Abstract
The thermodynamic parameters associated with the binding of several series of linear peptides to the third PDZ domain (PDZ3) of the postsynaptic density 95 protein (PSD-95) have been measured using isothermal titration calorimetry (ITC). Two strategies were pursued in developing these binding ligands: (1) systematic N-terminal truncation of sequences derived from the C-terminal regions of identified PDZ3-binding proteins (CRIPT, neuroligin-1 and citron); and (2) selective mutation of specific positions within a consensus hexapeptide (KKETEV) known to bind PDZ3. Each synthetically prepared peptide was used to titrate PDZ3, which yielded the changes in Gibbs free energy (ΔG), enthalpy (ΔH) and entropy (TΔS) for the binding event. Selected peptides were subjected to additional analysis, which entailed (1) measuring the change in heat capacity (ΔCp) upon association, to assess the character of the binding interface; and (2) constructing thermodynamic double-mutant cycles, to determine the presence of cooperative effects. From the first series, the CRIPT protein proved to be the better source for higher affinity sequences. From the second series, enhanced binding was associated with peptides that closely adhered to the established motif for class I PDZ domain C-termini, X-(T/S)-X-(V/I/L), and more specifically to a narrower motif of X-T-X-V. Further, in both series a length of six residues was necessary and sufficient to capture maximal affinity. In addition, there were significant influences upon binding by modifying the abutting ‘X’ positions. The cumulative results provide greater detail into the specific nature of ligand binding to PDZ3, and will assist in the development of selective molecular probes for the study of this and structurally homologous PDZ domains.
PDZ domains embody a genetically thrifty approach by which a variety of coexisting proteins can adopt specific yet parallel molecular recognition properties within densely populated cells (1, 2). As with other adaptor or scaffold protein domains (3, 4), the PDZ domain facilitates protein-protein interactions reliant upon discrete molecular patterns displayed by their endogenous binding partners. Often considered structurally and functionally semi-autonomous in temperament, PDZ domains are modular structures of about 100 amino acids that are singly or multiply present within longer polypeptides. One of the paradigmatic examples of the multidomain class is the mammalian neuronal postsynaptic density protein, 95 kDa, or PSD-95 (5).
PSD-95, also known as SAP90, is the marquee member of the membrane associated guanylate kinase (MAGUK) family (6), among the most scrutinized of the PDZ domain-containing proteins. While the full repertoire of its cellular functions has yet to be revealed, PSD-95 is already recognized as a hub for numerous protein-protein associations, variously involving one or more of its five domains, which consist of three non-identical PDZ (PDZ1, PDZ2, PDZ3), SH3 and GK domains. Viewed panoramically, PSD-95 appears to serve as an active scaffolding agent, fostering the assembly of transiently-formed protein complexes at the postsynaptic plasma membrane and the extracellular matrix (1). PSD-95 can affect clustering of NMDA receptors (7), AMPA receptor recruitment (8), synaptic organization, transmission and plasticity (9–11), and regulation of activity and trafficking of serotonin receptors (12). There may also exist therapeutic benefits to interfering with the normal binding activities of PSD-95 pertaining to ischemic brain damage (13), addiction (14) and pain management (15, 16). Given these varied biological entanglements involving PSD-95, membrane-permeable ligands selective for the PDZ domains of this protein could assist in deciphering their respective cellular roles.
Beyond that applied objective, there is also a biophysical interest in learning more about how protein structures like the PDZ domain recognize their molecular targets. In previous work, we reported both linear (17) and cyclic (18, 19) peptide ligands for PDZ3, in which isothermal titration calorimetry (ITC) was used to unearth the thermodynamic roots of the protein-binding events. The modest binding requirements of PDZ domains like PDZ3, with their predilection for relatively short sequences at the C-terminal region of protein partners, make them amenable to study with smaller ligands. PDZ3 has also been the subject of structural (20) and kinetic analysis (21). Collectively, inquiries such as these that seek to understand the fundamentals of molecular recognition can directly support the more utilitarian goal of molecular probe design and development. Our prior thermodynamic investigation with peptide macrocycles led to the discovery of a ligand with the ability to inhibit a PDZ domain-mediated interaction within human cells (22).
In this present study, we examine the binding behavior of PDZ3 with linear peptide ligands derived from two distinct sources: sequences from the C-terminal regions of proteins assigned as endogenous partners of PDZ3, and positional analogs of a consensus peptide sequence with known affinity for PDZ3. In the first case, we selected the proteins CRIPT (23), neuroligin-1 (24) and citron (25), and prepared peptides of varying length by removing residues from the N-terminal region. For the second series the hexapeptide KKETEV, which we had earlier reported as a good binding ligand for PDZ3 (17), was subjected to residue replacement at different positions. The binding to PDZ3 of each peptide from both series was measured by ITC to determine the basic thermodynamic parameters. These ‘standard’ titrations—at fixed temperature under the same pH and buffer conditions—provided the values for Kd (and thus ΔG), ΔH and TΔS. Selected peptides were then subjected to more advanced calorimetric titrations in which temperature was modified, in order to extract values for the change in heat capacity, ΔCp.
EXPERIMENTAL PROCEDURES
Materials
Plasmid encoding for GST-PDZ3 was provided by R. MacKinnon (Rockefeller Univ.). The following reagents were obtained from commercial sources: E. coli BLR-Gold (Stratagene); Coomassie Plus-200 Protein Assay (Pierce); human thrombin (Haematologic Technologies); TPCK trypsin and immobilized soybean trypsin inhibitor (Pierce), glutathione-bound agarose beads (Sigma).
GST-PDZ3 Fusion Protein Expression, Purification and Quantitation
The PDZ3 domain from PSD-95 (residues 302 – 402) was expressed in E. coli BLR-Gold as a glutathione-S-transferase (GST) fusion protein (the PDZ3 is fused—with an intervening thrombin recognition site—to the C-terminus of GST). Incubation of the cells took place in TB media at 37 °C until an OD600 of 1.2 was reached; this was followed by induction with IPTG (0.8 mM) and incubation at 30 °C for five hours. After centrifugation the cell pellet was either stored at −80 °C for later use or immediately resuspended in ice-cold PBS (with 1% Triton, 10 mM β-mercaptoethanol) then lysed by sonication (Branson 250 Sonifier). The supernatant was removed from the pelleted cellular debris after centrifugation of the lysed suspension, and maintained at 4 °C. Using a peristaltic pump (5 mL/min flow rate), the cellular lysate was loaded onto a glass column containing resuspended glutathione agarose beads in cold PBS buffer. After extensive column washing with ice-cold PBS buffer, GST-PDZ3 protein was eluted with Tris-HCl (50 mM, pH 8.0) containing reduced glutathione (5 mM). The extraction procedure was repeated until all the fusion protein was isolated from the cell lysate as determined by SDS-PAGE. The fusion protein samples were stored at 4 °C, and concentration determined by Coomassie Assay.
PDZ3 Domain Purification and Quantitation
Either human thrombin or TPCK trypsin was used to cleave the GST-PDZ3. A 1:50 (w/w) protease:fusion protein ratio was used for the thrombin cleavage reaction. The thrombin working solution was prepared by mixing reaction buffer (50 mM Tris-HCl (pH 8.0), 10% glycerol) with thrombin to obtain a 0.5 μg/mL final thrombin concentration. Thrombin working buffer (50 mM Tris-HCl, 150 mM NaCl, 2.5 mM CaCl2) was used to bring the final protein concentration to 0.4 mg/mL. The cleavage reaction was allowed to proceed for four hours at room temperature, after which time the solution was placed in dialysis tubing (3,500 MWCO, Pierce) and equilibrated with the ITC buffer (20 mM MES pH 6.0, 10 mM NaCl, 1 mM DTT).
For trypsin-mediated cleavage a preliminary screen, to determine optimized conditions, was designed in which small quantities of GST-PDZ3 were treated with varying amounts of the protease (trypsin:fusion (w/w) ratios ranging from 1:350 to 1:35,000) and incubated at room temperature for periods ranging from 30 min to five hours; the digests were then analyzed by SDS-PAGE. Optimized conditions (in which complete processing was observed without noticeable PDZ3 degradation) were found to be a 1:700 (w/w) trypsin:fusion protein ratio with a two-hour incubation. Trypsin working buffer (20 mM Tris-HCl, 200 mM NaCl, 1 mM EDTA, pH 7.4) was used to bring the final protease concentration to 1 μg/mL. A stock trypsin solution (1 mg/mL) was added to the fusion protein sample. To halt the trypsin cleavage reaction, immobilized soybean trypsin inhibitor on beaded agarose was added (2 mg, 50:50 v/v slurry for each 4.5 mg equivalent of trypsin used, twice the theoretical amount of inhibitor required). After mixing for one minute and standing at room temperature for five minutes, each cleavage reaction mixture was filtered (0.45 μm nylon syringe filter) to remove the immobilized inhibitor, then dialyzed into ITC buffer. The dialyzed cleavage mixture was loaded onto an ion-exchange column (Hi-trap Q, Amersham) attached to a peristaltic pump operating at 5 mL/min. The protein was eluted by continually washing with five column volumes of start buffer (20 mM MES pH 6.0, 10 mM NaCl, 1 mM DTT), followed by three column volumes each of increasing ionic strength buffer (25, 50, 75, 80, 85, 90, 95, 100, and 110 mM NaCl). The eluted fractions were collected according to their ionic strength and placed on ice, then analyzed by SDS-PAGE (20% gel). Elution fractions containing PDZ3 (usually between 80–85 mM NaCl) were combined and concentrated using a centrifugal filter device (MWCO 5000; Millipore). PDZ3 protein concentration was determined by Advanced Protein Assay and UV measurements at λ = 280 nm (ε280 = 2560 M−1 cm−1 for the PDZ3 domain was calculated (26) based on the protein sequence for the following conditions: 6.0 M guanidinium hydrochloride, 0.02 M phosphate buffer, pH 6.5). An SDS-PAGE gel depicting each stage of isolation and purification confirmed expected size and purity characteristics of each protein isolate (Figure 1).
Figure 1.
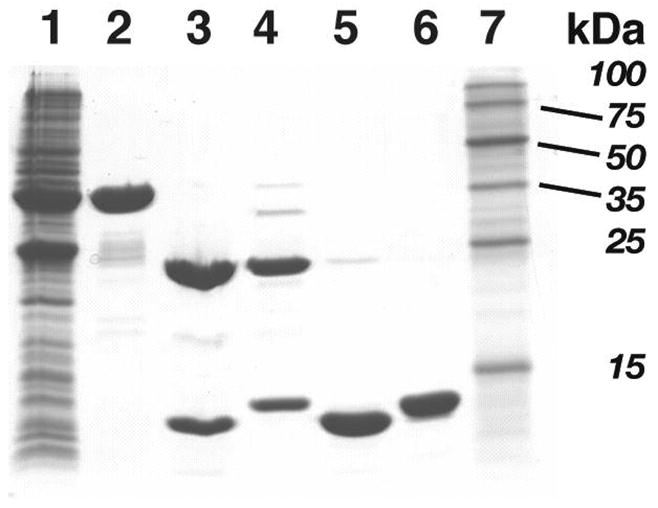
SDS-PAGE (20%) of expression and purification steps for PDZ3. Lane identification: 1, soluble bacterial lysate; 2, GST-PDZ3 fusion protein after affinity chromatography; 3, trypsin-cleaved GST-PDZ3; 4, thrombin-cleaved GST-PDZ3 fusion; 5, trypsin-cleaved PDZ3 (11.7 kDa) after ion exchange chromatography; 6, thrombin-cleaved PDZ3 (12.7 kDa) after ion exchange chromatography; 7, molecular weight markers (from top) 100, 75, 50, 35, 25, 15 kDa.
PDZ3 Domain Characterization
PDZ3 protein samples from trypsin or thrombin cleavage reactions were characterized by both mass spectrometry and circular dichroism (CD). PDZ3 was dialyzed into deionized water, 0.01% TFA and analyzed by electrospray ionization mass spectrometry (ESI-MS). PDZ3 solution conformation was examined by CD spectroscopy at room temperature (Jasco J-600 spectropolarimeter, wavelength range 200–260 nm; protein dialyzed into 5 mM MES (pH 6.0), 2.5 mM NaCl, 0.25 mM DTT buffer prior to the experiment). The CD spectrum of the dialysis buffer was also obtained and used to correct that of the protein.
