Abstract
Ribonuclease P (RNase P) is an essential endonuclease responsible for the 5′-end maturation of precursor tRNAs. Bacterial RNase P also processes precursor 4.5S RNA, tmRNA, 30S preribosomal RNA, and several reported protein-coding RNAs. Eukaryotic nuclear RNase P is far more complex than in the bacterial form, employing multiple essential protein subunits in addition to the catalytic RNA subunit. RNomic studies have shown that RNase P binds other RNAs in addition to tRNAs, but no non-tRNA substrates have previously been identified. Additional substrates were identified by using a multipronged approach in the budding yeast Saccharomyces cerevisiae. First, RNase P-dependant changes in RNA abundance were examined on whole-genome microarrays by using strains containing temperature sensitive (TS) mutations in two of the essential RNase P subunits, Pop1p and Rpr1r. Second, RNase P was rapidly affinity-purified, and copurified RNAs were identified by using a genome-wide microarray. Third, to identify RNAs that do not change abundance when RNase P is depleted but accumulate as larger precursors, >80 potential small RNA substrates were probed directly by Northern blot analysis with RNA from the RNase P TS mutants. Numerous potential substrates were identified, of which we characterized the box C/D intron-encoded small nucleolar RNAs (snoRNAs), because these both copurify with RNase P and accumulate larger forms in the RNase P temperature-sensitive mutants. It was previously known that two pathways existed for excising these snoRNAs, one using the pre-mRNA splicing path and the other that was independent of splicing. RNase P appears to participate in the splicing-independent path for the box C/D intron-encoded snoRNAs.
Keywords: RNA, biogenesis
Ribonuclease P (RNase P) is a conserved endoribonuclease responsible for removing the 5′ leader sequence from precursor transfer RNAs (pre-tRNAs) found in bacteria, archaea, eukarya (1, 2). In all cases, with the possible exception of some organelles, RNase P is composed of both RNA and protein subunits. Bacterial RNase P is the simplest form of the holoenzyme, with one large RNA subunit and a single small protein subunit (1). Although the RNA subunit of bacterial RNase P is sufficient for catalysis in vitro at high salt concentrations (3), both the RNA and protein subunits are required in vivo. The protein subunit appears to stabilize the catalytically active conformation of RNase P RNA and assist with substrate binding (4–7). In addition to pre-tRNAs, bacterial RNase P is known to process several substrates that are proposed to contain tRNA-like structures: 4.5S RNA, tmRNA, viral RNAs, mRNAs, ribo switches, ColE1 replication origin control RNAs, and C4 antisense RNA from phages P1 and P7 (8–16). The presence of the protein subunit in the RNase P holoenzyme increases the substrate versatility of the enzyme over the RNA enzyme alone (17).
The nature of the eukaryotic nuclear RNase P is much more complex. First, there are two very similar enzymes that are related to bacterial RNase P, termed RNase P and RNase MRP. RNase P is responsible for processing pre-tRNAs, whereas RNase MRP processes pre-rRNA, mitochondrial RNA primers and is required for the regulated turnover of a cell cycle mRNA (18–21). Both the eukaryotic RNase P and RNase MRP from nuclei are far more complex enzymes than bacterial RNase P (22). Each enzyme still employs a distinct but related RNA subunit and contains multiple required protein subunits for function in vivo. In yeast the two enzymes have eight identical proteins subunits, with RNase P having one unique protein and RNase MRP having two unique proteins (23, 24). Seven of the nine RNase P proteins are highly positively charged (pI 9.3–10.0), which could provide multiple substrate RNA binding sites in addition to the ones for pre-tRNAs that would be analogous to the bacterial enzyme. This might explain why yeast RNase P is much more susceptible to inhibition by single-stranded RNAs than bacterial RNase P (25). The additional protein components might provide the ability to hold other types of RNA in position to occupy the active cleavage site provided by the conserved, catalytic RNA subunit. Thus, given the number of non-tRNA substrates cleaved by even the bacterial enzymes, it seems possible that nuclear RNase P has been incorporated into the processing pathways for a number of different RNAs. Previous studies of the eukaryotic enzymes have suggested this (26, 27), and there is substantial evidence that the closely related RNase MRP participates in regulated turnover of specific mRNAs (18, 19).
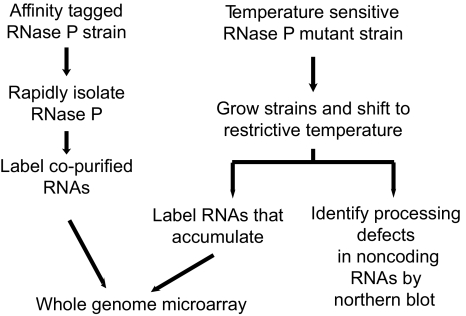
To search for physiologically relevant, previously unrecognized substrates for nuclear RNase P, we used three different approaches in Saccharomyces cerevisiae (Fig. 1). In the first, the RNase P holoenzyme was affinity purified, and RNAs that copurify with the enzyme were identified by using a whole-genome microarray. The second and third approaches utilize temperature-sensitive (TS) RNase P mutant strains. In the second approach, multiple TS mutant strains in multiple RNase P subunits were grown at the restrictive temperature and changes in the abundance of individual RNAs were measured by using a whole genome microarray. In the third approach, we examined the processing of possible small RNA substrates in TS mutant strains by Northern blot analysis to detect RNAs of altered size that accumulate in the absence of RNase P activity, even though they might not change in abundance. Here we report that this multipronged approach identified numerous potential substrates, and we focus on characterization of a particular class of RNAs that both copurify with RNase P and accumulate larger forms in the RNase P temperature-sensitive mutants. This class is the set of box C/D small nucleolar RNAs (snoRNAs) that are encoded in the introns of six pre-mRNA introns. It was previously known that two pathways existed for excising these snoRNAs, one using the pre-mRNA splicing path and the other that was independent of splicing (28). RNase P appears to participate in the splicing-independent path.
