for each data set, as branches tested. In accordance with our results (Table 3), we simulated nucleotide alignments with 84% of sites under purifying selection (ω0), 10% of sites under neutral evolution (ω1) and 6% of sites under positive selection on the foreground branch (ω2). Values of ω2 = 4 and ω2 = 9 were chosen, following, respectively, the method of Anisimova and Yang (2007), and the median value observed in our data.
Acknowledgments
We thank Darlene Goldstein, Jérôme Goudet, Tal Pupko, Nicolas Salamin, and Ken Wolfe for helpful discussions. We also thank Adam Eyre-Walker and anonymous reviewers for insightful remarks. R.S. and M.R.R. acknowledge funding from Etat de Vaud and Swiss National Science Foundation grant 116798, and L.D. from the Agence Nationale de la Recherche (GIP ANR JC05_49162). We thank the VITAL-IT project of the Swiss Institute of Bioinformatics for providing the computational resources.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.076992.108.
References
- Al-Shahrour F., Minguez P., Tarraga J., Medina I., Alloza E., Montaner D., Dopazo J., Minguez P., Tarraga J., Medina I., Alloza E., Montaner D., Dopazo J., Tarraga J., Medina I., Alloza E., Montaner D., Dopazo J., Medina I., Alloza E., Montaner D., Dopazo J., Alloza E., Montaner D., Dopazo J., Montaner D., Dopazo J., Dopazo J. FatiGO +: A functional profiling tool for genomic data. Integration of functional annotation, regulatory motifs and interaction data with microarray experiments. Nucleic Acids Res. 2007;35:W91–W96. doi: 10.1093/nar/gkm260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P. Hitchhiking effects of recurrent beneficial amino acid substitutions in the Drosophila melanogaster genome. Genome Res. 2007;17:1755–1762. doi: 10.1101/gr.6691007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M., Yang Z., Yang Z. Multiple hypothesis testing to detect lineages under positive selection that affects only a few sites. Mol. Biol. Evol. 2007;24:1219–1228. doi: 10.1093/molbev/msm042. [DOI] [PubMed] [Google Scholar]
- Anisimova M., Bielawski J.P., Yang Z., Bielawski J.P., Yang Z., Yang Z. Accuracy and power of Bayes prediction of amino acid sites under positive selection. Mol. Biol. Evol. 2002;19:950–958. doi: 10.1093/oxfordjournals.molbev.a004152. [DOI] [PubMed] [Google Scholar]
- Aparicio S., Chapman J., Stupka E., Putnam N., Chia J.-m., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Chapman J., Stupka E., Putnam N., Chia J.-m., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Stupka E., Putnam N., Chia J.-m., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Putnam N., Chia J.-m., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Chia J.-m., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Christoffels A., Rash S., Hoon S., Smit A., Rash S., Hoon S., Smit A., Hoon S., Smit A., Smit A., et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- Arbiza L., Dopazo J., Dopazo H., Dopazo J., Dopazo H., Dopazo H. Positive selection, relaxation, and acceleration in the evolution of the human and chimp genome. PLoS Comput. Biol. 2006;2:e38. doi: 10.1371/journal.pcbi.0020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakewell M.A., Shi P., Zhang J., Shi P., Zhang J., Zhang J. More genes underwent positive selection in chimpanzee evolution than in human evolution. Proc. Natl. Acad. Sci. 2007;104:7489–7494. doi: 10.1073/pnas.0701705104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. [Ser A] 1995;57:289–300. [Google Scholar]
- Benton M.J., Donoghue P.C.J., Donoghue P.C.J. Paleontological evidence to date the tree of life. Mol. Biol. Evol. 2006;24:26–53. doi: 10.1093/molbev/msl150. [DOI] [PubMed] [Google Scholar]