Peptide Synthesis and Purification
Peptide ligands were prepared manually using standard Fmoc-based solid-phase peptide synthesis protocols, in a manner commensurate with our previous synthetic work (17). Sequences for the partner proteins were based on the C-terminal region of the following proteins (with RefSeq protein accession numbers): CRIPT (NP 054890), neuroligin-1 (NP 055747), and citron (NP 009105). Peptides were purified to single peak homogeneity using reverse-phase HPLC and product masses confirmed by ESI-MS. Unless otherwise specified, all peptides were prepared with a free (non-acetylated) amino terminus.
Isothermal Titration Calorimetry (ITC)
Calorimetry experiments were performed with a VP-ITC microcalorimeter (Microcal, Inc.). With one exception, all ITC experiments involved direct titration of the peptide ligand into protein solution. PDZ3 samples were dialyzed for at least 24 hours in the ITC buffer prior to titrations. Peptides were dissolved in the same dialysis buffer as that of the protein samples in each experiment. The pH values for the separate solutions of PDZ3 and peptide were measured at room temperature, and the pH of the peptide sample was adjusted when necessary to match that of the protein to within 0.02 pH units. The ITC buffer for the calorimetry experiments was 20 mM MES, pH 6.0, 10 mM NaCl. No DTT was used in the buffers for calorimetry experiments with the isolated PDZ3, but was included for experiments with the PDZ3-GST fusion protein. The standard experimental temperature was 25 °C; additional experiments to determine ΔCp were performed at 15 °C and 35 °C. Solutions were degassed for 15–30 minutes under vacuum, without stirring, at a temperature a few degrees below that of the actual experiment.
For a standard titration experiment, PDZ3 (80–240 μM) in the ITC buffer was placed in the reaction cell (ca. 1.4 mL) while the peptide solution (1.8–4.0 mM) was loaded into the 250 μL injection syringe. Experiments were designed such that the protein and peptide concentrations used yielded c values (27) between 1 and 380. Each titration comprised an initial, single 1 or 2 μL injection (not used in data fitting) followed by 5 or 10 μL injections performed at 180-second intervals. The stirring speed used was 270 rpm and the reference power was 10 μcal/sec.
Heats of dilution were initially measured in blank titrations by injecting the peptides into the buffer used in the particular experiment, and subtracted from the observed ‘raw heat’ values. It was found that these values were comparable to those obtained from the final injections at the end of an actual peptide-into-protein titration, after saturation; this latter approach was used for the majority of the reported ITC experiments. Control experiments were also performed by injecting buffer into buffer and the PDZ3 protein into buffer, but these titrations afforded insignificant heats of dilution effects. The data from each titration experiment was collected by ORIGIN software (version 5.0, Microcal), which was also used to determine thermodynamic parameters through nonlinear least-squares fitting (assuming a single-site binding model). All thermodynamic parameters reported in this paper are the average values of at least two independent titration experiments, unless otherwise stated. Subsequent graphing and curve-fitting of data (for heat of ionization and change in heat capacity experiments) were performed with DeltaGraph (version 5, Red Rock Software).
Competition ITC experiments (28) were performed by titrating either one of two higher affinity peptides (KNYKQTSV and KNYKETSV, 1.2 mM) into the PDZ3 solution pre-incubated with the lower affinity peptide being tested (at a concentration of 5 mM). In these titrations, PDZ3 concentration varied from 65–85 μM.
Reverse titration experiments involved protein (1 mM) being injected into peptide (70–90 μM); all other aspects were identical to the protocols described above for standard direct titration ITC.
RESULTS
PDZ3 Preparation and Characterization
The overexpression and subsequent processing of GST-PDZ3 provided ample quantities of pure PDZ3 protein for the ITC experiments (Figure 1). The molecular weight of GST-PDZ3 was calculated as 39,520 Da based on sequence; this was verified by ESI-MS (39,520.85 ± 4.20 Da). Yields of the fusion protein ranging from 100–150 mg/L bacterial culture were typically achieved. Employed separately, thrombin and trypsin cleavage reactions of GST-PDZ3 generated two distinct forms of isolated PDZ3, with molecular masses of 12,725.64 ± 0.98 and 11,695.84 ± 0.43 Da, respectively, as determined by ESI-MS. In both forms of PDZ3 the N-terminal sequence comprised GSPEF, residues derived from the initial subcloning of PDZ3 into the pGEX vector. This was explicitly confirmed for the trypsin-generated PDZ3 by N-terminal protein sequencing of all five residues. From the combined data, it was calculated that PDZ3 processed by trypsin experienced an additional cleavage of ten residues from its carboxy terminus.
To determine whether the two forms of PDZ3 were functionally and structurally interchangeable, ITC and CD experiments were conducted. When each PDZ3 preparation was separately titrated with the C-terminal CRIPT pentapeptide KQTSV, the resulting thermodynamic parameters were comparable (PDZ3trypsin having Kd = 18.5 ± 2.7 μM, ΔH = −4.7 ± 0.5 kcal/mol, and TΔS = 1.7 ± 0.6 kcal/mol, versus PDZ3thrombin with Kd = 15.9 μM, ΔH = −3.9 kcal/mol, and TΔS = 2.6 kcal/mol. CD did not indicate any significant difference between the spectra taken of both forms of PDZ3 (data not shown). Given this similarity in structure and binding, all the reported ITC experiments used PDZ3 produced by trypsin treatment. Further, CD spectra were obtained for trypsin-prepared PDZ3 at variable temperatures, starting from 4 °C to 65 °C, in increments of 10 °C, then decreasing in the same intervals. The results of monitoring the ellipticity signal versus wavelength demonstrated that the PDZ3 maintained essentially unchanged structure throughout the range.
Standard ITC of CRIPT-based Peptides
Seven peptides comprising four to ten of the C-terminal residues of CRIPT were prepared and tested (Table 1). While insufficient heat from titration of PDZ3 with the tetrapeptide precluded the reliable determination of binding parameters, the addition of a single N-terminal residue led to modest binding (Kd ~18 μM). At the level of the corresponding hexapeptide, a dissociation constant of 0.76 μM was measured; thereafter, affinity values remained relatively consistent with the longer peptides. While in all cases changes in enthalpy and entropy favored association, the former parameter was the dominant contributor. Two N-acetylated CRIPT derivatives were also prepared and tested (Table 3), which exhibited lower affinity values than their corresponding non-acetylated sequences.
Table 1.
Thermodynamic binding parameters for PDZ3 and CRIPT-based peptides determined by ITC.a
| peptide | Kd (μM) | ΔG (kcal/mol) | ΔH (kcal/mol) | TΔS (kcal/mol) | |
|---|---|---|---|---|---|
| 1 | b QTSV | — | — | — | — |
| 2 | KQTSV | 18 ± 3 | −6.7 ± 0.1 | −4.7 ± 0.5 | 2.0 ± 0.6 |
| 3 | YKQTSV | 0.76 ± 0.04 | −8.4 ± 0.0 | −6.3 ± 0.2 | 2.1 ± 0.1 |
| 4 | NYKQTSV | 2.1 ± 0.1 | −7.8 ± 0.1 | −7.1 ± 0.1 | 0.7 ± 0.1 |
| 5 | KNYKQTSV | 0.47 ± 0.01 | −8.6 ± 0.1 | −6.6 ± 0.1 | 2.0 ± 0.1 |
| 6 | TKNYKQTSV | 0.80 ± 0.05 | −8.3 ± 0.1 | −6.6 ± 0.1 | 1.7 ± 0.1 |
| 7 | DTKNYKQTSV | 2.0 ± 0.1 | −7.8 ± 0.1 | −6.4 ± 0.1 | 1.4 ± 0.1 |
Values are the arithmetic mean of at least two independent experiments. All c values ranged between 5 and 162. Stoichiometric (n) values ranged from 0.90 to 1.06.
Insufficient heat detected to determine binding parameters.
Table 3.
Thermodynamic binding parameters for PDZ3 and N-acetylated CRIPT- and neuroligin-based peptides determined by ITC.a
| peptide | Kd (μM) | ΔG (kcal/mol) | ΔH (kcal/mol) | TΔS (kcal/mol) | |
|---|---|---|---|---|---|
| 1 | Ac-YKQTSV | 3.3 ± 0.1 | −7.5 ± 0.1 | −7.4 ± 0.2 | 0.1 ± 0.2 |
| 2 | Ac-NYKQTSV | 9.9 ± 1.4 | −6.8 ± 0.1 | −7.3 ± 0.1 | −0.5 ± 0.1 |
| 3 | Ac-HSTTRV | 55 ± 7 | −5.8 ± 0.1 | −4.5 ± 0.3 | 1.3 ± 0.4 |
| 4 | Ac-SHSTTRV | 70 ± 1 | −5.7 ± 0.5 | −4.5 ± 0.5 | 1.2 ± 0.1 |
Values are the arithmetic mean of at least two independent experiments. All c values measured between 5 and 65. Stoichiometric (n) values ranged from 0.91 to 1.12.
Except for a single instance, all titrations in this report were conducted in a ‘forward’ manner, wherein the smaller peptide ligand is injected into the protein sample until saturation is reached. The single reverse titration (protein injected into peptide) was performed as added confirmation that the correct model was used for data fitting, since the binding parameters should be the same irrespective of the direction of titration. This experiment, using the CRIPT-derived YKQTSV, yielded data (Kd = 0.62 μM; ΔG° = −8.5 kcal/mol; ΔH = −6.9 kcal/mol; TΔS = 1.6 kcal/mol; n = 1.2) that approximated that of the corresponding forward titration experiment (Table 1).
Standard ITC of Neuroligin-based Peptides
Seven peptides composed of four to ten of the C-terminal residues of neuroligin-1 were prepared and tested (Table 2). Whereas the tetrapeptide TTRV did not yield binding values, STTRV did, albeit with low affinity (Kd ~152 μM). STTRV was also the only peptide of the neuroligin-1 series that was performed as a competition; that is, the higher-affinity KNYKQTSV (Kd ~0.5 μM) was titrated into the PDZ3 solution preincubated with STTRV (5 mM). A further improvement in binding constant was observed with the hexapeptide (Kd ~18 μM), but thereafter affinity values remained relatively constant with additional elongation. Except for the pentapeptide, in which both enthalpy and entropy make significant contributions, all the peptides are driven by favorable ΔH and experience either a slight or small unfavorable TΔS. Two N-acetylated neuroligin-1 derivatives were also prepared and tested (Table 3), which exhibited lower affinity values than their corresponding non-acetylated sequences.
Table 2.
Thermodynamic binding parameters for PDZ3 and neuroligin-1-based peptides determined by ITC.a
| peptide | Kd (μM) | ΔG (kcal/mol) | ΔH (kcal/mol) | TΔS (kcal/mol) | |
|---|---|---|---|---|---|
| 1 | b TTRV | — | — | — | — |
| 2 | c STTRV | 152 ± 3 | −5.0 ± 0.2 | −3.2 ± 0.1 | 1.8 ± 0.2 |
| 3 | HSTTRV | 21 ± 1 | −6.4 ± 0.1 | −6.5 ± 0.1 | −0.1 ± 0.1 |
| 4 | SHSTTRV | 18 ± 1 | −6.5 ± 0.1 | −7.1 ± 0.2 | −0.6 ± 0.2 |
| 5 | HSHSTTRV | 15 ± 1 | −6.6 ± 0.1 | −6.9 ± 0.1 | −0.3 ± 0.1 |
| 6 | PHSHSTTRV | 11 ± 1 | −6.8 ± 0.1 | −7.4 ± 0.1 | −0.6 ± 0.1 |
| 7 | HPHSHSTTRV | 10 ± 1 | −6.8 ± 0.1 | −8.9 ± 0.1 | −2.1 ± 0.1 |
Values are the arithmetic mean of at least two independent experiments. All c values measured between 1 and 9. Stoichiometric (n) values ranged from 0.92 to 1.15.
Insufficient heat detected to determine binding parameters.
ITC was conducted as a competition experiment in which the displacement of preincubated STTRV was measured.
Standard ITC of Citron-based Peptides
Seven peptides consisting of four to ten of the C-terminal residues of citron were prepared and tested (Table 4). No meaningful binding was observed for peptides with six or fewer residues; the ligand with seven residues bound PDZ3 with an affinity of Kd ~70 μM. The longer peptides, from eight to ten residues, all possessed improved affinity but with binding constants only about half that of the heptapeptide.
