Abstract
We have characterized the molecular basis of the interaction between ASPP2 and Bcl-2, which are key proteins in the apoptotic pathway. The C-terminal ankyrin repeats and SH3 domain of ASPP2 (ASPP2Ank-SH3) mediate its interactions with the antiapoptotic protein Bcl-2. We used biophysical and computational methods to identify the interaction sites of Bcl-2 and its homologues with ASPP2. Using peptide array screening, we found that ASPP2Ank-SH3 binds two homologous sites in all three Bcl proteins tested: (i) the conserved BH4 motif, and (ii) a binding site for proapoptotic regulators. Quantitative binding studies revealed that binding of ASPP2Ank-SH3 to the Bcl-2 family members is selective at two levels: (i) interaction with Bcl-2-derived peptides is the tightest compared to peptides from the other family members, and (ii) within Bcl-2, binding of ASPP2Ank-SH3 to the BH4 domain is tightest. Sequence alignment of the ASPP2-binding peptides combined with binding studies of mutated peptides revealed that two nonconserved positions where only Bcl-2 contains positively charged residues account for its tighter binding. The experimental binding results served as a basis for docking analysis, by which we modeled the complexes of ASPP2Ank-SH3 with the full-length Bcl proteins. Using peptide arrays and quantitative binding studies, we found that Bcl-2 binds three loops in ASPP2Ank-SH3 with similar affinity, in agreement with our predicted model. Based on our results, we propose a mechanism in which ASPP2 induces apoptosis by inhibiting functional sites of the antiapoptotic Bcl-2 proteins.
Keywords: apoptosis, peptides, protein–protein interactions, peptide arrays, docking
Protein–protein interactions (PPI) are crucial for most processes critical for the proper function of living cells. Studying PPI is important for understanding the cell functionality and for designing drugs against diseases in which PPI are impaired. Peptides are excellent tools for studying PPI. They are good models for binding studies of protein domains because they often undergo induced fit upon ligand binding and gain their native structure. This was shown for peptides derived from the BH4 domain of Bcl-2 (1) and from Bak (2). Peptide arrays provide an efficient way for identifying binding sites (3). Peptide arrays were particularly valuable for epitope mapping and analyzing enzyme binding and PPI (4–7).
Impairment of regulation of apoptosis pathways is a key event in malignant transformation. Understanding how apoptotic pathways are regulated is essential to understanding the basics for developing anticancer therapies. Two key apoptotic pathways are the p53 pathway and the mitochondrial death pathway. We used the peptide approach to characterize the interaction between two key players in these apoptotic pathways: ASPP2 and the Bcl-2 family proteins.
Proteins from the Bcl-2 family play a major role in the mitochondrial death pathway (8). The Bcl-2 family consists of both proapoptotic (e.g., Bax and Bak) and antiapoptotic (e.g., Bcl-2, Bcl-XL, and Bcl-W) members, which cooperate through formation of homo-/heterodimers that maintain the balance between cell death and survival. The antiapoptotic proteins inhibit apoptosis in normal cells, but in response to apoptotic signals, the proapoptotic family members mediate the release of cytochrome C (reviewed in refs. 8 and 9). Inhibition of antiapoptotic Bcl-2 family members is a major target for anticancer therapies (10).
The ASPP2 protein specifically stimulates the apoptotic response mediated by the tumor suppressor p53, by enhancing the transactivation function of p53 on promoters of proapoptotic genes (11). Two ASPP2 variants were discovered as p53 and Bcl-2 binding proteins: 53BP2 and Bbp, respectively (12) [supporting information (SI) Fig. S1]. ASPP2 contains several structural and functional domains (Fig. S1) (13, 14). The C-terminal part of ASPP2 contains four ankyrin repeats and an SH3 domain (ASPP2Ank-SH3) (15), which mediate its interactions with numerous apoptosis-related partner proteins such as p53, NFκB, and Bcl-2 (13). ASPP2 and Bbp are predominantly localized in the cytoplasm, whereas Bbp is partly localized in the mitochondria and is involved in the mitochondrial death pathway (16). Expression of both Bcl-2 and Bcl-XL inhibited Bbp-induced cell death (17), suggesting that the ASPP2–Bcl complex may maintain the balance between the pro- and antiapoptotic activity of both proteins.
