Abstract
Background and purpose:
Long-term adaptations to pharmacological stimuli frequently originate from modulation of complex intracellular signalling pathways. We previously reported that HU210 and CP55940, two CB1 cannabinoid receptor agonists, induced opposite effects on TH expression. Herein, we characterized their influence on cAMP response element (CRE) and activator protein 1 (AP-1)-mediated regulation of gene transcription.
Experimental approach:
The activity of the agonists was examined on transfected N1E-115 cells in which expression of the luciferase reporter gene was controlled by transcription promoters consisting of repeats of either CRE or AP-1 elements. In addition, the implication of classical signalling pathways was investigated using a variety of kinase inhibitors.
Key results:
Consistent with the CB1-mediated reduction of cAMP accumulation, both ligands decreased CRE-driven luciferase expression with similar potencies. HU210 also exhibited a concentration-dependent reduction of luciferase activity in cells engineered to examine AP-1-controlled transcription, whereas such response was not obtained with CP55940. Responses were all inhibited by SR141716A and were modified in Pertussis toxin-treated cells, suggesting agonist-selective regulations of distinct Gi/o-dependent mechanisms through CB1 receptor activation. Finally, PKC inhibitors efficiently inhibited the paradoxical effect of HU210 on AP-1-mediated transcription, indicating selective regulation of PKC-dependent responses.
Conclusions and implications:
Together, our results demonstrate that two cannabinoid ligands, commonly used as reference agonists acting on the same receptor with similar affinities, differentially modulate gene transcription through distinct controls of AP-1. This could reflect activation of distinct subsets of Gi/o-proteins, supporting the concept of functional selectivity at CB1 receptors.
Keywords: luciferase, cannabinoid, agonist-selective trafficking of receptor signalling, transcription, CRE, AP-1, functional selectivity
Introduction
Besides radioligand-binding studies, pharmacological characterization of synthetic drugs acting at G-protein-coupled receptors consists of the study of their potency and intrinsic activity in functional assays. Considering the signalling pathways activated by these receptors, these assays are generally focused on the measure of conventional immediate and transient responses, such as production of second messengers, activation of kinases or alteration in ion equilibriums. Although these acute responses provide relevant details regarding the pharmacodynamic properties of the ligand, it is well documented that delayed or long-term modulations of cell functions contribute to the clinical outcome obtained with these ligands.
With respect to cannabinoids that are classically documented to signal through inhibition of adenylyl cyclase (AC), several reports indicate that activation of cannabinoid receptors leads to the regulation of several DNA-binding proteins, including activator protein 1 (AP-1) (Porcella et al., 1998) and cAMP response element-binding protein (CREB) (Herring et al., 1998). Little is presently known regarding the nature of cannabinoid-mediated cell signals putatively involved in these delayed responses. Hence, paradoxical results were sometimes observed regarding agonist-mediated alteration of second messenger production and modulation of transcriptional activity. Thus, 2-arachidonoylglycerol was shown to enhance cell transformation and carcinogenesis through the induction of AP-1 DNA binding (Zhao et al., 2005), whereas Δ9-tetrahydrocannabinol reduced interleukin-2 transcription through a decrease in nuclear factors binding to the AP-1 site of the interleukin-2 promoter (Condie et al., 1996; Faubert and Kaminski, 2000).
Apart from inhibition of AC via a Pertussis toxin (PTx)-sensitive Gi/o protein (Pertwee, 1999; Alexander et al., 2008), cannabinoid-induced accumulation of cAMP has also been reported (Glass and Felder, 1997). This response is likely to reflect a functional coupling with Gs-type G proteins, as it is preserved in recombinant (Bonhaus et al., 1998; Calandra et al., 1999) and native (Maneuf and Brotchie, 1997; Bash et al., 2003) systems after Gαi inactivation by PTx. Even more convincing are studies showing the functional switch of CB1 receptor coupling from Gαi to Gαs following the sequestration of Gi/o G-protein pool through dopamine D2 receptor coactivation (Glass and Felder, 1997) or overexpression (Jarrahian et al., 2004). In addition, both CB1 and CB2 receptors were reported to stimulate ERK1/2 (extracellular-signal regulated kinase) (Bouaboula et al., 1995) in a Gi/o-dependent manner, through the Gβγ subunit. The complexity of the intracellular signalling associated with the CB1 receptor extends to the activation of members of the mitogen-activated protein kinases (MAPK), including c-Jun N-terminal kinase and p38 MAPK (Rueda et al., 2000) and the modulation of calcium and potassium currents (Mackie and Hille, 1992; Mackie et al., 1995). In addition, CB1 receptors may induce rapid and transient elevations of intracellular free Ca2+ concentrations through Gβγ-mediated activation of PLCβ (Sugiura et al., 1997). Finally, regulation of PLC by CB1 receptors was proposed to require coupling with different α-subunits of the Gq/11 family (Ho et al., 1999). Hence, the selective coupling to Gq/11 protein in an agonist-specific manner was more recently demonstrated (Lauckner et al., 2005; McIntosh et al., 2007).
We recently reported cannabinoid-mediated regulation of tyrosine hydroxylase (TH) expression in the N1E-115 neuroblastoma cell line constitutively and selectively expressing the CB1 receptor subtype. Particularly, different classes of full CB1 agonists, belonging to different chemical families, were shown to induce opposite effects (Bosier et al., 2007), raising the question of functional selectivity at the CB1 receptor. In this context, the present study aims at further investigating CB1 receptor-mediated signalling pathways involved in the regulation of gene expression. In view of the essential roles of both CRE and AP-1 cis-enhancer elements in constitutive and induced TH gene expression, CRE- or AP-1-driven luciferase reporter gene assays were carried out to elucidate the CB1 receptor-mediated, agonist selective regulation of signalling pathways involved in long-term control of gene transcription.
Methods
Cell culture
All cell culture media and supplements were obtained from Invitrogen (Merelbeke, Belgium). Mouse neuroblastoma N1E-115 cells were grown in Dulbecco's MEM/NUT mix F-12 medium supplemented with 10% foetal calf serum, 100 UI mL−1 penicillin, 100 μg mL−1 streptomycin and 2 mM L-glutamine. All drug treatments were conducted in the same culture medium. At confluence, cells were trypsinized for dilutions. Cells were cultured at 37 °C in an atmosphere of humidified air and 5% CO2.
cAMP assay
N1E-115 cells were seeded in 96-well plates (104 cells per well), 24 h before cannabinoid treatments. Cells were pretreated for 30 min with 0.1 mM IBMX (3-isobutyl-1-methylxanthine). Then, cannabinoid agonists were added for another 30 min incubation treatment in the absence or presence of 1 μM forskolin. When N-(piperidin-1-yl)-5-(4-chlorophenyl)-1-(2,4-dichlorophenyl)-4-methyl-1H pyrazole-3-carboxamide hydrochloride (SR141716A) and N-((1S)-endo-1,3,3-trimethylbicyclo(2.2.1)heptan-2-yl)-5-(4-chloro-3-methylphenyl)-1-(4-methylbenzyl)-pyrazole-3-carboxamide (SR144528) were used, cells were pretreated for 5 min with the cannabinoid inverse agonists/antagonists at 1 μM before the addition of the cannabinoid agonist. Where indicated, cells were treated overnight with 100 ng mL−1 PTx. cAMP levels were measured using cAMP Biotrak enzyme immunoassay system (Amersham Pharmacia Biotech, Roosendaal, The Netherlands), according to the protocol provided by the manufacturer. Cells were incubated at 37 °C.
