Abstract
Spherical high density lipoproteins (HDL)† predominate in human plasma. However, little information exists on the structure of the most common HDL protein, apolipoprotein (apo) A-I, in spheres vs. better studied discoidal forms. We produced spherical HDL by incubating reconstituted discoidal HDL with physiological plasma-remodeling enzymes and compared apoA-I structure in discs and spheres of comparable diameter (79–80 and 93–96 Å). Using cross-linking chemistry and mass spectrometry, we determined that the general structural organization of apoA-I was overall similar between discs and spheres, regardless of diameter. This was the case despite the fact that the 93 Å spheres contained three molecules of apoA-I per particle compared with only two in the discs. Thus, apoA-I adopts a consistent general structural framework in HDL particles—irrespective of shape, size and the number of apoA-Is present. Furthermore, a similar cross-linking pattern was demonstrated in HDL particles isolated from human serum. We propose the first experiment-based molecular model of apoA-I in spherical HDL particles. This model provides a new foundation for understanding how apoA-I structure modulates HDL function and metabolism.
Keywords: sphere, disk
Given the inverse correlation between high density lipoprotein (HDL) levels and cardiovascular disease, a key question in vascular biology relates to how apolipoproteins modulate the metabolism and function of HDL. Significant evidence supports a role for HDL in the process of reverse cholesterol transport whereby lipids and cholesterol in the vessel wall are transported to the liver for catabolism. However, because of a lack of information on HDL structure and the molecular basis of its interactions with other proteins, our understanding of HDL metabolism and function is at a basic stage.
The “glue” that holds most HDL particles together is apolipoprotein (apo)A-I, a highly α-helical, 28-kDa polypeptide. It comprises some 70% of HDL protein mass, making it the clear starting point for deriving a basic understanding of HDL structure. In humans, apoA-I is primarily present in two major spherical HDL species, HDL2 (d = 1.063–1.125 g/ml) and HDL3 (d = 1.125–1.210 g/ml) with diameters ranging from 70 to 120 Å. More minor, but clearly important, HDL subspecies include lipid-poor apoA-I and nascent discoidal particles (reviewed in ref. 1). Highly reactive but low abundance discoidal HDLs are critical intermediates between lipid-poor apoA-I and mature spherical HDL. Easily produced in vitro, they have been heavily used for structural studies (2). Despite some debates on details of certain regions of apoA-I in discs, the majority of recent theoretical and experimental data supports the so-called “double belt” model (3). In this scheme, each of two ring-shaped apoA-I molecules wrap around a leaflet of a disk-like patch of lipid bilayer in an anti-parallel orientation.
Despite their abundance in plasma, much less is known about the structure of apoA-I in spherical particles. They contain a neutral lipid core composed of triglyceride and cholesteryl ester—encapsulated by a phospholipid and cholesterol monolayer—and are stabilized by surface apos (4). In the late 1990s, Borhani et al. (5) generated an x-ray crystal structure of a lipid-free apoA-I fragment, which they interpreted to represent a lipid-bound-like conformation. They hypothesized a “Faberge Egg” model for spherical HDL, suggesting that the same intermolecular contacts found in the crystal structure (and in the double belt model in discs) are also present in HDL spheres. This agrees with the predictions of Jonas et al. (6) who argued that spherical particles should have defined intermolecular protein interactions due to quantized diameter formation in vitro. However, other studies have shown differences in fluorescence emission (7), particle charge (8), and proteolytic sensitivity (9) between discs and spheres. It is not yet clear whether these effects are due to localized structural changes or whether they represent a global reorganization of apoA-I between the two shapes.
Because HDL particle shape can affect the activities of HDL remodeling factors such as lecithin cholesteryl acyl transferase (LCAT) (6) and cholesteryl ester transfer protein (CETP) (10), understanding apoA-I spatial arrangement in both discs and spheres is critical. Unfortunately, few published reports have experimentally addressed spherical HDL. We previously combined cross-linking chemistry with mass spectrometry (MS) to provide distance constraints for deriving and testing models for apos in apoA-I discs (11). Our results strongly supported the double belt model described above. Here, we applied the same approach to systematically compare experimental cross-link distance constraints between well-defined rHDL discs and spheres of similar diameter containing only apoA-I. The results indicate that the double belt model is a common organizational motif for apoA-I in both discs and spheres.
Results
Strategy.
