Abstract
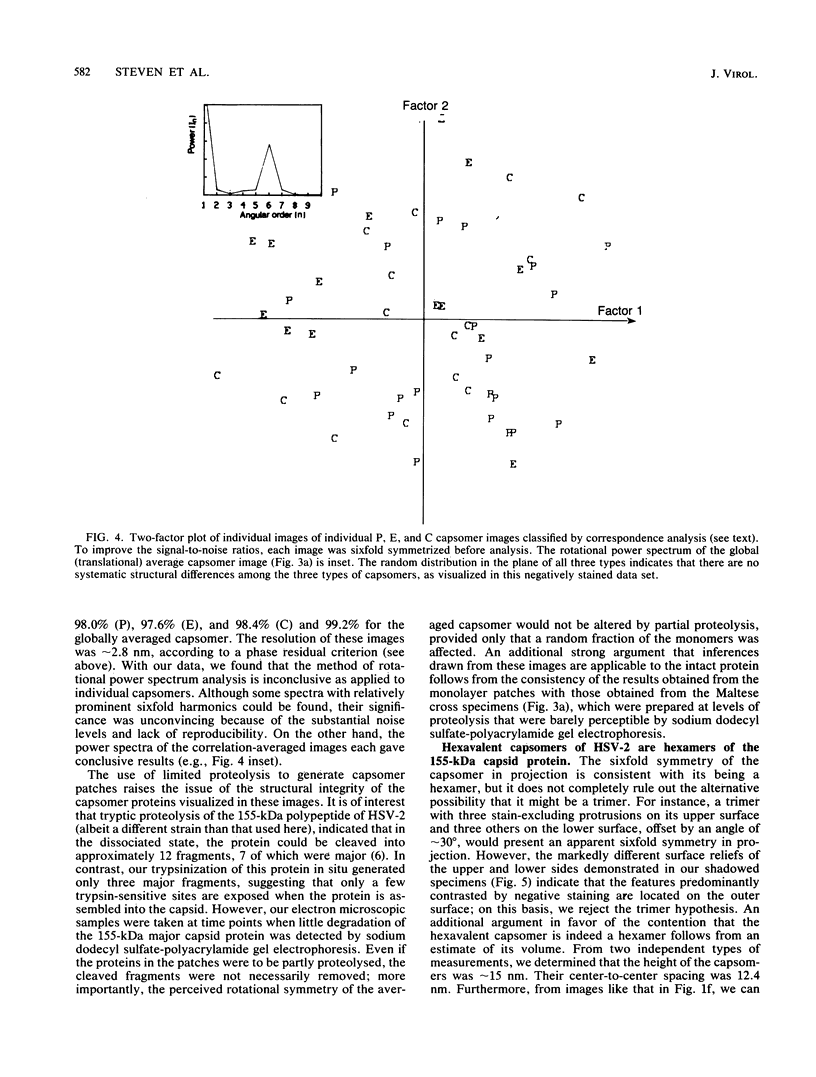
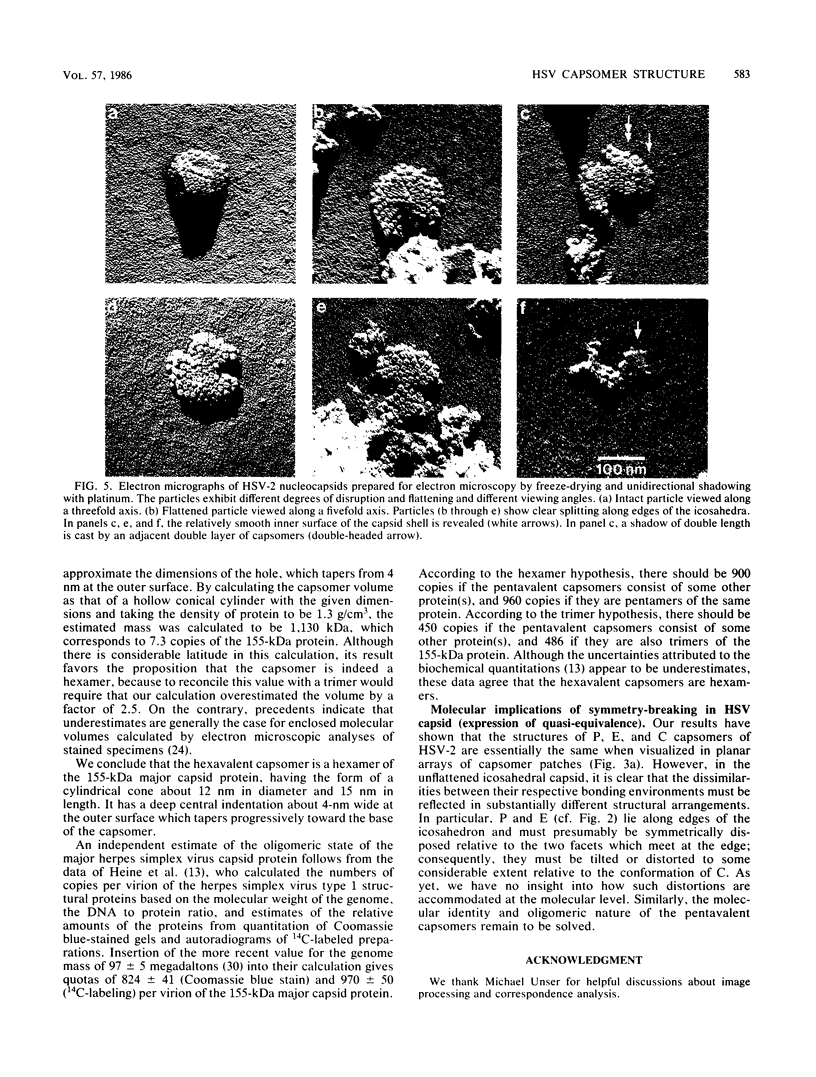
The structures of the hexavalent capsomers of herpes simplex virus type 2 were analyzed by negative staining electron microscopy of capsomer patches derived from partially disrupted nucleocapsids. Optimally computer-averaged images were formed for each of the three classes of capsomer distinguished by their respective positions on the surface of the icosahedral capsid with a triangulation number of 16; in projection, each capsomer exhibited unequivocal sixfold symmetry. According to correspondence analysis of our set of capsomer images, no significant structural differences were detected among the three classes of capsomers, as visualized under these conditions. Taking into account information from images of freeze-dried, platinum-shadowed nucleocapsid fragments, it was established that each hexavalent capsomer is a hexamer of the 155-kilodalton major capsid protein. The capsomer has the form of a sixfold hollow cone approximately 12 nm in diameter and approximately 15 nm in depth, whose axial channel tapers in width from the outside towards the inner capsid surface.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almeida J., Lang D., Talbot P. Herpesvirus morphology: visualization of a structural subunit. Intervirology. 1978;10(5):318–320. doi: 10.1159/000148994. [DOI] [PubMed] [Google Scholar]
- Baker T. S., Caspar D. L., Murakami W. T. Polyoma virus 'hexamer' tubes consist of paired pentamers. Nature. 1983 Jun 2;303(5916):446–448. doi: 10.1038/303446a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CASPAR D. L., KLUG A. Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- Cohen G. H., Ponce de Leon M., Diggelmann H., Lawrence W. C., Vernon S. K., Eisenberg R. J. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J Virol. 1980 May;34(2):521–531. doi: 10.1128/jvi.34.2.521-531.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther R. A., Franklin R. M. The structure of the groups of nine hexons from adenovirus. J Mol Biol. 1972 Jul 14;68(1):181–184. doi: 10.1016/0022-2836(72)90273-2. [DOI] [PubMed] [Google Scholar]
- Devaux C., Zulauf M., Boulanger P., Jacrot B. Molecular weight of adenovirus serotype 2 capsomers. A new characterization. J Mol Biol. 1982 Apr 25;156(4):927–939. doi: 10.1016/0022-2836(82)90148-6. [DOI] [PubMed] [Google Scholar]