Here, we used a combination of peptide arrays with biophysical and computational methods to gain insight into regulation of apoptosis by studying the molecular basis of the interactions between the antiapoptotic proteins from the Bcl-2 family and the proapoptotic protein ASPP2. We identified the sites that mediate the interactions in both the Bcl proteins and ASPP2Ank-SH3, and gained insight into the molecular mechanism of the interaction. Using docking studies supported by experimental data, we suggest a model for the complex between the full-length ASPP2Ank-SH3 and Bcl-2/Bcl-XL. We propose that ASPP2 binds and inhibits functional sites of antiapoptotic Bcl-2 proteins, promoting the release of proapoptotic proteins and the induction of apoptosis.
Results
Identification of the ASPP2 Binding Sites in Bcl-2 Family Proteins by Using Peptide Array Screening.
We designed an array comprising 59 overlapping peptides derived from the antiapoptotic proteins Bcl-2, Bcl-XL, and Bcl-W. Peptide length was between 10 and 30 residues (Table S1). Peptides were designed based on the secondary and tertiary structures of the parent proteins. ASPP2 893-1128, consisting of the Ankyrin and SH3 domains (ASPP2Ank-SH3), was expressed, purified, and screened for binding the peptide array (Fig. 1A and Table 1). Peptides from Yes-associated protein 1, which are known to bind ASPP2 (18), served as positive control (data not shown). ASPP2Ank-SH3-bound peptides were derived from two homologous sites at all three tested antiapoptotic Bcl-2 proteins (Fig. 1 B–D and Table 1). The first site (“BH4 site”) is the N-terminal α-helix, represented by peptides Bcl-W 8–26, Bcl-2 7–24, and Bcl-XL 6–26. This site corresponds to the BH4 domain, which is conserved among antiapoptotic Bcl-2 family members, and is essential for apoptosis inhibition (19). The second ASPP2 binding site (“proapoptotic site”) consists of parts of helix 2 and helix 3, represented by the binding peptides Bcl-2 45–108, Bcl-XL 104–123, and Bcl-W 41–67. The common motif for all of these peptides is the sequence R-aromatic-R-R-X-F. This site overlaps with a known binding site for proapoptotic members from Bcl-2 family such as Bak and Bad (2, 20).
Fig. 1.
Analysis of the ASPP2 binding sites in the Bcl-2 family proteins. (A) An array consisting of overlapping peptides derived from the Bcl-2 family proteins Bcl-2, Bcl-W, and Bcl-XL was screened for binding ASPP2Ank-SH3. Each dark spot represents binding of ASPP2Ank-SH3 to a specific peptide (Table 1). (B–D) The ASPP2 binding sites, as discovered in the peptide array screening, are highlighted on the known 3D structures of the Bcl family proteins. The BH4 site is in magenta and the proapoptotic site is in green. See Table 1 for peptides details. (B) Bcl-2 [PDB entry 1YSW (23)]. (C) Bcl-XL [PDB entry 1G5J (20)]. (D) Bcl-W [PDB entry 1O0l (31)]. Figures were generated by using PyMOL (32). (E–G) Quantitative analysis of the interaction between ASPP2Ank-SH3 and Bcl-2 BH4 peptide (7–24) using three independent methods. (E) Fluorescence spectroscopy. ASPP2Ank-SH3 was titrated into fluorescein-labeled Bcl-2 7–24, and the binding curve was fit to 1:1 binding model. Kd was found to be 4.7 ± 0.2 μM. (F) SPR. Biotinylated Bcl-2 7–24 peptide was captured on a ProteOn NLC sensor chip, followed by the association of 150 μl of ASPP2Ank-SH3, simultaneously injected at concentrations of 40, 30, 15, 10, and 7.5 μM (from top to bottom). Kd was found to be 1.6 μM. (G) ELISA. ASPP2Ank-SH3 was adsorbed on the ELISA plate, and binding of the peptide was studied as described in Materials and Methods. The apparent Kd is at the low micromolar range, in agreement with the two other methods.
