Abstract
Mitochondria are essential for cellular energy production in most eukaryotic organisms. However, when glucose is abundant, yeast species that underwent whole-genome duplication (WGD) mostly conduct fermentation even under aerobic conditions, and most can survive without a functional mitochondrial genome. In this study, we show that the rate of evolution for the nuclear-encoded mitochondrial genes was greater in post-WGD species than pre-WGD species. Furthermore, codon usage bias was relaxed for these genes in post-WGD yeast species. The codon usage pattern and the distribution of a particular transcription regulatory element suggest that the change to an efficient aerobic fermentation lifestyle in this lineage might have emerged after WGD between the divergence of Kluyveromyces polysporus and Saccharomyces castellii from their common ancestor. This new energy production strategy could have led to the relaxation of mitochondrial function in the relevant yeast species.
Two major processes generate cellular energy in eukaryotic organisms: fermentation, which occurs anaerobically, and respiration, which requires oxygen (Freeman 2005). Respiration that is carried out in mitochondria produces ∼80% of the energy needed by most eukaryotic organisms under aerobic conditions (Rolfe and Brown 1997). Whole-genome duplication (WGD) in yeast, estimated to have occurred in the ancestor of the Saccharomyces sensu stricto species complex around 100 million years ago (Mya) (Wolfe and Shields 1997; Dietrich et al. 2004; Dujon et al. 2004; Kellis et al. 2004), has been proposed to have led to the evolution of an efficient fermentation system in this lineage (Piskur 2001; Piskur and Langkjaer 2004; Piskur et al. 2006). This hypothesis is based on the observation that with plentiful glucose, most post-WGD yeast species primarily carry out fermentation to generate energy even under aerobic conditions (Crabtree-positive) (Merico et al. 2007). In contrast, pre-WGD yeast species mostly respire under such conditions.
Although the above hypothesis is yet to be proven, efficient fermentation is likely to have had a profound impact on the evolution of the relevant yeast species because cellular energy is an important factor in determining growth and hence fitness of organisms in their environments. With the introduction of aerobic fermentation in post-WGD yeast species, the role of mitochondria in generating energy for cellular growth appeared to have become attenuated. Indeed, most post-WGD yeast species can live even without a functional mitochondrial genome (Petite-positive) (Merico et al. 2007). In this study, we investigated the impact of this metabolic strategy change on mitochondrial evolution. Protein evolution rates and codon usage patterns indicate that a relaxation of function may have occurred for genes that act in the mitochondria (called “mitochondrial genes” in this study) for post-WGD species. By examining the codon usage bias and evolution pattern for a specific regulatory motif, we concluded that the efficient aerobic fermentation system in this lineage might have emerged between the divergence of Kluyveromyces polysporus and Saccharomyces castellii from their common ancestor after WGD.
Results
Evolutionary rates of mitochondrial genes increased in post-WGD species
Rates of amino acid substitution are predominantly used to gauge functional constraints on gene products (Nei 2005). To investigate whether WGD led to a relaxation of functional constraints on genes that act in the mitochondria, we compared the evolutionary trajectory of orthologous genes in six post-WGD yeast species and three closely related pre-WGD yeast species in the clade Saccharomycotina. A total of 2603 one-to-one orthologous genes, including 296 mitochondrial genes, were identified. Nonsynonymous distances (dN) were calculated for each orthologous gene between individual pre-WGD or post-WGD species pairs. Because different species diverged at different time points during evolution, and in order to compare the gene evolutionary rate between different species pairs, we normalized the dN value of each gene in a specific species pair by the median dN of all the studied orthologous genes for the same species pair. The latter measurements are hypothesized to represent rough estimates for the divergence time between species (Supplemental Fig. 1).
