Surface plasmon resonance (SPR) measurements provide a highly sensitive means for detecting biomolecular interactions in a label-free manner. Numerous studies of biomolecular interactions have been performed with fixed-angle SPR imaging (SPRi) on surfaces patterned with a variety of biomolecules such as DNA, RNA, proteins, and peptides.1 Arrays increase the information obtainable in a single experiment as multiple reactions can be monitored in parallel.
A current and significant limitation of SPR is that the substrate must be a metal thin film. A number of metal thin films are capable of supporting surface plasmons in the near-infrared and visible regions of the electromagnetic spectrum.2,3 Gold surfaces have been the substrate of choice for SPR measurements for two reasons: gold is relatively stable in the aqueous environments needed for monitoring biomolecular interactions and a versatile chemistry based on the attachment of sulfur-containing ligands to the gold surface has been developed and well-characterized. The readily formed gold–sulfur bond enables the direct attachment of ligands to the gold surface4 as well as attachment via an intermediate self-assembled monolayer (SAM).5–7 This gold–thiol chemistry has made possible the routine analysis of aqueous binding processes to immobilized molecules at near-neutral pH values and moderate temperatures. The susceptibility of the gold–sulfur bond to oxidation and photodecomposition has prevented SPR sensing from finding utility in areas such as on-surface combinatorial chemistry (due to the harsh chemical conditions employed) and photolithography (due to the adverse effects of ultraviolet radiation on the gold–sulfur bond).8
Here we describe the development of a lamellar structure in which a thin layer of amorphous carbon is deposited onto a surface plasmon-active gold thin film (Figure 1a). Carbon-based surfaces are readily modified with biomolecules of interest using a well-developed and robust chemistry, based upon the attachment of alkene-containing molecules to the substrate through the UV light-mediated formation of carbon–carbon bonds.9 Recently, a similar lamellar structure utilizing a thin silicate overlayer was used to fabricate and monitor supported bilayer membranes with SPR.10
Figure 1.
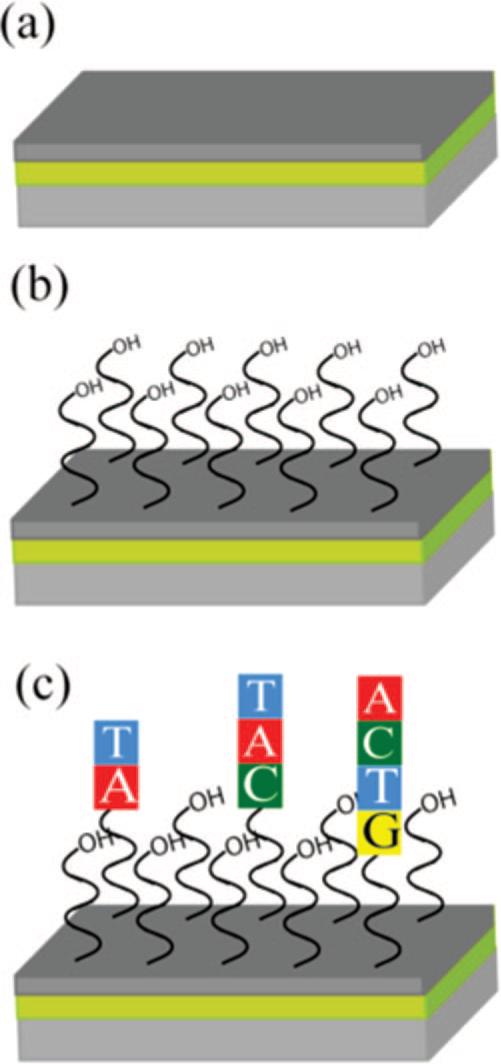
The carbon-on-gold substrates used in this work consist of a lamellar structure (a) in which an SPR conducting gold thin film was evaporated onto a high-index glass substrate and then an amorphous carbon overlayer was sputtered onto the gold. The amorphous carbon was then hydrogen terminated in a 13.65 MHz inductively coupled hydrogen plasma and UV photofunctionalized with 9-decene-1-ol to yield (b) a hydroxyl-terminated surface. An (c) oligonucleotide array was then synthesized on the substrate in a base-by-base fashion using photolithographic chemical methods.