Table 1.
Binding between ASPP2Ank-SH3 and peptides from Bcl-2 family proteins: SPR studies
| Parent protein | Peptide no. in array | Residues* | Sequence* | Kd, μM |
|---|---|---|---|---|
| Bcl-2 | 2–4 | 7–24 | YDNREIVMKYIHYKLSQR | 1.6 |
| 8 | 45–108† | TESEVVHLTLRQAGDDFSRRYRRD | 10 | |
| 103–120 | RRYRRDFAEMSSQLHLTP | 13 | ||
| Bcl-XL | 42–45 | 6–26 | SQSNRELVVDFLSYKLSQKGY | 23 |
| 103–111 | LRYRRAFSD | 20 | ||
| 50 | 104–123 | RYRRAFSDLTSQLHITPGTA | 26 | |
| 89–111 | AVKQALREAGDEFELRYRRAFSD | No binding | ||
| Bcl-W | 20–22 | 6–28 | PDTRALVADFVGYKLRQKG | Too weak to quantify |
| 54–67 | WETRFRRTFSDLAAQ | Too weak to quantify | ||
| 26–27 | 41–67 | WPLHQAMRAAGDEFETRFRRTFSDLAAQ | Too weak to quantify |
*The combined sequence of overlapping peptides from the same region that bound ASPP2Ank-SH3 in the array. Trp was added at the N terminus of several peptides for UV spectroscopy.
†Bcl-2 45–108 lacks residues 49–88, an unstructured loop that lacks at the crystal structure (23).
Quantitative Analysis of the Interactions Between ASPP2 and the Bcl-2 Family Proteins: Selectivity Among Proteins and Among Binding Sites.
The peptide array method is semiquantitative and served for initial screening of the binding sites. To gain quantitative information about the binding of the Bcl peptides to ASPP2, the binding peptides discovered in the array screening were synthesized and their binding to ASPP2Ank-SH3 was studied by using three independent quantitative methods: surface plasmon resonance (SPR), fluorescence spectroscopy, and ELISA. The peptides from the proapoptotic site were overlapping but not completely homologous, so we also tested the exact homologous peptides Bcl-2 103–120 and Bcl-XL 89–111, as well as short peptides containing the conserved R-aromatic-R-R-X-F motif: Bcl-2 103–120, Bcl-XL 103–111, and Bcl-W 54–67 (Table 1).
The binding of all of the peptides to ASPP2Ank-SH3 was quantified by SPR experiments because for some peptides binding was too weak to quantify by using fluorescence spectroscopy, which required much higher protein concentrations. The SPR results verified that all of the peptides selected in the peptide array indeed bind ASPP2Ank-SH3. Quantitative analysis of the SPR binding data (Table 1 and Fig. 1F) revealed that the binding of ASPP2Ank-SH3 to the Bcl-2 family proteins is selective at two levels. (i) Selectivity between proteins: Binding of ASPP2Ank-SH3 to Bcl-2 peptides is the tightest, binding of Bcl-XL is an order of magnitude weaker, and binding of Bcl-W is very weak. Fluorescence and ELISA binding studies confirmed that the Bcl-2 BH4 peptide (Bcl-2 7–24) bound tightest to ASPP2Ank-SH3, with affinity at the low-micromolar range (4.7 ± 0.2 μM; Fig. 1 E–G). (ii) Selectivity between the two binding sites within Bcl-2: Binding to the BH4 domain (Fig. 1B, magenta) is an order of magnitude tighter than binding to the proapoptotic site (Fig. 1B, green, and Table 1).