Fig. 1.
Multipronged approach to identify additional RNase P substrates. Three complementary approaches were taken to discover previously unrecognized in vivo substrates for yeast RNase P. RNAs that physically associate with RNase P were identified by copurification with affinity-tagged holoenzyme. Functional relationships were identified in temperature-sensitive mutant strains by examining changes in abundance by microarray or accumulation of aberrant-size processing products by Northern blot. Microarray data are summarized in Table S1.
Results
Identifying RNAs That Copurify with RNase P.
Potential RNase P substrates were determined by identifying RNAs that copurify with RNase P. RNase P was purified by using either a small RNA affinity tag (aptamer) in the RNA subunit (Rpr1r) that binds to streptavidin (29, 30) or a TAP tag (31) on the protein subunit that is unique to RNase P, Rpr2p. Untagged wild-type RNase P was subjected to the same purification steps to establish a background for RNA contaminants in the purification process. The coisolated RNA was then reverse transcribed into fluorescently labeled cDNA. The cDNA was used to probe a microarray containing oligos to the entire yeast genome: ORFs, known noncoding RNAs, and intergenic regions (32, 33).
Comparison of results from independent purifications indicated that data from the RNA subunit aptamer tag purification (R2 = 0.708) were much more consistent than the protein subunit (Rpr2p) TAP purification (R2 = 0.196). It would be expected that more transient interactions would be lost during the more protracted dual-column purification of the Rpr2p-TAP tag than the single-column RPR1-aptamer tag. Additionally, the RNA aptamer purification involves mild elution from the affinity resin with only the addition of a small molecule (biotin). Therefore, we focused on data obtained from the RNA subunit tag purification.
Numerous RNAs were detected as copurifying with RNase P. The 250 most abundantly copurified RNAs mapped predominantly to ORFs encoding components of the translation machinery [Table 1; full listing can be found in supporting information (SI) Table S1]. The prevalence of these mRNAs in the copurification does not correlate to their abundance in the cell (R2 = 0.263), referenced to the yeast transcriptome (34) (Fig. S1), consistent with selective association with RNase P. The correlation drops even further (R2 = 0.125) when limited to the 250 most abundantly copurified RNAs. It is interesting to note that tRNAs are not identified in this isolation, possibly because of the transient binding of tRNA substrates and products to RNase P. However, it is also possible that cDNA probe production is inefficient due to difficulties reverse transcribing tRNAs because of nucleotide modifications or tertiary structure.
Table 1.
Nuclear-encoded RNAs that copurify with RNase P
| mRNAs with intron encoded snoRNAs | ASC1 (3.67), EFB1 (3.01), IMD4 (3.14), RPL7A (4.36), RPL7B (3.18), TEF4 (2.90) |
| Ribosomal small subunit mRNAs | ASC1 (3.67), RPS0A (3.66), RPS0B (3.05), RPS1A (4.04), RPS1B (4.24), RPS2 (3.61), RPS3 (2.76), RPS4A (4.74), RPS4B (3.36), RPS5 (3.88), RPS6A (3.59), RPS7A (4.62), RPS7B (4.24), RPS8A (3.95), RPS9A (2.74), RPS10A (3.72), RPS10B (2.71), RPS11A (4.17), RPS11B (3.67), RPS12 (4.90), RPS13 (3.10), RPS14A (3.75), RPS15 (2.63), RPS16B (3.72), RPS17A (3.87), RPS17B (4.15), RPS18A (3.94), RPS18B (3.19), RPS19A (4.20), RPS19B (4.18), RPS20 (2.80), RPS21A (2.91), RPS21B (3.44), RPS22B (4.84), RPS23A (2.58), RPS23B (4.00), RPS24A (3.06), RPS24B (3.28), RPS25A (4.71), RPS25B (4.51), RPS27A (3.61), RPS27B (2.86), RPS28A (4.33), RPS28B (2.71), RPS29A (3.67), RPS29B (3.84), RPS30A (2.76), RPS31 (3.23) |
| Ribosomal large subunit mRNAs | RPL1A (3.04), RPL1B (4.54), RPP1B (3.01), RPL2B (3.97), RPP2A (4.18), RPP2B (3.24), RPL3 (2.90), RPL4B (2.98), RPL5 (6.05), RPL6A (2.84), RPL6B (4.48), RPL7A (4.36), RPL7B (3.18), RPL8A (3.34), RPL9A (5.61), RPL9B (3.60), RPL11A (4.32), RPL11B (4.62), RPL12A (4.47), RPL13A (7.24), RPL13B (2.89), RPL14A (3.62), RPL14B (3.78), RPL15B (4.94), RPL16B (2.91), RPL17B (3.13), RPL18A (4.73), RPL18B (4.11), RPL19A (5.01), RPL19B (5.01), RPL20A (3.81), RPL20B (3.54), RPL21B (3.94), RPL22A (2.57), RPL23A (3.78), RPL23B (3.77), RPL24A (3.05), RPL24B (2.90), RPL26A (3.47), RPL26B (2.67), RPL27A (3.17), RPL27B (4.45), RPL28 (3.86), RPL29 (3.55), RPL30 (4.65), RPL31A (3.74), RPL33A (3.80), RPL33B (3.55), RPL34A (5.11), RPL34B (5.72), RPL35B (2.86), RPL36A (2.99), RPL36B (3.57), RPL37B (2.67), RPL38 (4.56), RPL40A (3.43), RPL40B (3.69), RPL41A (3.57), RPL41B (3.73), RPL42A (3.78), RPL42B (3.39), RPL43A (2.70) |
| RNA polymerase I subunit mRNAs | RPB8 (2.70), RPA135 (3.10), RPC40 (3.06), RPO26 (3.66) |
| Translation Initiation mRNAs | PAB1 (2.86), TIF1 (2.67), TIF11 (3.20), NIP1 (2.82), TIF3 (3.79) |
| Translation elongation mRNAs | EFB1 (3.01), TEF4 (2.90), EFT1 (3.27) |
| Noncoding RNAs | RUF5 (3.43) |
Shown is enrichment with aptamer-tagged RPR1 listed in parentheses; bolded genes accumulate an RNase P ts strain.