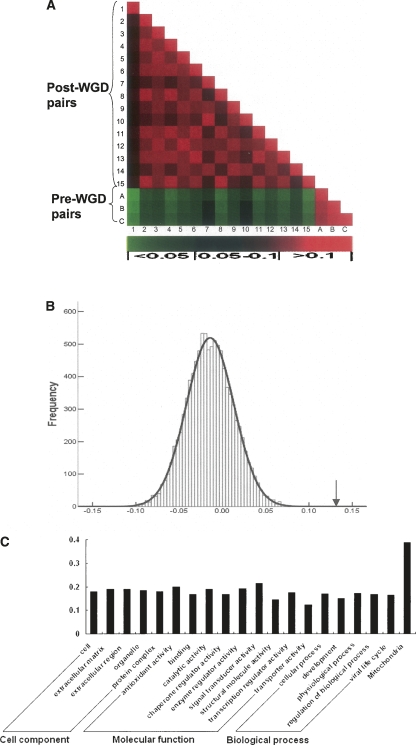
As shown in Figure 1A, the normalized evolutionary distances for all the mitochondrial genes are similar within the pre-WGD and post-WGD species groups. They are significantly different, however, between the pre-WGD and post-WGD groups: 41 of 45 cross-WGD comparisons (there are 15 post-WGD and three pre-WGD species pairs, respectively) show significant differences (P < 0.05). Only two comparisons are expected to show significant difference under the same threshold for P-values. The other four comparisons have P-values between 0.05 and 0.07 (Fig. 1A). The average mitochondrial gene evolutionary distance after normalization (by evolutionary rates of all the studied genes in the whole genome) is about 0.75 for the pre-WGD species pairs, but 0.85 for the post-WGD species pairs. To see whether the 13% [(0.85 − 0.75)/0.75] increase in mitochondrial gene evolutionary rate in the post-WGD species is statistically significant, we randomly picked 296 non-mitochondrial genes from the orthologous matrix, calculated their overall evolutionary rate increase in the post-WGD species, and iterated the process 10,000 times. None of the random gene sets show a greater evolutionary rate increase than that seen for the mitochondrial genes (P < 10−4;Fig. 1B).
Figure 1.
Mitochondrial genes evolve faster in post-WGD yeast species. (A) Pairwise comparison of the normalized dN values for mitochondrial genes. Numbers and letters to the left and at the bottom of the figure indicate species pairs. (1–15, post-WGD species pairs; A–C, pre-WGD species pairs. 1, Sces-Spar; 2, Sces-Smik; 3, Sces-Sbay; 4, Sces-Scas; 5, Sces-Kpol; 6, Spar-Smik; 7, Spar-Sbay; 8, Spar-Scas; 9, Spar-Kpol; 10, Smik-Sbay; 11, Smik-Scas; 12, Smik-Kpol; 13, Sbay-Scas; 14, Sbay-Kpol; 15, Scas-Kpol; A, Kwal-Sklu; B, Kwal-Klac; C, Sklu-Klac [see Methods “Sequence Data” section for abbreviation descriptions]). The color in each intersecting cell represents the P-value of the comparison between mitochondrial gene dN values for the two species pairs in that row and column, respectively. The scale of P-values is shown at the bottom of the figure. (B) Distribution of the evolutionary rate increase in the post-WGD species. The distribution was obtained by randomly sampling 296 genes, calculating their evolution rate increase in the post-WGD species, and iterating the process for 10,000 times. The X-axis is the average evolutionary rate increase in the post-WGD species. The curve represents the normal distribution estimated from the simulated data, and the arrow indicates the observed evolutionary rate increase for the mitochondrial genes (P < 1.0 × 10−4). (C) Proportions of genes that have increased evolutionary rates in the post-WGD species. Non-mitochondrial genes were grouped into 19 different categories based on their Gene Ontology (GO) annotations. The Y-axis indicates the percentage of genes with increased rates of evolution in the post-WGD species. The similar results were obtained using a nonredundant set of more specific GO categories for biological function (Myers et al. 2006) or subcellular location data (Supplemental Fig. 2A,B).
The evolutionary distances of individual genes were also compared between the pre-WGD and post-WGD species. Out of the 296 studied mitochondrial genes, 115 (38.9%) have significantly increased evolutionary rates in the post-WGD yeast species. In comparison, only 18.5% (427 out of 2307) of non-mitochondrial genes show a similar trend (Fisher’s exact test P = 3 × 10−14) (Supplemental Table 1). The proportion of non-mitochondrial genes showing increased evolutionary rates in the post-WGD yeast species for different functional categories are also significantly smaller than that of the mitochondrial genes (Z test, P = 0) (Fig. 1C; Supplemental Fig. 2A,B).
Genes with increased evolutionary rates in the post-WGD species are enriched with mitochondrial translation machine components (42 out of 70, P = 4.0 × 10−5). Because protein translation is crucial for mitochondrial functions, the observed relaxation could have profound impacts on organelle evolution. Indeed, a majority (∼65%) of the mitochondrial proteome is composed of genes that have evolutionary rate increases in the post-WGD species and their physical interaction partners (Supplemental Table 2). Among the 31 studied genes that are directly involved in aerobic respiration, 14 genes show evolutionary rate increases in the post-WGD species. This proportion (14/31 = 45.2%) is not statistically different from that of the mitochondrial genes (38.9%), but it is much larger than that of the non-mitochondrial genes in the whole genome (18.5%).