Plasmids, transfection and dual luciferase assay
pCRE-Luc and pAP-1-Luc plasmids were purchased from Stratagene (La Jolla, CA, USA) and included 4 and 7 copies of CRE and AP-1 cis-enhancer elements, respectively, fused to the firefly luciferase gene. pRL 138 obtained from Dr E Pierreux (UCL, ICP, Brussels, Belgium) was used as an internal control to normalize for transfection variability. pRL 138 was constructed by the introduction of a 225 bp sequence of the PFK-2 promoter in the pRL null vector from Promega (Mannheim, Germany) encoding for Renilla luciferase, as described previously (Pierreux et al., 1998).
Cells were plated at a density of 105 cells per well in 24-well plates. Culture medium was changed after 24 h, and the cells were co-transfected with a reporter plasmid (3 μg per well and 5 μg per well for pCRE-Luc and pAP-1-Luc, respectively) and the pRL 138 plasmid at 0.25 μg per well. Transfection was performed overnight using the calcium phosphate co-precipitation method. After transfection, cells were washed three times with PBS buffer (137 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.47 mM KH2PO4, pH 7.4) and fresh medium was added 24 h before the 5 h treatment with the appropriate amounts of cannabinoid agonists. When indicated, SR141716A and SR144528 were added 5 min before applying the agonist. When PTx was used, cells were treated overnight with a 100 ng mL−1 concentration. To stop the reaction, cells were washed three times with PBS buffer before lysis by addition of 100 μL passive lysis buffer supplied with the Dual Luciferase Reporter Assay System (Promega). Firefly luciferase reporter activity was normalized for Renilla luciferase activity. Respective measurement of light emission was determined according to the manufacturer's instructions. Luminescence was detected by a TD20/20 luminometer (Turner design, Sunnyvale, CA, USA).
Data analysis and statistical procedures
Data presented in the text and in the figures were expressed as percentages of the corresponding values obtained with cells treated with vehicle (dimethyl sulphoxide diluted in culture medium). pEC50 values were determined from at least three separate experiments by nonlinear regression analysis performed using Graph Pad prism software (San Diego, CA, USA). Statistical analysis was performed on the log-transformed value of relative luminescence or on the net cAMP level by ANOVA with repeated measurements or Student's t-test. A post hoc analysis was performed by Scheffe test, using the SPSS software.
Drugs, chemical reagents and other materials
(6,R)-trans-3-(1,1-dimethylheptyl)-6,7,10,10,-tetrahydro-1-hy-droxy-6,6-dimethyl-6H-dibenzo(b,d)pyran-9-methanol (HU210) and (1R,3R,4R)-3-(2-hydroxy-4-(1,1-dimethylheptyl) phenyl)-4-(3-hydroxypropyl)cyclohexan-1-ol (CP55940) were purchased from Tocris Cookson (Bristol, UK). The CB1 receptor inverse agonist/antagonist, SR141716A, and the CB2 receptor inverse agonist/antagonist, SR144528, were generously provided by Dr Barth and Dr Mossé, respectively (Sanofi-Synthélabo Research, Montpellier, France). Forskolin, IBMX, phorbol 12-myristate 13-acetate and PTx were purchased from Sigma (Boornem, Belgium). Stock solutions of drugs were prepared in dimethyl sulphoxide at 0.01 M and stored in aliquots at −80 °C. The final dimethyl sulphoxide concentration never exceeded 0.1%, which had no significant effect on assays.
Results
Modulation of cAMP accumulation
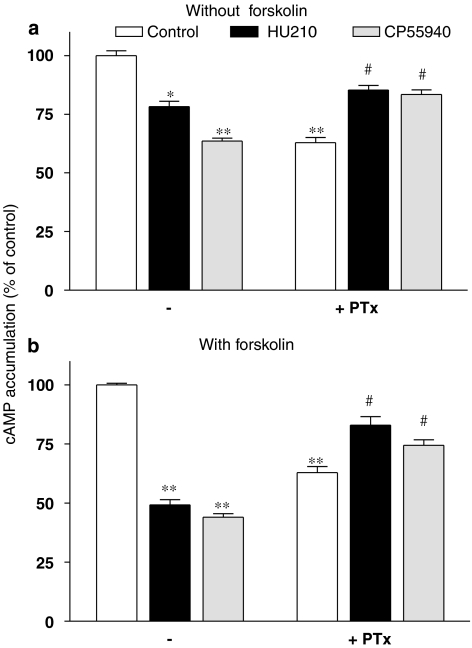
In agreement with several studies indicating the functional coupling of the cannabinoid receptors with Gi/o proteins, both HU210 (0.1 μM) and CP55940 (0.1 μM), two typical cannabinoid full agonists, elicited a significant reduction in cAMP accumulation in N1E-115 neuroblastoma cells (21.8 and 36.5% reduction relative to control cells, respectively) (Figure 1a). To facilitate the detection of Gi/o-mediated responses, basal AC activity was enhanced by the addition of 1 μM forskolin (leading to a robust 725±56% induction of cAMP accumulation, relative to control). At the tested concentration, maximal response to forskolin is certainly not reached, as the EC50 value of forskolin in mediating cAMP accumulation in N1E-115 cells is close to 10 μM (Stenstrom et al., 1985; Murphy and Byczko, 1989). In these conditions, both HU210 and CP55940 exhibited a more pronounced inhibition of cAMP formation (50.9 and 56.0% reduction relative to forskolin treated cells, respectively) (Figure 1b). Confirming the specific involvement of CB1 receptors, these responses were totally prevented by SR141716A, a selective CB1 inverse agonist/antagonist, whereas SR144528, a selective CB2 inverse agonist/antagonist, had no influence on the agonist-mediated reduction of cAMP levels (data not shown). Although basal cAMP level was significantly decreased in PTx-treated cells, HU210 (0.1 μM) and CP55940 (0.1 μM) elicited a significant increase in cAMP synthesis either in the absence (30.0 and 28.4%, respectively) or presence of forskolin (32.0 and 18.4%, respectively) (Figure 1). Increased cAMP level induced by cannabinoids was already reported, when tested in the absence of functional Gi/o proteins, and is currently attributed to a functional coupling of the CB1 receptor with Gs proteins.
Figure 1.
Regulation of cAMP accumulation in N1E-115 neuroblastoma cells. Cells were exposed to HU210 or CP55940 (0.1 μM), in the absence (a) or presence (b) of 1 μM forskolin. The responses to both agonists were also measured after overnight treatment of the cells with PTx (100 ng mL−1). Results are expressed as percentage of responses in control cells (exposed to vehicle or forskolin, which caused a sevenfold increase in basal cAMP accumulation). Data are mean values with s.e.mean from three separate experiments performed in triplicate. **P<0.01, *P<0.05 relative to control condition; ##P<0.01, #P<0.05 relative to responses in the presence of PTx.