Table 4.
Thermodynamic binding parameters for PDZ3 and citron-based peptides determined by ITC.a
| peptide | Kd (μM) | ΔG (kcal/mol) | ΔH (kcal/mol) | TΔS (kcal/mol) | |
|---|---|---|---|---|---|
| 1 | b QSSV | — | — | — | — |
| 2 | b DQSSV | — | — | — | — |
| 3 | b WDQSSV | — | — | — | — |
| 4 | VWDQSSV | 701 ± 3 | −5.7 ± 0.1 | −5.1 ± 0.1 | 0.6 ± 0.1 |
| 5 | KVWDQSSV | 30 ± 1 | −6.2 ± 0.1 | −5.4 ± 0.4 | 0.8 ± 0.4 |
| 6 | NKVWDQSSV | 30 ± 1 | −6.2 ± 0.1 | −5.8 ± 0.2 | 0.4 ± 0.1 |
| 7 | VNKVWDQSSV | 36 ± 1 | −5.8 ± 0.3 | −6.1 ± 0.3 | −0.3 ± 0.1 |
Values are the arithmetic mean of at least two independent. All c values measured between 1 and 5. Stoichiometric (n) values ranged from 0.92 to 1.15.
Insufficient heat detected to determine binding parameters.
Standard ITC of Consensus Sequence Peptides
Thirty peptides comprising the parent sequence KKETEV and positionally-substituted analogs were individually prepared, and binding to PDZ3 analyzed by ITC (Table 5). In all but three cases, a sufficient heat signal was detected that allowed for direct binding measurement and the construction of a titration curve, which together provide for the determination of the remaining thermodynamic parameters (Figures 3 and 4). For the three recalcitrant peptides it was necessary to use competition binding experiments, as the affinity of the interaction was too low to be detected directly. With respect to the parent peptide KKETEV, Kd values were larger for all derivatives except for that of KKETAV, which ranked as the highest affinity ligand of all the peptides in this study. The observed values for the thermodynamic parameters for each ligand varied, but aside from a few exceptions, they generally exhibited favorable changes in enthalpy and entropy, with the former often dominating.
Table 5.
ITC-determined thermodynamic binding parameters for PDZ3 and KKETEV-derived peptides. a The underlined residues indicate changes with respect to the parent peptide, KKETEV.
| peptide | Kd (μM) | ΔG (kcal/mol) | ΔH (kcal/mol) | TΔS (kcal/mol) | |
|---|---|---|---|---|---|
| 1 | KETEV | 18 ± 1 | −6.5 ± 0.1 | −4.3 ± 0.3 | 2.2 ± 0.3 |
| 2 | KKETEV | 1.9 ± 0.1 | −7.8 ± 0.1 | −6.2 ± 0.1 | 1.6 ± 0.1 |
| 3 | KKKETEV | 1.3 ± 0.1 | −8.0 ± 0.1 | −5.9 ± 0.3 | 2.1 ± 0.2 |
| 4 | KKETEA | 91 ± 2 | −5.5 ± 0.1 | −4.6 ± 0.2 | 0.9 ± 0.2 |
| 5 | KKETEL | 7.9 ± 1.3 | −7.0 ± 0.1 | −4.1 ± 0.3 | 2.9 ± 0.2 |
| 6 | KKETEI | 7.7 ± 1.2 | −7.0 ± 0.1 | −4.3 ± 0.2 | 2.7 ± 0.1 |
| 7 | KKETEM | 21 ± 2 | −6.4 ± 0.1 | −6.8 ±± 0.2 | −0.4 ± 0.1 |
| 8 | b KKETEF | 57 ± 2 | −5.8 ± 0.1 | −4.4 ± 0.4 | 1.4 ± 0.4 |
| 9 | KKETET | 105 ± 6 | −5.4 ± 0.1 | −5.9 ± 0.2 | −0.5 ± 0.2 |
| 10 | KKESEV | 6.6 ± 0.9 | −7.1 ± 0.1 | −4.8 ± 0.1 | 2.3 ± 0.2 |
| 11 | KKECEV | 72 ± 7 | −5.7 ± 0.1 | −1.7 ± 0.1 | 4.0 ± 0.2 |
| 12 | KKESEL | 33 ± 2 | −6.1 ± 0.1 | −4.0 ± 0.1 | 2.1 ± 0.1 |
| 13 | KKESEI | 24 ± 6 | −6.3 ± 0.2 | −5.0 ± 0.2 | 1.3 ± 0.4 |
| 14 | b KKESEF | 98 ± 16 | −5.5 ± 0.1 | −3.1 ± 0.1 | 2.4 ± 0.1 |
| 15 | KKETGV | 2.4 ± 0.0 | −7.7 ± 0.1 | −5.7 ± 0.2 | 2.0 ± 0.2 |
| 16 | KKETAV | 0.45 ± 0.10 | −8.7± 0.1 | −5.3 ± 0.4 | 3.4 ± 0.4 |
| 17 | KKETVV | 1.3 ± 0.2 | −8.1 ± 0.1 | −5.9 ± 0.1 | 2.2 ± 0.1 |
| 18 | KKETLV | 1.8 ± 0.3 | −7.8 ± 0.1 | −3.7 ± 0.4 | 4.1 ± 0.3 |
| 19 | KKETPV | 0.95 ± 0.15 | −8.2 ± 0.1 | −4.3 ± 0.1 | 3.9 ± 0.2 |
| 20 | KKETWV | 2.8 ± 0.4 | −7.6 ± 0.1 | −3.5 ± 0.2 | 4.1 ± 0.1 |
| 21 | KKETDV | 20 ± 2 | −6.4 ± 0.1 | −4.1 ± 0.3 | 2.3 ± 0.3 |
| 22 | KKETKV | 1.2 ± 0.0 | −8.1 ± 0.1 | −5.6 ± 0.6 | 2.5 ± 0.6 |
| 23 | KKGTEV | 80 ± 3 | −5.6 ± 0.1 | −2.7 ± 0.1 | 2.9 ± 0.1 |
| 24 | KKATEV | 21 ± 4 | −6.4 ± 0.1 | −2.4 ± 0.1 | 4.0 ± 0.2 |
| 25 | KKQTEV | 4.0 ± 0.0 | −7.4 ± 0.1 | −4.9 ± 0.3 | 2.5 ± 0.3 |
| 26 | KKDTEV | 85 ± 12 | −5.6 ± 0.1 | −3.9 ± 0.3 | 1.7 ± 0.2 |
| 27 | KKKTEV | 27 ± 4 | −6.2 ± 0.1 | −2.7 ± 0.3 | 3.5 ± 0.4 |
| 28 | b KKGTGV | 273 ± 30 | −4.9 ± 0.1 | −2.6 ± 0.3 | 2.3 ± 0.2 |
| 29 | KKATAV | 8.3 ± 1.5 | −6.9 ± 0.1 | −3.0 ± 0.1 | 3.9 ± 0.2 |
| 30 | YKETEV | 1.2 ± 0.1 | −8.1 ± 0.1 | −6.9 ± 0.1 | 1.2 ± 0.2 |
Values are the arithmetic mean of at least two independent experiments. All c values measured between 1 and 237. Stoichiometric (n) values ranged from 0.85 to 1.21.
ITC was conducted as a competitive experiment.
Figure 3.
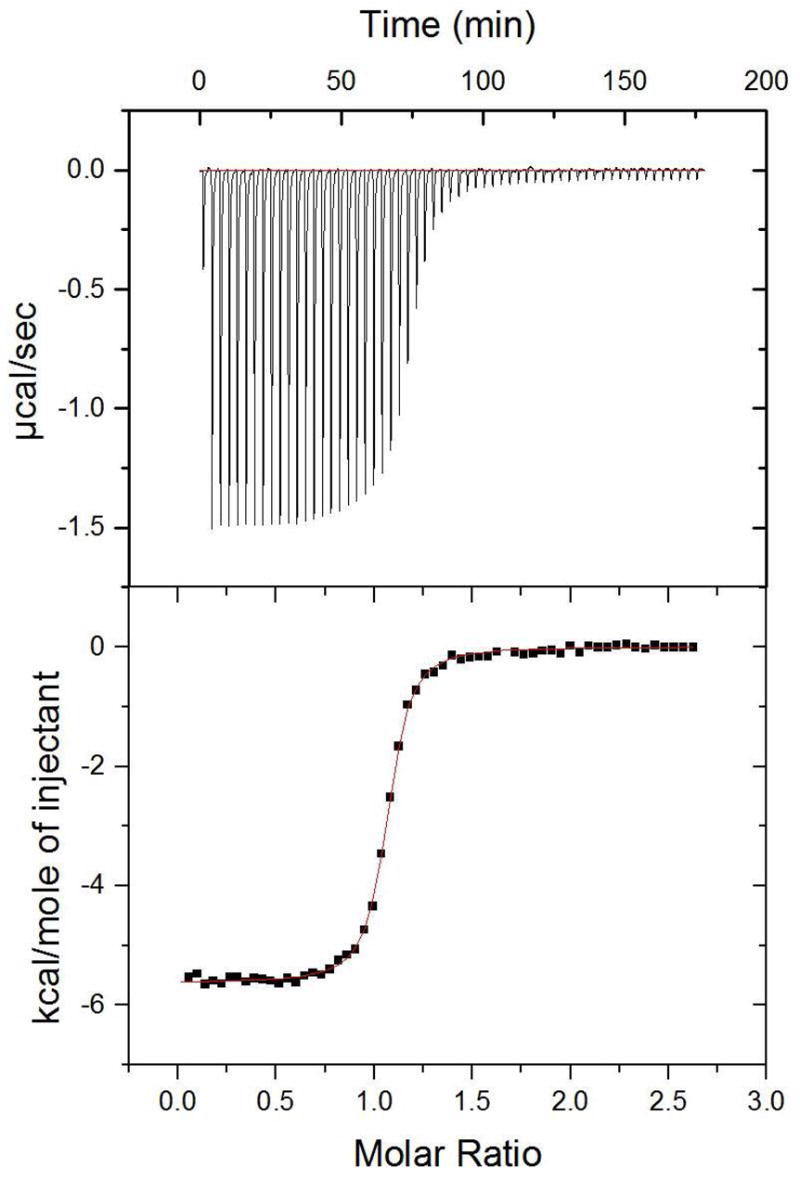
ITC data for higher-affinity peptide ligand. Raw thermogram (top) and integrated titration curve (bottom) for KKETAV and PDZ3 at 25 °C.
Figure 4.
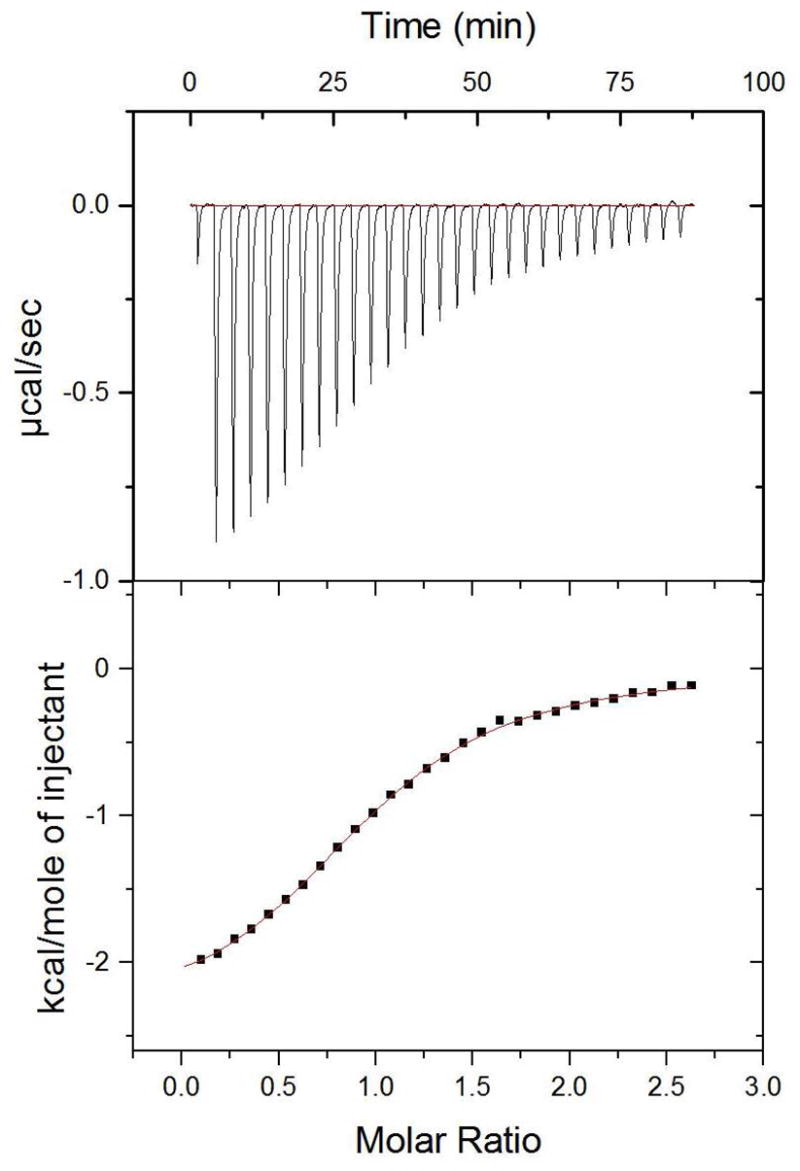
ITC data for lower-affinity peptide ligand. Raw thermogram (top) and integrated titration curve (bottom) for KKATEV and PDZ3 at 25 °C.