*Full listing of individual genes can be found in Table S1.
Identifying RNAs That Accumulate in Temperature-Sensitive Mutants.
The next approach to identifying previously unrecognized RNase P substrates was to identify RNAs that change in abundance in the TS RNase P mutant strains. Temperature-sensitive RNase P mutations were available in two subunits of yeast RNase P: the unique RNA subunit, Rpr1r (35, 36), and the largest protein subunit, Pop1p, that is also a component of RNase MRP (37).
The RNAs affected in the temperature-sensitive strains are vastly different between RPR1 TS and the two POP1 TS strains (Table S1). This could be because of the dual role of Pop1p in RNases P and MRP or the different time courses of the temperature shift (2 h to see growth inhibition in RPR1 TS compared with 6 h for POP1 TS strains). However, there is an interesting general preference for the RNAs that coisolate with the RNase P and accumulate in response to the TS mutations (Table 2) in that they tend to be components of the translation machinery out of proportion to their abundance in the cell (Fig. S1). This carries through when considering only the RNAs that both coisolate and accumulate in response to TS mutation. Of these RNAs, 16 are mRNAs encoding protein subunits of the ribosome. The remaining RNAs that both copurify with RNase P and accumulate in the mutant strain include mRNAs for two translation initiation factors (TIF11, SUI13), a box C/D snoRNA binding protein (SNU13), a common subunit in RNA polymerases I, II, and III (RPO26), the CUP1-1/RUF5 locus, and six intergenic regions. Signal from three of the intergenic regions neighboring ribosomal protein genes (RPL42B, RPL41A, RPL38) were also identified, although signal from the coding regions of the genes themselves was not found and no characterized RNA is made from these intergenic regions. We note that tRNAs do not accumulate substantially in this microarray analysis, but this is not unexpected in that the amount of uncut pre-tRNAs that accumulates before the cells stop growing is small compared with the stable population of mature tRNAs.
Table 2.
Nuclear-encoded RNAs that accumulate with RNase P
| RNAs that accumulate in RPR1 TS strain (from top 250)* | |
| Ribosomal small subunit mRNAs | RPS30A (2.05), RPS17A (1.87), RPS18B (2.05), RPS16A (1.88), RPS10B (1.93), RPS28A (2.19), RPS10A (2.17) |
| Ribosomal large subunit mRNAs | RPL31A (1.98), RPL41B (2.79), RPL41A (3.05), RPL37B (2.60), RPL34A (2.02), RPL30 (2.01), RPL34B (1.89), RPL38 (3.92), RPL36A (2.17) |
| RNA polymerase I subunit mRNAs | RPC10 (2.53), RPA34 (1.88), RPC19 (2.00), RPO26 (2.04) |
| RNA polymerase III subunit mRNAs | RPC10 (2.53), RPC19 (2.00), RPO26 (2.04) |
| Mitochondrial Ribosome | MRPL37 (1.86), MRPL49 (2.09), MRP49 (1.82), MRPL3 (2.09), MRPL24 (1.85), MRPL40 (2.16) |
| Noncoding RNAs | RUF5 (2.08) |
| RNAs that accumulate in POP1660D6 TS strain (from top 250)* | |
| Ribosomal large subunit mRNAs | RPL41A (1.98), RLP24 (2.03), RPL38 (1.97) |
| RNA polymerase I subunit mRNAs | RPC10 (1.93), RPA34 (2.43), RPC19 (1.93) |
| Noncoding RNAs | RUF5 (2.92) |
| RNAs that accumulate in POP1233E11 TS strain (from top 250)* | |
| Ribosomal large subunit mRNAs | RPL41A (2.04), RPL27B (2.23), RPL37B (2.93), RPL40B (2.10), RPL38 (2.34), RPL36A (2.39), RPL33B (2.34) |
| Ribosomal small subunit mRNAs | RPS28A (6.51), RPS31 (2.12), RPS26A (2.51), RPS20 (2.05), RPS27A (2.08) |
| Noncoding RNAs | RUF5 (2.10) |
| RNAs that accumulate larger forms (possible precursors) in RPR1 TS (identified by northern blot) | |
| Intron-encoded snoRNAs | snR24, snR18, snR54, snR39, snR38 |
Accumulation values are included in parenthesis following the gene name, and bolded gene products copurify with RNase P RNA affinity tag.
*Full listing of individual genes can be found in Table S1.
Intron-Encoded snoRNAs.