Arrays prepared on functionalized carbon-based substrates such as diamond thin films,11,12 glassy carbon,12 and amorphous carbon thin films13 have superior stability to analogous arrays prepared on functionalized glass, silicon, and gold substrates. Amorphous carbon is of particular interest as it can be deposited at room temperature, allowing it to be integrated with other materials14 such as quartz crystal microbalances,13 electrodes,15 and metal thin films without perturbing their structure. The utility of a multilayered substrate containing a metal thin film and an amorphous carbon overlayer is shown here by their use for in situ synthesis of oligonucleotide arrays, which are then employed in the analysis of biomolecule binding processes with SPRi (Figure 1). The in situ fabrication of oligonucleotide arrays utilizing photochemically protected oligonucleotide building blocks16 is not possible with traditional gold SPR substrates because of the extended exposure to ultraviolet light and oxidizing chemical conditions. To illustrate the possibilities offered by this approach, we use these DNA arrays to analyze biomolecular interactions with DNA and proteins.
To determine the effect of the carbon overlayer on the ability of the gold film to support surface plasmons, a series of scanning angle SPR and SPRi measurements were made for substrates with varying thicknesses of amorphous carbon (0 − 20 nm). The experimentally measured scanning angle SPR curves were fit to n-phase Fresnel models (Figure 2a). The agreement with theory indicates that the amorphous carbon overlayer does not alter the gold thin film and can also be used to confirm the thickness of the amorphous carbon on the gold surface.17 Decreases in the photon-plasmon coupling efficiency as well as a broadening of the angle scanning SPR curves are ascribed to the complex dielectric function of amorphous carbon.13,18 The broadening of the SPR curves leads to a loss in surface sensitivity. A series of fixed-angle SPR imaging (SPRi) experiments were performed to quantify this decrease in sensitivity. In these experiments, changes in reflectivity resulting from a 3.0 × 10−4 change in the bulk solution index of refraction (caused by replacing pure water, n = 1.3333, with a 1% ethanol solution, n = 1.3336) were monitored for the surfaces described above. The changes in reflectivity for each substrate were compared to that for bare gold substrate and normalized accordingly (Figure 2b legend). Next, these surfaces were used to determine the minimum thickness of functionalized amorphous carbon needed to support the chemical requirements of in situ oligonucleotide array synthesis. Each surface was functionalized with 9-decene-1-ol and then exposed to the reaction conditions needed to synthesize an 18-nucleotide sequence array by light-directed synthesis.19 The minimum thickness of the amorphous carbon needed to consistently support the array fabrication process is 7.5 nm. This results in a 42% loss of sensitivity relative to a bare gold film (Figure 2b), where sensitivity is defined as the maximum change in reflectivity as a function of refractive index change. Amorphous carbon films less than 7.5 nm sometimes delaminated during array synthesis, rendering the substrates unusable.
Figure 2.
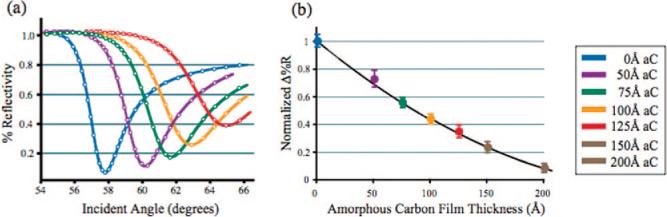
Varying thicknesses of amorphous carbon were applied to 42.5 nm gold thin films and monitored with both scanning angle SPR and SPRi. (a) Scanning angle reflectivity curves (8994 cm−1) of gold substrates containing 5, 7.5, 10, and 12.5 nm of amorphous carbon were compared to a bare gold substrate. Experimentally obtained values (circles) were compared to theoretically calculated reflectivity curves for each substrate (lines). The theoretical values were determined with a n-phase Fresnel equation. (b) A series of fixed angle SPRi measurements were made to determine the effect of the amorphous carbon overlayer on binding sensitivity. Each substrate was exposed to two solutions with a 3.4 × 10−4 index of refraction difference and the change in reflectivity measured. The total change in reflectivity obtained for the bare gold substrate was used to define a range from 0 to 1 (0 = minimum reflectivity; 1 = maximum reflectivity); the total change in reflectivity obtained for the carbon–gold substrates is shown as a fraction of that value.