Codon usage biases of mitochondrial genes were relaxed in most post-WGD species
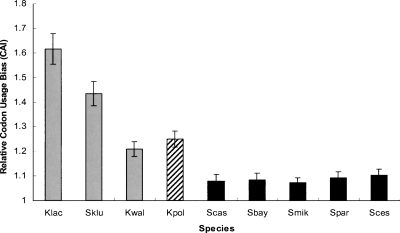
Highly expressed genes tend to have strong codon usage bias, most likely to ensure efficient and accurate translation (Ikemura 1981, 1982). A reduction in functional constraint on a gene product could lead to a reduction in its codon usage bias (Akashi 1997; Gu et al. 2005). Therefore, if mitochondrial energy production became less important to the post-WGD species, a decreased codon usage bias would be expected for the mitochondrial genes of these species. To test this hypothesis, we calculated the Codon Adaptation Index (CAI) (Sharp and Li 1987; Sharp and Matassi 1994) value for individual genes in each species. A Student’s t-test was used to compare codon usage bias for mitochondrial and non-mitochondrial genes within each species. Interestingly, mitochondrial genes displayed significantly stronger codon usage bias than non-mitochondrial genes in all three studied pre-WGD species (P-values < 0.001 for each case). In contrast, there are no significant differences in codon usage bias between mitochondrial and non-mitochondrial genes (P-values > 0.05) for all the studied post-WGD species except for K. polysporus, which was the first species among those studied to have diverged from a common ancestor after WGD (Scannell et al. 2007).
In order to compare gene codon usage bias between species, we normalized the CAI value of each gene by the median CAI value of all studied genes in the same species. This was done because codon usage bias can be affected by many species-specific factors such as mutation bias, tRNA pools, and RNA stabilities (Ermolaeva 2001). As shown in Figure 2, with the exception of K. polysporus, the relative CAI values for mitochondrial genes in all post-WGD species are significantly lower than those in pre-WGD species. In addition, the levels of mitochondrial gene codon usage bias in pre-WGD species show an interesting relationship with respect to fermentation: the least efficient fermenter (Kluyveromyces lactis) has the strongest codon usage bias, while the most efficient (Kluyveromyces waltii) has the weakest codon usage bias (Fig. 2) (the fermentation efficiency was inferred from Merico et al. 2007). These observations substantiate the hypothesis that codon usage bias, and thus functional constraints on mitochondrial genes, are less severe when fermentation plays a greater role in cellular energy production (Andersson et al. 2003).
Figure 2.
Average codon usage bias for mitochondrial genes in each species. The means and standard errors were calculated from the normalized CAI values. (Shaded bars) Three pre-WGD species; (black bars) the post-WGD species except for K. polysporus (striped bar).
K. polysporus might be an intermediate between pre-WGD to post-WGD species regarding evolution of fermentation
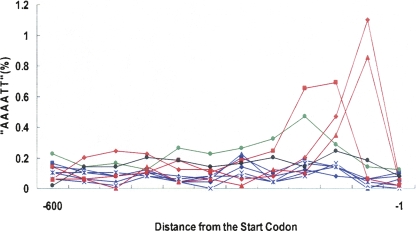
Although K. polysporus is a post-WGD species, it shows a different pattern of mitochondrial gene codon usage bias from the other post-WGD species (Fig. 2). Because the mitochondrial gene evolutionary rate also increased in this species (Fig. 1), we suspect that this species represents a transitional stage for the evolution of fermentation in this lineage. To test this, we investigated an important regulatory motif (“AAAATT,” rapid growth element [RGE]) that is present in the mitochondrial ribosomal protein genes in pre-WGD species, but was claimed to be missing in all post-WGD species (Ihmels et al. 2005). The motif responds to fast organism growth and was also found in the regulatory regions for rRNA and ribosomal protein genes. Compared to all three studied pre-WGD and other post-WGD species, K. polysporus has an intermediate number of mitochondrial ribosomal genes that retain the motif. This holds true even after correcting for high AT content in the genome of this species (Fig. 3). A recent work on yeast physiology by Fekete et al. (2007) revealed that all of the pre-WGD species that we studied as well as K. polysporus are petite negative, whereas the other post-WGD species in our study are petite positive. In addition, the species that are closely related to K. polysporus during evolution have a mixture of petite positive/negative phenotypes (Fekete et al. 2007). These data support a transitional role of K. polysporus during yeast fermentation evolution in this lineage.