We elected to study two sets of HDL particles with diameters of approximately 79–80 Å and 93–96 Å, a size range comparable to human plasma HDL (4). A large body of work from the Jonas (12) and Rye (13) laboratories shows that discoidal and spherical apoA-I particles can be generated with these two diameters. Discs can be generated by varying the ratio of synthetic phospholipids to apoA-I, using a modification of the cholate dialysis technique (11, 14). To generate HDL spheres, we felt it was important to use methods that are as physiological as possible. Bypassing commonly used sonication methods (12), we incubated a 96-Å reconstituted disk with human LCAT and low density lipoprotein (LDL) as a phospholipid donor (15) — the same reaction that matures nascent HDL in human plasma (1). This resulted in a spherical particle 93 Å in diameter. In turn, this large sphere was converted to a smaller sphere of 80 Å by incubating with CETP and Intralipid (15). These particles represent a viable compromise between a physiological mode of production and the necessary control over apo composition required for detailed structural studies. The characterized HDL particles were chemically cross-linked on near-neighbor Lys residues under native conditions in buffer. After lipid extraction and trypsinization, cross-linked peptides were identified by high resolution MS. Throughout the manuscript, “small” discs and spheres of ≈79–80 Å are designated D79 and S80, respectively; “large” discs and spheres of 93–96 Å are signified as D96 and S93.
Comparing Small Discs and Spheres.
The physical characteristics of the particles generated for this study are listed in Table 1. By polyacrylamide gradient gel electrophoresis (PAGGE), the discs exhibited a hydrodynamic diameter of 79 Å (11, 14) (Fig. 1A). They contained phospholipid and apoA-I only. Without neutral lipids, the particles demonstrated clear rouleaux formation when analyzed by negative stain electron microscopy (EM) (Fig. 1B), suggesting that they are discoidal in solution. EM histograms showed diameter distributions of 79 ± 11 Å consistent with the native PAGGE estimation [supporting information (SI) Fig. S1]. D79 have exactly two apoA-I molecules per particle (11).
Table 1.
Characterization of discoidal and spherical HDL particles studied
| Particle | Diameter, Å (± 3)* | Particle shape/size by EM (Å)† | No: of apoA-I per particle‡ | Composition PC/FC/CE/TG/A-I§ | Helicity¶ |
|---|---|---|---|---|---|
| D79 | 79 | 79 ± 11 | 2 | 30/0/0/0/1 | 79.8 |
| S80 | 80 | 76 ± 11 | 2 | 43/3/6/5/1 | 76.2 |
| D96 | 96 | 97 ± 16 | 2 | 78/0/0/0/1 | 78.0 |
| S93 | 93 | 92 ± 13 | 3 | 34/1/17/0/1 | 77.0 |
*Average apparent particle hydrodynamic diameters were calculated based on the particle migration distance on native PAGGE (Figs 1 and 2).
†The diameters calculated by EM were based on averaging 200 randomly chosen particles (SI Methods).
‡Number of apoA-I molecules per particle was determined by particle cross linking followed by PAGGE analysis. Each value represents averaged data from at least two independent particle preparations with the exception of S80 (n = 1).
§Particle compositions were calculated as described in the Methods after particle purification by gel filtration (SI Methods).
¶Helicity was calculated by subjecting particles solutions to circular dichroism spectroscopy followed by analysis using the SELCON 3 algorithm (SI Methods).
Fig. 1.
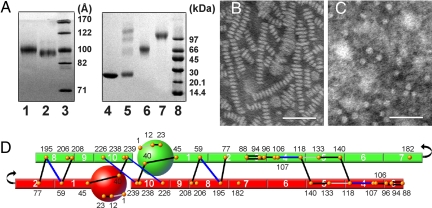
Structural analysis of D79 and S80 particles. (A) Native PAGGE (8–25%) of D79 (lane 1) and S80 (lane 3) particles. An 8–25% SDS/PAGGE analysis of cross-linked S80 after separation into monomeric (lane 5) and dimeric (lane 6) forms by gel filtration chromatography is also shown. Cross-linking was carried out at a 1:10 molar ratio of apoA-I: BS3 at 1 mg/ml apoA-I concentration. All gels were stained with Coomassie blue. (B and C) Negative stain electron micrographs of D79 and S80 particles. (Scale bars: 50 nm.) (D) Experimental cross-links compatible with 5/5 molecular registry of the double-belt model are shown as solid lines. The apoA-I molecules have been drawn as if they have been peeled off the edge of an HDL disk and laid flat. Cross-links in black are common to both D79 and S80 particles; those in blue are found in D79 only and those in gray are found in S80 only. Putative apoA-I helical segments are numbered according to Roberts et al. (31). Locations of the 21 Lys residues in apoA-I are identified as orange dots. All experimental cross-links found in D79 and S80 are listed in Table S1.