The loss in sensitivity caused by the amorphous carbon can be reduced by altering the thickness of the surface plasmon conducting gold underlayer. Theoretical calculations of the SPR scanning angle response indicate that a decrease in gold thickness will result in sensitivity gain.
Oligonucleotide arrays employed in this work were synthesized on substrates containing a 42.5 nm gold thin film and a 7.5 nm amorphous carbon overlayer (Figure 1). Prior to use, the amorphous carbon substrate was hydrogen-terminated in a 13.65 MHz inductively coupled hydrogen plasma and then photofunctionalized with 9-decene-1-ol to yield an alcohol-terminated substrate. The oligonucleotide arrays were fabricated using a previously described light-directed photolithographic synthesis method,16 allowing the oligonucleotide sequences to be synthesized in a base-by-base manner.
An array consisting of 420 individual oligonucleotide features was fabricated (Figure 3). The array contained 34 copies of two oligonucleotide probes (probe 1 and probe 2, 18 nt each) placed throughout the array.20 The remaining oligonucleotide features were randomly generated 18 nt sequences. Probes 1 and 2 were then hybridized with their complements (100 nM) and monitored with SPRi. A difference image was calculated for each hybridization event (image after binding minus image before binding) and the intensities of each feature determined (Figure 2c,d). The relative standard deviation of each set of hybridized features was 2.5%, indicative of substrate uniformity.
Figure 3.
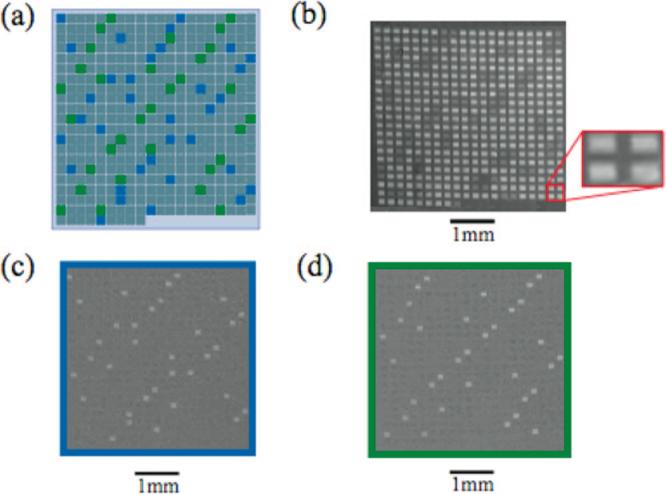
DNA-DNA binding experiment. An array containing 420 individual oligonucleotide features (128 μm × 128 μm per feature, with a 96 μm spacing between each feature) was fabricated on a substrate containing 7.5 nm of amorphous carbon thin film applied to 42.5 nm of gold. Each oligonucleotide feature was synthesized using light-directed synthetic methods previously described. (a) Diagram of the array used in the SPRi experiments. The array contained 34 copies of 18 nt oligonucleotide probe sequences placed throughout the array, probe 1 (blue) and probe 2 (green). (b) Image of the array before binding. A portion of the array is enlarged to show the relative sizes of the oligonucleotide features and interfeature spacing. (c,d) Difference images of the oligonucleotide array after exposure to a 100 nM solution of the complementary oligonucleotide sequence of probe 1 (c) or probe 2 (d).