Figure 3.
Frequency of the “AAAATT” motif occurrence for the mitochondrial ribosomal protein genes. The X-axis indicates the location of the motif (−1 denotes the first nucleotide upstream of the start codon). (Red) The pre-WGD species (▴, K. waltii; ◆, S. kluyveri; ◼, K. lactis); (blue) the post-WGD species (◆, S. cerevisiae; ◼, S. paradoxus; ▴, S. mikatae; ×, S. bayanus; and *, S. castellii), except for K. polysporus (green ●). (Black ●) Motif occurrence in random sequences that was generated according to the GC content of K. polysporus.
Discussion
In aerobic conditions with ample carbon sources such as glucose, pre-WGD yeast species predominantly conduct respiration to generate energy for fast cellular growth. Under the same conditions, however, most post-WGD yeast species undergo aerobic fermentation due to glucose repression of respiration pathways (Wang et al. 2004; Santangelo 2006). Mitochondria are essential for eukaryotic organisms to carry out respiration. Our results show that the evolutionary rate of genes that function in the organelle increased in post-WGD yeast species. During aerobic fermentation, yeast produce ethanol in the presence of glucose, and use ethanol as an energy resource when glucose is exhausted. Since ethanol is toxic to most microorganisms, the capacity to produce, accumulate, tolerate, and consume ethanol must have provided these yeast species selective advantages over other organisms. We hypothesize that this initial stage of adaptive evolution could have shaped the evolution of certain mitochondrial genes in post-WGD species.
Adaptive evolution, however, might not play a major role in the increase of evolutionary rate seen in mitochondrial genes in post-WGD yeast species. With the advent of aerobic fermentation in these species, mitochondria have less of a role in generating energy that was otherwise essential to pre-WGD yeast species. Our results are consistent with a relaxed functional role of this organelle in post-WGD species. Reduced codon usage bias for the mitochondrial genes, which is not expected under adaptive evolution, also supports this conclusion. The rewiring of a regulatory network for mitochondrial ribosomal protein genes in post-WGD yeast species due to the loss of a regulatory motif might be a consequence of a functional relaxation of these genes (Ihmels, et al. 2005). A recent study showed that translational efficiency of mitochondrial genes is higher in aerobic compared with anaerobic yeasts (Man and Pilpel 2007). This observation is also consistent with a relaxed codon usage bias for the mitochondrial genes in anaerobic yeasts because relaxed codon usage bias in a gene decreases its translation efficiency. Interestingly, genes involved in glycolysis, which are important for fermentative growth, displayed stronger codon usage bias in anaerobic than those in aerobic yeast species (Man and Pilpel 2007). This is expected because of the increased usage of fermentation in these anaerobic species. Because mitochondria are also important for various functions unrelated to energy production (McBride et al. 2006), and respiration is essential for post-WGD yeast species when glucose is depleted, a total degeneration of mitochondria and loss of genes that function in this organelle is not expected in post-WGD species.
Our study indirectly tested the relationship between the WGD and the evolution of efficient fermentation in the lineage leading to the baker’s yeast, Saccharomyces cerevisiae. The importance of WGD to the evolution of efficient fermentation in this lineage is supported by various observations (Wolfe 2004; Blank et al. 2005; Conant and Wolfe 2007). Nevertheless, the intermediate evolutionary status of K. polysporus inferred from our results, which was substantiated by others (Fekete et al. 2007), indicates that the WGD alone was not sufficient for the emergence of efficient fermentation. Relaxation of mitochondrial function is more likely linked to the advent of aerobic fermentation. Because a new fermentative metabolic strategy could have taken a long time to become optimal, speciation events leading to K. polysporus could have happened during this process. It will be interesting to investigate whether K. polysporus is a better fermenter than the studied pre-WGD species despite being petite-negative.
The speciation events leading to K. polysporus and S. castellii occurred ∼80–90 Mya (Scannell et al. 2007). Efficient fermentation in this lineage might have emerged after the divergence of K. polysporus but before the divergence of S. castellii from the post-WGD common ancestor. Remarkably, fermentable fruits started a wide geographic expansion around the same time (Benner et al. 2002), thus providing an environment in which efficient fermentation could flourish. By comparing the genomic differences—particularly for the relevant pathways—between the post-WGD species with and without efficient fermentation ability, we hope to gain a clearer picture of how this novel lifestyle developed during evolution.