The Tighter Binding of Bcl2 to ASPP2 Is Due to Extra Positive Charge at the Conserved Binding Interfaces.
To reveal the basis for the selectivity in ASPP2 binding to the Bcl proteins, we aligned their ASPP2 binding sequences. Most amino acids are conserved in all three Bcl proteins (Fig. 2A). However, there are some nonconserved amino acids. Within the aligned sequences of the BH4 domain, two positively charged amino acids exist in Bcl-2 and are missing from its homologues: (i) Lys-15 is replaced by Asp in the Bcl-W and Bcl-XL, and (ii) His-18 is replaced by Gly in Bcl-W and by Ser in Bcl-XL. In the proapoptotic site, instead of Bcl-2 Arg-103, Bcl-XL has Leu and Bcl-W has Thr. In all cases the Bcl-2 sequence is more positively charged than the others, which may explain its tighter binding to the negatively charged ASPP2Ank-SH3 (pI = 4.72). To examine the contribution of the positively charged residues to the tighter binding of the Bcl-2-derived peptide, we synthesized a series of Bcl peptides modified at the positively charged residues and compared their binding affinities to ASPP2Ank-SH3 using ELISA (Table S2). We tested peptides where the positive charge from the Bcl-2 sequence was eliminated or replaced by the corresponding residue from Bcl-XL (Bcl-2 7–24 K15A, H18A, K15D, H18S) and peptides where positive charges were introduced into the relevant positions at the Bcl-XL homologous peptide (Bcl-XL 6–26 D15K, S18H). Bcl-2 7–24 K15A and H18A showed significant reduction in binding affinity to ASPP2Ank-SH3 compared to the WT peptide (Fig. 2B). Introduction of a negative charge in Bcl-2 7–24 K15D abolished the binding almost completely (Fig. 2B). The Bcl-XL-derived mutated peptides bearing the additional positively charged residues—e.g., Bcl-XL 6–26 D15K—showed significant increase in the binding affinity, whereas Bcl-XL 6–26 S18H did not (Fig. 2C). However, introducing both residues into the Bcl-XL peptide had a synergistic effect that raised the binding to the level of the WT Bcl-2 peptide (Fig 2 B and C). In the mutated peptides derived from the proapoptotic domain, the modification Bcl-XL L103R (the corresponding residue in Bcl-2) did not result in a significant change in the binding to ASPP2Ank-SH3 (data not shown). The contribution of the Arg residues to the binding of ASPP2Ank-SH3 to the proapoptotic site was tested using NMR. The TOCSY spectrum of Bcl-XL 103–111 was compared to its spectrum when incubated with ASPP2Ank-SH3 under identical conditions. The differences between the bound and nonbound peptide were extremely pronounced (Fig. S2A and Table S3) in both amide and Hα proton chemical shifts. Changes in the chemical shifts of Arg-106 and -107 and S110 upon binding indicate their involvement in the interaction with ASPP2Ank-SH3 (Fig. S2B). The Arg-104 amide was unresolved in the free state, but the Hα proton did not show a significant deviation in the bound state.
Fig. 2.
Positively charged residues in Bcl-2 account for its tighter binding To ASPP2. (A) Sequence alignment of ASPP2 binding peptides: The ASPP2 binding peptides derived from the three Bcl proteins are bold and colored by amino acid type. (B and C) ELISA studies of ASPP2Ank-SH3 binding to Bcl-2 and Bcl-XL peptides modified at the nonconserved amino acid K15 and H18 of Bcl-2. (B) Peptides in which the positively charged residues from the Bcl-2 peptides were replaced by alanine or by the corresponding residues from Bcl-XL. (C) Peptides in which positive charges were introduced into the relevant positions at the Bcl-XL peptide.