The ability to fabricate a large number of unique oligonucleotide features on a single, SPR-compatible array provides a means for probing biomolecular interactions such as DNA–RNA, DNA–protein, and DNA–small molecule interactions. Two protein:DNA SPRi binding experiments were explored as models. First is the binding of thrombin, a protein involved in the coagulation cascade, to a single-stranded DNA aptamer, 5′-GGTTGGTGTGGTTGG-3′ which has been shown to have nanomolar binding affinity.21 Second, the binding of the VFR (virulence factor regulator) transcription regulatory protein to a known double-stranded DNA recognition sequence was monitored. The VFR protein modulates quorum sensing in Pseudomonas aeruginosa.22
An oligonucleotide array containing the thrombin-specific 15-nucleotide single-stranded aptamer and a 52-nucleotide oligomer encoding the VFR-binding site was photolithographically fabricated on an amorphous carbon–gold substrate.23 The VFR-binding oligomer employed is self-complementary, such that it forms a hairpin structure upon self-annealing. A four-nucleotide sequence (TCCT) was inserted to form the hairpin loop-structure. Hairpin formation was achieved by heating the array to 90 °C and slowly cooling to allow for proper annealing. Figure 3a contains a schematic of the array once the hairpins have formed along with the SPRi image of the array. The surface was first exposed to a 10.0 μg/mL thrombin solution and a difference image was obtained after 2 min of binding (Figure 3b). The thrombin only binds to the aptamer sequence and not to the VFR binding site. Next, a 1.0 μg/mL solution of VFR was added, and after 2 min a difference image was obtained showing specific binding to the double-stranded oligonucleotide sequence (Figure 3c). In both protein-binding experiments, nonspecific adsorption across the substrate was observed. The sequence-specific binding resulted in a signal increase of approximately 10:1 while the adsorption to non-target oligonucleotide sequences was equal to that of the background, demonstrating the ability to analyze specific DNA–protein interactions by SPRi on photolithographically fabricated DNA arrays.
In summary, we describe here a novel carbon-on-metal thin film substrate architecture, which enables surface plasmon resonance detection with photolithographically fabricated DNA arrays for the analysis of biomolecular interactions. The utility of the technology is shown in the analysis of specific DNA–DNA and DNA–protein binding interactions. Presently, our group is utilizing these new substrates to determine the secondary structure of RNA molecules, to probe the sequence-specific binding kinetics and affinity of proteins and small molecules, and exploring the use of these substrates as possible small-molecule combinatorial chemistry platforms for drug discovery applications. This new material opens many such possibilities for the fabrication of molecular arrays and large-scale analysis of their binding interactions utilizing SPR spectroscopic methods.
Supplemental Material
Figure 4.
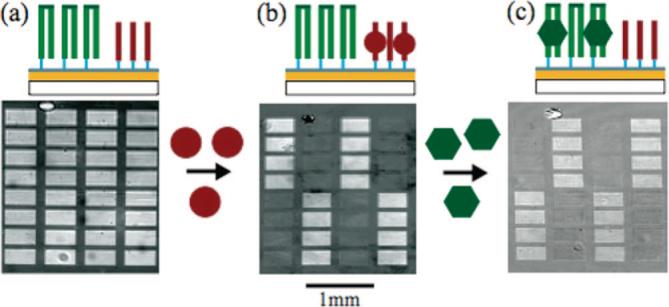
Protein-DNA binding experiment. (a) SPRi image of an oligonucleotide array consisting of two oligonucleotide sequences photolithographically synthesized on an amorphous carbon substrate. The double-stranded oligonucleotide sequences are hairpin structures (green) formed by synthesizing a self-complementary oligonucleotide sequence containing a TCCT hairpin loop sequence. The second oligonucleotide sequence (red) had no self- and/or interstrand complementary sequences, such that it remained single-stranded throughout the experiment. Hairpin structures were formed by incubating the surface at 90 °C followed by a slow cooling process. (b) First, the surface was exposed to a 10.0 μg/mL thrombin solution and binding to the single-stranded target oligonucleotide sequence was monitored with SPRi. The difference image shown was obtained by subtracting the image after thrombin binding from the reference image taken prior to binding. (c) Next a 1.0 μg/mL solution of the VFR protein was exposed to the oligonucleotide array and binding was monitored. The difference image was obtained by subtracting the image after VFR binding from the image after thrombin binding.
Acknowledgment
The authors would like to thank Professor Aseem Ansari for providing the VFR protein. This work was funded by NIH Grants R01HG002298, R01HG003275, R33DK070297, and the University of Wisconsin Technology Innovation Fund. Authors S.C.W., R.M.C., and L.M.S. have a financial interest in GWC Technologies.