KKETEV was also tested against the uncleaved fusion protein, GST-PDZ3, using peptide and protein concentrations comparable to those of the experiments with isolated PDZ3. Without DTT present, the values obtained were Kd = 4.5 μM, ΔH = −5.8 kcal/mol, and TΔS = 1.5 kcal/mol; with DTT (1 mM), the parameters measured differed only slightly, with Kd = 3.9 μM, ΔH = −5.9 kcal/mol, and TΔS = 1.4 kcal/mol. In both instances a cloudy appearance of the sample was noted after removal upon completion of the titration.
Temperature-Dependent ITC and Change in Heat Capacity (ΔCp)
Two peptides from the CRIPT series and two from the neuroligin-1 series were used to directly titrate PDZ3 at two additional temperatures (15 and 35 °C) under the same experimental conditions as initially conducted at 25 °C. The ΔH° values for each were plotted against all three temperature points, which were then fit with linear curves (Figure 5). The slopes from each were extracted to yield the ΔCp values for YKQTSV (−163 cal/mol K), DTKNYKQTSV (−160 cal/mol K), HSTTRV (−131 cal/mol K) and HPHSHSTTRV (−128 cal/mol K).
Figure 5.
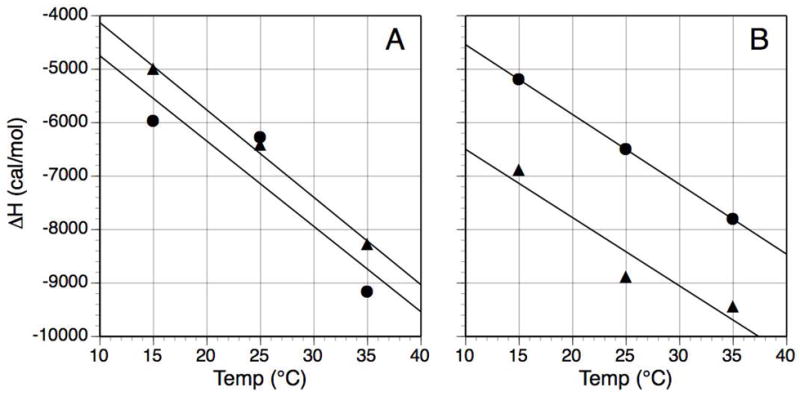
Determination of ΔCp for the binding of four peptides to PDZ3. Temperature-dependent values for ΔH were fit with linear curves, where the value for the slope is ΔCp. (A) CRIPT-derived peptides: YKQTSV (λ), curve: y = −163x – 2497; DTKNYKQTSV (σ) y = −160x – 3154; (B) Neuroligin-derived peptides: HSTTRV (λ), y = −131x – 3235; HPHSHSTTRV (σ), y = −128x – 5221. Error ranges, when graphed, are about the dimension of the data point symbols; for visual clarity, error bars are not shown.
Double Mutant Cycles and Ligand Residue Coupling
Using the existing data from the standard ITC determinations, three different double-mutant cycles constructed from four peptide-PDZ3 interactions apiece were assembled to probe for coupling effects between specific positions within the consensus hexapeptide. The Po/P-2 pair of positions was examined with peptides that reflect the sequential mutation of Val and Thr in KKETEV into Leu/Ile and Ser, respectively (Figure 10). The P-1/P-3 pair relationship employed KKETEV, in which each Glu was similarly replaced with Ala (Figure 11). In each case the observed free energy of coupling, ΔGc, was negligible, with values well below 0.5 kcal/mol. The values for enthalpy of coupling, ΔH°c, however, were energetically significant, ranging from 1–2 kcal/mol.
Figure 10.
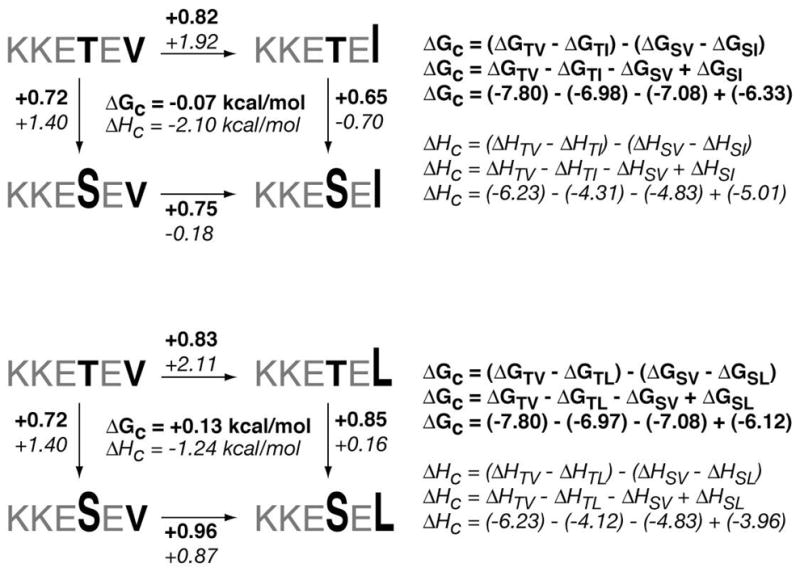
Double-mutant cycles for analysis of P0 and P-2 positions in peptide ligands.
Figure 11.
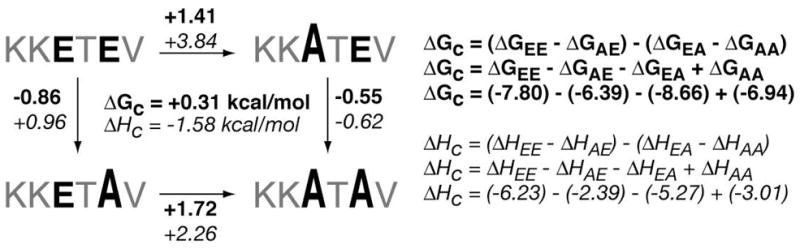
Double-mutant cycles for analysis of P-1 and P-3 positions in peptide ligands.
DISCUSSION
A Calorimetric Approach to PDZ Binding Analysis
Many experimental methods are available for the quantitative evaluation of protein-ligand interactions, each with its attendant strengths and weaknesses (29). Approaches taken to examine the binding properties of varying members of the PDZ domain family have included surface plasmon resonance (30, 31), fluorescence perturbation (32, 33), fluorescence anisotropy/polarization (25, 34–36), chemiluminescence (37), NMR titration (36, 38) and ELISA (39). While these investigations have provided insights into ligand-binding behavior of PDZ domains, fewer are the studies that plumb the depths of enthalpy and entropy contributions, parameters necessary to fully characterize the noncovalent molecular interactions involved in any molecular association.
To reliably access these thermodynamic values in practice demands the use of calorimetry as the analytical technique.2 With isothermal titration calorimetry (ITC), a single titration experiment of peptide into protein (or vice versa) immediately affords not only the binding constant, but the individual contributions to ΔG made by ΔH and ΔS (40–42). The key parameters are all interconnected through the following expression:
| (1) |
The application of ITC to characterize ligand binding to PDZ domains is now still in a stage of infancy. In addition to our calorimetric explorations of PSD-95 PDZ3 binding properties (17–19), others have utilized this technique to study the PDZ domains of syntenin (43, 44), Erbin (45), and DegS protease (46).
Characterization of PDZ3
Given the larger material requirements of ITC when compared to other assay techniques, optimization efforts were undertaken to increase the bacterial expression of soluble GST fusion protein and recover an ample amount of functional PDZ3. The efficiency of the step connecting those processes—the proteolytic cleavage of GST-PDZ3—was considerably enhanced by identifying conditions for using the more rapid and economical trypsin over thrombin, which facilitated the processing of large quantities of the fusion protein.
Temperature-dependent CD spectra demonstrated that PDZ3 maintained its structural characteristics throughout the range from 4 °C up to 65 °C, the upper value being much higher than that employed in any of the ITC experiments. Since the titrations performed to determine ΔCp require different temperatures, it was useful to confirm that PDZ3 would maintain a stable conformation when heated. The CD experiments themselves, however, were more of an added confirmation and not strictly necessary, since ITC can serve as its own internal quality control check by way of the stoichiometry value (n). In the case of PDZ3, which has an established 1:1 binding stoichiometry with peptides, an n value of unity indicates that the protein sample is fully folded and functional. Although deviations of up to 20% from n = 1 were occasionally observed, the majority of titrations resulted in stochiometry values that varied well within a range of 10%.
The isolated PDZ domain was used rather than the fusion protein to avoid possible thermodynamic effects associated with the dimerization of GST (47, 48) or the potential for fusion protein precipitation. This latter point is a particular concern with ITC since protein concentrations are relatively high compared to other assay formats. We were curious, however, to determine to what extent the GST fusion might alter the binding behavior, and a single example using the CRIPT-derived peptide KKETEV (discussed below) against the GST-PDZ3 fusion was tested. As reported in the Results section, affinity of KKETEV for GST-PDZ3 is only somewhat weaker than for PDZ3. This provides a degree of validation for the design of future non-ITC based, higher throughput assays that might utilize the more easily accessible GST-PDZ3.
Developing Ligands for PDZ3
Strategies to generate peptide ligands for PDZ domains can be roughly divided into two camps: identification of putative binding sequences from partner proteins known to associate with the desired domain, or target-based affinity selection of peptides from libraries of biological (39) or chemical (30) origin. In this report, ligand design was based on both approaches, using identified protein partners and sequence information provided by an oriented peptide library (30).
PDZ3-binding Partner Peptides: Standard ITC
Three proteins identified as endogenous binding partners for PDZ3 were selected for the ligand design: Cysteine-rich interactor of PDZ3, or CRIPT (23); neuroligin-1 (24); and citron (25). All three are designated as class I PDZ domains, with a C-terminus that adheres to the X-(Ser/Thr)-X-(Val)-CO2H sequence (49, 50). The exact C-terminal sequences from each were used to prepare the corresponding peptides in varying length, and each used in ITC experiments with PDZ3.
CRIPT, a postsynaptic protein and cytosolic protein, was selected as a sequence source based on evidence that it is a cellular binding partner of PDZ3. Yeast two-hybrid data showed that CRIPT bound strongly to PDZ3 with weak or no association to PDZ1 or PDZ2 of PSD-95 (51). Fluorescence polarization experiments measured the affinity of a C-terminal CRIPT-derived nine-residue peptide for PDZ3 at Kd ~1 μM (51), and the binding of a CRIPT peptide to PDZ3 has also been characterized by X-ray crystallography (20).
On a per-residue basis, the CRIPT C-terminal sequences possessed the highest affinities of the partner-derived peptides, and the binding of all ligands where favored both enthalpically and entropically (Table 1). While no parameters could be calorimetrically determined for the tetrapeptide, KQTSV did exhibit reasonable affinity with Kd = 18 μM. Extension to the hexapeptide resulted in a jump in binding strength by over an order of magnitude, with Kd ~0.8 μM. Further elongation was relatively inconsequential to affinity, with dissociation constant values fluctuating slightly about a mean of approximately one micromolar. This slight bobbling of ΔG could be attributable to the ‘floating’ amino terminus; the positively charged unit might therefore experience different binding interactions than a non-charged amide moiety occurring at the same location. When viewing the PDZ3-peptide structure determined by X-ray crystallography, the Lys at P-4 is only partially resolved, and the remaining N-terminal residues are completely absent (20). Taken together, all the preceding suggests that PDZ3 reserves the greater share of its recognition powers for canonical binding at six residues; longer sequences carry residues that are not favorably (nor unfavorably) accommodated by the PDZ3 surface, while a shorter sequence of five captures only a portion of the available binding energy.