- Bielawski J.P., Yang Z., Yang Z. A maximum likelihood method for detecting functional divergence at individual codon sites, with application to gene family evolution. J. Mol. Evol. 2004;59:121–132. doi: 10.1007/s00239-004-2597-8. [DOI] [PubMed] [Google Scholar]
- Biswas S., Akey J.M., Akey J.M. Genomic insights into positive selection. Trends Genet. 2006;22:437–446. doi: 10.1016/j.tig.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Boussau B., Gouy M., Gouy M. Efficient likelihood computations with nonreversible models of evolution. Syst. Biol. 2006;55:756–768. doi: 10.1080/10635150600975218. [DOI] [PubMed] [Google Scholar]
- Brunet F.G., Crollius H.R., Paris M., Aury J.M., Gibert P., Jaillon O., Laudet V., Robinson-Rechavi M., Crollius H.R., Paris M., Aury J.M., Gibert P., Jaillon O., Laudet V., Robinson-Rechavi M., Paris M., Aury J.M., Gibert P., Jaillon O., Laudet V., Robinson-Rechavi M., Aury J.M., Gibert P., Jaillon O., Laudet V., Robinson-Rechavi M., Gibert P., Jaillon O., Laudet V., Robinson-Rechavi M., Jaillon O., Laudet V., Robinson-Rechavi M., Laudet V., Robinson-Rechavi M., Robinson-Rechavi M. Gene loss and evolutionary rates following whole-genome duplication in teleost fishes. Mol. Biol. Evol. 2006;23:1808–1816. doi: 10.1093/molbev/msl049. [DOI] [PubMed] [Google Scholar]
- Bustamante C.D., Fledel-Alon A., Williamson S., Nielsen R., Todd Hubisz M., Glanowski S., Tanenbaum D.M., White T.J., Sninsky J.J., Hernandez R.D., Fledel-Alon A., Williamson S., Nielsen R., Todd Hubisz M., Glanowski S., Tanenbaum D.M., White T.J., Sninsky J.J., Hernandez R.D., Williamson S., Nielsen R., Todd Hubisz M., Glanowski S., Tanenbaum D.M., White T.J., Sninsky J.J., Hernandez R.D., Nielsen R., Todd Hubisz M., Glanowski S., Tanenbaum D.M., White T.J., Sninsky J.J., Hernandez R.D., Todd Hubisz M., Glanowski S., Tanenbaum D.M., White T.J., Sninsky J.J., Hernandez R.D., Glanowski S., Tanenbaum D.M., White T.J., Sninsky J.J., Hernandez R.D., Tanenbaum D.M., White T.J., Sninsky J.J., Hernandez R.D., White T.J., Sninsky J.J., Hernandez R.D., Sninsky J.J., Hernandez R.D., Hernandez R.D., et al. Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- Byrne K.P., Wolfe K.H., Wolfe K.H. Consistent patterns of rate asymmetry and gene loss indicate widespread neofunctionalization of yeast genes after whole-genome duplication. Genetics. 2006;175:1341–1350. doi: 10.1534/genetics.106.066951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Chamary J.V., Parmley J.L., Hurst L.D., Parmley J.L., Hurst L.D., Hurst L.D. Hearing silence: Non-neutral evolution at synonymous sites in mammals. Nat. Rev. Genet. 2006;7:98–108. doi: 10.1038/nrg1770. [DOI] [PubMed] [Google Scholar]
- The Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Clamp M., Cuff J., Searle S.M., Barton G.J., Cuff J., Searle S.M., Barton G.J., Searle S.M., Barton G.J., Barton G.J. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Clark A.G., Glanowski S., Nielsen R., Thomas P.D., Kejariwal A., Todd M.A., Tanenbaum D.M., Civello D., Lu F., Murphy B., Glanowski S., Nielsen R., Thomas P.D., Kejariwal A., Todd M.A., Tanenbaum D.M., Civello D., Lu F., Murphy B., Nielsen R., Thomas P.D., Kejariwal A., Todd M.A., Tanenbaum D.M., Civello D., Lu F., Murphy B., Thomas P.D., Kejariwal A., Todd M.A., Tanenbaum D.M., Civello D., Lu F., Murphy B., Kejariwal A., Todd M.A., Tanenbaum D.M., Civello D., Lu F., Murphy B., Todd M.A., Tanenbaum D.M., Civello D., Lu F., Murphy B., Tanenbaum D.M., Civello D., Lu F., Murphy B., Civello D., Lu F., Murphy B., Lu F., Murphy B., Murphy B., et al. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science. 2003;302:1960–1963. doi: 10.1126/science.1088821. [DOI] [PubMed] [Google Scholar]