The spherical particles, S80, exhibited a nearly identical migration distance to D79 by native PAGGE (Fig. 1A). Unlike the discs, they contained free cholesterol, triglyceride and cholesteryl ester in addition to phospholipid and apoA-I (Table 1). As a result, they appeared as discrete circular structures by EM. There was no evidence of discoid or rectangular shapes that would indicate discs, even when visualized at high concentrations (Fig. 1C), suggesting a spherical shape in bulk solution. S80 particles were distributed about a mean of 76 ± 11 Å (Fig. S1). Cross-linking of S80 with BS3 resulted in a diffuse 56-kDa apoA-I dimer band by SDS/PAGGE (Fig. 1A, lane 6), indicating two molecules of apoA-I like the D79 particles.
The particles were cross-linked and analyzed by MS. All resulting cross-linked peptide pairs and involved Lys residues are listed in Table S1. The most informative for determining the spatial arrangement of apoA-I molecules on a particle are the intermolecular cross-links. We have previously proposed that the majority of observed intermolecular D79 cross-links are consistent with the 5/5 LL double belt model (11) as described by Segrest (3). Fig. 1D shows a two-dimensional representation of two anti-parallel apoA-I molecules in this configuration with these cross-links in D79 and S80 compared. Interestingly, the majority of the identified intermolecular cross-links were found in both the discs and spheres. The few that were unique to either the sphere or the disk seemed to be clustered either near the termini or near the junction of helices 4 and 5. These are the same regions that displayed cross-link differences between 96Å and 78 Å discoidal particles in our previous studies (11, 14). Overall, these data indicate that apoA-I molecules interact similarly when present on a disk or a sphere with a diameter of ≈80 Å.
Comparing Large Discs and Spheres.
The larger D96 discs exhibited the expected hydrodynamic diameter of 96 Å as well known (Table 1 and Fig. 2A) (14, 16, 17). Containing only phospholipid and apoA-I, they produced clear rouleaux by negative stain EM (Fig. 2B). Image analysis showed a normal distribution about a mean of ≈97 ± 16 Å, consistent with native PAGGE (Fig. S1).
Fig. 2.
Structural analysis of D96 and S93 particles. (A) Native PAGGE (8–25%) of D96 (lane 1) and S93 (lane 2). An 8–25% SDS/PAGGE analysis of cross-linked D96 (lane 6) and S93 (lane 7) is also shown. The particles were cross-linked at a 1:100 molar ratio of apoA-I: BS3 at 1 mg/ml apoA-I for PAGGE analysis to avoid partial cross-linking and to detect the highest MW bands. Lipid-free apoA-I (lane 4) and lipid-free apoA-I cross-linked under the same conditions (lane 5) is shown for comparison. All gels were stained with Coomassie Blue. (B and C) Negative stain EM images of D96 and S93 particles, respectively. (Scale bars: 50 nm.) (D) Experimental cross-links compatible with 5/5 molecular registry of the double-belt model are shown as for Fig. 1. Cross-linker color coding is the same as in Fig. 1.
The large spherical particles migrated slightly faster than the discs in a native gel (Fig. 2A), corresponding to a diameter of ≈93 Å. The S93 particles contained a cholesteryl ester rich core of ≈20% of the particle volume, which is comparable with the denser fractions of human HDL3 (18) (Table 1). They appeared to be uniformly spherical with no discoidal shapes by negative stain EM (Fig. 2C Left) and were normally distributed about a mean of 92 ± 13 Å, consistent with native PAGGE. To further confirm the particle shape, we used electron Cryomicroscopy (Cryo-EM) to visualize S93 particles in a frozen-hydrated native state. Imaging was performed at three different tilt angles (−45°, 0° and + 45°), allowing different views of the same field of particles (Fig. S2). The particles were clearly spherical or slightly oval with no evidence of swollen discs or prolate shapes. Cross-linking with BS3 resulted in diffuse band at ≈100 kDa by SDS/PAGGE (Fig. 2A, lane 7). Comparing this band to oligomeric lipid-free apoA-I cross-linked under similar conditions (Fig. 2A, lane 5), it is clear that the S93 particles contained three molecules of apoA-I. This is in contrast to the D96 and D78 particles, which contained only two (Fig. 2A, lane 6) (6, 7).
Fig. 2D compares the cross-linking patterns between S93 and D96 particles. Again, most of the diagnostic intermolecular cross-links were shared between the two particle shapes, suggesting that the apoA-I spatial arrangements were generally similar between the D96 and S93 particles, despite the additional apoA-I molecule in the S93 particles.
To translate our observations to human plasma HDL particles, we isolated the human HDL3a (d = 1.123 g/ml) subfraction from the plasma of a normal human donor by isopycnic density ultracentrifugation (18) and cross-linked as with S80 and S93. Particles in this fraction ranged from ≈85 to 110 nm in diameter by native PAGE (data not shown) and contained an average of 23% CE and 3% TG by weight. Of the 23 cross-links identified in the in vitro produced S93 particles, 21 were observed in the in vivo produced HDL3a preparation (Table S1).