With a non-modified amino terminus, these ligands will have an additional site of protonation under the buffer conditions used. From an experimental perspective, this can be beneficial by improving aqueous solubility, an important consideration since the peptides are at high concentration in the injecting syringe prior to titration. After the binding profile of the CRIPT peptides had been established, the two peptides that represent the major transition points in affinity were prepared with N-terminal acetylation, so as to more closely mimic the corresponding protein sequence. For both of the resultant peptides, Ac-KQTSV and Ac-YKQTSV, the affinity was approximately four times weaker than the corresponding non-acetylated peptides (Table 3). This demonstrated that the positively-charged terminal amine group actually contributes favorably to the binding free energy of association.
Neuroligin-1 was the source for the second series of peptides tested against PDZ3. Neuroligins are proteins embodied in the membrane of neuronal cells that expose an extracellular N-terminal domain on the outside and project a highly conserved C-terminal tail within the cell (53), and are involved in synapse development and function (54). Yeast two-hybrid data indicate that the neuroligin-1 cytosolic C-terminal region—which displays the canonical class I PDZ domain residues—binds tightly to PDZ3 of PSD-95 (24).
The neuroligin-1 peptides exhibited a similar profile as the CRIPT series, although values for the free energy changes were close to an order of magnitude weaker. This, despite the fact that both series possess the same optimal Thr/Val pair at P-2/P0, indicates the importance of proximal residues. The large seven-fold binding enhancement from penta- to hexapeptide is, as before, followed by a tapering off of the affinity, here settling at a modest Kd ~10 μM. As with the CRIPT-derived peptides, in addition to the large affinity enhancement, the most significant favorable ΔΔH (of −3.3 kcal/mol) occurs with the transition from five to six residues. These last few observations reinforce the notion that a minimally ideal ligand length for PDZ3 comprises six residues. Neuroligin-1 penta- and hexapeptides acetylated at the N-terminal for this series show, as with the CRIPT-derived ligands, a reduction of binding affinity by at least a factor of three (Table 3).
Citron provided the sequence for the third and last series of peptides replicating the C-terminal end of an assigned protein partner of PDZ3. Cytosolic citron was first identified in a yeast two-hybrid screen for proteins that interact with the activated form of Rho GTPase (55). In GABAergic neurons from the hippocampus, citron associates with PSD-95 through its PDZ3 domain and is concentrated at the postsynaptic side of glutamatergic synapses (25). The binding pattern for the citron peptides differed markedly from the other two protein partner series, exhibiting the lowest affinity when compared to equal-length ligands from CRIPT and neuroligin-1 (Table 4). Association was only detectable with sequences of seven residues or longer, and binding strength did not improve beyond Kd ~30 μM.
Reviewing the data from all the protein-partner peptides prompts the first of several questions: how would the PDZ domain-binding behavior of these peptides compare with that of the whole partner proteins from which they derive? Since such PDZ3-protein interactions have not yet been measured, this is currently a matter of conjecture. It is conceivable that the C-terminal region of the partner protein is in some way conformationally biased or restricted, without the full flexibility and pliancy of the linear peptide models, and thus preorganized for association. This argument is especially important for neuroligin proteins, which are membrane associated and perhaps more restricted, and locally more concentrated, than the cytosolic CRIPT and citron proteins. Alternatively, the C-terminus of the PDZ3-binding protein might be part of a larger molecular recognition motif, in which additional residues on the surface of either the partner or PDZ3 participate in supplementary binding interactions.
Even if the values of their affinities differ from their corresponding peptide models, one wonders whether the parent proteins would themselves exhibit a parallel pattern of differential binding strengths. That is, would the preferred order for PDZ3 association—CRIPT > neuroligin-1 > citron—as seen with the peptides in vitro likewise be observed with the actual partner proteins in vivo?
The previous speculations inevitably lead to one that questions the biological significance of such thermodynamic values, and that is whether they ultimately correlate with the biological services provided by these associations within the cell. Since PDZ domains are generally believed to form transient complexes, many considered regulatory in nature, variability in the strength with which PDZ3 binds to endogenous partners may be a function of its cellular roles. A spectrum of binding affinity values might therefore be reasonably expected between a given PDZ domain, such as PDZ3, and its partners.
PDZ3-binding Partner Peptides: Change in Heat Capacity (ΔCp)
The change in heat capacity (ΔCp) is considered a fundamental thermodynamic parameter, and its specific value places a limit on the magnitude of the changes in enthalpy and entropy:
| (2) |
The value for ΔCp may also be temperature dependent if it is not a truly “rigid body” binding event. Additionally, and particularly relevant to fostering insight into the structural nature of the interaction, ΔCp may be correlated with the surface area buried upon complex formation (58, 59), With this type of interpretation comes the monition that other molecular behaviors may influence the ΔCp value, such as changes in protein conformation or solvation patterns that are not part of the demarcated binding interface.
One of the open questions regarding the in vivo binding of PDZ domains to their endogenous protein partners is how much of the latter is involved. Many in vitro appraisals of PDZ domains function have demonstrated that relatively short peptides can bind them with moderate to high affinity, but it is not clear whether the actual cellular event is marked with additional points of contact involving regions of the partner protein distal to its C-terminus. One way to address this using simple peptide models is to inquire into the magnitude of the corresponding ΔCp values, and whether a length dependence is present. Simply measuring affinity would, a priori, not be sufficient, since it is possible through enthalpy-entropy compensation for ligands of differing lengths to possess comparable binding strengths.
In our studies, ΔCp values were determined for peptide ligands of six and ten residues in length from both the CRIPT- and neuroligin-derived series (Tables 1 and 2). The ΔCp values for the peptides were modest (between −127 and −163 cal/mol K) (Figure 5), which advocates a rigid body type of interaction that does not involve any substantial conformational changes in the protein upon binding (59). These results are consistent with the structural studies with the PDZ3 domain in the free and complexed forms that show very limited conformational changes between these two forms (20). The ΔCp values were almost identical for the peptides of each series with six and ten residues, implying that the addition of residues beyond those that fit into the canonical PDZ3 binding pocket does not affect the heat capacity changes. This seems to be the case for the CRIPT-derived ligands, whose binding is also characterized at the structural level, and for which the ΔΔH value of the six- to ten-residue transition is negligible, within experimental error. For the neuroligin-1-derived series of ligands the situation is different, since the ΔΔH value calculated for the transition from hexa- to decapeptide is −2.3 kcal/mol, suggestive of net bond formation. These new bonds might be formed with polar residues on the surface of the PDZ3 domain. If this is correct, then it will affect the ΔCp values in an opposite direction and less significantly compared with the burial of non-polar surface area that is characteristic of a short peptide binding event.
Consensus Sequence Peptides: Standard ITC
Based on the established class I binding motif, coupled to reported information on residue preferences for assorted PDZ domains determined by an oriented peptide library (30), a consensus sequence—KKETEV—was posited as a potential ligand for PDZ3. This hexapeptide was prepared and tested in a study reported earlier (17), and shown to exhibit good affinity for PDZ3 with Kd = 1.9 μM (Table 5). Further truncation to KETEV yielded a binding constant almost an order of magnitude weaker. Elongation to form the KKKETEV only improved affinity for PDZ3 marginally. Based on the preceding results, KKETEV was chosen as the parent peptide for further residue substitution studies. The hexapeptide length was also the smallest that embodied near-peak affinity for the ligands derived from CRIPT and neuroligin.
After establishing KKETEV as a benchmark consensus ligand, we subdivided the analogs to be prepared into categories based on which residues would be replaced (i.e., P0, P-1, etc; Figure 2). The approach was to minimize the magnitude of the changes, starting first with single mutations then moving to double substitutions, such that the binding contribution of specific positions within a sequence of fixed residue length can be evaluated. In this manner, any thermodynamic comparisons to be made are on firmer footing, since the structural changes are conservative in scope and set within the context of unvarying neighbors. Juxtaposing such data for analysis can be especially fruitful when relatively small, controlled structural changes are accompanied by dramatically altered thermodynamic parameters. This lays the groundwork for constructing binding hypotheses that rely on comparisons between ligands exhibiting energetically significant differences in their thermodynamic parameters (≥1 kcal/mol for ΔΔG or ΔΔH, and ≥2 kcal/mol for TΔΔS (41)). Although the possible structural and conformation changes resulting from replacing a residue side chain might be notable, they are fewer in number and magnitude than those that result from more dramatic alterations, such as the addition or deletion of entire residues as performed on the protein partner series of ligands described earlier.
Figure 2.
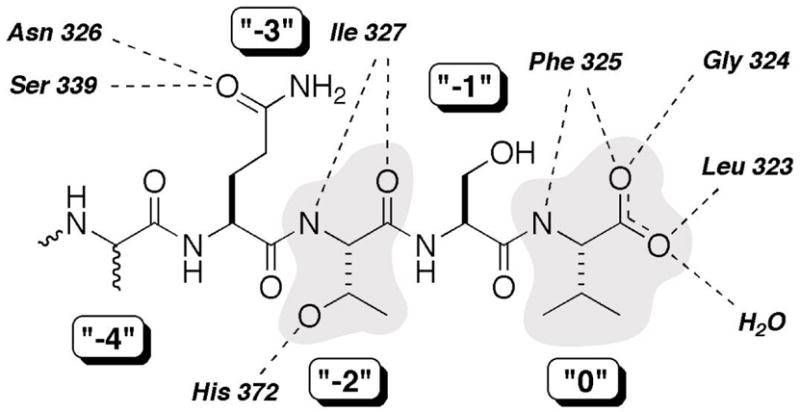
Binding interactions and labeling of a PDZ-peptide complex. Figure based on structural data from the X-ray crystallographic study of PDZ3 with a CRIPT-derived peptide reported by Doyle et al (20). Shaded are the “0” (Val, at the C-terminus), or “P0”, and “−2” (Thr), or “P-2”, residues, the core positions that largely define the class I PDZ domains. Dashed lines denote assigned hydrogen bonds derived from the crystal structure. Of note is that the peptide structure was not fully resolved at, and beyond, the P-4 position.
The order and priority of substitutions were based on the canonical residues that define a class I PDZ domain such as PDZ3. Since the definition of this category is predicated on the near-invariant nature of the P0 (C-terminus) and P-2 positions, substitutions were initially made to those residues, both as single (Table 5, 4–11) and double modifications (Table 5, 12–14). A similar approach was taken to study the vicinal P-1 and P-3 sites, singly (Table 5, 15–27) and in tandem (Table 5, 28 and 29), followed by an isolated example of a P-5 substitution (Table 5, 30).