We used ELISA to compare the binding affinities of Bcl-2 7–24 to ASPP2Ank-SH3 at different ionic strengths and found that the ionic strength had only a modest effect on binding (Fig. S3).
Docking Studies Suggest a Model for ASPP2Ank-SH3 Binding with Full-Length Bcl-2 and Bcl-XL.
To move from the peptide to the protein level and suggest structural models for the complexes between the full-length Bcl proteins and ASPP2Ank-SH3, we performed docking studies. We used the algorithms PatchDock (21) and RosettaDock (22) to dock the structure of ASPP2Ank-SH3 (15) to structures of Bcl-2 (23, 24) and Bcl-XL (10, 25). We defined a biologically valid docking model as one that its protein–protein interface (within 5 Å) contains at least four residues from each experimentally detected ASPP2 binding peptide from Bcl-2/Bcl-XL (Table 1). The 5-Å and four-residue limits were set based on known PPI (26, 27). From the top 200 models generated, 19–29 models were valid according to our binding peptides data. To further select fitting models, we examined the sequence alignment of the Bcl proteins (Fig. 2A) in the binding peptides regions. We looked for models that involve contacts of specific amino acids that are surface-exposed and conserved, as well as those that contribute to the tighter binding affinity of Bcl-2 to ASPP2Ank-SH3 (Bcl-2 numbering; PDB entry 1YSW). (i) From the BH4 site: Y19, K20, and R24 are conserved, and K15 and H18 are unique to Bcl-2. (ii) From the proapoptotic site: R104, Y105, R106, and R107 are conserved, and R103 is unique to Bcl-2 (Fig. 3A). Our docking results suggested models for the complexes of ASPP2Ank-SH3 with both Bcl-2 and Bcl-XL (Fig. 3B). The SH3 domain of ASPP2 interacts with the BH4 domain of Bcl, and the Ank domain of ASPP2 interacts with the proapoptotic site of Bcl. The conserved interactions in both models involve mainly D/E from ASPP2Ank-SH3 with conserved K/R of Bcl-2/Bcl-XL.
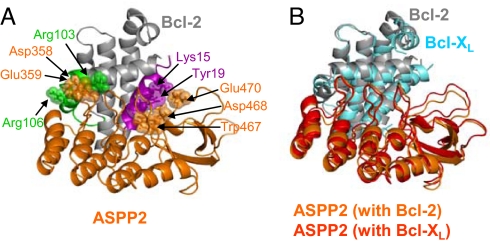
Fig. 3.
Docking model for the interaction between the ASPP2Ank-SH3 and Bcl-2/Bcl-XL proteins. (A) Bcl-2 binding to ASPP2Ank-SH3. Bcl-2 is colored gray, the BH4 site is in magenta, and the proapoptotic site is in green. ASPP2Ank-SH3 is colored orange. (B) Two aligned models of Bcl-2 and Bcl-XL complexes with ASPP2Ank-SH3. Bcl-XL is colored cyan, and Bcl-2 is in gray. ASPP2Ank-SH3 is colored red and orange for its complex models with Bcl-XL and Bcl-2, respectively. Representative binding residues from both partners are depicted in sticks and spheres. The conserved interactions between Bcl-2 (PDB entry 1YSW) and ASPP2 (PDB entry 1ycs) are as follows: K15–D468 and E470; H18–D468 and E481; Y19–W467; K20–D468 and E481; R24–D352; R103–D358 and D359; Y105–P356; and R106–D333, D358, and E359. The homologous amino acids in Bcl-XL show the same interactions with ASPP2. For consistency with the PDB file, numbering of the ASPP2 residues is according to the sequence of its truncated form 53BP2.
Identification of the Bcl-2 Binding Sites in ASPP2.