In the top 250 copurified RNAs, we observed seven intron-encoded small nucleolar RNAs (snoRNAs). There are 75 known snoRNAs in the yeast genome. The majority of the snoRNAs in yeast are independently transcribed, however there are also examples of polycistronic transcripts and intron-encoded snoRNAs (38). There are eight total intron-encoded snoRNAs, six of which are box C/D. RNA from all six box C/D snoRNA ORFs were in the top 250 RNAs copurifying with RNase P. Of the two box H/ACA snoRNAs, only the snR44/RPS22B locus was in the top 250 copurifying RNAs, whereas snR191/NOG2 was the 319th-ranked RNA.
Box C/D Intron-Encoded snoRNAs Accumulate Known Processing Intermediates in an RNase P Temperature-Sensitive Mutant.
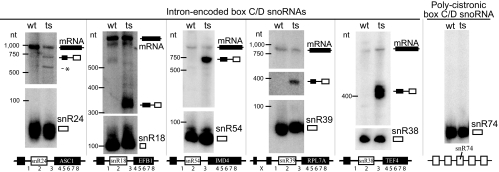
As part of our screen for possible substrates for RNase P, we performed Northern blot analysis on RNA from RNase P TS strains to see whether some processing intermediates of some small RNAs might accumulate, even though the overall amount of RNA from that transcription unit did not change significantly. Accumulation of pre-tRNAs in these mutants was previously demonstrated (36, 37), and we opted to use the TS in the RPR1 RNA subunit because it, unlike Pop1 protein, is unique to RNase P and not found in MRP. Here, 79 additional noncoding RNAs were examined by Northern blot, representing all classes of small nuclear RNAs, small cytoplasmic RNA (SCR1), and box H/ACA and C/D small nucleolar RNAs from independently transcribed, polycistronic, and intron-encoded genes (Table S2). In most cases RNAs other than tRNAs did not accumulate larger (or smaller) forms in the RNase P mutant. A subset of the box C/D snoRNAs was the notable exception. All six of the intron-encoded box C/D snoRNAs accumulate a processing intermediate in the RNase P TS mutant that is larger than the mature snoRNA (Fig. 2). There are also two intron-encoded box H/ACA snoRNAs, but no aberrant forms of these were observed.
Fig. 2.
Box C/D intron-encoded snoRNAs accumulate processing intermediate in temperature-sensitive RNase P strain. Northern blot analysis of RNA from wild-type (WT) and temperature-sensitive (TS) RNase P mutants reveals an accumulation of an unusual processing form of the box C/D intron-encoded snoRNAs in only the RNase P-deficient strain. Because RPL7A and RPL7B are highly homologous, only 7A and its snoRNA are shown. The identity of the accumulated transcripts is indicated schematically to the right of the panels and was determined by which oligo probes detected them and by the length of the primer extension product to the 5′ end of the RNA. (Asterisk indicates a breakdown intermediate with an undetermined 5′ terminus.) Probes 1 and 2 hybridize to the processing intermediate. Oligo probes 3 and X did not give a signal. Probes 4–8 hybridize to the mRNA. Probe 2 hybridizes to the mature snoRNA. Noncoding RNAs (79) were examined for altered forms by Northern blot (Table S1). A blot of a snoRNA from a polycistronic transcript is shown for contrast.
Probing Northern blots with oligonucleotides spaced at the indicated positions (Fig. 2) showed that each accumulated RNA is a 5′-extended pre-snoRNA that contains the 5′ exon, the intron 5′ of the snoRNA, and the snoRNA itself. Primer extension confirms the 5′ end is the expected primary transcript for the pre-mRNA containing the intron (data not shown). The intron sequence 3′ of the snoRNA and 3′ exons are absent in the accumulated RNA, suggesting the RNase P defect causes loss of an essential cleavage that initiates processing on the 5′ side of these snoRNAs. Unfortunately, attempts to identify the sites of cleavage in vitro or in vivo failed to identify specific RNase P cleavage sites (see below). However, the nature of these 5′-extended pre-snoRNAs is compatible with predicted processing intermediates in one maturation pathway for intron-encoded box C/D snoRNAs (28, 39).
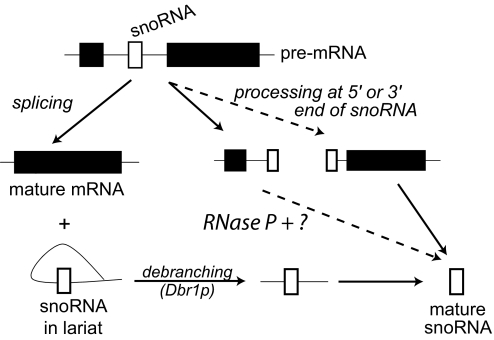
Maturation of box C/D snoRNAs can proceed through two pathways in yeast (28, 39): the primary pathway depends on splicing the pre-mRNA to free a snoRNA-containing lariat, and the minor pathway is independent of the splicing reaction (Fig. 3). The primary splicing-dependent pathway requires linearization of the intron lariat product by debranching enzyme (Dbr1p), allowing subsequent release of the snoRNA by endonucleases and exonucleases. In the minor, the snoRNA is excised from the intron of the pre-mRNA directly, leading to the destruction of the mRNA. Although this process is thought to involve the Rnt1 endonuclease (40), the enzymatic path is not well defined and our data suggest that in vivo it also involves cleavage by RNase P upstream of the snoRNA. The unusual 5′-extended pre-snoRNAs that accumulate in the RNase P mutant strain have already undergone 3′ maturation of the snoRNA and require one or more 5′ cleavages. Multiple attempts to delete DBR1 in the presence of the TS RPR1 mutation were unsuccessful (data not shown), suggesting the combination is synthetically lethal and strengthening the interpretation that RNase P is required in the alternative pathway.
Fig. 3.