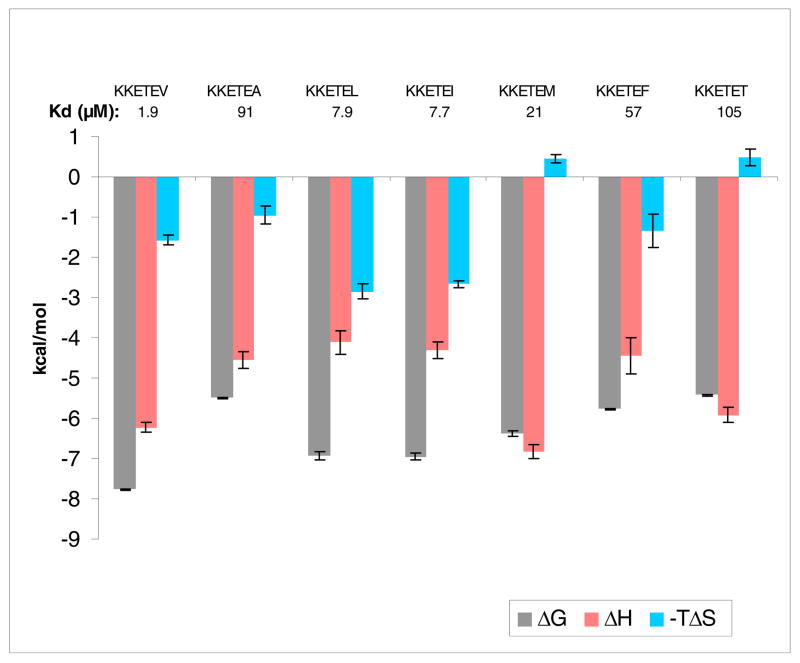
At P0, PDZ3 optimally accommodates the hydrophobic Val side chain, and adjustments to decrease (P0 = Ala) or increase (P0 = Leu, Ile, Phe) the side chain by increasing the hydrocarbon content resulted in diminished affinity (Figure 6). With its lone methyl side chain at P0, alanine is a violation of the class I PDZ motif and understandably loses affinity through suboptimal occupancy of the larger hydrophobic pocket. But there is measurable if weak binding, and perhaps even more notably (and inexplicably), the ΔH value of −4.6 kcal/mol compares to that of the Leu and Ile analogs. Those two isosteric peptides gain a binding edge over the Ala derivative with TΔΔS values that approach ~ 2 kcal/mol; if pressed for a rationale, this might be due to an entropically favored dehydration that accompanies binding of those larger alkyl moieties at P0 over that of Ala. The Leu and Ile ligands display Kd values only four-fold weaker than that of Val, and with Phe, an upper limit to allowable size appears to be reached and affinity weakens to Kd ~57 μM. This is in keeping with the outcome of our earlier report that included residues at P0 with nonproteinogenic alkyl side chains (17).
Figure 6.
Bar graph of thermodynamic data for binding of consensus sequence peptides mutated at P0 to PDZ3. Note that TΔS is plotted as the negative value.
Placing methionine at P0 resulted in a lower affinity peptide with Kd ~21 μM. With a more polarizable sulfur in lieu of carbon, a useful comparison can be made with the isosteric analog (where SΠCH2, using Ahx (2-aminohexanoic acid, or norleucine) in place of Met). A hexapeptide with a nonstandard residue at P0 we had reported earlier, KKETE(Ahx) has moderate affinity with Kd ~7 μM for PDZ3 (17). Both the Met and Ahx derivatives possess values for TΔS that are close to zero, the lowest among all the consensus sequence peptides. The major structural commonality is a linear, flexible side chain, a feature associated with low or unfavorable changes in entropy in other related analogs that employed nonstandard amino acids at the C-terminal residue (17).
Finally, an isosteric threonine replacement for the preferred valine at P0 to yield KKETET resulted in, unsurprisingly, weak affinity, with Kd ~105 μM. And yet, perhaps some surprise is warranted, considering that such a violent departure from the canonical class I binding sequence does not completely abolish association. The thermodynamic reason is not simply a worsening in enthalpy—the value of ΔH actually equals that of KKKETEV and bests those of the other KKETEX derivatives—but rather a dramatic shift in TΔΔS of ~ −2 kcal/mol with respect to the parent KKETEV. It may follow from an interpretation that the desolvation of the isopropyl group of Val is entropically quite favorable, whereas this is not so with the Thr side chain. The uncomplexed Thr may also lose hydrogen-bonding partners that are not replaced when bound at P0. With the barnase protein, it has been reported that the burial of the Thr hydroxyl moiety in a site intended for the γ-methyl group of Val costs 2.5 kcal/mol, in the range expected for the loss of two hydrogen bonds (60). With PDZ3, the ValΠThr change in the ligand yields a similar value, with ΔΔG ~2.4 kcal/mol.
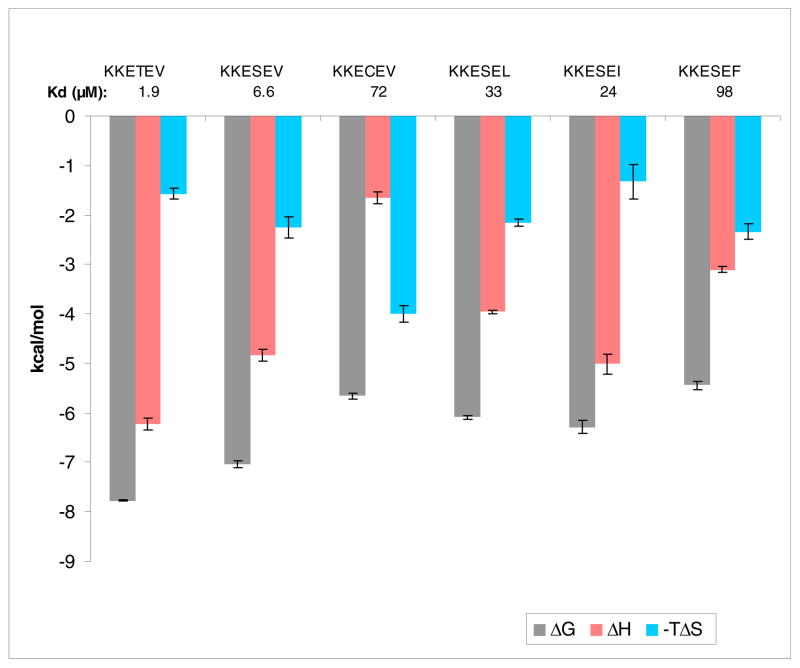
The next substitutions considered were those of the P-2 position. Although class I PDZ domains recognize both Thr and Ser at P-2, there is an approximate 3- to 4-fold preference for the more branched residue by PDZ3 (Figure 7). This may be the consequence of a strengthened hydrogen bond to His372 of PDZ3, in which the methyl of the Thr side chain restricts the rotation of the hydroxyl group in an optimal orientation. The methyl group might additionally impart a small amount of favorable van der Waals interaction. In keeping with this observed residue preference, both the CRIPT and neuroligin-1 sequences have Thr at P-2. Placement of cysteine at P-2 results in a Kd of ~73 μM. The 10-fold drop in affinity when converting SerΠCys is characterized by a sizeable loss in binding enthalpy, with ΔΔH = 3.2 kcal/mol, although this is softened somewhat by an entropic improvement of TΔΔS = 1.8 kcal/mol. One hypothetical scenario to explain this is a weakened or ineffectual hydrogen bond of the thiol group to His372 when compared to the hydroxyl side chain of serine. But this type of assessment has to also consider the strength of the thiol hydrogen bond to water when the peptide is in the uncomplexed state, as compared to the parallel interaction with the serine analog.
Figure 7.
Bar graph of thermodynamic data for binding of consensus sequence peptides mutated at P-2 to PDZ3. Note that TΔS is plotted as the negative value.
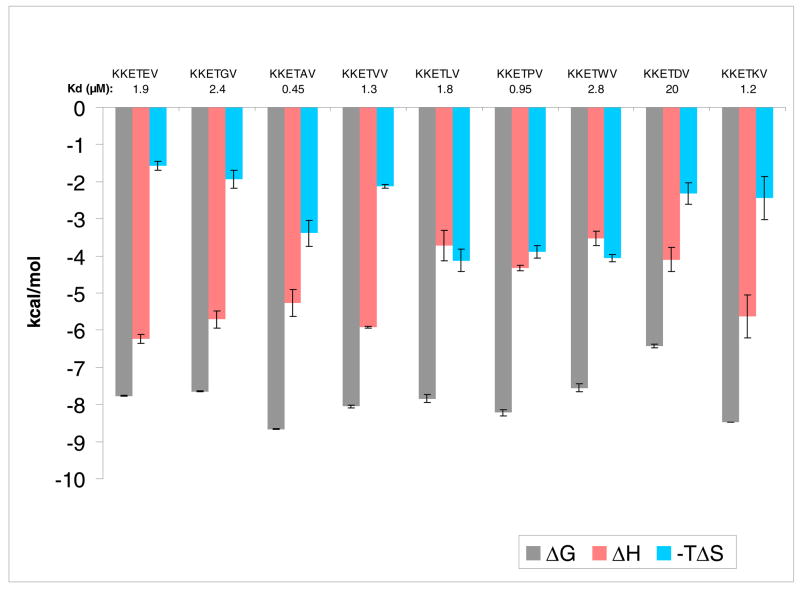
Although the P0 and P-2 subsites have generally been considered the primary determinants for PDZ3 binding, we also examined the contributions of the neighboring P-1 and P--3 positions. Previously we exploited the exposed nature of these residues by modifying both side chains and covalently imposing a bridging unit to form macrocyclic ligands (18, 19). Here, both glutamates of the KKETEV consensus sequence were replaced, singly and in tandem, with either alanine or glycine. Interestingly, the GluΠAla mutation at P-1 resulted in the ligand with the highest affinity for PDZ3 of the entire consensus-modified collection, with Kd = 0.45 μM. The submicromolar affinity of KKETAV is attributable to its favorable entropic increase (TΔΔS = 1.8 kcal/mol), since the enthalpy change was in fact unfavorable (ΔΔH = 1.0 kcal/mol).
In general, the P-1 position of PDZ3 appears quite tolerant to substitution, with most derivatives maintaining Kd values in the range of 1–3 μM (Figure 8). With respect to hydrocarbon size, accommodated residues span the gamut from the minimalist Gly, to Ala, Val, Leu, Pro, and up to the very large Trp. From the standpoint of conformation, the glycine and proline derivatives represent the two ends of flexibility and constraint, respectively. Even the cationic Lys is accepted, but the line is drawn when negatively-charged aspartate is introduced, and affinity drops by an order of magnitude. The development of certain PDZ domain-selective molecular probes might take advantage of this observation. Further derivitization at this position, for example, might allow the production of ligands that maintain affinity for PDZ3 of PSD-95, but disfavor binding to the PDZ3 domains of other MAGUK proteins.
Figure 8.
Bar graph of thermodynamic data for binding of consensus sequence peptides mutated at P-1 to PDZ3. Note that TΔS is plotted as the negative value.
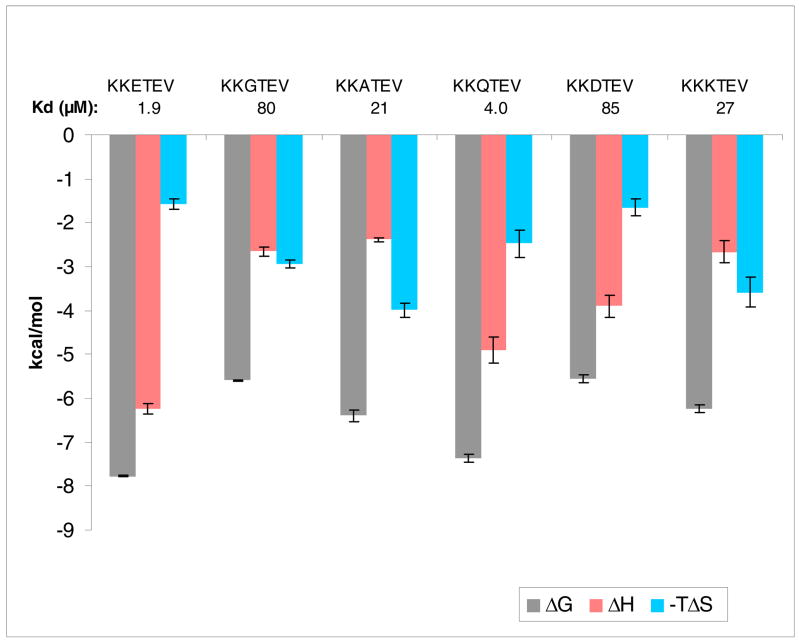
Moving further out to the other relatively solvent-exposed exposed position, P-3, the consensus residue Glu was replaced with a few select amino acids (Figure 9). In the crystal structure of PDZ3 with a peptide bearing glutamine at P-3, there appears to be a protein surface-mediated hydrogen bond with the amide side chain. Replacing Glu with Gln resulted in a two-fold reduction in affinity, and significant towards that was an enthalpic drop of ΔΔH = 1.3 kcal/mol. This could reflect an improved hydrogen bond of the Glu carboxylate over that of the Gln amide, or viewed from the perspective of glutamine, correlates with the loss of one or two H-bonds with PDZ3 (20).
Figure 9.
Bar graph of thermodynamic data for binding of consensus sequence peptides mutated at P-3 to PDZ3. Note that TΔS is plotted as the negative value.