Figs. 1D and 2D highlight cross-links that we have previously interpreted to be consistent with the 5/5 double belt model. Throughout our studies over the past 5 years, we have consistently observed a cross-link population that may be compatible with a alternate model in which helix 5 of one molecule aligns with helix 2 of the second (LL5/2). Although these tend to be of lower intensity, we found cross-links compatible with this registry in all four particle preparations in this study (Fig. S3) and in the human HDL3a particles (Table S1). Moreover, we found 3 cross-links in S80 and S93 that involved the N-terminal Asp of apoA-I. The structural significance of these is unclear, but they may indicate an increased flexibility of the apoA-I N terminus in spherical particles.
Discussion
An HDL disk is essentially a circular patch of phospholipid (PL) bilayer stabilized by amphipathic apolipoproteins protecting fatty acyl side-chains from water. Conversion to a sphere occurs when neutral lipid esters, accumulated principally via LCAT and CETP, intercalate between the bilayer leaflets. As this occurs, the particle surface becomes a continuous phospholipid monolayer without an “edge” for the apos to protect. Thus, it is tempting to imagine that the amphipathic helices of apoA-I might spread out across the sphere, perhaps disengaging from the protein:protein contacts observed in discs. However, our current data show overall similar apoA-I intermolecular cross-linking patterns in discs and spheres of matched diameter and in human plasma HDL. A straightforward interpretation is that the double belt model represents a common structural framework for apoA-I in both discs and spheres.
Although potentially surprising, we believe that this conclusion is supported by three major arguments. First, the “spreading out” concept described above presupposes that it is the lipid that forces apoA-I molecules together at the disk edge. However, there is evidence that protein:protein attractions may be more important for maintaining the apoA-I spatial arrangements in HDL. In the original double belt theoretical analysis by Segrest (3), up to 21 intermolecular salt bridges were possible between adjacent amphipathic helices contributed by two apoA-I molecules. This registry exhibited the highest salt bridge potential of all theoretically possible registries. Although salt bridges are not usually considered strong molecular forces for maintaining protein structure, Segrest argued that the lower dielectric at the lipid interface should enhance intermolecular attraction. There are also likely to be significant hydrophobic interactions between the helices. Cross-linking studies (11, 17) and resonance coupling experiments (16) clearly support a general 5/5 registry between apoA-I molecules on a disk. Thus, protein:protein interactions can maintain specific molecular registries between apoA-I belts on HDL discs.
Second, there is little evidence that apoA-I helices interact differently with phospholipids in a disk vs. a sphere. ApoA-I contains repeating amphipathic helices, most of which are class A. They have a hydrophilic face with acidic residues arrayed opposite a hydrophobic face and Arg and Lys residues at the non-polar/polar interface (19). These basic residues may interact both with the aliphatic acyl chains and the phospholipid headgroups (20). This “snorkeling” should occur regardless of whether a given amphipathic helix is on a disk edge or floating in a continuous PL surface on a sphere (21). Therefore, if the lipid:protein contacts are similar in discs and spheres, it is difficult to imagine that protein:protein contacts should be disrupted when particle shape changes.
Third, recent studies have blurred the traditional distinction between HDL discs and spheres. Segrest et al. (22) performed an in silico analysis in which a 96-Å disk (similar to D96) was gradually stripped of PL. As they shrunk, the particles lost their disk morphology and became twisted pseudospheroids. However, the basic protein:protein contacts were essentially preserved, as were the lipid:protein contacts. This is consistent with our cross-linking studies of ≈78Å discs showing a similar double belt organization as 96-Å discs (11). More recent simulations addressed spherical particles with a neutral lipid core. Again, it was theorized that double belt-like contacts could exist in spheres, although apoA-I might twist across the sphere like the seams of a baseball (23). Also, Weisgraber et al. demonstrated convincingly that apoE/DPPC particles, which by composition should be discoidal, are clearly spheroidal by x-ray scattering (24). Thus, there may be little fundamental difference in the forces mediating apoA-I intermolecular contacts in a particle containing PL only and one with a neutral lipid core.
Given these arguments, our observations may not be so surprising. We propose that the double belt model represents a general structural framework adopted by all lipidated forms of apoA-I, regardless of shape (with the possible exception of certain lipid-poor forms (25)). We further propose that localized changes within this framework, changes that might be difficult to detect by the relatively low-resolution cross-linking methodology, could be responsible for observed differences in LCAT and CETP activation between discs and spheres. For example, a recent study by Curtiss et al. showed that the manipulation of the TG to CE ratio of the HDL neutral lipid core can affect the expression of monoclonal antibody epitopes on apoA-I (26). Antibody binding, although not able to define intermolecular contacts, is likely to be more sensitive to such localized conformational changes that may be critical for modulating interactions with plasma enzymes and transfer proteins.