Intron-encoded snoRNA processing pathways. Two distinct processing pathways exist for intron-encoded snoRNAs. The splicing-dependent pathway produces the mature mRNA and snoRNA after the intron lariat form has been opened by Dbr1p and further processing. The splicing-independent pathway produces only the mature snoRNA. The dashed lines indicate the step affected by RNase P.
Although computer folding of the pre-snoRNAs does not predict tRNA-like structures, we have previously shown that highly purified, yeast nuclear RNase P binds tightly to single-stranded RNAs (25) and cuts efficiently at highly preferred sites in pre-rRNA that do not have obvious tRNA-like structures (35). We therefore tested whether the pre-snoRNA alone (without protein) was a highly selective substrate for yeast nuclear RNase P. Purified RNase P cuts T7-transcribed, 5′-extended pre-snoRNAs in multiple places in vitro (Fig. S2). As previously observed for other non-tRNA substrates (35), there was not a clear correlation to specific sequences or structures upstream of the mature 5′ end of the snoRNAs. The major cleavage site in pre-snR14 is at the 5′ end of the mature snoRNA, with a minor cut site 38 nucleotides upstream in the intron. In pre-snR18, the major cleavage site corresponds to ≈43 nucleotides upstream of the 5′ end of the mature snoRNA, which is seven nucleotides upstream of a proposed stem essential for splicing-independent snoRNA maturation (39, 40). RNase P makes multiple cuts in pre-snR38, the strongest is 58 nucleotides upstream of the mature snoRNA, with additional cuts at the 5′ end of the mature snoRNA and 17 nucleotides and 21 nucleotides upstream. Because these naked RNA cleavages did not form a detectable pattern, we refrain from suggesting correlations to physiologically relevant sites.
Because the in vivo pre-snoRNA substrates are likely to be ribonucleoprotein complexes (28, 38–40), we also tried to cleave accumulated RNPs in soluble extracts of the RNase P TS mutant after accumulation of precursor. A large excess of purified RNase P was added to such extracts under conditions where the longer forms were not cleaved by the endogenous temperature-sensitive activity (37°C). Although the longer snoRNAs degraded slowly with time (data not shown), no RNase P-specific preferred cleavage locations were identified, including those shown to be sensitive in the naked RNA. Thus, if correct precursor small nucleolar RNPs (snoRNPs) for RNase P cleavage can be reconstituted, it will require more extended biochemical analysis.
Discussion
Recent studies by other groups have suggested various substrates for eukaryotic RNase P. Although it has been shown that RNase P can cut the noncoding HRA1 RNA in vitro (27), we see no evidence of an in vivo association or function, because HRA1 neither copurifies with RNase P nor does it accumulate in any of the three temperature-sensitive mutants. Another recent study depleted one of the protein subunits, Rpp1p, found in both RNases P and MRP (26). This identified 74 RNAs that accumulate with the Rpp1p depletion. Of these 74 noncoding RNAs, two copurify with RNase P in our hands, MAN7 and TLN1, and another one, TLN20, accumulates in the RPR1 TS mutant strain (Table S3). Interestingly, MAN7 and TLN1 are both found antisense from genes encoding ribosomal proteins. Thus, a future direction of interest might be to investigate participation of RNase P in processing and turnover of antisense RNAs.
In this study we focused on the intron-encoded snoRNAs, because transcripts encoding them copurify with RNase P. We find that 5′-extended pre-snoRNAs for each of the box C/D intron-encoded snoRNAs, but not the H/ACA snoRNAs, accumulated in the TS RNase P strains. The link between the intron-encoded box C/D snoRNAs and the other RNAs that copurify with affinity-purified RNase P is strengthened by the identities of the host mRNAs of the intron-encoded snoRNA: seven of eight host mRNAs encoded proteins are involved in translation. Although the abundance of the pre-mRNAs from this pathway did not increase significantly in the RNase P TS mutants, this is not unexpected for a maturation, rather than turnover defect. The 5′-extended pre-snoRNA is a known processing intermediate in the splicing-independent, intron-encoded snoRNA maturation pathway (Fig. 3). This splicing-independent pathway requires endonucleolytic cuts both 5′ and 3′ of the snoRNA and leads to the destruction of the mRNA (28). The pre-snoRNA that accumulates in the RNase P mutant strain has already been trimmed at the 3′ end of the snoRNA, but it still contains the full transcript 5′ to the snoRNA. It is possible that RNase P cuts at the 5′ end of these snoRNAs in vivo, but it is also possible that it cuts one or more sites upstream of the snoRNA and the 5′ maturation is subsequently performed by other nucleases. This would resemble the case for 5′ maturation of 5.8S rRNA by RNase MRP cleavage followed by further trimming.
In vitro cleavage assays with purified RNase P and pre-snoRNAs did not produce a consistent model for the nature of the RNase P cleavage site in the intron-encoded snoRNAs. Although this is likely because of a lack of appropriate additional components in the purified system (e.g., snoRNA structure), it leaves open the possibility that the RNase P effects on the intronic pre-snoRNAs is indirect. However, direct participation by RNase P is strongly suggested by the combination of (i) the physical interaction demonstrated by copurification of snoRNAs with RNase P, (ii) the functional relationship seen by the accumulation of known processing intermediates in the RNase P mutant strain, and (iii) the previously demonstrated ability of yeast nuclear RNase P to cleave non-tRNA substrates at high rates and in a partially sequence-specific fashion (35). The analysis of the sequences and RNP structures that lead to RNase P recognition will presumably be an extended undertaking, especially because the highly complex 10-subunit RNase P RNP structure could hypothetically recognize a relatively large number of signals.