Methods
Sequence data
According to the phylogenetic tree constructed by Kurtzman and Robnett (2003), there are three pre-WGD and eight post-WGD yeast species whose whole genome sequences are available in clades 1–11. We did not use Saccharomyces kudriavzevii because of its low DNA sequencing coverage. Candida glabrata was also not included because this species showed an accelerated evolutionary rate at the whole genome level after speciation (Supplemental Fig. 3). Since we were comparing evolutionary rates of mitochondrial genes that are normalized by the median rate of all genes in the genome, the global evolutionary rate increase in C. glabrata made it difficult to delineate the evolutionary rate acceleration for mitochondrial genes in this lineage. Hence, three pre-WGD (Saccharomyces kluyveri [Sklu], K. waltii [Kwal], and K. lactis [Klac]) and six post-WGD species (K. polysporus [Kpol], S. cerevisiae [Sces], Saccharomyces paradoxus [Spar], Saccharomyces mikatae [Smik], Saccharomyces bayanus [Sbay], and S. castellii [Scas]) were used in our study. The protein, coding, and genomic sequences for the recently sequenced post-WGD species, K. polysporus, are from Scannell et al. (2007). Sequences for the other species were downloaded from the Fungal Comparative Genomics database (http://fungal.genome.duke.edu/) (Fitzpatrick et al. 2006) and the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/).
Orthologous gene definition and evolutionary distance estimation
Using the INPARANOID software package (Remm et al. 2001), orthologs between any two species were identified. Orthologous clusters among all species were generated by MultiParanoid (Alexeyenko et al. 2006) according to the results of INPARANOID. We removed all duplicated genes such as Ohnologs (Byrne and Wolfe 2005) that belong to the same orthologous cluster and retained the clusters that had only one member in each species. A one-to-one orthologous matrix with 2603 rows (genes) and nine columns (species) was created. The mitochondrial genes were downloaded from the manually curated MitoP2 database (Andreoli et al. 2004; http://www.mitop.de:8080/mitop2/). In the database, 528 S. cerevisiae genes are annotated as mitochondrial genes based on published literature. There are 296 such mitochondrial gene entries in our defined orthologous matrix.
For each orthologous gene, we did 15 pairwise alignments for the post-WGD species and three pairwise alignments for the pre-WGD species pairs using GeneWise (Birney et al. 2004). The software YN00 in the package PAML was used to estimate the nonsynonymous distance (dN) for each of the 18 alignments (Yang and Nielsen 2000). Because different species diverged at different times, the dN value of each gene in a species pair was normalized by the median dN value for all studied orthologous genes of the same species pair. In Supplemental Figure 1, we used the median dN values between each species pair to reconstruct phylogenetic trees among the pre-WGD and post-WGD species. The trees are consistent with the previous results (Kurtzman and Robnett 2003), indicating that the median dN could represent the divergence time between species and provide a calibrating scale for evolutionary rate normalization. Student’s t-test was used to compare evolutionary rates of mitochondrial genes between any two species pairs.
Gene Ontology (GO) analysis
The GO annotation file of S. cerevisiae was downloaded from the Gene Ontology website (http://www.geneontology.org/GO.current.annotations.shtml) in July 2007. Using WEGO (Ye et al. 2006), we assigned all studied non-mitochondrial genes into 19 categories based on their GO items. The proportions of genes with increased evolutionary rates in the post-WGD species were calculated in each category and also for the mitochondrial genes (Fig. 1C). Similar results for more specific functional and sublocalization categories were obtained (Supplemental Fig. 2A,B).
GO items of the mitochondrial genes with increased evolutionary rates in the post-WGD species were compared with those for the studied mitochondrial genes. Gene enrichment for each GO item was analyzed by software GeneMerge1.2 (Castillo-Davis and Hartl 2003).
Codon usage bias (codon adaptation index [CAI]) calculation
According to the MultiParanoid results, we identified all the orthologous clusters that contained the known cytoplasmic ribosomal protein genes in S. cerevisiae regardless of gene copy number in each species. These ribosomal protein genes in each species were used as reference to calculate the CAI value for individual genes in the same species by CodonW (Sharp and Matassi 1994; http://codonw.sourceforge.net/).
Regulatory motif analysis
There are 49 mitochondrial ribosomal protein genes in our orthologous matrix. For each species, we scanned 600 bp of DNA sequence upstream of all mitochondrial ribosomal protein genes to discern the occurrence of regulatory motif “AAAATT” and its reverse complement (Ihmels et al. 2005). The percentage of motif occurrence in each 50-bp window was calculated. Multiple occurrences of the motif in the same sliding window were regarded as independent events. To see whether the motif occurrence in K. polysporus is different from random expectation, we calculated the GC content of the upstream sequences for these 49 mitochondrial ribosomal protein genes in K. polysporus. Based on the observed GC content, 49 random sequences were generated by RANDNA (Piva and Principato 2006) software, and the occurrence of “AAAATT” and its reverse complement was calculated.