Cannabinoid-mediated regulation of CRE and AP-1-dependent transcriptional activity
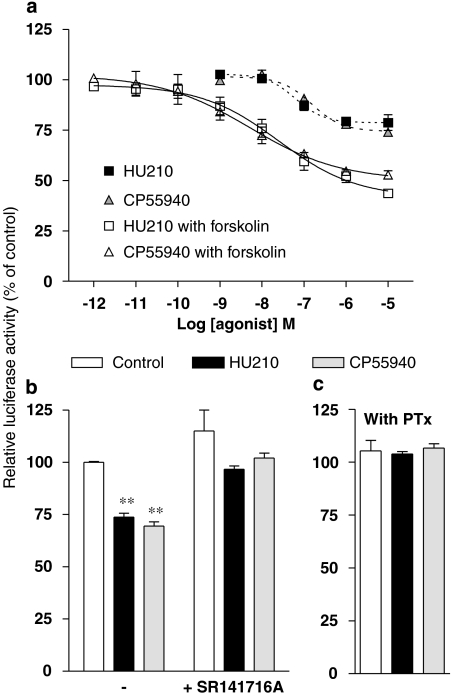
To examine the effects of CB1 receptor activation on CRE and AP-1-dependent transcriptional activities, N1E-115 cells were transiently transfected with plasmid constructs containing the luciferase gene under the control of cis-enhancer element repeats of either CRE (pCRE-Luc) or AP-1 (pAP-1-Luc). As expected, cells transfected with pCRE-Luc constructs displayed a robust basal luciferase activity (not shown), confirming a constitutive activity of CRE-dependent transcription in N1E-115 cells. In addition, the strong increase in luciferase activity (up to 850%) observed with forskolin 1 μM confirmed that the luciferase reporter gene is a reliable indicator of cAMP levels in this model. Both HU210 and CP55940 were found to modestly inhibit CRE-mediated transcriptional activity (estimated pEC50 values of 7.21±0.01 and 6.82±0.09, respectively) (Figure 2a). More convincingly, when the agonists were tested in cells exposed to forskolin, both markedly reduced luciferase expression in a concentration-dependent manner, with pEC50 values of 7.60±0.01 and 8.07±0.04 for HU210 and CP55940, respectively. In this assay, the analysis of concentration–response curves indicated Hill slopes distinct from unity (0.47±0.04 and 0.41±0.02 for HU210 and CP55940, respectively). Nevertheless, the responses to both agonists were markedly impaired using SR141716A (Figure 2b), suggesting that inhibition of CRE activity was specifically driven through CB1 receptor-dependent signalling pathways.
Figure 2.
Cannabinoid-mediated regulation of CRE-dependent transcriptional activity in N1E-115 neuroblastoma cells. Luciferase activity was measured in transiently transfected N1E-115 cells carrying the pCRE-Luc construct. Cells were exposed to increasing concentrations of HU210 or CP55940 in the absence or presence of 1 μM forskolin (a) and luciferase activity was monitored 5 h later. The responses to both agonists (used at 0.1 μM) were also measured in the presence of 1 μM SR141716A (b). The role of Gi/o-type G proteins was investigated by repeating these measures in the presence of forskolin on cells treated overnight with PTx (100 ng mL−1) (c). Results (mean values with s.e.mean from at least three separate experiments performed in triplicate) are given as the percentages of relative luciferase activity (firefly luciferase relative to Renilla luciferase activity) compared with untreated or forskolin-treated cells. Forskolin increased the basal level of luciferase by 8.5-fold. **P<0.01 and *P<0.05 denote significant difference as compared with control conditions.
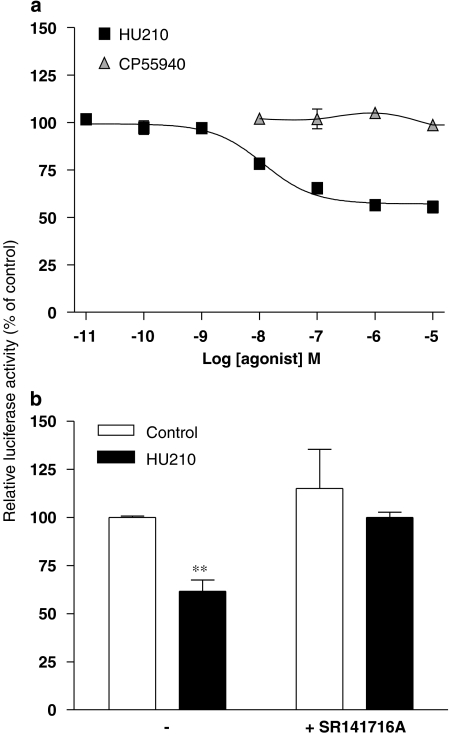
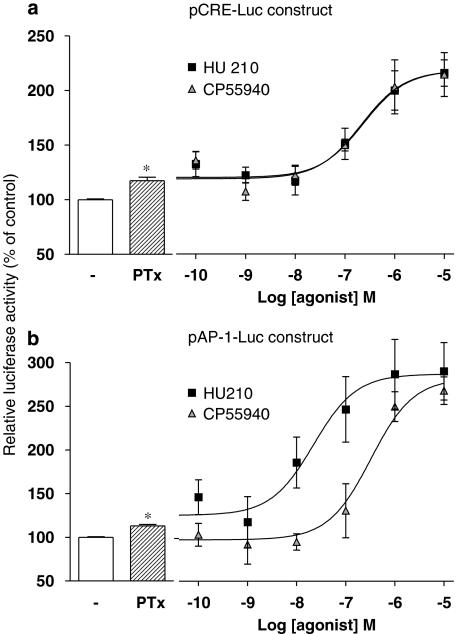
Experiments were repeated in cells engineered to examine AP-1-controlled luciferase activity. As with the CRE construct, a noticeable constitutive AP-1-driven luciferase activity was detected. The efficiency of this system was validated using 0.08 μM phorbol 12-myristate 13-acetate, a PKC activator known to regulate AP-1 transcription, which produced a modest but significant increase in luciferase activity (124.8±2.4%, relative to control; P<0.01). In this assay, HU210 elicited a concentration-dependent reduction of AP-1 activity (pEC50 of 7.95±0.11) (Figure 3a) and experimental data best fitted with a standard sigmoidal curve (Hill slope close to 1). Again, SR141716A abolished the HU210-mediated response (Figure 3b), confirming that the regulation of AP-1-dependent transcription was operating through CB1 receptor activation.
Figure 3.
HU210-mediated regulation of AP-1-dependent transcriptional activity in N1E-115 neuroblastoma cells. Luciferase activity was measured in N1E-115 cells transiently transfected with pAP-1-Luc construct. (a) Cells were exposed to increasing concentrations of HU210 or CP55940 and luciferase activity was monitored 5 h later. The responses to both agonists (used at 0.1 μM) were also measured in the presence of 1 μM SR141716A (b). Results shown are mean values with s.e.mean from at least three separate experiments performed in triplicate and are given as the percentages of relative luciferase activity (firefly luciferase relative to Renilla luciferase activity) compared with untreated cells. **P<0.01 and *P<0.05 denote significant difference as compared with control conditions.
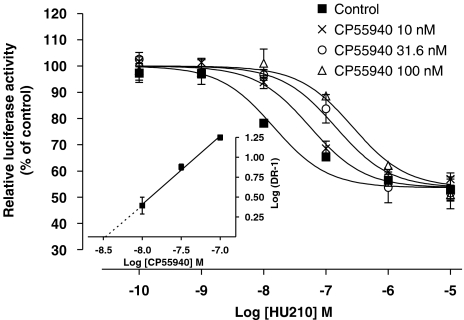
Surprisingly, no significant influence on AP-1 activity was observed using CP55940, indicating that this commonly used agonist of the CB1 receptor behaves as a neutral ligand on the cell-signalling pathway regulating AP-1-dependent transcriptional activity. The pharmacological profile of CP55940 was further examined by analysing its ability to antagonize cannabinoid-mediated regulation of AP-1 activity. As shown in Figure 4, CP55940 caused a concentration-dependent rightward shift of the sigmoidal curve for HU210-induced luciferase activation. This clearly identifies CP55940 as a competitive antagonist at the CB1 receptor in this functional assay and Schild plot transformation indicated a pA2 value of 8.48 (95% confidence interval of 8.22–8.92).
Figure 4.
CP55940 competitively antagonizes HU210-induced inhibition of AP-1-dependent luciferase activity. Luciferase activity was measured in N1E-115 cells transiently transfected with pAP-1-Luc construct. The apparent potency of HU210 in decreasing luciferase activity was evaluated in the presence of different concentrations of CP55940. Inset: Schild plot analysis allowing estimation of the pA2 value of CP55940 at inhibiting the response to HU210. Results shown are mean values with s.e.mean from at least three separate experiments performed in triplicate and are given as the percentage of relative luciferase activity (firefly luciferase relative to Renilla luciferase activity) as compared with untreated cells.