It is especially interesting that one of the intron-encoded snoRNA processing pathways is compromised, because this suggests the pathway might be nucleolar. Not only is the final destination of the snoRNPs the nucleolus, but RNase P is also found primarily in the nucleolus in yeast (41). Thus, RNase P might provide a link between production of both tRNAs and ribosomes, the two most abundant RNA components of the translational machinery.
Materials and Methods
Yeast Strains.
Affinity purification.
In a yeast strain containing a C-terminal tandem affinity purification (TAP) tag on RPR2 (Open Biosystems; YSC1178-7501110), chromosomal RPR1 was disrupted with HIS3 and replaced with a plasmid, pRS315, containing RPR1 with RNA affinity tags for streptavidin and Sephadex (29, 30).
Temperature-sensitive mutations.
Previously described strains with defined RNase P mutations were used in this study: G207G211 RPR1 (42), R233K POP1, and R626L/P628K POP1 (37).
Yeast Growth.
Yeast were grown in standard synthetic media containing dextrose and lacking histidine (SDC-H). For temperature-sensitive (TS) assays, yeast were grown at 30°C into log phase (OD600 of 0.6–0.8) and then diluted into SDC-H media prewarmed to 37°C. The strain-specific time period was determined from growth curves for wild-type (WT) and TS RNase P strains, using the earliest time after the growth curve of the WT and TS strains diverged. G207G211 RPR1 was grown at 37°C for 2 h, and the POP1 TS strains were grown at 37°C for 6 h.
RNase P Purification.
Two yeast strains were subjected to RNase P purification: (i) a control strain that expresses WT RPR1 and WT RPR2 and (ii) a tagged strain that expresses TAP-tagged RPR2 and aptamer-tagged RPR1.
RNase P was purified by using either a single-column aptamer-streptavidin affinity purification or a two-column TAP purification. Sequential TAP then aptamer-streptavidin purifications were also attempted but did not yield sufficient amounts of RNase P for analysis. Briefly, 8 liters of yeast were grown in yeast extract/peptone/dextrose media to an OD600 of 0.8–1.0. Yeast were lysed in lysis buffer [50 mM Hepes, pH 7.5/10% glycerol/0.5 mM EDTA/150 mM NaCl/1 mM DTT/0.1% Nonidet P-40/complete, EDTA-free protease inhibitor (Roche Diagnostics Corporation)] with a model 110-Y Microfluidizer (Microfluidics), using five passes each through a 200-μm chamber and a 100-μm chamber. Cell extract was cleared by centrifugation for 10 min at 17,000 × g, followed by a 100-min spin at 105,000 × g. Twenty five microliters of a 50% slurry of IgG Sepharose was added per 1 ml of cell extract and incubated on a rotating drum at 4°C for 2 h. The IgG Sepharose was washed with 25 column volumes of lysis buffer. RNase P was eluted overnight with excess tobacco etch virus (TEV) protease. The elution was adjusted to 2 mM CaCl2 and bound for 2 h to 25 μl of a 50% slurry of Calmodulin Affinity Resin (CAR) per milliliter of starting extract. The CAR was washed five times with five volumes of lysis buffer and 2 mM CaCl2. RNase P was eluted with five volumes of lysis buffer and 10 mM EGTA.
Microarray Preparation.
Associated RNAs were reverse transcribed to cDNAs and fluorescently labeled with either Cy3 or Cy5 dyes (32). Labeled cDNAs were then hybridized to a yeast whole-genome microarray, which contains >13,000 features corresponding to both known ORFs and intergenic regions. The design is accessible at the Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo/) under platform accession no. GPL6424 (43). The ratio of Cy3 to Cy5 fluorescence indicates the relative amounts of RNA coming through the purification of the tagged to untagged yeast strains. The resulting data were analyzed as previously described (32). All data are accessible at the GEO (43) database under provisional series accession no. GSE10514.
RNAs were detected by microarray analysis as described (32). Briefly, RNAs were reverse transcribed into cDNA in the presence of aminoallyl-dUTP using random nonamers as primers. The cDNA was then labeled with either Cy3 or Cy5 (Amersham Biosciences). Labeled cDNA was then hybridized to a yeast whole-genome microarray (44).
Northern Blotting of RNAs.
Hot acid phenol (45) was used to extract total RNA from yeast cells harvested at 30°C and 37°C, and concentrations were determined by UV absorbance. Ten micrograms of total yeast RNA per lane was electrophoresed on denaturing 8% polyacrylamide gels. The RNA was then electrotransferred to a Nytran SuperCharge membrane (Schleicher & Schuell Bioscience).
Specific oligodeoxynucleotide probes were designed to the majority of yeast small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs). Probes were radiolabeled with [γ-32P]ATP. Labeled probes were hybridized and washed according to instructions accompanying the Nytran SuperCharge membrane. Signals on the Northern blots were determined with a PhosphorImager (Molecular Dynamics; model no. 445 SI).
In Vitro Cleavage Reactions and Primer Extensions.
Templates for T7 transcription were made from genomic S. cerevisiae DNA by PCR using primers to the region 100 nucleotides 5′ of the mRNA (to include any essential 5′-untranslated region that may be structurally significant) and a primer complementary to the 3′ end of the intron-encoded snoRNA. These templates were then used for T7 transcription. The in vitro-transcribed RNA was gel purified on an 8% polyacrylamide gel. RNase P purified using the tandem affinity purification (TAP) tag on the unique protein subunit Rpr1p was added to the gel-purified 5′-extended pre-snoRNA in RNase P assay buffer (10 mM Hepes, pH 7.9/100 mM KCl/10 mM MgCl2) and incubated for 15 min at 37°C. The reaction was then treated with proteinase K, and the RNA was extracted by using phenol-chloroform. Primers complementary to the snoRNA were kinased with [γ-32P]ATP, which was then gel isolated on a 12% polyacrylamide gel. The labeled primers were hybridized with the treated and untreated pre-snoRNA for 1 h at 42°C and then extended by using SuperScript II reverse transcriptase according to manufacturers instructions (Invitrogen). The cDNA was then electrophoresed onto an 8% polyacrylamide gel along with respective dideoxy sequencing ladders (46).