Acknowledgments
We thank Kenneth Wolfe, Andrew Clark, Patrick Stover, Thomas Fox, and Paul Soloway for discussions; Orna Man for technical help with INPARANOID and MultiParanoid; Guojie Zhang for technical help with WEGO; and Eric Alani for reading of the manuscript. This work was supported by a NSFC key grant (no. 30430400) and a 973 Program of China (no. 2007CB815703-5) to W.W. and startup funds from Cornell University awarded to Z.G.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.074674.107.
References
- Akashi H. Codon bias evolution in Drosophila. Population genetics of mutation-selection drift. Gene. 1997;205:269–278. doi: 10.1016/s0378-1119(97)00400-9. [DOI] [PubMed] [Google Scholar]
- Alexeyenko A., Tamas I., Liu G., Sonnhammer E.L., Tamas I., Liu G., Sonnhammer E.L., Liu G., Sonnhammer E.L., Sonnhammer E.L. Automatic clustering of orthologs and inparalogs shared by multiple proteomes. Bioinformatics. 2006;22:e9–e15. doi: 10.1093/bioinformatics/btl213. [DOI] [PubMed] [Google Scholar]
- Andersson S.G., Karlberg O., Canbäck B., Kurland C.G., Karlberg O., Canbäck B., Kurland C.G., Canbäck B., Kurland C.G., Kurland C.G. On the origin of mitochondria: A genomics perspective. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2003;358:165–177. doi: 10.1098/rstb.2002.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreoli C., Prokisch H., Hörtnagel K., Mueller J.C., Münsterkötter M., Scharfe C., Meitinger T., Prokisch H., Hörtnagel K., Mueller J.C., Münsterkötter M., Scharfe C., Meitinger T., Hörtnagel K., Mueller J.C., Münsterkötter M., Scharfe C., Meitinger T., Mueller J.C., Münsterkötter M., Scharfe C., Meitinger T., Münsterkötter M., Scharfe C., Meitinger T., Scharfe C., Meitinger T., Meitinger T. MitoP2, an integrated database on mitochondrial proteins in yeast and man. Nucleic Acids Res. 2004;32:D459–D462. doi: 10.1093/nar/gkh137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benner S.A., Caraco M.D., Thomson J.M., Gaucher E.A., Caraco M.D., Thomson J.M., Gaucher E.A., Thomson J.M., Gaucher E.A., Gaucher E.A. Planetary biology—Paleontological, geological, and molecular histories of life. Science. 2002;296:864–868. doi: 10.1126/science.1069863. [DOI] [PubMed] [Google Scholar]
- Birney E., Clamp M., Durbin R., Clamp M., Durbin R., Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank L.M., Lehmbeck F., Sauer U., Lehmbeck F., Sauer U., Sauer U. Metabolic-flux and network analysis of fourteen hemiascomycetous yeasts. FEMS Yeast Res. 2005;5:545–558. doi: 10.1016/j.femsyr.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Byrne K.P., Wolfe K.H., Wolfe K.H. The Yeast Gene Order Browser: Combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Davis C.I., Hartl D.L., Hartl D.L. GeneMerge–post-genomic analysis, data mining, and hypothesis testing. Bioinformatics. 2003;19:891–892. doi: 10.1093/bioinformatics/btg114. [DOI] [PubMed] [Google Scholar]
- Conant G.C., Wolfe K.H., Wolfe K.H. Increased glycolytic flux as an outcome of whole-genome duplication in yeast. Mol. Syst. Biol. 2007;3:129. doi: 10.1038/msb4100170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich F.S., Voegeli S., Brachat S., Lerch A., Gates K., Steiner S., Mohr C., Pöhlmann R., Luedi P., Choi S., Voegeli S., Brachat S., Lerch A., Gates K., Steiner S., Mohr C., Pöhlmann R., Luedi P., Choi S., Brachat S., Lerch A., Gates K., Steiner S., Mohr C., Pöhlmann R., Luedi P., Choi S., Lerch A., Gates K., Steiner S., Mohr C., Pöhlmann R., Luedi P., Choi S., Gates K., Steiner S., Mohr C., Pöhlmann R., Luedi P., Choi S., Steiner S., Mohr C., Pöhlmann R., Luedi P., Choi S., Mohr C., Pöhlmann R., Luedi P., Choi S., Pöhlmann R., Luedi P., Choi S., Luedi P., Choi S., Choi S., et al. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- Dujon B., Sherman D., Fischer G., Durrens P., Casaregola S., Lafontaine I., De Montigny J., Souciet J.L., Sherman D., Fischer G., Durrens P., Casaregola S., Lafontaine I., De Montigny J., Souciet J.L., Fischer G., Durrens P., Casaregola S., Lafontaine I., De Montigny J., Souciet J.L., Durrens P., Casaregola S., Lafontaine I., De Montigny J., Souciet J.L., Casaregola S., Lafontaine I., De Montigny J., Souciet J.L., Lafontaine I., De Montigny J., Souciet J.L., De Montigny J., Souciet J.L., Souciet J.L. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- Ermolaeva M.D. Synonymous codon usage in bacteria. Curr. Issues Mol. Biol. 2001;3:91–97. [PubMed] [Google Scholar]