Signalling pathways involved in CRE and AP-1-dependent transcriptional activity
The cellular mechanisms involved in the regulation of CRE and AP-1-dependent transcriptional activity by cannabinoid agonists were examined using inhibitors of intracellular signalling cascades putatively associated with CB1 receptor functions. As this receptor appears predominantly associated with modulation of cAMP levels, we first studied the effect of the PKA inhibitors, H89 and KT5720, on both CRE and AP-1-driven luciferase activities. Also, as cannabinoid responses on CRE activity were more pronounced when the basal activity of AC was boosted, the effect of inhibitors on CRE-controlled transcription was evaluated in the presence of 1 μM forskolin. As expected, incubation of N1E-115 cells with H89 (10 μM) or KT5720 (5 μM) significantly reduced pCRE-Luc activity in the presence of forskolin (Table 1). In the same conditions, these PKA inhibitors completely abolished the modulation of pCRE-Luc activity caused by the cannabinoid agonists HU210 and CP55940.
Table 1.
Biochemical characterization of the signalling pathways involved in the modulation of pCRE-Luc activity by cannabinoid ligands
| Treatments | No agonist | HU210 | CP55940 |
|---|---|---|---|
| No inhibitor | 100.0±0.2 | 75.0±0.9** | 65.7±2.3** |
| PKA inhibitors | |||
| H89 (10 μM) | 45.7±7.2* | 48.9±9.3 | 52.1±10.2 |
| 100.0±15 | 106.4±8.5 | 112.2±21.1 | |
| KT5720 (5 μM) | 62.9±1.8* | 67.7±5.5 | 65.5±2.7 |
| 100.0±2.9 | 107.9±12.1 | 102.6±7.2 | |
| MEK/MAPKK inhibitors | |||
| U0126 (5 μM) | 83.6±4.1* | 64.7±7.2 | 62.1±7.1 |
| 100.0±4.9 | 76.6±4.8# | 74.1±6.7# | |
| PD98059 (25 μM) | 78.2±4.3** | 61.7±6.9 | 56.0±5.3 |
| 100.0±5.5 | 78.6±4.4# | 71.4±2.8# | |
| PI3K inhibitor | |||
| Wortmannin (0.2 μM) | 114.3±5.4 | 85.9±1.8 | 70.7±6.5 |
| 100.0±4.7 | 74.9±2.8## | 61.9±5.1## | |
| PKC inhibitors | |||
| Chelerythrine (10 μM) | 101.8±3.7 | 73.2±1.9 | 67.9±4.7 |
| 100.0±3.7 | 75.4±2.1## | 68.7±2.1## | |
| GF109203X (0.25 μM) | 124.7±12.1 | 108.2±11.0 | 79.7±3.2 |
| 100.0±9.7 | 80.6±2.4# | 65.0±3.3## | |
Transfected N1E-115 cells carrying pCRE-Luc construct were pretreated for 1 h in the absence or presence of inhibitors. Thereafter, cells were exposed to a vehicle, HU210 (0.1 μM) or CP55940 (0.1 μM), in the presence of forskolin 1 μM, and the luciferase activity was monitored 5 h later. Results are given as the percentages of luciferase activity compared with control (forskolin only). Numbers in italics show the same data normalized to the values obtained in the presence of the inhibitor (set at 100%). Shown are mean values with s.e.mean from at least three independent experiments performed in triplicate.
**P<0.01, *P<0.05 relative to forskolin-induced response; ##P<0.01, #P<0.05 relative to the cells treated with the inhibitor alone.
The effects of MAPK kinase (MEK) and PI3K inhibitors were also investigated, as cannabinoids are known to regulate MAPK activity, possibly through PI3K activation. The MEK inhibitors U0126 (5 μM) and PD98059 (25 μM) were found to decrease the effect of forskolin on CRE-driven luciferase transcription. Similar reduction in CRE activity was observed in the absence of forskolin (74.5±7.9 and 67.1±2.3% relative to control cells, for U0126 and PD98059, respectively), suggesting that MAPKs are involved in the basal activity of CRE cis-enhancer elements. In contrast, these MEK inhibitors as well as the PI3K inhibitor wortmannin (0.2 μM) failed to reduce the effects of HU210 or CP55940. Finally, the PKC inhibitors chelerythrine (10 μM) or GF 109203X (0.25 μM), which were tested on the basis of the key role of this kinase in the modulation of AP-1-dependent transcriptional activity, were unable to influence the regulation of CRE activity by the cannabinoid agonists.
In contrast to the responses obtained with the pCRE-Luc construct, the PKA inhibitors were without influence on the HU210-mediated regulation of AP-1-driven luciferase expression, whereas efficient blockade was obtained with the PKC inhibitors (Table 2). Compound Ro-31-6045, an analogue of GF 109203X, generally used as a negative control for inhibition of PKC activity, failed to influence the response to HU210, validating the specificity of the PKC inhibition. The failure of CP55940 to efficiently regulate AP-1 element was confirmed by our finding that none of the inhibitors tested here affected the response to this agonist. When used alone, inhibitors of MEK, PI3K or PKC were also found to affect the basal AP-1-mediated luciferase activity, suggesting that these kinases are involved in the constitutive AP-1-dependent transcriptional activity. Finally, neither MEK nor PI3K inhibitors appeared to influence the response to HU210.
Table 2.
Biochemical characterization of the signalling pathways involved in the modulation of pAP-1-Luc activity by cannabinoid ligands
| Treatments | No agonist | HU210 | CP55940 |
|---|---|---|---|
| No inhibitor | 100.0±0.76 | 65.3±5.8** | 101.1±4.4 |
| PKA inhibitors | |||
| H89 (10 μM) | 98.5±11.2 | 71.5±6.5 | 104.8±7.1 |
| 100.0±11.1 | 73.0±2.2# | 105.6±6.0 | |
| KT5720 (5 μM) | 94.3±1.0 | 70.1±2.3 | 96.9±2.5 |
| 100.0±1.1 | 74.3±3.1## | 101.4±2.4 | |
| MEK/MAPKK inhibitors | |||
| U0126 (5 μM) | 94.9±12 | 58.1±9.2 | 104.6±9.6 |
| 100.0±13.0 | 60.7±2.7# | 107.3±7.4 | |
| PD98059 (25 μM) | 74.3±1.0** | 52.8±1.5 | 74.3±1.8 |
| 100.0±1.3 | 71.0±3.0## | 100.1±3.9 | |
| PI3K inhibitor | |||
| Wortmannin (0.2 μM) | 300.4±68.1* | 212.7±58.3 | 315.9±38.3 |
| 100.0±22.2 | 68.7±5.0 | 107.4±5.5 | |
| PKC inhibitors | |||
| Chelerythrine (10 μM) | 70.1±2.8** | 70.9±4.9 | 72.8±6.9 |
| 100.0±3.9 | 101.1±3.1 | 103.6±5.8 | |
| GF109203X (0.25 μM) | 65.6±11.0** | 75.3±25.0 | 73.6±19.1 |
| 100.0±16.1 | 108.7±17.1 | 113.4±8.6 | |
| Negative control for PKC inhibition | |||
| Ro-31-6045 (0.25 μM) | 100.0±1.5 | 61.2±1.6 | 98.9±0.4 |
| 100.0±1.5 | 61.2±1.6## | 98.9±0.4 | |
MEK, mitogen-activated kinase kinase; MAPK, mitogen-activated protein kinase.