Supplementary Material
Acknowledgments.
We thank Felicia H. Scott for advice on RNase P purification and Christine Guthrie for helpful discussions and help from her laboratory in carrying out microarray analysis. This work was funded by National Institutes of Health Grant GM034869 (to D.R.E.) and National Institutes of Health Pharmacological Sciences Predoctoral Training Grant T32 GM007767 Fellowship (to D.J.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE10514).
This article contains supporting information online at www.pnas.org/cgi/content/full/0801906105/DCSupplemental.
References
- 1.Frank DN, Pace NR. Ribonuclease P: Unity and diversity in a tRNA processing ribozyme. Annu Rev Biochem. 1998;67:153–180. doi: 10.1146/annurev.biochem.67.1.153. [DOI] [PubMed] [Google Scholar]
- 2.Xiao S, Scott F, Fierke CA, Engelke DR. Eukaryotic ribonuclease P: A plurality of ribonucleoprotein enzymes. Annu Rev Biochem. 2002;71:165–189. doi: 10.1146/annurev.biochem.71.110601.135352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 4.Niranjanakumari S, Stams T, Crary SM, Christianson DW, Fierke CA. Protein component of the ribozyme ribonuclease P alters substrate recognition by directly contacting precursor tRNA. Proc Natl Acad Sci USA. 1998;95:15212–15217. doi: 10.1073/pnas.95.26.15212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang XW, Yang XJ, Littrell K, Niranjanakumari S, Thiyagarajan P, et al. The Bacillus subtilis RNase P holoenzyme contains two RNase P RNA and two RNase P protein subunits. RNA. 2001;7:233–241. doi: 10.1017/s1355838201001352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurz JC, Niranjanakumari S, Fierke CA. Protein component of Bacillus subtilis RNase P specifically enhances the affinity for precursor-tRNAAsp. Biochemistry. 1998;37:2393–2400. doi: 10.1021/bi972530m. [DOI] [PubMed] [Google Scholar]
- 7.Rueda D, Hsieh J, Day-Storms JJ, Fierke CA, Walter NG. The 5′ leader of precursor tRNAAsp bound to the Bacillus subtilis RNase P holoenzyme has an extended conformation. Biochemistry. 2005;44:16130–16139. doi: 10.1021/bi0519093. [DOI] [PubMed] [Google Scholar]
- 8.Alifano P, et al. Ribonuclease E provides substrates for ribonuclease P-dependent processing of a polycistronic mRNA. Genes Devel. 1994;8:3021–3031. doi: 10.1101/gad.8.24.3021. [DOI] [PubMed] [Google Scholar]
- 9.Altman S, Wesolowski D, Guerrier-Takada C, Li Y. RNase P cleaves transient structures in some riboswitches. Proc Natl Acad Sci USA. 2005;102:11284–11289. doi: 10.1073/pnas.0505271102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giege R, Florentz C, Dreher TW. The TYMV tRNA-like structure. Biochimie. 1993;75:569–582. doi: 10.1016/0300-9084(93)90063-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gimple O, Schon A. In vitro and in vivo processing of cyanelle tmRNA by RNase P. Biol Chem. 2001;382:1421–1429. doi: 10.1515/BC.2001.175. [DOI] [PubMed] [Google Scholar]
- 12.Hartmann RK, Heinrich J, Schlegl J, Schuster H. Precursor of C4 antisense RNA of bacteriophages P1 and P7 is a substrate for RNase P of Escherichia coli. Proc Natl Acad Sci USA. 1995;92:5822–5826. doi: 10.1073/pnas.92.13.5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komine Y, Kitabatake M, Yokogawa T, Nishikawa K, Inokuchi H. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc Natl Acad Sci USA. 1994;91:9223–9227. doi: 10.1073/pnas.91.20.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Altman S. A specific endoribonuclease, RNase P, affects gene expression of polycistronic operon mRNAs. Proc Natl Acad Sci USA. 2003;100:13213–13218. doi: 10.1073/pnas.2235589100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Cole K, Altman S. The effect of a single, temperature-sensitive mutation on global gene expression in Escherichia coli. RNA. 2003;9:518–532. doi: 10.1261/rna.2198203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peck-Miller KA, Altman S. Kinetics of the processing of the precursor to 4.5 S RNA, a naturally occurring substrate for RNase P from Escherichia coli. J Mol Biol. 1991;221:1–5. doi: 10.1016/0022-2836(91)80194-y. [DOI] [PubMed] [Google Scholar]
- 17.Liu F, Altman S. Differential evolution of substrates for an RNA enzyme in the presence and absence of its protein cofactor. Cell. 1994;77:1093–1100. doi: 10.1016/0092-8674(94)90448-0. [DOI] [PubMed] [Google Scholar]
- 18.Cai T, Aulds J, Gill T, Cerio M, Schmitt ME. The Saccharomyces cerevisiae RNase mitochondrial RNA processing is critical for cell cycle progression at the end of mitosis. Genetics. 2002;161:1029–1042. doi: 10.1093/genetics/161.3.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gill T, Cai T, Aulds J, Wierzbicki S, Schmitt ME. RNase MRP cleaves the CLB2 mRNA to promote cell cycle progression: Novel method of mRNA degradation. Mol Cell Biol. 2004;24:945–953. doi: 10.1128/MCB.24.3.945-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang DD, Clayton DA. A novel endoribonuclease cleaves at a priming site of mouse mitochondrial DNA replication. EMBO J. 1987;6:409–417. doi: 10.1002/j.1460-2075.1987.tb04770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee DY, Clayton DA. Initiation of mitochondrial DNA replication by transcription and R-loop processing. J Biol Chem. 1998;273:30614–30621. doi: 10.1074/jbc.273.46.30614. [DOI] [PubMed] [Google Scholar]