- Fekete V., Cierna M., Poláková S., Piskur J., Sulo P., Cierna M., Poláková S., Piskur J., Sulo P., Poláková S., Piskur J., Sulo P., Piskur J., Sulo P., Sulo P. Transition of the ability to generate petites in the Saccharomyces/Kluyveromyces complex. FEMS Yeast Res. 2007;7:1237–1247. doi: 10.1111/j.1567-1364.2007.00287.x. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick D.A., Logue M.E., Stajich J.E., Butler G., Logue M.E., Stajich J.E., Butler G., Stajich J.E., Butler G., Butler G. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol. Biol. 2006;6:99. doi: 10.1186/1471-2148-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman S. Biological science. 2d ed. Prentice Hall; Upper Saddle River, New Jersey: 2005. [Google Scholar]
- Gu Z., David L., Petrov D., Jones T., Davis R.W., Steinmetz L.M., David L., Petrov D., Jones T., Davis R.W., Steinmetz L.M., Petrov D., Jones T., Davis R.W., Steinmetz L.M., Jones T., Davis R.W., Steinmetz L.M., Davis R.W., Steinmetz L.M., Steinmetz L.M. Elevated evolutionary rates in the laboratory strain of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 2005;102:1092–1097. doi: 10.1073/pnas.0409159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihmels J., Bergmann S., Gerami-Nejad M., Yanai I., McClellan M., Berman J., Barkai N., Bergmann S., Gerami-Nejad M., Yanai I., McClellan M., Berman J., Barkai N., Gerami-Nejad M., Yanai I., McClellan M., Berman J., Barkai N., Yanai I., McClellan M., Berman J., Barkai N., McClellan M., Berman J., Barkai N., Berman J., Barkai N., Barkai N. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science. 2005;309:938–940. doi: 10.1126/science.1113833. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J. Mol. Biol. 1981;15:389–409. doi: 10.1016/0022-2836(81)90363-6. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs. J. Mol. Biol. 1982;158:573–597. doi: 10.1016/0022-2836(82)90250-9. [DOI] [PubMed] [Google Scholar]
- Kellis M., Birren B.W., Lander E., Birren B.W., Lander E., Lander E. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- Kurtzman C.P., Robnett C.J., Robnett C.J. Phylogenetic relationships among yeasts of the ‘Saccharomyces complex’ determined from multigene sequence analyses. FEMS Yeast Res. 2003;3:417–432. doi: 10.1016/S1567-1356(03)00012-6. [DOI] [PubMed] [Google Scholar]
- Man O., Pilpel Y., Pilpel Y. Differential translation efficiency of orthologous genes is involved in phenotypic divergence of yeast species. Nat. Genet. 2007;39:415–421. doi: 10.1038/ng1967. [DOI] [PubMed] [Google Scholar]
- McBride H.M., Neuspiel M., Wasiak S., Neuspiel M., Wasiak S., Wasiak S. Mitochondria: More than just a powerhouse. Curr. Biol. 2006;16:R551–R560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- Merico A., Sulo P., Piskur J., Compagno C., Sulo P., Piskur J., Compagno C., Piskur J., Compagno C., Compagno C. Fermentative lifestyle in yeasts belonging to the Saccharomyces complex. FEBS J. 2007;274:976–989. doi: 10.1111/j.1742-4658.2007.05645.x. [DOI] [PubMed] [Google Scholar]