Transfected N1E-115 cells carrying pAP-1-Luc construct were pretreated for 1 h in the absence or presence of inhibitors. Thereafter, cells were exposed to a vehicle, HU210 (0.1 μM) or CP55940 (0.1 μM), and the luciferase activity was monitored 5 h later. Results are given as the percentages of luciferase activity compared with control. Numbers in italics show the same data normalized to the values obtained in the presence of the inhibitor (set at 100%). Shown are mean values with s.e.mean from at least three independent experiments performed in triplicate.
**P<0.01, *P<0.05 relative to control; ##P<0.01, #P<0.05 relative to the cells treated with the inhibitor alone.
Cannabinoid-mediated regulation of CRE and AP-1-dependent transcriptional activity is directed by Gi/o proteins
The role of Gi/o-type G proteins in the modulation of gene transcription by cannabinoid agonists was examined by repeating the luciferase reporter assays in cells treated overnight with PTx (100 ng mL−1). In cells expressing either the pCRE-Luc or pAP-1-Luc constructs, the basal luciferase activities appeared to be substantially increased after treatment with PTx (17.4 and 12.4% relative to control, respectively), indicating that both transcription promoters were negatively influenced by a constitutive activation of Gi/o-type G proteins.
As shown in Figure 2c, the inhibition of CRE-dependent luciferase activity induced by both HU210 and CP55940 in the presence of forskolin was abolished by PTx pretreatment (Figure 2c). In contrast, when the assay was conducted in the absence of forskolin, the effect of cannabinoid agonists was switched from inhibition to induction of CRE-dependent transcription, and concentration–response curves revealed pEC50 values of 6.64±0.26 and 6.67±0.27 for HU210 and CP55940, respectively (Figure 5a). These data are consistent with the above-mentioned increase in cAMP levels in PTx-treated cells, demonstrating the dual coupling of the CB1 receptor with the Gi/o and Gs-type G proteins.
Figure 5.
Involvement of Gi/o-type G proteins in the modulation of CRE and AP-1-dependent transcriptional activities by cannabinoid ligands. Luciferase activity was measured in N1E-115 cells transiently transfected with either pCRE (a) or pAP-1-Luc (b) constructs. Cells were treated overnight with 100 ng mL−1 PTx. Thereafter, cells were exposed to increasing concentrations of HU210 or CP55940 and luciferase activity was monitored after 5 h. Results shown are mean values with s.e.mean from at least three separate experiments performed in triplicate and are given as the percentage of relative luciferase activity (firefly luciferase relative to Renilla luciferase activity) compared with control. *P<0.05 denotes significant difference as compared with control conditions.
Such reversal of the functional response to HU210 in PTx-treated cells was also observed when studying the modulation of luciferase under the control of AP-1. In these conditions, HU210 concentration-dependently stimulated luciferase activity with pEC50 values of 7.66±0.38. More surprisingly, in this model of PTx-treated cells, CP55940 appeared to be effective in inducing AP-1-dependent transcription with a pEC50 of 6.48±0.23 (Figure 5b). Together, these data indicate that the regulation of AP-1 activity by cannabinoid agonists was also operating through the activation of members of the Gi/o protein family.
Discussion and conclusions
The effects of cannabinoids on CNS activity are frequently related to the CB1 receptor-mediated regulation of signalling cascades, controlling the release of other neurotransmitters. Beside these acute responses, several reports indicate that activation of the CB1 receptor modulates transcription factors, including CREB, nuclear factor kappa B, nuclear factor-activated T cells and AP-1 (Ouyang et al., 1998; Herring et al., 2001). In this study, using N1E-115 neuroblastoma cells that constitutively express CB1 receptors, we report that the cannabinoid agonist HU210 concentration-dependently inhibits both CRE and AP-1-mediated transcriptional activities. In contrast, the unrelated cannabinoid agonist CP55940 fails to influence AP-1-controlled gene expression while efficiently regulating CRE activity. Furthermore, these responses are abolished and/or reversed in the cells treated with PTx, suggesting the involvement of Gi/o-type G proteins.
Although the involvement of cannabinoid receptors was not definitely established, cAMP-dependent inhibition of CREB DNA binding after cannabinoid treatment has been previously demonstrated (Koh et al., 1997; Herring et al., 1998, 2001). More recently, CB1 receptor-mediated increase in CREB activation through ERK-dependent signalling was demonstrated in discrete regions of the rat brain after administration of cannabinoid agonists (Casu et al., 2005; Rubino et al., 2007). Consistent with these studies, the use of a specific luciferase-based reporter assay revealed that both HU210 and CP55940 concentration-dependently inhibited CRE-dependent transcriptional activity through activation of CB1 receptors, as indicated by the efficient blockade of the responses by SR141716A. The detection of this response was facilitated in cells exposed to forskolin, which boosts basal CRE-dependent transcriptional activity, suggesting that agonists were acting through inhibition of AC. This is consistent with our data showing a decreased cAMP accumulation in cells exposed to HU210 and CP55940. Indeed, the use of specific inhibitors confirmed that PKA was required for the forskolin-mediated induction of basal CRE activity. Accordingly, the PKA inhibitors totally abolished HU210 and CP55940-mediated regulation of CRE-dependent luciferase activity. Together, these data strongly support the general concept that agonists of CB1 receptors influence gene transcription through inhibition of cAMP production and modulation of PKA activity. Nevertheless, concentration–response curves were characterized by a Hill coefficient <1, indicating that the regulation of CRE-dependent activity by the cannabinoid agonists could involve the combination of distinct but converging signalling pathways. Although cannabinoid receptors have been shown to regulate several types of G proteins, this complexity could merely involve different members of the family of Gi/o proteins.
The AP-1 transcription factor is best characterized as a family of protein heterodimers encoded by defined immediate early genes, such as c-Fos and JunB. Therefore, AP-1 activity is dependent on the level of immediate early gene expression and the phosphorylation of the different subunits by PKC, ERK or c-Jun N-terminal kinase. As cannabinoid receptors are known to activate ERK1/2 and c-Jun N-terminal kinase, and regulate the expression of immediate early genes, the effects of cannabinoid agonists on AP-1-dependent transcriptional activity were characterized. Several studies have already shown cannabinoid-mediated regulations of AP-1 activity with inconsistent responses reflecting either increased or decreased in AP-1 DNA binding and/or AP-1-controlled transcription (Porcella et al., 1998; Faubert and Kaminski, 2000; Zhao et al., 2005; Giuliano et al., 2006). Here, the direct measure of AP-1-dependent activity revealed a reduction of AP-1-mediated transcription in neuroblastoma cells, consecutively to HU210 stimulation. Accordingly, anandamide-mediated inhibition of AP-1 activation in a CB1 receptor- and PKC-dependent mechanism was previously demonstrated (Maccarrone et al., 2003). Likewise, HU210-mediated inhibition of AP-1-driven luciferase activity was efficiently inhibited by PKC inhibitors and antagonized using SR141716A. Although regulation of PKC-related pathways by cannabinoids remains poorly documented, several studies have already suggested a role of this kinase in cannabinoid receptor-mediated reduction of potassium currents (Hampson et al., 2000) and regulation of L-type voltage dependent calcium channels (Rubovitch et al., 2002). In addition, demonstrations that CB1 receptors may functionally interact with Gq/11-type G protein in response to (R)-(+)-(2,3-dihydro-5-methyl-3-(4-morpholinylmethyl)-pyrrolo-(1,2,3-de)-1,4-benzoxazin-6-yl)-1-naphthalenylmethanone mesylate (WIN55212-2) (Lauckner et al., 2005) and efficiently regulate PLC activity (Sugiura et al., 1996, 1997; Ho et al., 1999) are compelling indications for a CB1 receptor-mediated regulation of PKC.