- 22.Walker SC, Engelke DR. Ribonuclease P: The evolution of an ancient RNA enzyme. Crit Rev Biochem Mol Biol. 2006;41:77–102. doi: 10.1080/10409230600602634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmitt ME, Clayton DA. Characterization of a unique protein component of yeast RNase MRP: An RNA-binding protein with a zinc-cluster domain. Genes Dev. 1994;8:2617–2628. doi: 10.1101/gad.8.21.2617. [DOI] [PubMed] [Google Scholar]
- 24.Salinas K, Wierzbicki S, Zhou L, Schmitt ME. Characterization and purification of Saccharomyces cerevisiae RNase MRP reveals a new unique protein component. J Biol Chem. 2005;280:11352–11360. doi: 10.1074/jbc.M409568200. [DOI] [PubMed] [Google Scholar]
- 25.Ziehler WA, Day JJ, Fierke CA, Engelke DR. Effects of 5′ leader and 3′ trailer structures on pre-tRNA processing by nuclear RNase P. Biochemistry. 2000;39:9909–9916. doi: 10.1021/bi000603n. [DOI] [PubMed] [Google Scholar]
- 26.Samanta MP, Tongprasit W, Sethi H, Chin CS, Stolc V. Global identification of noncoding RNAs in Saccharomyces cerevisiae by modulating an essential RNA processing pathway. Proc Natl Acad Sci USA. 2006;103:4192–4197. doi: 10.1073/pnas.0507669103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang L, Altman S. A noncoding RNA in Saccharomyces cerevisiae is an RNase P substrate. RNA. 2007;13:682–690. doi: 10.1261/rna.460607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Villa T, Ceradini F, Presutti C, Bozzoni I. Processing of the intron-encoded U18 small nucleolar RNA in the yeast Saccharomyces cerevisiae relies on both exo- and endonucleolytic activities. Mol Cell Biol. 1998;18:3376–3383. doi: 10.1128/mcb.18.6.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srisawat C, Engelke DR. Streptavidin aptamers: Affinity tags for the study of RNAs and ribonucleoproteins. RNA. 2001;7:632–641. doi: 10.1017/s135583820100245x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Srisawat C, Engelke DR. RNA affinity tags for purification of RNAs and ribonucleoprotein complexes. Methods. 2002;26:156–161. doi: 10.1016/S1046-2023(02)00018-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rigaut G, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 32.Inada M, Guthrie C. Identification of Lhp1p-associated RNAs by microarray analysis in Saccharomyces cerevisiae reveals association with coding and noncoding RNAs. Proc Natl Acad Sci USA. 2004;101:434–439. doi: 10.1073/pnas.0307425100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iyer VR, et al. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- 34.Holstege FC, et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 35.Chamberlain JR, Pagan R, Kindelberger DW, Engelke DR. An RNase P RNA subunit mutation affects ribosomal RNA processing. Nucleic Acids Res. 1996;24:3158–3166. doi: 10.1093/nar/24.16.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pagan-Ramos E, Lee Y, Engelke DR. Mutational analysis of Saccharomyces cerevisiae nuclear RNase P: Randomization of universally conserved positions in the RNA subunit. RNA. 1996;2:441–451. [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao S, et al. Functional characterization of the conserved amino acids in Pop1p, the largest common protein subunit of yeast RNases P and MRP. RNA. 2006;12:1023–1037. doi: 10.1261/rna.23206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petfalski E, Dandekar T, Henry Y, Tollervey D. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol Cell Biol. 1998;18:1181–1189. doi: 10.1128/mcb.18.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Villa T, Ceradini F, Bozzoni I. Identification of a novel element required for processing of intron-encoded box C/D small nucleolar RNAs in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:1311–1320. doi: 10.1128/mcb.20.4.1311-1320.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giorgi C, Fatica A, Nagel R, Bozzoni I. Release of U18 snoRNA from its host intron requires interaction of Nop1p with the Rnt1p endonuclease. EMBO J. 2001;20:6856–6865. doi: 10.1093/emboj/20.23.6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bertrand E, Houser-Scott F, Kendall A, Singer RH, Engelke DR. Nucleolar localization of early tRNA processing. Genes Dev. 1998;12:2463–2468. doi: 10.1101/gad.12.16.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pagan-Ramos E, Lee Y, Engelke DR. A conserved RNA motif involved in divalent cation utilization by nuclear RNase P. RNA. 1996;2:1100–1109. [PMC free article] [PubMed] [Google Scholar]
- 43.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 45.Kohrer K, Domdey H. Preparation of high molecular weight RNA. Methods Enzymol. 1991;194:398–405. doi: 10.1016/0076-6879(91)94030-g. [DOI] [PubMed] [Google Scholar]
- 46.Hull MW, Thomas G, Huibregtse JM, Engelke DR. Protein-DNA interactions in vivo–examining genes in Saccharomyces cerevisiae and Drosophila melanogaster by chromatin footprinting. Methods Cell Biol. 1991;35:383–415. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.