- Conant G.C., Wagner G.P., Stadler P.F., Wagner G.P., Stadler P.F., Stadler P.F. Modeling amino acid substitution patterns in orthologous and paralogous genes. Mol. Phylogenet. Evol. 2007;42:298–307. doi: 10.1016/j.ympev.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Davis J.C., Petrov D.A., Petrov D.A. Preferential duplication of conserved proteins in eukaryotic genomes. PLoS Biol. 2004;2:e55. doi: 10.1371/journal.pbio.0020055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Dufayard J.F., Duret L., Penel S., Gouy M., Rechenmann F., Perriere G., Duret L., Penel S., Gouy M., Rechenmann F., Perriere G., Penel S., Gouy M., Rechenmann F., Perriere G., Gouy M., Rechenmann F., Perriere G., Rechenmann F., Perriere G., Perriere G. Tree pattern matching in phylogenetic trees: Automatic search for orthologs or paralogs in homologous gene sequence databases. Bioinformatics. 2005;21:2596–2603. doi: 10.1093/bioinformatics/bti325. [DOI] [PubMed] [Google Scholar]
- Duret L., Mouchiroud D., Gouy M., Mouchiroud D., Gouy M., Gouy M. HOVERGEN: A database of homologous vertebrate genes. Nucleic Acids Res. 1994;22:2360–2365. doi: 10.1093/nar/22.12.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T., Ikeo K., Gojobori T., Ikeo K., Gojobori T., Gojobori T. Large-scale search for genes on which positive selection may operate. Mol. Biol. Evol. 1996;13:685–690. doi: 10.1093/oxfordjournals.molbev.a025629. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A. The genomic rate of adaptive evolution. Trends Ecol. Evol. 2006;21:569–575. doi: 10.1016/j.tree.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Force A., Lynch M., Pickett F.B., Amores A., Yan Y.L., Postlethwait J., Lynch M., Pickett F.B., Amores A., Yan Y.L., Postlethwait J., Pickett F.B., Amores A., Yan Y.L., Postlethwait J., Amores A., Yan Y.L., Postlethwait J., Yan Y.L., Postlethwait J., Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman R., Hughes A.L., Hughes A.L. Likelihood-ratio tests for positive selection of human and mouse duplicate genes reveal nonconservative and anomalous properties of widely used methods. Mol. Phylogenet. Evol. 2007;42:388–393. doi: 10.1016/j.ympev.2006.07.015. [DOI] [PubMed] [Google Scholar]
- Gibbs R.A., Rogers J., Katze M.G., Bumgarner R., Weinstock G.M., Mardis E.R., Remington K.A., Strausberg R.L., Venter J.C., Wilson R.K., Rogers J., Katze M.G., Bumgarner R., Weinstock G.M., Mardis E.R., Remington K.A., Strausberg R.L., Venter J.C., Wilson R.K., Katze M.G., Bumgarner R., Weinstock G.M., Mardis E.R., Remington K.A., Strausberg R.L., Venter J.C., Wilson R.K., Bumgarner R., Weinstock G.M., Mardis E.R., Remington K.A., Strausberg R.L., Venter J.C., Wilson R.K., Weinstock G.M., Mardis E.R., Remington K.A., Strausberg R.L., Venter J.C., Wilson R.K., Mardis E.R., Remington K.A., Strausberg R.L., Venter J.C., Wilson R.K., Remington K.A., Strausberg R.L., Venter J.C., Wilson R.K., Strausberg R.L., Venter J.C., Wilson R.K., Venter J.C., Wilson R.K., Wilson R.K., et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Gojobori J., Tang H., Akey J.M., Wu C.-I., Tang H., Akey J.M., Wu C.-I., Akey J.M., Wu C.-I., Wu C.-I. Adaptive evolution in humans revealed by the negative correlation between the polymorphism and fixation phases of evolution. Proc. Natl. Acad. Sci. 2007;104:3907–3912. doi: 10.1073/pnas.0605565104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Gascuel O., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- He X., Zhang J., Zhang J. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics. 2005;169:1157–1164. doi: 10.1534/genetics.104.037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellsten U., Khokha M.K., Grammer T.C., Harland R.M., Richardson P., Rokhsar D.S., Khokha M.K., Grammer T.C., Harland R.M., Richardson P., Rokhsar D.S., Grammer T.C., Harland R.M., Richardson P., Rokhsar D.S., Harland R.M., Richardson P., Rokhsar D.S., Richardson P., Rokhsar D.S., Rokhsar D.S. Accelerated gene evolution and subfunctionalization in the pseudotetraploid frog Xenopus laevis. BMC Biol. 2007;5:31. doi: 10.1186/1741-7007-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra H.E., Coyne J.A., Coyne J.A. The locus of evolution: Evo-Devo and the genetics of adaptation. Evolution Int. J. Org. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- Hubbard T.J., Aken B.L., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Aken B.L., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Clarke L., Coates G., Cunningham F., Cutts T., Coates G., Cunningham F., Cutts T., Cunningham F., Cutts T., Cutts T., et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A.L. Looking for Darwin in all the wrong places: The misguided quest for positive selection at the nucleotide sequence level. Heredity. 2007;99:364–373. doi: 10.1038/sj.hdy.6801031. [DOI] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Jaillon O., Aury J.M., Brunet F., Petit J.L., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Aury J.M., Brunet F., Petit J.L., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Brunet F., Petit J.L., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Petit J.L., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Fischer C., Ozouf-Costaz C., Bernot A., Ozouf-Costaz C., Bernot A., Bernot A., et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Johnston C.R., O'Dushlaine C., Fitzpatrick D.A., Edwards R.J., Shields D.C., O'Dushlaine C., Fitzpatrick D.A., Edwards R.J., Shields D.C., Fitzpatrick D.A., Edwards R.J., Shields D.C., Edwards R.J., Shields D.C., Shields D.C. Evaluation of whether accelerated protein evolution in chordates has occurred before, after, or simultaneously with gene duplication. Mol. Biol. Evol. 2007;24:315–323. doi: 10.1093/molbev/msl162. [DOI] [PubMed] [Google Scholar]
- Jordan I.K., Wolf Y.I., Koonin E.V., Wolf Y.I., Koonin E.V., Koonin E.V. Duplicated genes evolve slower than singletons despite the initial rate increase. BMC Evol. Biol. 2004;4:22. doi: 10.1186/1471-2148-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen F., Hobolth A., Hornshoj H., Bendixen C., Fredholm M., Schierup M., Hobolth A., Hornshoj H., Bendixen C., Fredholm M., Schierup M., Hornshoj H., Bendixen C., Fredholm M., Schierup M., Bendixen C., Fredholm M., Schierup M., Fredholm M., Schierup M., Schierup M. Comparative analysis of protein coding sequences from human, mouse and the domesticated pig. BMC Biol. 2005;3:2. doi: 10.1186/1741-7007-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M., Naruse K., Sasaki S., Nakatani Y., Qu W., Ahsan B., Yamada T., Nagayasu Y., Doi K., Kasai Y., Naruse K., Sasaki S., Nakatani Y., Qu W., Ahsan B., Yamada T., Nagayasu Y., Doi K., Kasai Y., Sasaki S., Nakatani Y., Qu W., Ahsan B., Yamada T., Nagayasu Y., Doi K., Kasai Y., Nakatani Y., Qu W., Ahsan B., Yamada T., Nagayasu Y., Doi K., Kasai Y., Qu W., Ahsan B., Yamada T., Nagayasu Y., Doi K., Kasai Y., Ahsan B., Yamada T., Nagayasu Y., Doi K., Kasai Y., Yamada T., Nagayasu Y., Doi K., Kasai Y., Nagayasu Y., Doi K., Kasai Y., Doi K., Kasai Y., Kasai Y., et al. The medaka draft genome and insights into vertebrate genome evolution. Nature. 2007;447:714–719. doi: 10.1038/nature05846. [DOI] [PubMed] [Google Scholar]