A GluΠAla mutation at P-3 resulted in an opposing effect to that seen when the same change was effected at P-1. KKATEV experiences a near 10-fold drop in affinity with respect to KKETEV. Glycine at P-3 fares poorly as well, as do the analogs with oppositely charged Asp and Lys residues. The tolerance of P-1 stands in contrast to the more restrictive requirements of the P-3 position, although this cannot be stated definitely since fewer analogs were tested for the latter. Doubly-substituted ligands were prepared at P-1 and P-3 with alanine (KKATAV) and glycine (KKGTGV). Both displayed a decrease in the binding affinity compared to the parent KKETEV, with widely divergent Kd values of 8.3 μM and 272 μM, respectively.
A single exchange at P-5 concluded the standard ITC investigation of the consensus peptide. Looking at the CRIPT hexapeptide YKQTSV from the protein partner series, the Tyr residue represents a distinctly different character than the Lys found in the corresponding position of KKETEV. The transfer of that aromatic residue in place of the positively charged one led to YKETEV, which was only marginally improved in affinity over the original consensus peptide, and which exhibited a similar thermodynamic enthalpy-entropy profile. The ability to accept two functionally distinct side chains implies a possible opening for exploring diverse substitution patterns that may also display equal or enhanced affinity, as was seen with the P-1 position.
Consensus Sequence Peptides: Residue Coupling in Ligand Binding
Considering structural, conformational, electronic and solvation effects, that proximal residues in proteins are coupled and interact cooperatively—favorably or unfavorably—would seem probable. It has been stated that for proteins, in general, residues separated by less than 7 Å interact cooperatively; at greater separations the effects of mutation are additive, and the energetics of the interactions are independent of each other (61). To the extent that this holds for intermolecular binding events as well as intramolecular folding behavior requires empirical thermodynamic evaluation.
PDZ domains have to date been insufficiently examined as to whether there are cooperative effects within the binding ligand upon protein association. To address this question, we employed the tactic of a double mutant cycle analysis (61–63). This method has been used to calculate the degree of energetic coupling or cooperativity between different mutants. Cooperative interactions between amino acids are thought to be important in determining the specificity of the macromolecular interactions (64), and might be a widespread phenomena in molecular recognition events as supported by different investigations (65–67). To probe whether one residue influences the binding propensity of another within the same ligand, single and double mutations at the desired positions are prepared, necessitating the preparation and binding evaluation of a total of four constructs and then close the thermodynamic cycle with double mutants at the same positions.
Here, the two main binding determinant positions of the PDZ3 binding sequence, P0 and P-2, were first examined. To test the level of cooperativity, if any, between these two positions, we assembled double mutant cycles to calculate the free energy of coupling, ΔGc, and the free enthalpy of coupling, ΔHc, In such an analysis, if ΔGc is negative then the two residues are positively coupled (the binding of the first amino acid favors binding of the second); if ΔGc is positive, the two positions are negatively coupled (the presence of the first amino acid disfavors binding of the second). A value of zero for ΔGc indicates that the there is no cross interaction of the two residues upon binding. A corresponding interpretation applies to ΔHc.
The analysis in this case indicates that there is no substantial ΔGc between the P0 and P-2 residues when transforming from either a Val/Thr to Ile/Ser or a Val/Thr to Leu/Ser state (Figure 10). However, there appears to be a significant coupling effect at the level of enthalpy; the first residue is coupled to the second with a ΔHc = −2.1 kcal/mol for the Ile/Ser cycle, and ΔHc = −1.2 kcal/mol for the Leu/Ser cycle. The finding of significant ΔHc values in protein interactions has been reported in prior studies (65, 68, 69). In these investigations, the residues exhibited significant enthalpic coupling energetics while ΔGc remained small or insignificant. In the case of another modular protein domain, that of SH2 (65), the values of ΔHc are very similar to that observed for our PDZ3-peptide interactions. The magnitude of this value is arguably comparable with the worth of a single hydrogen bond, but what does enthalpic coupling signify if it is masked via enthalpy-entropy compensation such that the free energy change itself is nominal? A clear mechanistic rationale is not evident. Certainly some form of interaction between residues at P0 and P-2 is anticipated since these sites are linchpins for PDZ domain recognition. What function or purpose—if any—is served by enthalpically-coupled residues is an open question. The presence of enthalpic coupling may represent a way in which PDZ domains bias their recognition of residues on their binding partners. Perhaps it is a mode whereby binding affinities are maintained at a certain level—not too tight, not too weak, as befitting a transient, regulatory function—even as the PDZ domains and their protein partners evolve and undergo residue mutation that promote new binding selectivities (that is, eliminating old, or creating new, PDZ domain-mediated partnerships.
Similarly, we examined the vicinal P-1 and P-3 sites for evidence of cooperativity (Figure 11). As seen for P-0 and P-2, the results indicate that while ΔGc is not significant, these two positions experience a meaningful ΔHc. Thus, while the P0 and P-2 residues still dictate the fundamental class or mode of PDZ domain recognition, the occupants of the vicinal P-1 and P-3 positions may be fundamentally important in how PDZ domains evolve over time to form transient in vivo partnerships. From a structural perspective, this is particularly interesting since those residues are apparently more surface-oriented and not involved in deeply buried, extensive interactions with the PDZ3 domain.
In conclusion, our accumulated results from an array of calorimetrically-based experiments with distinctly different series of peptide ligands have revealed a number of features pertaining to the molecular recognition of a well-characterized and reasonably representative PDZ domain. The analysis of thermodynamic binding parameters is made all the more meaningful by comparing between a variety of analogs in which modifications are made systematically and at fixed points. In addition, the determination of specific, short sequences with good affinity for PDZ3 of PSD-95 provides a foundation for the development of cellular probes to investigate the biological consequences of disrupting the endogenous protein-protein interactions mediated by this PDZ domain.
Acknowledgments
We thank L. Hryhorczuk (WSU) for mass spectrometry services, and Ms. Amy Griffin for the preparation of selected peptides.
Footnotes
This research was supported by the National Institutes of Health (GM63021)
Abbreviations: CD, circular dichroism; DTT, dithiothreitol; ESI-MS, electrospray ionization mass spectrometry; GST, glutathione-S-transferase; ITC, isothermal titration calorimetry; MAGUK, membrane associated guanylate kinase; PSD-95, postsynaptic density 95 kDa.
Another approach to determining thermodynamic parameters is the van’t Hoff analysis of temperature-dependent Kd values obtained from non-calorimetric assays. There has been debate as to what extent this is appropriate: see Mizoue and Tellinghuisen, Biophys Chem (2004) 110, 15-24, and references therein.
References
- 1.Sheng M, Sala C. PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci. 2001;24:1–29. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Harris BZ, Lim WA. Mechanism and role of PDZ domains in signaling complex assembly. J Cell Sci. 2001;114:3219–31. doi: 10.1242/jcs.114.18.3219. [DOI] [PubMed] [Google Scholar]
- 3.Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–80. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 4.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–52. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 5.Cho KO, Hunt CA, Kennedy MB. The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron. 1992;9:929–42. doi: 10.1016/0896-6273(92)90245-9. [DOI] [PubMed] [Google Scholar]
- 6.Montgomery JM, Zamorano PL, Garner CC. MAGUKs in synapse assembly and function: an emerging view. Cell Mol Life Sci. 2004;61:911–29. doi: 10.1007/s00018-003-3364-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morabito MA, Sheng M, Tsai LH. Cyclin-dependent kinase 5 phosphorylates the N-terminal domain of the postsynaptic density protein PSD-95 in neurons. J Neurosci. 2004;24:865–76. doi: 10.1523/JNEUROSCI.4582-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schnell E, Sizemore M, Karimzadegan S, Chen L, Bredt DS, Nicoll RA. Direct interactions between PSD-95 and stargazin control synaptic AMPA receptor number. Proc Natl Acad Sci U S A. 2002;99:13902–7. doi: 10.1073/pnas.172511199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hata Y, Takai Y. Roles of postsynaptic density-95/synapse-associated protein 90 and its interacting proteins in the organization of synapses. Cell Mol Life Sci. 1999;56:461–72. doi: 10.1007/s000180050445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beique JC, Andrade R. PSD-95 regulates synaptic transmission and plasticity in rat cerebral cortex. J Physiol. 2003;546:859–67. doi: 10.1113/jphysiol.2002.031369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yao WD, Gainetdinov RR, Arbuckle MI, Sotnikova TD, Cyr M, Beaulieu JM, Torres GE, Grant SG, Caron MG. Identification of PSD-95 as a regulator of dopamine-mediated synaptic and behavioral plasticity. Neuron. 2004;41:625–38. doi: 10.1016/s0896-6273(04)00048-0. [DOI] [PubMed] [Google Scholar]
- 12.Xia Z, Gray JA, Compton-Toth BA, Roth BL. A direct interaction of PSD-95 with 5-HT2A serotonin receptors regulates receptor trafficking and signal transduction. J Biol Chem. 2003;278:21901–8. doi: 10.1074/jbc.M301905200. [DOI] [PubMed] [Google Scholar]
- 13.Aarts M, Liu Y, Liu L, Besshoh S, Arundine M, Gurd JW, Wang YT, Salter MW, Tymianski M. Treatment of ischemic brain damage by perturbing NMDA receptor- PSD-95 protein interactions. Science. 2002;298:846–50. doi: 10.1126/science.1072873. [DOI] [PubMed] [Google Scholar]
- 14.Roche KW. The expanding role of PSD-95: a new link to addiction. Trends Neurosci. 2004;27:699–700. doi: 10.1016/j.tins.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Tao F, Tao YX, Gonzalez JA, Fang M, Mao P, Johns RA. Knockdown of PSD-95/SAP90 delays the development of neuropathic pain in rats. Neuroreport. 2001;12:3251–5. doi: 10.1097/00001756-200110290-00022. [DOI] [PubMed] [Google Scholar]
- 16.Tao YX, Raja SN. Are synaptic MAGUK proteins involved in chronic pain? Trends Pharmacol Sci. 2004;25:397–400. doi: 10.1016/j.tips.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Saro D, Klosi E, Paredes A, Spaller MR. Thermodynamic analysis of a hydrophobic binding site: probing the PDZ domain with nonproteinogenic peptide ligands. Org Lett. 2004;6:3429–32. doi: 10.1021/ol049181q. [DOI] [PubMed] [Google Scholar]
- 18.Li T, Saro D, Spaller MR. Thermodynamic profiling of conformationally constrained cyclic ligands for the PDZ domain. Bioorg Med Chem Lett. 2004;14:1385–8. doi: 10.1016/j.bmcl.2003.09.103. [DOI] [PubMed] [Google Scholar]
- 19.Udugamasooriya G, Saro D, Spaller MR. Bridged peptide macrocycles as ligands for PDZ domain proteins. Org Lett. 2005;7:1203–6. doi: 10.1021/ol0475966. [DOI] [PubMed] [Google Scholar]