The absence of CP55940-mediated regulation of AP-1-dependent transcription clearly shows that distinct signalling cascades contribute to the modulation of CRE- and AP-1-dependent activities. In addition, these data support the concept of functional selectivity of agonists acting at the CB1 receptor. Indeed, functional selectivity of receptor signalling is characterized by the ability of distinct ligands acting at a single G-protein-coupled receptor to differentially control intracellular signal transduction pathways. Thus, functionally selective ligands activate or inhibit multiple intracellular cascades with different efficacies or potencies (Bosier and Hermans, 2007). Such selectivity at the level of regulation of AP-1-dependent transcriptional activity by cannabinoid agonists is an unprecedented observation. Although the expression of the CB1 receptor in this model of neuroblastoma cells remains poorly characterized, we have accumulated convincing pharmacological data showing that both agonists tested act on a single molecular target identified as the CB1 receptor. It is noteworthy that both agonists promote reduction of CRE-dependent luciferase activity with similar potencies and decrease cAMP. This is also consistent with the data showing the possibility to competitively antagonize the HU210-mediated regulation of AP-1-dependent response by CP55940. Importantly, the pA2 value determined for CP55940 in this competition assay is consistent with its published nanomolar affinity for the CB1 receptor. The detection of signalling specificity with some drugs, as shown here for CP55940, could be purely related to their partial agonist profile. For these drugs, the density of receptors and the availability of defined signalling partners could influence the apparent efficacy in functional assays. However, to our knowledge, the vast majority of pharmacological studies have identified CP55940 as a full agonist of the CB1 receptor. In spite of repeated attempts, we failed to find cannabinoid agonist-induced [35S]-GTPγS binding to G proteins in this model of neuroblastoma cells, suggesting that the expression of the CB1 receptor is not sufficient to allow the detection of significant responses. Therefore, we were unable to characterize the potencies and efficacies of CP55940 and HU210 at the earliest steps of the signalling cascade in this model. Nevertheless, in our hands, both ligands were characterized as full agonists of the CB1 receptor in [35S]-GTPγS-binding assays conducted on mouse cerebellar membrane preparations (pEC50: 7.77±0.72 and 7.80±0.18; Emax: 173±1 and 176±2% stimulation over basal, for HU210 and CP55940, respectively; Bosier et al., unpublished data).
Although functional selectivity at G-protein-coupled receptors is extensively documented in the literature, little is known regarding the CB1 receptor, and most available studies have been focused on coupling with different members of the Gi/o protein family (Glass and Northup, 1999; Prather et al., 2000; Mukhopadhyay and Howlett, 2005). Nevertheless, the ability of different CB1 receptor agonists to differently activate Gi/o or Gs-dependent signalling pathways was described in recombinant systems (Bonhaus et al., 1998). To our knowledge, this study provides the first evidence for functional selectivity at the native CB1 receptor in a neural cell line, reflected by an agonist-selective response monitored at the transcriptional level. In a closely related study conducted in the same cell line, we recently reported the complex regulation of TH by CB1 receptor agonists (Bosier et al., 2007). Importantly, HU210 or CP55940 was shown to decrease or increase the expression of TH, respectively, raising the question of an agonist-selective trafficking of CB1 receptor signalling. Earlier studies demonstrated that AP-1 and CRE motifs present in the rat TH gene promoter were involved in the pharmacological modulation of transcription (Kim et al., 1993; Najimi et al., 2002; Lewis-Tuffin et al., 2004). Therefore, identifying an agonist-dependent differential regulation of TH expression in a catecholamine-producing cell line, highlights the putative relevance of functional selectivity.
Inhibition or reversal of the responses to the cannabinoid agonists in PTx-treated cells suggests that both CRE and AP-1 activities are regulated through activation of Gi/o-type G proteins, in accordance with the documented predominant coupling of the CB1 receptor. The unexpected reduction of basal cAMP level observed after overnight treatment with PTx could reveal some influence of this toxin on the metabolism or proliferation of N1E-115 cells. However, forskolin was found to induce equivalent increases in cAMP accumulation in cells with or without exposure to PTx (725.08±55.97 and 728.58±41.15% stimulation, relative to control), suggesting that the pharmacological modulation of AC and associated signalling were not affected. Taking advantage of an internal control plasmid (encoding for Renilla luciferase) that allowed us to normalize for cell density, an enhanced basal luciferase activity in PTx-treated cells was detected, confirming the stimulation of both CRE and AP-1-dependent transcription by some constitutive activation of Gi/o-type G proteins in N1E-115 cells. In addition, the inverse agonist, SR141716A, also increased luciferase activity (not shown), indicating that a constitutive coupling of the CB1 receptor participates in the activation of CRE and AP-1 in the absence of agonist.
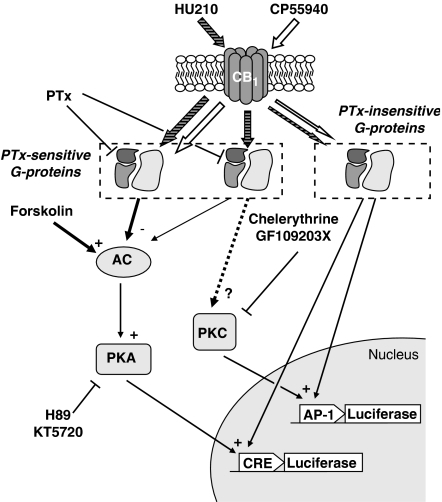
The putative interaction of the CB1 receptor with several isoforms of Gi/o proteins could certainly hold clues for the diversity in intracellular signalling triggered by cannabinoid ligands and explain the results observed with HU210 and CP55940 (Figure 6). On the other hand, although the aim of the study was not to examine the diversity of couplings with different types of G proteins, our data suggest that in the absence of functional Gi/o protein, the CB1 receptor may operate atypical coupling with other G proteins. Indeed, increased cAMP accumulation and stimulation of CRE-driven luciferase activity in PTx-treated cells indicate that CB1 receptors also exert a positive influence on AC through a putative coupling with Gs-type G protein. Besides, considering the consequence of PTx pretreatment on HU210 and CP55940-mediated regulation of AP-1 activity, the possibility of coupling with members of the Gq/11 proteins family cannot be excluded.
Figure 6.
Putative signalling pathways associated with regulation of CRE and AP-1-controlled transcription by the CB1 receptor. Through stabilization of different active conformations of the receptor, HU210 and CP55940 could selectively promote coupling with several types of G proteins, supporting activation/inhibition of distinct signalling pathways. On the one hand, the high affinity agonists HU210 (striped arrow) and CP55940 (open arrow) mediate inhibition of CRE-dependent transcription through activation of Pertussis toxin-sensitive G proteins and related inhibition of AC and PKA activity, as confirmed by the use of PKA inhibitors. On the other hand, only HU210 regulates AP-1-dependent transcription via an unresolved mechanism involving the activity of PKC, as suggested by the use of appropriate inhibitors. In addition, experimental observations demonstrate the possible interaction of CB1 receptors with PTx-insensitive G proteins. Although these alternative couplings exert a modest influence with comparison to the response mediated by Gi/o proteins, activation of PTx-insensitive G proteins by both HU210 and CP55940 is thought to equally promote CRE and AP-1-controlled transcription.
In conclusion, the present results indicate that two cannabinoid ligands, commonly used as reference agonists at the CB1 receptor, may differentially influence gene transcription through distinct controls on AP-1 cis-enhancer DNA element. Supporting the concept of functional selectivity, these data suggest that the choice of appropriate ligands acting on a given receptor should help to selectively modulate defined cellular responses. In addition, the present observations clearly indicate that ligands, with similar pharmacodynamic properties and eliciting similar regulation of classical immediate effectors, may trigger divergent delayed responses. This should encourage the systematic study of a wide variety of early and late responses to drugs to better define their therapeutic potential or explain the differences in their clinical profiles.