- Myers C.L., Barrett D.R., Hibbs M.A., Huttenhower C., Troyanskaya O.G., Barrett D.R., Hibbs M.A., Huttenhower C., Troyanskaya O.G., Hibbs M.A., Huttenhower C., Troyanskaya O.G., Huttenhower C., Troyanskaya O.G., Troyanskaya O.G. Finding function: Evaluation methods for functional genomic data. BMC Genomics. 2006;7:187. doi: 10.1186/1471-2164-7-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Selectionism and neutralism in molecular evolution. Mol. Biol. Evol. 2005;22:2318–2342. doi: 10.1093/molbev/msi242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskur J. Origin of the duplicated regions in the yeast genomes. Trends Genet. 2001;17:302–303. doi: 10.1016/s0168-9525(01)02308-3. [DOI] [PubMed] [Google Scholar]
- Piskur J., Langkjaer R.B., Langkjaer R.B. Yeast genome sequencing: The power of comparative genomics. Mol. Microbiol. 2004;52:381–389. doi: 10.1111/j.1365-2958.2004.04182.x. [DOI] [PubMed] [Google Scholar]
- Piskur J., Rozpedowska E., Polakova S., Merico A., Compagno C., Rozpedowska E., Polakova S., Merico A., Compagno C., Polakova S., Merico A., Compagno C., Merico A., Compagno C., Compagno C. How did Saccharomyces evolve to become a good brewer? Trends Genet. 2006;22:183–186. doi: 10.1016/j.tig.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Piva F., Principato G., Principato G. RANDNA: A random DNA sequence generator. In Silico Biol. 2006;6:253–258. [PubMed] [Google Scholar]
- Remm M., Storm C.E., Sonnhammer E.L., Storm C.E., Sonnhammer E.L., Sonnhammer E.L. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- Rolfe D.F., Brown G.C., Brown G.C. Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiol. Rev. 1997;77:731–758. doi: 10.1152/physrev.1997.77.3.731. [DOI] [PubMed] [Google Scholar]
- Santangelo G. Glucose signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2006;70:253–282. doi: 10.1128/MMBR.70.1.253-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell D.R., Frank A.C., Conant G.C., Byrne K.P., Woolfit M., Wolfe K.H., Frank A.C., Conant G.C., Byrne K.P., Woolfit M., Wolfe K.H., Conant G.C., Byrne K.P., Woolfit M., Wolfe K.H., Byrne K.P., Woolfit M., Wolfe K.H., Woolfit M., Wolfe K.H., Wolfe K.H. Independent sorting-out of thousands of duplicated gene pairs in two yeast species descended from a whole-genome duplication. Proc. Natl. Acad. Sci. 2007;104:8397–8402. doi: 10.1073/pnas.0608218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P.M., Li W.H., Li W.H. The codon Adaptation Index—A measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P.M., Matassi G., Matassi G. Codon usage and genome evolution. Curr. Opin. Genet. Dev. 1994;4:851–860. doi: 10.1016/0959-437x(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Wang Y., Pierce M., Schneper L., Güldal C., Zhang X., Tavazoie S., Broach J., Pierce M., Schneper L., Güldal C., Zhang X., Tavazoie S., Broach J., Schneper L., Güldal C., Zhang X., Tavazoie S., Broach J., Güldal C., Zhang X., Tavazoie S., Broach J., Zhang X., Tavazoie S., Broach J., Tavazoie S., Broach J., Broach J. Ras and Gpa2 mediate one branch of a redundant glucose signaling pathway in yeast. PLoS Biol. 2004;2:e128. doi: 10.1371/journal.pbio.0020128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe K.H. Evolutionary genomics: Yeasts accelerate beyond BLAST. Curr. Biol. 2004;14:R392–R394. doi: 10.1016/j.cub.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Wolfe K.H., Shields D.C., Shields D.C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- Yang Z., Nielsen R., Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- Ye J., Fang L., Zheng H., Zhang Y., Chen J., Zhang Z., Wang J., Li S., Li R., Bolund L., Fang L., Zheng H., Zhang Y., Chen J., Zhang Z., Wang J., Li S., Li R., Bolund L., Zheng H., Zhang Y., Chen J., Zhang Z., Wang J., Li S., Li R., Bolund L., Zhang Y., Chen J., Zhang Z., Wang J., Li S., Li R., Bolund L., Chen J., Zhang Z., Wang J., Li S., Li R., Bolund L., Zhang Z., Wang J., Li S., Li R., Bolund L., Wang J., Li S., Li R., Bolund L., Li S., Li R., Bolund L., Li R., Bolund L., Bolund L., et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006;34:W293–W297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]