- Katoh K., Toh H., Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- Landan G., Graur D., Graur D. Heads or tails: A simple reliability check for multiple sequence alignments. Mol. Biol. Evol. 2007;24:1380–1383. doi: 10.1093/molbev/msm060. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Chang J.L., Kulbokas E.J., Zody M.C., Kulbokas E.J., Zody M.C., Zody M.C., et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Lynch M., O’Hely M., Walsh B., Force A., O’Hely M., Walsh B., Force A., Walsh B., Force A., Force A. The probability of preservation of a newly arisen gene duplicate. Genetics. 2001;159:1789–1804. doi: 10.1093/genetics/159.4.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayrose I., Doron-Faigenboim A., Bacharach E., Pupko T., Doron-Faigenboim A., Bacharach E., Pupko T., Bacharach E., Pupko T., Pupko T. Towards realistic codon models: Among site variability and dependency of synonymous and non-synonymous rates. Bioinformatics. 2007;23:i319–i327. doi: 10.1093/bioinformatics/btm176. [DOI] [PubMed] [Google Scholar]
- Mikkelsen T.S., Wakefield M.J., Aken B., Amemiya C.T., Chang J.L., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Wakefield M.J., Aken B., Amemiya C.T., Chang J.L., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Aken B., Amemiya C.T., Chang J.L., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Amemiya C.T., Chang J.L., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Chang J.L., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Garber M., Gentles A.J., Goodstadt L., Heger A., Gentles A.J., Goodstadt L., Heger A., Goodstadt L., Heger A., Heger A., et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- Nei M. The new mutation theory of phenotypic evolution. Proc. Natl. Acad. Sci. 2007;104:12235–12242. doi: 10.1073/pnas.0703349104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., Yang Z., Yang Z. Estimating the distribution of selection coefficients from phylogenetic data with applications to mitochondrial and viral DNA. Mol. Biol. Evol. 2003;20:1231–1239. doi: 10.1093/molbev/msg147. [DOI] [PubMed] [Google Scholar]
- Nielsen R., Bustamante C., Clark A.G., Glanowski S., Sackton T.B., Hubisz M.J., Fledel-Alon A., Tanenbaum D.M., Civello D., White T.J., Bustamante C., Clark A.G., Glanowski S., Sackton T.B., Hubisz M.J., Fledel-Alon A., Tanenbaum D.M., Civello D., White T.J., Clark A.G., Glanowski S., Sackton T.B., Hubisz M.J., Fledel-Alon A., Tanenbaum D.M., Civello D., White T.J., Glanowski S., Sackton T.B., Hubisz M.J., Fledel-Alon A., Tanenbaum D.M., Civello D., White T.J., Sackton T.B., Hubisz M.J., Fledel-Alon A., Tanenbaum D.M., Civello D., White T.J., Hubisz M.J., Fledel-Alon A., Tanenbaum D.M., Civello D., White T.J., Fledel-Alon A., Tanenbaum D.M., Civello D., White T.J., Tanenbaum D.M., Civello D., White T.J., Civello D., White T.J., White T.J., et al. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:e170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E., Claude J., Strimmer K., Claude J., Strimmer K., Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Parmley J., Hurst L., Hurst L. How common are intragene windows with KA > KS owing to purifying selection on synonymous mutations? J. Mol. Evol. 2007;64:646–655. doi: 10.1007/s00239-006-0207-7. [DOI] [PubMed] [Google Scholar]