Acknowledgments
This study was supported by grants from the National Fund for Scientific Research (FNRS, Crédit au chercheur 1.5303.04) and from UPSA (Institut de la douleur). EH is the Research Director of the FNRS.
Abbreviations
- AC
adenylyl cyclase
- AP-1
activator protein 1
- CP55940
(1R,3R,4R)-3-(2-hydroxy-4-(1,1-dimethylheptyl) phenyl)-4-(3-hydroxypropyl)cyclohexan-1-ol
- CRE
cAMP response element
- CREB
cAMP response element-binding protein
- ERK
extracellular-signal regulated kinase
- HU210
(6,R)-trans-3-(1,1-dimethylheptyl)-6,7,10,10,-tetrahydro-1-hydroxy-6,6-dimethyl-6H-dibenzo(b,d)pyran-9-methanol
- MAPK
mitogen-activated protein kinase
- MEK
mitogen-activated kinase kinase
- PTx
Pertussis toxin
- SR141716A
N-(piperidin-1-yl)-5-(4-chlorophenyl)-1-(2,4-dichlorophenyl)-4-methyl-1H pyrazole-3-carboxamide hydrochloride
- SR144528
N-((1S)-endo-1,3,3-trimethylbicyclo(2.2.1)heptan-2-yl)-5-(4-chloro-3-methylphenyl)-1-(4-methylbenzyl)-pyrazole-3-carboxamide
- TH
tyrosine hydroxylase
Conflict of interest
The authors state no conflict of interest.
References
- Alexander SPH, Mathie A, Peters JA.Guide to receptors and channels (GRAC) Br J Pharmacol 2008153(Suppl 2)S1–S209.3rd edn. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bash R, Rubovitch V, Gafni M, Sarne Y. The stimulatory effect of cannabinoids on calcium uptake is mediated by Gs GTP-binding proteins and cAMP formation. Neurosignals. 2003;12:39–44. doi: 10.1159/000068915. [DOI] [PubMed] [Google Scholar]
- Bonhaus DW, Chang LK, Kwan J, Martin GR. Dual activation and inhibition of adenylyl cyclase by cannabinoid receptor agonists: evidence for agonist-specific trafficking of intracellular responses. J Pharmacol Exp Ther. 1998;287:884–888. [PubMed] [Google Scholar]
- Bosier B, Hermans E. Versatility of GPCR recognition by drugs: from biological implications to therapeutic relevance. Trends Pharmacol Sci. 2007;28:438–446. doi: 10.1016/j.tips.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Bosier B, Tilleux S, Najimi M, Lambert DM, Hermans E. Agonist selective modulation of tyrosine hydroxylase expression by cannabinoid ligands in a murine neuroblastoma cell line. J Neurochem. 2007;102:1996–2007. doi: 10.1111/j.1471-4159.2007.04679.x. [DOI] [PubMed] [Google Scholar]
- Bouaboula M, Poinot-Chazel C, Bourrie B, Canat X, Calandra B, Rinaldi-Carmona M, et al. Activation of mitogen-activated protein kinases by stimulation of the central cannabinoid receptor CB1. Biochem J. 1995;312 (Part 2):637–641. doi: 10.1042/bj3120637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calandra B, Portier M, Kerneis A, Delpech M, Carillon C, Le Fur G, et al. Dual intracellular signaling pathways mediated by the human cannabinoid CB1 receptor. Eur J Pharmacol. 1999;374:445–455. doi: 10.1016/s0014-2999(99)00349-0. [DOI] [PubMed] [Google Scholar]
- Casu MA, Pisu C, Sanna A, Tambaro S, Spada GP, Mongeau R, et al. Effect of delta9-tetrahydrocannabinol on phosphorylated CREB in rat cerebellum: an immunohistochemical study. Brain Res. 2005;1048:41–47. doi: 10.1016/j.brainres.2005.04.053. [DOI] [PubMed] [Google Scholar]
- Condie R, Herring A, Koh WS, Lee M, Kaminski NE. Cannabinoid inhibition of adenylate cyclase-mediated signal transduction and interleukin 2 (IL-2) expression in the murine T-cell line, EL4.IL-2. J Biol Chem. 1996;271:13175–13183. doi: 10.1074/jbc.271.22.13175. [DOI] [PubMed] [Google Scholar]
- Faubert BL, Kaminski NE. AP-1 activity is negatively regulated by cannabinol through inhibition of its protein components, c-fos and c-jun. J Leukoc Biol. 2000;67:259–266. doi: 10.1002/jlb.67.2.259. [DOI] [PubMed] [Google Scholar]
- Giuliano M, Calvaruso G, Pellerito O, Portanova P, Carlisi D, Vento R, et al. Anandamide-induced apoptosis in Chang liver cells involves ceramide and JNK/AP-1 pathway. Int J Mol Med. 2006;17:811–819. [PubMed] [Google Scholar]
- Glass M, Felder CC. Concurrent stimulation of cannabinoid CB1 and dopamine D2 receptors augments cAMP accumulation in striatal neurons: evidence for a Gs linkage to the CB1 receptor. J Neurosci. 1997;17:5327–5333. doi: 10.1523/JNEUROSCI.17-14-05327.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass M, Northup JK. Agonist selective regulation of G proteins by cannabinoid CB(1) and CB(2) receptors. Mol Pharmacol. 1999;56:1362–1369. doi: 10.1124/mol.56.6.1362. [DOI] [PubMed] [Google Scholar]
- Hampson RE, Mu J, Deadwyler SA. Cannabinoid and kappa opioid receptors reduce potassium K current via activation of G(s) proteins in cultured hippocampal neurons. J Neurophysiol. 2000;84:2356–2364. doi: 10.1152/jn.2000.84.5.2356. [DOI] [PubMed] [Google Scholar]
- Herring AC, Faubert Kaplan BL, Kaminski NE. Modulation of CREB and NF-kappaB signal transduction by cannabinol in activated thymocytes. Cell Signal. 2001;13:241–250. doi: 10.1016/s0898-6568(01)00145-0. [DOI] [PubMed] [Google Scholar]
- Herring AC, Koh WS, Kaminski NE. Inhibition of the cyclic AMP signaling cascade and nuclear factor binding to CRE and kappaB elements by cannabinol, a minimally CNS-active cannabinoid. Biochem Pharmacol. 1998;55:1013–1023. doi: 10.1016/s0006-2952(97)00630-8. [DOI] [PubMed] [Google Scholar]
- Ho BY, Uezono Y, Takada S, Takase I, Izumi F. Coupling of the expressed cannabinoid CB1 and CB2 receptors to phospholipase C and G protein-coupled inwardly rectifying K+ channels. Receptors Channels. 1999;6:363–374. [PubMed] [Google Scholar]
- Jarrahian A, Watts VJ, Barker EL. D2 dopamine receptors modulate Gα-subunit coupling of the CB1 cannabinoid receptor. J Pharmacol Exp Ther. 2004;308:880–886. doi: 10.1124/jpet.103.057620. [DOI] [PubMed] [Google Scholar]
- Kim KS, Park DH, Wessel TC, Song B, Wagner JA, Joh TH. A dual role for the cAMP-dependent protein kinase in tyrosine hydroxylase gene expression. Proc Natl Acad Sci USA. 1993;90:3471–3475. doi: 10.1073/pnas.90.8.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh WS, Crawford RB, Kaminski NE. Inhibition of protein kinase A and cyclic AMP response element (CRE)-specific transcription factor binding by delta9-tetrahydrocannabinol (delta9-THC): a putative mechanism of cannabinoid-induced immune modulation. Biochem Pharmacol. 1997;53:1477–1484. doi: 10.1016/s0006-2952(97)82441-0. [DOI] [PubMed] [Google Scholar]