Footnotes
Supporting Information Available: Details of materials and methods used in this study. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Wark AW, Lee HJ, Corn RM. Handbook of Surface Plasmon Resonance. RSC Publishing; 2008. Advanced Methods for SPR Biosensing. pp. 251–280. [Google Scholar]
- 2.Mitsushio M, Miyashita K, Higo M. Sens. Actuators, A. 2006;125:296–303. [Google Scholar]
- 3.Ordal MA, Long LL, Bell RJ, Bell SE, Bell RR, Alexander RW, Ward CA. Appl. Opt. 1983;22:1099–1119. doi: 10.1364/ao.22.001099. [DOI] [PubMed] [Google Scholar]
- 4.Rabkeclemmer CE, Leavitt AJ, Beebe TP. Langmuir. 1994;10:1796–1800. [Google Scholar]
- 5.Bain CD, Whitesides GM. Science. 1988;240:62–63. doi: 10.1126/science.240.4848.62. [DOI] [PubMed] [Google Scholar]
- 6.Brockman JM, Frutos AG, Corn RM. J. Am. Chem. Soc. 1999;121:8044–8051. [Google Scholar]
- 7.Frey BL, Corn RM. Anal. Chem. 1996;68:3187–3193. [Google Scholar]
- 8.Ulman A. An Introduction to Ultrathin Organic Films: From Langmuir-Blodgett to Self-Assembly. Academic Press; Boston, MA: 1991. p. 442. [Google Scholar]
- 9.Strother T, Knickerbocker T, Russell JN, Butler JE, Smith LM, Hamers RJ. Langmuir. 2002;18:968–971. [Google Scholar]
- 10.Phillips KS, Han JH, Martinez M, Wang ZZ, Carter D, Cheng Q. Anal. Chem. 2006;78:596–603. doi: 10.1021/ac051644y. [DOI] [PubMed] [Google Scholar]
- 11.Yang WS, Auciello O, Butler JE, Cai W, Carlisle JA, Gerbi J, Gruen DM, Knickerbocker T, Lasseter TL, Russell JN, Smith LM, Hamers RJ. Nat. Mater. 2003;2:63–63. doi: 10.1038/nmat779. [DOI] [PubMed] [Google Scholar]
- 12.Phillips MF, Lockett MR, Rodesch MJ, Shortreed MR, Cerrina F, Smith LM. Nucleic Acids Res. 2008;36:e7. doi: 10.1093/nar/gkm1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun B, Colavita PE, Kim H, Lockett M, Marcus MS, Smith LM, Hamers RJ. Langmuir. 2006;22:9598–9605. doi: 10.1021/la061749b. [DOI] [PubMed] [Google Scholar]
- 14.Hauert R. Diamond Relat. Mater. 2003;12:583–589. [Google Scholar]
- 15.Fiaccabrino GC, Tang XM, Skinner N, deRooij NF, KoudelkaHep M. Sens. Actuators, B. 1996;35:247–254. [Google Scholar]
- 16.Singh-Gasson S, Green RD, Yue YJ, Nelson C, Blattner F, Sussman MR, Cerrina F. Nat. Biotechnol. 1999;17:974–978. doi: 10.1038/13664. [DOI] [PubMed] [Google Scholar]
- 17.The thickness of each amorphous carbon layer was independently confirmed with an AFM stepping experiment.
- 18.Robertson J. Adv. Phys. 1986;35:317–374. [Google Scholar]
- 19.See Supporting Information for a detailed outline of the chemical processes used in array synthesis.
- 20.Oligonucleotide sequences and experimental details are located in the Supporting Information.
- 21.Macaya R, Schultze P, Smith F, Roe J, Feigon J. Proc. Natl. Acad. Sci. U.S.A. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Albus AM, Pesci EC, Runyen-Janecky LJ, West SEH, Iglewski BH. J. Bacteriol. 1997;179:3928–3935. doi: 10.1128/jb.179.12.3928-3935.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oligonucleotide sequences and experimental details are located in the Supporting Information.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