ApoA-I on Small Discs and Spheres.
Consistent with the arguments of Segrest, we suggest that apoA-I probably twists itself into a conformation that allows a smaller diameter in the D79 vs. the D96 particles, likely involving an atypical PL bilayer structure. With the addition of neutral lipid in the S80 particles, a filling of the core might relax strain on the phospholipids, but apoA-I registry appears to remain similar. Alternatively, one could invoke one or more hinge domains in both the D79 and S80 particles to account for the reduced diameter vs. the 96 Å particles (14, 16, 27). We should also note that the S80 particles contained ≈12% neutral lipid core lipids by mass, less than the 17% measured in human HDL3c particles of similar diameter (18). Therefore, it is possible that these represent pseudospheroids. However, the similar cross-link pattern between S80 and S93 supports our assertion that the double-belt contacts are a common theme for apoA-I organization, despite the shape of the host HDL particle.
Trefoil Model for Three apoA-I Molecules on Large Spheres.
The similarity of the cross-linking pattern between D96 with two apoA-I molecules, and S93 with three was intriguing. It follows that all three molecules in the sphere must make contacts that are similar to those made by two molecules in the double belt. How can this happen? We envisioned three potential models that can account for this observation (Fig. 3). In the first scenario, two apoA-I's are arranged in a double belt around the equator of the sphere with the third arranged separately as a hairpin (Fig. 3B). Although we cannot distinguish between intra- and intermolecular cross-links on particles with >2 apoA-I molecules, we feel that this model is unlikely. During cross-linking, the kinetics of trimer formation for S93 particles were similar to the appearance of the dimer in D96 by SDS/PAGGE (data not shown). This suggests that all three molecules exist in the same spatial relationship, rather than one sitting off on its own. In a second scenario, all three apoA-I molecules wrap around the equator in a stacked anti-parallel arrangement, i.e., a triple belt (Fig. 3C). Although we cannot rule this out, we feel that this is less likely because the third molecule must interact in a different docking interface (3) than the other two.
Fig. 3.
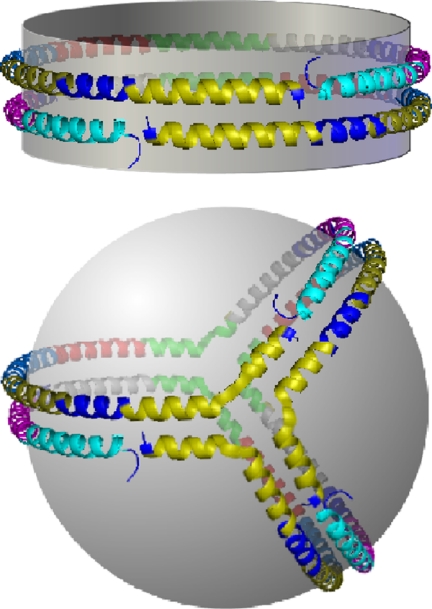
Potential molecular arrangements of three apoA-I molecules on the larger HDL spherical particles. The classical 5/5 double belt model proposed for D96 is shown first with two molecules of apoA-I shown in red and green (model A). The small spheres represent the N-terminal 44 aa of each molecule. Model B shows two molecules of apoA-I arranged in a double belt as in the discs with a third molecule arranged as a hairpin on one hemisphere. Model C has all three molecules arranged in an antiparallel fashion at the particle equator, i.e., triple belt. Model D, called the trefoil model, was generated by splitting the right hand half of two molecules of apoA-I in the double belt 60° out of the plane of the disk. Then a third molecule, bent the same way, was inserted.
We favor a third scenario, which we call the trefoil model (Fig. 3D). This arrangement can be generated a priori from the basic disk double belt model. If an inflection point is introduced in helices 5 and 10 of both apoA-I molecules, then half of each ring can be bent 60° out of the plane of the particle. A third molecule, bent the same way, can be placed between the original two. This breaks the PL surface into three equal slices subtending 120° of the sphere surface. In the vernacular of sphere geometry, these are called lunes or biangles. This model offers a degree of parsimony in that all three molecules exist in identical conformations. Importantly, each of the cross-links and/or salt bridge interactions found in discs with two apoA-I molecules are still possible in the trefoil (Fig. 4), accounting for the similar cross-linking pattern between the D96 and S93 particles. Also, the equal PL lunes provide a rationale for how apoA-I can maintain such high lipid surface curvature in HDL sized particles. We have built a theoretical computer model of the Trefoil and verified the plausibility of the intermolecular cross-links discussed above.
Fig. 4.