- Perriere G., Duret L., Gouy M., Duret L., Gouy M., Gouy M. HOBACGEN: Database system for comparative genomics in bacteria. Genome Res. 2000;10:379–385. doi: 10.1101/gr.10.3.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen L., Bollback J.P., Dimmic M., Hubisz M., Nielsen R., Bollback J.P., Dimmic M., Hubisz M., Nielsen R., Dimmic M., Hubisz M., Nielsen R., Hubisz M., Nielsen R., Nielsen R. Genes under positive selection in Escherichia coli. Genome Res. 2007;17:1336–1343. doi: 10.1101/gr.6254707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond S.K., Muse S.V., Muse S.V. Site-to-site variation of synonymous substitution rates. Mol. Biol. Evol. 2005;22:2375–2385. doi: 10.1093/molbev/msi232. [DOI] [PubMed] [Google Scholar]
- Prud’homme B., Gompel N., Carroll S.B., Gompel N., Carroll S.B., Carroll S.B. Emerging principles of regulatory evolution. Proc. Natl. Acad. Sci. 2007;104:8605–8612. doi: 10.1073/pnas.0700488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam N.H., Hellsten U., Yu J.S., Pennachio L., Blow M., Shoguchi E., Robinson-Rechavi M., Butts T., Ferrier D.E.K., Garcia-Fernàndez J., Hellsten U., Yu J.S., Pennachio L., Blow M., Shoguchi E., Robinson-Rechavi M., Butts T., Ferrier D.E.K., Garcia-Fernàndez J., Yu J.S., Pennachio L., Blow M., Shoguchi E., Robinson-Rechavi M., Butts T., Ferrier D.E.K., Garcia-Fernàndez J., Pennachio L., Blow M., Shoguchi E., Robinson-Rechavi M., Butts T., Ferrier D.E.K., Garcia-Fernàndez J., Blow M., Shoguchi E., Robinson-Rechavi M., Butts T., Ferrier D.E.K., Garcia-Fernàndez J., Shoguchi E., Robinson-Rechavi M., Butts T., Ferrier D.E.K., Garcia-Fernàndez J., Robinson-Rechavi M., Butts T., Ferrier D.E.K., Garcia-Fernàndez J., Butts T., Ferrier D.E.K., Garcia-Fernàndez J., Ferrier D.E.K., Garcia-Fernàndez J., Garcia-Fernàndez J., et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- R Development Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2007. [Google Scholar]
- Rat Genome Sequencing Project Consortium Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Roth C., Liberles D., Liberles D. A systematic search for positive selection in higher plants (Embryophytes) BMC Plant Biol. 2006;6:12. doi: 10.1186/1471-2229-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer S.A., Parsch J., Zhang Z., Hartl D.L., Parsch J., Zhang Z., Hartl D.L., Zhang Z., Hartl D.L., Hartl D.L. Prevalence of positive selection among nearly neutral amino acid replacements in Drosophila. Proc. Natl. Acad. Sci. 2007;104:6504–6510. doi: 10.1073/pnas.0701572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell D.R., Wolfe K.H., Wolfe K.H. A burst of protein sequence evolution and a prolonged period of asymmetric evolution follow gene duplication in yeast. Genome Res. 2007;18:137–147. doi: 10.1101/gr.6341207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner P., Diekhans M., Diekhans M. Regions of extreme synonymous codon selection in mammalian genes. Nucleic Acids Res. 2006;34:1700–1710. doi: 10.1093/nar/gkl095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoighe C., Johnston C.R., Shields D.C., Johnston C.R., Shields D.C., Shields D.C. Significantly different patterns of amino acid replacement after gene duplication as compared to after speciation. Mol. Biol. Evol. 2003;20:484–490. doi: 10.1093/molbev/msg059. [DOI] [PubMed] [Google Scholar]
- Shapiro J.A., Huang W., Zhang C., Hubisz M.J., Lu J., Turissini D.A., Fang S., Wang H.-Y., Hudson R.R., Nielsen R., Huang W., Zhang C., Hubisz M.J., Lu J., Turissini D.A., Fang S., Wang H.-Y., Hudson R.R., Nielsen R., Zhang C., Hubisz M.J., Lu J., Turissini D.A., Fang S., Wang H.-Y., Hudson R.R., Nielsen R., Hubisz M.J., Lu J., Turissini D.A., Fang S., Wang H.-Y., Hudson R.R., Nielsen R., Lu J., Turissini D.A., Fang S., Wang H.-Y., Hudson R.R., Nielsen R., Turissini D.A., Fang S., Wang H.-Y., Hudson R.R., Nielsen R., Fang S., Wang H.-Y., Hudson R.R., Nielsen R., Wang H.