- 20.Doyle DA, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Crystal structures of a complexed and peptide-free membrane protein- binding domain: molecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–76. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 21.Gianni S, Engstrom A, Larsson M, Calosci N, Malatesta F, Eklund L, Ngang CC, Travaglini-Allocatelli C, Jemth P. The kinetics of PDZ domain-ligand interactions and implications for the binding mechanism. J Biol Chem. 2005;280:34805–12. doi: 10.1074/jbc.M506017200. [DOI] [PubMed] [Google Scholar]
- 22.Piserchio A, Salinas GD, Li T, Marshall J, Spaller MR, Mierke DF. Targeting specific PDZ domains of PSD-95; structural basis for enhanced affinity and enzymatic stability of a cyclic peptide. Chem Biol. 2004;11:469–73. doi: 10.1016/j.chembiol.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 23.Niethammer M, Valtschanoff JG, Kapoor TM, Allison DW, Weinberg TM, Craig AM, Sheng M. CRIPT, a novel postsynaptic protein that binds to the third PDZ domain of PSD-95/SAP90. Neuron. 1998;20:693–707. doi: 10.1016/s0896-6273(00)81009-0. [DOI] [PubMed] [Google Scholar]
- 24.Irie M, Hata Y, Takeuchi M, Ichtchenko K, Toyoda A, Hirao K, Takai Y, Rosahl TW, Sudhof TC. Binding of neuroligins to PSD-95. Science. 1997;277:1511–5. doi: 10.1126/science.277.5331.1511. [DOI] [PubMed] [Google Scholar]
- 25.Zhang W, Vazquez L, Apperson M, Kennedy MB. Citron binds to PSD-95 at glutamatergic synapses on inhibitory neurons in the hippocampus. J Neurosci. 1999;19:96–108. doi: 10.1523/JNEUROSCI.19-01-00096.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gasteiger EHC, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein Identification and Analysis Tools on the ExPASy Server. In: Walker JM, editor. The Proteomics Protocols Handbook. Humana Press; 2005. pp. 571–607. [Google Scholar]
- 27.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–7. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 28.Zhang YL, Zhang ZY. Low-affinity binding determined by titration calorimetry using a high- affinity coupling ligand: a thermodynamic study of ligand binding to protein tyrosine phosphatase 1B. Anal Biochem. 1998;261:139–48. doi: 10.1006/abio.1998.2738. [DOI] [PubMed] [Google Scholar]
- 29.Chowdhry BZ, Harding SE. In: Protein-Ligand Interactions: Hydrodynamics and Calorimetry. Harding SE, Chowdhry BZ, editors. Oxford University Press; New York: 2001. pp. 1–17. [Google Scholar]
- 30.Songyang Z, Fanning AS, Fu C, Xu J, Marfatia SM, Chishti AH, Crompton A, Chan AC, Anderson JM, Cantley LC. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–7. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 31.Grootjans JJ, Zimmermann P, Reekmans G, Smets A, Degeest G, Durr J, David G. Syntenin, a PDZ protein that binds syndecan cytoplasmic domains. Proc Natl Acad Sci U S A. 1997;94:13683–8. doi: 10.1073/pnas.94.25.13683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris BZ, Hillier BJ, Lim WA. Energetic determinants of internal motif recognition by PDZ domains. Biochemistry. 2001;40:5921–30. doi: 10.1021/bi010142l. [DOI] [PubMed] [Google Scholar]
- 33.Harris BZ, Lau FW, Fujii N, Guy RK, Lim WA. Role of electrostatic interactions in PDZ domain ligand recognition. Biochemistry. 2003;42:2797–805. doi: 10.1021/bi027061p. [DOI] [PubMed] [Google Scholar]
- 34.Novak KA, Fujii N, Guy RK. Investigation of the PDZ domain ligand binding site using chemically modified peptides. Bioorg Med Chem Lett. 2002;12:2471–4. doi: 10.1016/s0960-894x(02)00345-1. [DOI] [PubMed] [Google Scholar]
- 35.Wang L, Piserchio A, Mierke DF. Structural characterization of the intermolecular interactions of synapse-associated protein-97 with the NR2B subunit of N-methyl-D-aspartate receptors. J Biol Chem. 2005;280:26992–6. doi: 10.1074/jbc.M503555200. [DOI] [PubMed] [Google Scholar]
- 36.Piserchio A, Pellegrini M, Mehta S, Blackman SM, Garcia EP, Marshall J, Mierke DF. The PDZ1 domain of SAP90. Characterization of structure and binding. J Biol Chem. 2002;277:6967–73. doi: 10.1074/jbc.M109453200. [DOI] [PubMed] [Google Scholar]
- 37.Skelton NJ, Koehler MF, Zobel K, Wong WL, Yeh S, Pisabarro MT, Yin JP, Lasky LA, Sidhu SS. Origins of PDZ domain ligand specificity. Structure determination and mutagenesis of the Erbin PDZ domain. J Biol Chem. 2003;278:7645–54. doi: 10.1074/jbc.M209751200. [DOI] [PubMed] [Google Scholar]
- 38.Kachel N, Erdmann KS, Kremer W, Wolff P, Gronwald W, Heumann R, Kalbitzer HR. Structure determination and ligand interactions of the PDZ2b domain of PTP-Bas (hPTP1E): splicing-induced modulation of ligand specificity. J Mol Biol. 2003;334:143–55. doi: 10.1016/j.jmb.2003.09.026. [DOI] [PubMed] [Google Scholar]
- 39.Fuh G, Pisabarro MT, Li Y, Quan C, Lasky LA, Sidhu SS. Analysis of PDZ domain-ligand interactions using carboxyl-terminal phage display. J Biol Chem. 2000;275:21486–91. doi: 10.1074/jbc.275.28.21486. [DOI] [PubMed] [Google Scholar]
- 40.Jelesarov I, Bosshard HR. Isothermal titration calorimetry and differential scanning calorimetry as complementary tools to investigate the energetics of biomolecular recognition. J Mol Recognit. 1999;12:3–18. doi: 10.1002/(SICI)1099-1352(199901/02)12:1<3::AID-JMR441>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 41.Ward WH, Holdgate GA. Isothermal titration calorimetry in drug discovery. Prog Med Chem. 2001;38:309–76. doi: 10.1016/s0079-6468(08)70097-3. [DOI] [PubMed] [Google Scholar]
- 42.Velazquez Campoy A, Freire E. ITC in the post-genomic era…? Priceless. Biophys Chem. 2005;115:115–24. doi: 10.1016/j.bpc.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 43.Grootjans JJ, Reekmans G, Ceulemans H, David G. Syntenin-syndecan binding requires syndecan-synteny and the co-operation of both PDZ domains of syntenin. J Biol Chem. 2000;275:19933–41. doi: 10.1074/jbc.M002459200. [DOI] [PubMed] [Google Scholar]
- 44.Kang BS, Cooper DR, Jelen F, Devedjiev Y, Derewenda U, Dauter Z, Otlewski J, Derewenda ZS. PDZ tandem of human syntenin: crystal structure and functional properties. Structure. 2003;11:459–68. doi: 10.1016/s0969-2126(03)00052-2. [DOI] [PubMed] [Google Scholar]
- 45.Birrane G, Chung J, Ladias JA. Novel mode of ligand recognition by the Erbin PDZ domain. J Biol Chem. 2003;278:1399–402. doi: 10.1074/jbc.C200571200. [DOI] [PubMed] [Google Scholar]
- 46.Walsh NP, Alba BM, Bose B, Gross CA, Sauer RT. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell. 2003;113:61–71. doi: 10.1016/s0092-8674(03)00203-4. [DOI] [PubMed] [Google Scholar]
- 47.Tudyka T, Skerra A. Glutathione S-transferase can be used as a C-terminal, enzymatically active dimerization module for a recombinant protease inhibitor, and functionally secreted into the periplasm of Escherichia coli. Protein Sci. 1997;6:2180–7. doi: 10.1002/pro.5560061012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niedziela-Majka A, Rymarczyk G, Kochman M, Ozyhar A. GST-Induced dimerization of DNA-binding domains alters characteristics of their interaction with DNA. Protein Expr Purif. 1998;14:208–20. doi: 10.1006/prep.1998.0932. [DOI] [PubMed] [Google Scholar]
- 49.Vaccaro P, Dente L. PDZ domains: troubles in classification. FEBS Lett. 2002;512:345–9. doi: 10.1016/s0014-5793(02)02220-2. [DOI] [PubMed] [Google Scholar]
- 50.Bezprozvanny I, Maximov A. Classification of PDZ domains. FEBS Lett. 2001;509:457–62. doi: 10.1016/s0014-5793(01)03214-8. [DOI] [PubMed] [Google Scholar]
- 51.Niethammer M, Valtschanoff JG, Kapoor TM, Allison DW, Weinberg TM, Craig AM, Sheng M. CRIPT, a novel postsynaptic protein that binds to the third PDZ domain of PSD-95/SAP90. Neuron. 1998;20:693–707. doi: 10.1016/s0896-6273(00)81009-0. [DOI] [PubMed] [Google Scholar]
- 52.Passafaro M, Sala C, Niethammer M, Sheng M. Microtubule binding by CRIPT and its potential role in the synaptic clustering of PSD-95. Nat Neurosci. 1999;2:1063–9. doi: 10.1038/15990. [DOI] [PubMed] [Google Scholar]
- 53.Ichtchenko K, Hata Y, Nguyen T, Ullrich B, Missler M, Moomaw C, Sudhof TC. Neuroligin 1: a splice site-specific ligand for beta-neurexins. Cell. 1995;81:435–43. doi: 10.1016/0092-8674(95)90396-8. [DOI] [PubMed] [Google Scholar]
- 54.Lise MF, El-Husseini A. The neuroligin and neurexin families: from structure to function at the synapse. Cell Mol Life Sci. 2006;63:1833–49. doi: 10.1007/s00018-006-6061-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Madaule P, Furuyashiki T, Reid T, Ishizaki T, Watanabe G, Morii N, Narumiya S. A novel partner for the GTP-bound forms of rho and rac. FEBS Lett. 1995;377:243–8. doi: 10.1016/0014-5793(95)01351-2. [DOI] [PubMed] [Google Scholar]
- 56.Madaule P, Eda M, Watanabe N, Fujisawa K, Matsuoka T, Bito H, Ishizaki T, Narumiya S. Role of citron kinase as a target of the small GTPase Rho in cytokinesis. Nature. 1998;394:491–4. doi: 10.1038/28873. [DOI] [PubMed] [Google Scholar]
- 57.Zhang W, Benson DL. Targeting and clustering citron to synapses. Mol Cell Neurosci. 2006;31:26–36. doi: 10.1016/j.mcn.2005.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Connelly PR, Varadarajan R, Sturtevant JM, Richards FM. Thermodynamics of protein-peptide interactions in the ribonuclease S system studied by titration calorimetry. Biochemistry. 1990;29:6108–14. doi: 10.1021/bi00477a031. [DOI] [PubMed] [Google Scholar]
- 59.Spolar RS, Record MT., Jr Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–84. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 60.Serrano L, Kellis JT, Jr, Cann P, Matouschek A, Fersht AR. The folding of an enzyme. II. Substructure of barnase and the contribution of different interactions to protein stability. J Mol Biol. 1992;224:783–804. doi: 10.1016/0022-2836(92)90562-x. [DOI] [PubMed] [Google Scholar]
- 61.Schreiber G, Fersht AR. Energetics of protein-protein interactions: analysis of the barnase- barstar interface by single mutations and double mutant cycles. J Mol Biol. 1995;248:478–86. doi: 10.1016/s0022-2836(95)80064-6. [DOI] [PubMed] [Google Scholar]
- 62.Carter PJ, Winter G, Wilkinson AJ, Fersht AR. The use of double mutants to detect structural changes in the active site of the tyrosyl-tRNA synthetase (Bacillus stearothermophilus) Cell. 1984;38:835–40. doi: 10.1016/0092-8674(84)90278-2. [DOI] [PubMed] [Google Scholar]
- 63.Horovitz A. Double-mutant cycles: a powerful tool for analyzing protein structure and function. Folding & Design. 1996;1:R121–R126. doi: 10.1016/S1359-0278(96)00056-9. [DOI] [PubMed] [Google Scholar]
- 64.Di Cera E. Site-Specific Thermodynamics: Understanding Cooperativity in Molecular Recognition. Chem Rev. 1998;98:1563–1592. doi: 10.1021/cr960135g. [DOI] [PubMed] [Google Scholar]
- 65.Bradshaw JM, Waksman G. Calorimetric examination of high-affinity Src SH2 domain-tyrosyl phosphopeptide binding: dissection of the phosphopeptide sequence specificity and coupling energetics. Biochemistry. 1999;38:5147–54. doi: 10.1021/bi982974y. [DOI] [PubMed] [Google Scholar]
- 66.Hunter CA, Tomas S. Cooperativity, partially bound states, and enthalpy-entropy compensation. Chem Biol. 2003;10:1023–32. doi: 10.1016/j.chembiol.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 67.Kiel C, Serrano L, Herrmann C. A detailed thermodynamic analysis of ras/effector complex interfaces. J Mol Biol. 2004;340:1039–58. doi: 10.1016/j.jmb.2004.05.050. [DOI] [PubMed] [Google Scholar]
- 68.Richieri GV, Low PJ, Ogata RT, Kleinfeld AM. Mutants of rat intestinal fatty acid-binding protein illustrate the critical role played by enthalpy-entropy compensation in ligand binding. J Biol Chem. 1997;272:16737–40. doi: 10.1074/jbc.272.27.16737. [DOI] [PubMed] [Google Scholar]
- 69.Frisch C, Schreiber G, Johnson CM, Fersht AR. Thermodynamics of the interaction of barnase and barstar: changes in free energy versus changes in enthalpy on mutation. J Mol Biol. 1997;267:696–706. doi: 10.1006/jmbi.1997.0892. [DOI] [PubMed] [Google Scholar]