Molecular comparison of the apoA-I interactions in a disk double belt vs. the spherical trefoil. Each putative helical domain shown in Fig. 1 is represented as a separate color; helix 1, teal; helix 2 purple; helix 3, dark blue; helix 4, gray; helix 5, green; helix 6, red; helix 7, light blue; helix 8, dark yellow; helix 9, navy blue; helix 10, yellow. Notice that all helix to helix interactions present in the double belt between two molecules of apoA-I in the disk are also present between three apoA-I molecules in the trefoil. For example, helix 10 of all three molecules in the trefoil (the yellow one in front) can interact with helix 10 of the other molecules in the same way as the double belt. Another example is the similar arrangements of helix 9 (navy blue) and helix 1 (teal) in both models.
Human Plasma HDL.
The fact that HDL3a isolated from human plasma showed cross-linking patterns that were highly consistent with S93 is strong evidence that the structural information gleaned from synthetic particles is relevant to native HDL. However, despite the tight density cut used to isolate HDL3a, this fraction is still compositionally heterogeneous, containing uncertain apoA-I stoichiometries and neutral lipid core compositions on a per particle basis. Although we believe our data indicate that the intermolecular registries found in the double belt model represent a general organizing motif that applies also to native plasma HDL, we leave open the possibility of lower abundance conformations with potentially important biological functions. More detailed applications of this methodology to the study of native plasma HDL are clearly warranted.
Relevance to HDL Biology.
Although this topic is surprisingly understudied, common wisdom holds that human plasma HDL2 and HDL3 particles contain between 2 and 5 molecules of apoA-I per particle. A satisfying feature of the trefoil model is that it allows for the intercalation of additional apoA-I molecules with a corresponding increase in the bend angle for each resident apoA-I—each maintaining the same intramolecular interactions. Furthermore, recent proteomics studies indicate that up to 50 additional proteins can associate with HDL in widely varying abundances (28). Our models suggest that apoA-I acts as an organizing scaffold that maintains stable PL surfaces for the attachment of some of these proteins. The symmetric nature of the trefoil model may allow for optimal interactions of these proteins with apoA-I itself, perhaps to modulate HDL interplay with plasma remodeling factors or cell surface proteins. An interesting topic for future work is how the relatively abundant apoA-II impacts apoA-I organization on HDL. Finally we would point out that the protein dynamics described within the framework of the double belt such as looped regions (16), hinge domains (14), and “solar flares” (29) should work in the trefoil sphere exactly as in discs. This may explain how spherical HDL particles modulate their diameter in response to variations in lipid cargo as they circulate.
In conclusion, we suggest that the molecular contacts inherent to the original double belt model dictate a general organizational motif for apoA-I in most of its lipidated states. The proposed models, particularly the trefoil, provide a solid basis for additional experimentation aimed toward a detailed understanding of apoA-I structure and how it modulates HDL metabolism.
Methods
rHDL Particle Preparation.
Human apoA-I isolation and purification for particle preparation was carried as reported (14). The Bio-bead cholate removal method was used for the preparation of reconstituted HDL (rHDL) discs (11, 30). An initial ratio of 78:1 and 35:1 1-palmitoyl-2-oleoyl-sn-glycero-3 phosphatidylcholine (POPC):apoA-I was used for ≈96 Å and ≈79 Å disk reconstitutions, respectively. Particle purification by gel filtration chromatography was done essentially as before (11) (SI Methods). S93 spherical particles were prepared by incubating discoidal D96 rHDL with LCAT and LDL at 37°C for 24 h (6). Small, S80 spherical particles were generated by incubating S93 with CETP and intralipid (6).
rHDL Particle Characterization.
Chemical composition of the particles was determined using established assays (SI Methods). Particle hydrodynamic diameter and diameter distributions were calculated based on PAGGE and negative stain electron microscopy. The secondary structure of apoA-I in each particle was determined in solution by circular dichroism followed by SELCON 3 software, a component of CDPro software package.
Cross-Linking and MS Measurements and Data Analysis.
All rHDL particles were cross-linked at 1 mg/ml apoA-I with bis(sulfosuccinimidyl) suberate (BS3) at a molar ratio of 1:10 for MS analysis. To determine the number of apoA-I molecules per particle, the larger particles D96 and S93 were cross-linked at a 1:100 molar ratio of apoA-I:BS3 for PAGGE analysis. Cross-linked particle process for MS analysis was carried out using optimized protocols in the laboratory (SI Methods). MS measurements were performed on an Sciex/Applied Biosystems QSTAR XL. The MS analysis were carried out using commercially available and home built software.
Supplementary Material
Acknowledgments.