- Frank J. Averaging of low exposure electron micrographs of non-periodic objects. Ultramicroscopy. 1975 Dec;1(2):159–162. doi: 10.1016/s0304-3991(75)80020-9. [DOI] [PubMed] [Google Scholar]
- Frank J., Verschoor A., Boublik M. Computer averaging of electron micrographs of 40S ribosomal subunits. Science. 1981 Dec 18;214(4527):1353–1355. doi: 10.1126/science.7313694. [DOI] [PubMed] [Google Scholar]
- Furlong D. Direct evidence for 6-fold symmetry of the herpesvirus hexon capsomere. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2764–2766. doi: 10.1073/pnas.75.6.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson W., Roizman B. Proteins specified by herpes simplex virus. Staining and radiolabeling properties of B capsid and virion proteins in polyacrylamide gels. J Virol. 1974 Jan;13(1):155–165. doi: 10.1128/jvi.13.1.155-165.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine J. W., Honess R. W., Cassai E., Roizman B. Proteins specified by herpes simplex virus. XII. The virion polypeptides of type 1 strains. J Virol. 1974 Sep;14(3):640–651. doi: 10.1128/jvi.14.3.640-651.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler J., Aebi U., Kellenberger E. Freeze drying and shadowing a two-dimensional periodic specimen. J Ultrastruct Res. 1977 Apr;59(1):76–86. doi: 10.1016/s0022-5320(77)80030-0. [DOI] [PubMed] [Google Scholar]
- Labaw L. W., Davies D. R. The molecular outline of human gamma G1 immunoglobulin from an EM study of crystals. J Ultrastruct Res. 1972 Aug;40(3):349–365. doi: 10.1016/s0022-5320(72)90106-2. [DOI] [PubMed] [Google Scholar]
- Maizel J. V., Jr, White D. O., Scharff M. D. The polypeptides of adenovirus. II. Soluble proteins, cores, top components and the structure of the virion. Virology. 1968 Sep;36(1):126–136. doi: 10.1016/0042-6822(68)90122-0. [DOI] [PubMed] [Google Scholar]
- Palmer E. L., Martin M. L., Gary G. W., Jr The ultrastructure of disrupted herpesvirus nucleocapsids. Virology. 1975 May;65(1):260–265. doi: 10.1016/0042-6822(75)90026-4. [DOI] [PubMed] [Google Scholar]
- RUSSELL W. C. A sensitive and precise plaque assay for herpes virus. Nature. 1962 Sep 8;195:1028–1029. doi: 10.1038/1951028a0. [DOI] [PubMed] [Google Scholar]