-Y., Hudson R.R., Nielsen R., Hudson R.R., Nielsen R., Nielsen R., et al. Adaptive genic evolution in the Drosophila genomes. Proc. Natl. Acad. Sci. 2007;104:2271–2276. doi: 10.1073/pnas.0610385104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J.D., Tibshirani R., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P.D., Campbell M.J., Kejariwal A., Mi H., Karlak B., Daverman R., Diemer K., Muruganujan A., Narechania A., Campbell M.J., Kejariwal A., Mi H., Karlak B., Daverman R., Diemer K., Muruganujan A., Narechania A., Kejariwal A., Mi H., Karlak B., Daverman R., Diemer K., Muruganujan A., Narechania A., Mi H., Karlak B., Daverman R., Diemer K., Muruganujan A., Narechania A., Karlak B., Daverman R., Diemer K., Muruganujan A., Narechania A., Daverman R., Diemer K., Muruganujan A., Narechania A., Diemer K., Muruganujan A., Narechania A., Muruganujan A., Narechania A., Narechania A. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallender E.J., Lahn B.T., Lahn B.T. Positive selection on the human genome. Hum. Mol. Genet. 2004;13:R245–R254. doi: 10.1093/hmg/ddh253. [DOI] [PubMed] [Google Scholar]
- Voight B.F., Kudaravalli S., Wen X., Pritchard J.K., Kudaravalli S., Wen X., Pritchard J.K., Wen X., Pritchard J.K., Pritchard J.K. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E.T., Kodama G., Baldi P., Moyzis R.K., Kodama G., Baldi P., Moyzis R.K., Baldi P., Moyzis R.K., Moyzis R.K. Global landscape of recent inferred Darwinian selection for Homo sapiens. Proc. Natl. Acad. Sci. 2006;103:135–140. doi: 10.1073/pnas.0509691102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski I., Pfeffer A., Friedman N., Regev A., Pfeffer A., Friedman N., Regev A., Friedman N., Regev A., Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449:54–61. doi: 10.1038/nature06107. [DOI] [PubMed] [Google Scholar]
- Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwala R., Ainscough R., Alexandersson M., An P., Ainscough R., Alexandersson M., An P., Alexandersson M., An P., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wernersson R., Pedersen A.G., Pedersen A.G. RevTrans: Multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res. 2003;31:3537–3539. doi: 10.1093/nar/gkg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K.M., Suchard M.A., Huelsenbeck J.P., Suchard M.A., Huelsenbeck J.P., Huelsenbeck J.P. Alignment uncertainty and genomic analysis. Science. 2008;319:473–476. doi: 10.1126/science.1151532. [DOI] [PubMed] [Google Scholar]
- Wyckoff G.J., Malcom C.M., Vallender E.J., Lahn B.T., Malcom C.M., Vallender E.J., Lahn B.T., Vallender E.J., Lahn B.T., Lahn B.T. A highly unexpected strong correlation between fixation probability of nonsynonymous mutations and mutation rate. Trends Genet. 2005;21:381–385. doi: 10.1016/j.tig.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- Yang Z. Computational molecular evolution. Oxford University Press; Oxford, UK: 2006. [Google Scholar]
- Yang Z., Nielsen R., Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- Yang Z., Nielsen R., Goldman N., Pedersen A.M., Nielsen R., Goldman N., Pedersen A.M., Goldman N., Pedersen A.M., Pedersen A.M. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Frequent false detection of positive selection by the likelihood method with branch-site models. Mol. Biol. Evol. 2004;21:1332–1339. doi: 10.1093/molbev/msh117. [DOI] [PubMed] [Google Scholar]
- Zhang L., Li W.-H., Li W.-H. Human SNPs reveal no evidence of frequent positive selection. Mol. Biol. Evol. 2005;22:2504–2507. doi: 10.1093/molbev/msi240. [DOI] [PubMed] [Google Scholar]
- Zhang J., Nielsen R., Yang Z., Nielsen R., Yang Z., Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]