We thank Dr. Timothy A. Keiderling (University of Illinois at Chicago) for assistance with CD measurements and Matt Wortman for initial consultations on building the trefoil model. We also thank Cali Smith for excellent administrative assistance. This work was supported by a RO1 (HL67093) grant to W.S.D., a RO1 (HL48148) to W.G.J. and a K99/R00 (HL1004925) award to R.A.G.D.S. from the NHLBI. K.-A.R. was supported by National Health and Medical Research Council of Australia Grant 222722. Negative stain EM was carried out in the Vanderbilt University Research Electron Microscopy Resource of the Cell Imaging Core. This resource is partially supported by National Institutes of Health grants CA68485, DK20593, and DK58404. J.F. was supported by P20RR16475. G.R. was supported by W. M. Keck foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The trefoil model reported in this paper has been deposited in the Protein Model database, http://mi.caspur.it/PMDB/main.php (accession no. PM0075240).
This article contains supporting information online at www.pnas.org/cgi/content/full/0803626105/DCSupplemental.
References
- 1.Curtiss LK, Valenta DT, Hime NJ, Rye KA. What is so special about apolipoprotein AI in reverse cholesterol transport? Arterioscler Thromb Vasc Biol. 2006;26:12–19. doi: 10.1161/01.ATV.0000194291.94269.5a. [DOI] [PubMed] [Google Scholar]
- 2.Brouillette CG, Anantharamaiah GM, Engler JA, Borhani DW. Structural models of human apolipoprotein A-I: A critical analysis and review. Biochim Biophys Acta. 2001;1531:4–46. doi: 10.1016/s1388-1981(01)00081-6. [DOI] [PubMed] [Google Scholar]
- 3.Segrest JP, et al. A detailed molecular belt model for apolipoprotein A-I in discoidal high density lipoprotein. J Biol Chem. 1999;274:31755–31758. doi: 10.1074/jbc.274.45.31755. [DOI] [PubMed] [Google Scholar]
- 4.Jonas A. In: Biochemistry of Lipids, Lipoproteins and Membranes. Vance DE, Vance JE, editors. Vol. 483. Elsevier Science; 2002. p. 504. [Google Scholar]
- 5.Borhani DW, Rogers DP, Engler JA, Brouillette CG. Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. Proc Natl Acad Sci USA. 1997;94:12291–12296. doi: 10.1073/pnas.94.23.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jonas A, Wald JH, Toohill KL, Krul ES, Kezdy KE. Apolipoprotein A-I structure and lipid properties in homogeneous, reconstituted spherical and discoidal high density lipoproteins. J Biol Chem. 1990;265:22123–22129. [PubMed] [Google Scholar]
- 7.Li HH, et al. ApoA-I structure on discs and spheres. Variable helix registry and conformational states. J Biol Chem. 2002;277:39093–39101. doi: 10.1074/jbc.M206770200. [DOI] [PubMed] [Google Scholar]
- 8.Davidson WS, Sparks DL, Lund-Katz S, Phillips MC. The molecular basis for the difference in charge between pre- beta- and alpha-migrating high density lipoproteins. J Biol Chem. 1994;269:8959–8965. [PubMed] [Google Scholar]
- 9.Dalton MB, Swaney JB. Structural and functional domains of apolipoprotein A-I within high density lipoproteins. J Biol Chem. 1993;268:19274–19283. [PubMed] [Google Scholar]
- 10.Bruce C, et al. Molecular determinants of plasma cholesteryl ester transfer protein binding to high density lipoproteins. J Biol Chem. 1995;270:11532–11542. doi: 10.1074/jbc.270.19.11532. [DOI] [PubMed] [Google Scholar]
- 11.Silva RA, Hilliard GM, Li L, Segrest JP, Davidson WS. A Mass Spectrometric Determination of the Conformation of Dimeric Apolipoprotein A-I in Discoidal High Density Lipoproteins. Biochemistry. 2005;44:8600–8607. doi: 10.1021/bi050421z. [DOI] [PubMed] [Google Scholar]
- 12.Jonas A. Reconstitution of high-density lipoproteins. Methods Enzymol. 1986;128:553–582. doi: 10.1016/0076-6879(86)28092-1. [DOI] [PubMed] [Google Scholar]
- 13.Rye KA, Barter PJ. Formation and metabolism of prebeta-migrating, lipid-poor apolipoprotein A-I. Arterioscler Thromb Vasc Biol. 2004;24:421–428. doi: 10.1161/01.ATV.0000104029.74961.f5. [DOI] [PubMed] [Google Scholar]
- 14.Maiorano JN, Jandacek RJ, Horace EM, Davidson WS. Identification and structural ramifications of a hinge domain in apolipoprotein A-I discoidal high-density lipoproteins of different size. Biochemistry. 2004;43:11717–11726. doi: 10.1021/bi0496642. [DOI] [PubMed] [Google Scholar]