- Lauckner JE, Hille B, Mackie K. The cannabinoid agonist WIN55212-2 increases intracellular calcium via CB1 receptor coupling to Gq/11 G proteins. Proc Natl Acad Sci USA. 2005;102:19144–19149. doi: 10.1073/pnas.0509588102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis-Tuffin LJ, Quinn PG, Chikaraishi DM. Tyrosine hydroxylase transcription depends primarily on cAMP response element activity, regardless of the type of inducing stimulus. Mol Cell Neurosci. 2004;25:536–547. doi: 10.1016/j.mcn.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Maccarrone M, Di Rienzo M, Battista N, Gasperi V, Guerrieri P, Rossi A, et al. The endocannabinoid system in human keratinocytes. Evidence that anandamide inhibits epidermal differentiation through CB1 receptor-dependent inhibition of protein kinase C, activation protein-1, and transglutaminase. J Biol Chem. 2003;278:33896–33903. doi: 10.1074/jbc.M303994200. [DOI] [PubMed] [Google Scholar]
- Mackie K, Hille B. Cannabinoids inhibit N-type calcium channels in neuroblastoma-glioma cells. Proc Natl Acad Sci USA. 1992;89:3825–3829. doi: 10.1073/pnas.89.9.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie K, Lai Y, Westenbroek R, Mitchell R. Cannabinoids activate an inwardly rectifying potassium conductance and inhibit Q-type calcium currents in AtT20 cells transfected with rat brain cannabinoid receptor. J Neurosci. 1995;15:6552–6561. doi: 10.1523/JNEUROSCI.15-10-06552.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maneuf YP, Brotchie JM. Paradoxical action of the cannabinoid WIN 55,212-2 in stimulated and basal cyclic AMP accumulation in rat globus pallidus slices. Br J Pharmacol. 1997;120:1397–1398. doi: 10.1038/sj.bjp.0701101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh BT, Hudson B, Yegorova S, Jollimore CA, Kelly ME. Agonist-dependent cannabinoid receptor signalling in human trabecular meshwork cells. Br J Pharmacol. 2007;152:1111–1120. doi: 10.1038/sj.bjp.0707495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Howlett AC. Chemically distinct ligands promote differential CB1 cannabinoid receptor–Gi protein interactions. Mol Pharmacol. 2005;67:2016–2024. doi: 10.1124/mol.104.003558. [DOI] [PubMed] [Google Scholar]
- Murphy MG, Byczko Z. Effects of adenosine analogues on basal, prostaglandin E1- and forskolin-stimulated cyclic AMP formation in intact neuroblastoma cells. Biochem Pharmacol. 1989;38:3289–3295. doi: 10.1016/0006-2952(89)90627-8. [DOI] [PubMed] [Google Scholar]
- Najimi M, Robert JJ, Mallet J, Rostene W, Forgez P. Neurotensin induces tyrosine hydroxylase gene activation through nitric oxide and protein kinase C signaling pathways. Mol Pharmacol. 2002;62:647–653. doi: 10.1124/mol.62.3.647. [DOI] [PubMed] [Google Scholar]
- Ouyang Y, Hwang SG, Han SH, Kaminski NE. Suppression of interleukin-2 by the putative endogenous cannabinoid 2-arachidonyl-glycerol is mediated through down-regulation of the nuclear factor of activated T cells. Mol Pharmacol. 1998;53:676–683. doi: 10.1124/mol.53.4.676. [DOI] [PubMed] [Google Scholar]
- Pertwee RG. Pharmacology of cannabinoid receptor ligands. Curr Med Chem. 1999;6:635–664. [PubMed] [Google Scholar]
- Pierreux CE, Urso B, De Meyts P, Rousseau GG, Lemaigre FP. Inhibition by insulin of glucocorticoid-induced gene transcription: involvement of the ligand-binding domain of the glucocorticoid receptor and independence from the phosphatidylinositol 3-kinase and mitogen-activated protein kinase pathways. Mol Endocrinol. 1998;12:1343–1354. doi: 10.1210/mend.12.9.0172. [DOI] [PubMed] [Google Scholar]
- Porcella A, Gessa GL, Pani L. Delta9-tetrahydrocannabinol increases sequence-specific AP-1 DNA-binding activity and Fos-related antigens in the rat brain. Eur J Neurosci. 1998;10:1743–1751. doi: 10.1046/j.1460-9568.1998.00175.x. [DOI] [PubMed] [Google Scholar]
- Prather PL, Martin NA, Breivogel CS, Childers SR. Activation of cannabinoid receptors in rat brain by WIN 55212-2 produces coupling to multiple G protein alpha-subunits with different potencies. Mol Pharmacol. 2000;57:1000–1010. [PubMed] [Google Scholar]
- Rubino T, Sala M, Vigano D, Braida D, Castiglioni C, Limonta V, et al. Cellular mechanisms underlying the anxiolytic effect of low doses of peripheral Delta9-tetrahydrocannabinol in rats. Neuropsychopharmacology. 2007;32:2036–2045. doi: 10.1038/sj.npp.1301330. [DOI] [PubMed] [Google Scholar]
- Rubovitch V, Gafni M, Sarne Y. The cannabinoid agonist DALN positively modulates L-type voltage-dependent calcium-channels in N18TG2 neuroblastoma cells. Brain Res Mol Brain Res. 2002;101:93–102. doi: 10.1016/s0169-328x(02)00174-2. [DOI] [PubMed] [Google Scholar]
- Rueda D, Galve-Roperh I, Haro A, Guzman M. The CB(1) cannabinoid receptor is coupled to the activation of c-Jun N-terminal kinase. Mol Pharmacol. 2000;58:814–820. doi: 10.1124/mol.58.4.814. [DOI] [PubMed] [Google Scholar]
- Stenstrom S, Seppala M, Pfenning M, Richelson E. Inhibition by ethanol of forskolin-stimulated adenylate cyclase in a murine neuroblastoma clone (N1E-115) Biochem Pharmacol. 1985;34:3655–3659. doi: 10.1016/0006-2952(85)90226-6. [DOI] [PubMed] [Google Scholar]
- Sugiura T, Kodaka T, Kondo S, Nakane S, Kondo H, Waku K, et al. Is the cannabinoid CB1 receptor a 2-arachidonoylglycerol receptor? Structural requirements for triggering a Ca2+ transient in NG108-15 cells. J Biochem (Tokyo) 1997;122:890–895. doi: 10.1093/oxfordjournals.jbchem.a021838. [DOI] [PubMed] [Google Scholar]
- Sugiura T, Kodaka T, Kondo S, Tonegawa T, Nakane S, Kishimoto S, et al. 2-Arachidonoylglycerol, a putative endogenous cannabinoid receptor ligand, induces rapid, transient elevation of intracellular free Ca2+ in neuroblastoma × glioma hybrid NG108-15 cells. Biochem Biophys Res Commun. 1996;229:58–64. doi: 10.1006/bbrc.1996.1757. [DOI] [PubMed] [Google Scholar]
- Zhao Q, He Z, Chen N, Cho YY, Zhu F, Lu C, et al. 2-Arachidonoylglycerol stimulates activator protein-1-dependent transcriptional activity and enhances epidermal growth factor-induced cell transformation in JB6 P+ cells. J Biol Chem. 2005;280:26735–26742. doi: 10.1074/jbc.M412828200. [DOI] [PMC free article] [PubMed] [Google Scholar]