- Rayment I., Baker T. S., Caspar D. L., Murakami W. T. Polyoma virus capsid structure at 22.5 A resolution. Nature. 1982 Jan 14;295(5845):110–115. doi: 10.1038/295110a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann M. G. Constraints on the assembly of spherical virus particles. Virology. 1984 Apr 15;134(1):1–11. doi: 10.1016/0042-6822(84)90267-8. [DOI] [PubMed] [Google Scholar]
- Smith P. R. An integrated set of computer programs for processing electron micrographs of biological structures. Ultramicroscopy. 1978;3(2):153–160. doi: 10.1016/s0304-3991(78)80021-7. [DOI] [PubMed] [Google Scholar]
- Smith P. R., Fowler W. E., Pollard T. D., Aebi U. Structure of the actin molecule determined from electron micrographs of crystalline actin sheets with a tentative alignment of the molecule in the actin filament. J Mol Biol. 1983 Jul 5;167(3):641–660. doi: 10.1016/s0022-2836(83)80103-x. [DOI] [PubMed] [Google Scholar]
- Straus S. E., Aulakh H. S., Ruyechan W. T., Hay J., Casey T. A., Vande Woude G. F., Owens J., Smith H. A. Structure of varicella-zoster virus DNA. J Virol. 1981 Nov;40(2):516–525. doi: 10.1128/jvi.40.2.516-525.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timbury M. C. Temperature-sensitive mutants of herpes simplex virus type 2. J Gen Virol. 1971 Nov;13(2):373–376. doi: 10.1099/0022-1317-13-2-373. [DOI] [PubMed] [Google Scholar]
- Vernon S. K., Lawrence W. C., Cohen G. H. Morphological components of herpesvirus. I. Intercapsomeric fibrils and the geometry of the capsid. Intervirology. 1974;4(4):237–248. doi: 10.1159/000149968. [DOI] [PubMed] [Google Scholar]
- WILDY P., RUSSELL W. C., HORNE R. W. The morphology of herpes virus. Virology. 1960 Oct;12:204–222. doi: 10.1016/0042-6822(60)90195-1. [DOI] [PubMed] [Google Scholar]
- Wadsworth S., Jacob R. J., Roizman B. Anatomy of herpes simplex virus DNA. II. Size, composition, and arrangement of inverted terminal repetitions. J Virol. 1975 Jun;15(6):1487–1497. doi: 10.1128/jvi.15.6.1487-1497.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrigley N. G. An electron microscope study of the structure of Sericesthis iridescent virus. J Gen Virol. 1969 Jul;5(1):123–134. doi: 10.1099/0022-1317-5-1-123. [DOI] [PubMed] [Google Scholar]
- van Heel M., Frank J. Use of multivariate statistics in analysing the images of biological macromolecules. Ultramicroscopy. 1981;6(2):187–194. doi: 10.1016/0304-3991(81)90059-0. [DOI] [PubMed] [Google Scholar]