- 15.Rye KA, Hime NJ, Barter PJ. The influence of cholesteryl ester transfer protein on the composition, size, and structure of spherical, reconstituted high density lipoproteins. J Biol Chem. 1995;270:189–196. doi: 10.1074/jbc.270.1.189. [DOI] [PubMed] [Google Scholar]
- 16.Martin DD, Budamagunta MS, Ryan RO, Voss JC, Oda MN. Apolipoprotein A-I assumes a “looped belt” conformation on reconstituted high density lipoprotein. J Biol Chem. 2006;281:20418–20426. doi: 10.1074/jbc.M602077200. [DOI] [PubMed] [Google Scholar]
- 17.Bhat S, Sorci-Thomas MG, Alexander ET, Samuel MP, Thomas MJ. Intermolecular contact between globular N-terminal fold and C-terminal domain of ApoA-I stabilizes its lipid-bound conformation: Studies employing chemical cross-linking and mass spectrometry. J Biol Chem. 2005;280:33015–33025. doi: 10.1074/jbc.M505081200. [DOI] [PubMed] [Google Scholar]
- 18.Kontush A, Chantepie S, Chapman MJ. Small, dense HDL particles exert potent protection of atherogenic LDL against oxidative stress. Arterioscler Thromb Vasc Biol. 2003;23:1881–1888. doi: 10.1161/01.ATV.0000091338.93223.E8. [DOI] [PubMed] [Google Scholar]
- 19.Segrest JP. Amphipathic helixes and plasma lipoproteins: Thermodynamic and geometric considerations. Chem Phys Lipids. 1977;18:7–22. doi: 10.1016/0009-3084(77)90023-8. [DOI] [PubMed] [Google Scholar]
- 20.Mishra VK, Palgunachari MN, Segrest JP, Anantharamaiah GM. Interactions of synthetic peptide analogs of the class A amphipathic helix with lipids. Evidence for the snorkel hypothesis. J Biol Chem. 1994;269:7185–7191. [PubMed] [Google Scholar]
- 21.Segrest JP, Garber DW, Brouillette CG, Harvey SC, Anantharamaiah GM. The amphipathic alpha helix: A multifunctional structural motif in plasma apolipoproteins [Review] Adv Protein Chem. 1994;45:303–369. doi: 10.1016/s0065-3233(08)60643-9. [DOI] [PubMed] [Google Scholar]
- 22.Catte A, et al. Novel changes in discoidal high density lipoprotein morphology: A molecular dynamics study. Biophys J. 2006;90:4345–4360. doi: 10.1529/biophysj.105.071456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Catte A, et al. Structure of spheroidal HDL particles revealed by combined atomistic and coarse-grained simulations. Biophys J. 2008;94:2306–2319. doi: 10.1529/biophysj.107.115857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peters-Libeu CA, Newhouse Y, Hall SC, Witkowska HE, Weisgraber KH. Apolipoprotein E*dipalmitoylphosphatidylcholine particles are ellipsoidal in solution. J Lipid Res. 2007;48:1035–1044. doi: 10.1194/jlr.M600545-JLR200. [DOI] [PubMed] [Google Scholar]
- 25.Gillard BK, Courtney HS, Massey JB, Pownall HJ. Serum opacity factor unmasks human plasma high-density lipoprotein instability via selective delipidation and apolipoprotein A-I desorption. Biochemistry. 2007;46:12968–12978. doi: 10.1021/bi701525w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Curtiss LK, Bonnet DJ, Rye KA. The conformation of apolipoprotein A-I in high-density lipoproteins is influenced by core lipid composition and particle size: A surface plasmon resonance study. Biochemistry. 2000;39:5712–5721. doi: 10.1021/bi992902m. [DOI] [PubMed] [Google Scholar]
- 27.Li L, et al. Double belt structure of discoidal high density lipoproteins: Molecular basis for size heterogeneity. J Mol Biol. 2004;343:1293–1311. doi: 10.1016/j.jmb.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 28.Vaisar T, et al. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J Clin Invest. 2007;117:746–756. doi: 10.1172/JCI26206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Z, et al. The refined structure of nascent HDL reveals a key functional domain for particle maturation and dysfunction. Nat Struct Mol Biol. 2007;14:861–868. doi: 10.1038/nsmb1284. [DOI] [PubMed] [Google Scholar]
- 30.Maiorano JN, Davidson WS. The orientation of helix 4 in apolipoprotein A-I-containing reconstituted high density lipoproteins. J Biol Chem. 2000;275:17374–17380. doi: 10.1074/jbc.M000044200. [DOI] [PubMed] [Google Scholar]
- 31.Roberts LM, et al. Structural analysis of apolipoprotein A-I: Limited proteolysis of methionine-reduced and -oxidized lipid-free and lipid-bound human apo A-I. Biochemistry. 1997;36:7615–7624. doi: 10.1021/bi962952g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.