To identify the Bcl-2 binding sites in ASPP2 and further validate our docking model, we designed an array comprising 41 overlapping peptides derived from the Ank-SH3 domains of ASPP2 sequence (Table S4). Bcl-2 1–217 was expressed, purified, and screened for binding the peptide array (Fig. 4A and Table 2). Peptides from the Bak protein, which are known to bind Bcl-2 (2), served as positive control (not shown). Bcl-2 bound several peptides derived from the ASPP2 Ank-SH3 domains (Fig. 4A and Table 2), which were in agreement with our docking model. They are derived from three major binding sites that form a continuous binding interface (Fig. 4B): (i) an unstructured loop at the beginning of the SH3 domain, represented by ASPP2 1065–1082; (ii) a loop following the first ankyrin repeat, represented by ASPP2 945–961 and ASPP2 931–942; and (iii) the loop following the second ankyrin repeat, represented by ASPP2 978–988, ASPP2 988-1002, and ASPP2 993-1004. The binding peptides discovered in the array screening were synthesized, and their bindings to Bcl-2 1–217 were quantitatively studied by using fluorescence spectroscopy. Five of six peptides derived from the ASPP2 binding interface, as was revealed by our docking model, bound Bcl-2 with nanomolar affinity (Fig. 4 C–E and Table 2). ASPP2 1039–1053, the only selected peptide derived from the other interface of the protein that is not involved in Bcl-2 binding according to our model, did not show any binding. We assume that the positive signal in the peptide array is an artifact in this case.
Fig. 4.
Bcl-2 binding sites in ASPP2. (A) An array consisting of overlapping peptides derived from ASPP2 was screened for binding Bcl-2. Each dark spot represents binding of Bcl-2 to a specific peptide (Table 2). (B) The Bcl-2 binding sites, as discovered in the peptide array screening, are colored blue on the 3D structure of ASPP2Ank-SH3 (PDB entry 1YCS). (C–E) Quantitative analysis. Shown are representative binding curves of the interaction between Bcl-2 and the peptides derived from ASPP2Ank-SH3 as indicated. Binding was quantified by using fluorescence spectroscopy as described in Materials and Methods. For binding affinity, see Table 2.
Table 2.
Binding of Bcl-2 protein to peptides from ASPP2Ank-SH3: Fluorescence spectroscopy studies
| Parent protein | Peptide no. in array | Residues | Sequence | Kd, nM |
|---|---|---|---|---|
| ASPP2 | 14 | 931–942 | LDSSLEGEFDLV | 142 ± 38 |
| 15 | 945–961 | IIYEVDDPSLPNDEGIT | 210 ± 36 | |
| 23 | 988–1002 | ADSDGWTPLHCAASC | 158 ± 17 | |
| 24 | 993–1004 | *WTPLHCAASCNN | 476 ± 40 | |
| 32 | 1039–1053 | GYTQCSQFLYGVQEK | No binding | |
| 37 | 1065–1082 | LWDYEPQNDDELPMKEGD | 96 ± 17 |
*Trp was added at the N terminus of some peptides for UV spectroscopy.
†ASPP2 978–988 was not tested because we were unable to label it with fluorescein.
Discussion
Using a combination of peptide array screening, biophysical methods, and docking studies, we characterized the interaction between ASPP2 and the Bcl-2 family proteins. We present evidence for a direct interaction of ASPP2 with the Bcl-2 family proteins Bcl-XL and Bcl-W. We identified the binding sites in both ASPP2 and Bcl-2 family proteins, demonstrated that binding of ASPP2 to the Bcl-2 family proteins is selective with preference to the BH4 domain of Bcl-2, and proposed a model for the structure of the complex between the proteins. Our computational model was in excellent agreement with our experimental data from the peptide array screening and quantitative binding studies.
Our results show that peptides are a reliable model for the ASPP2–Bcl interaction. (i) For each Bcl protein tested, ASPP2 bound only some peptides but not all of them, and most of the binding peptides were overlapping, indicating specific binding of particular Bcl sequences. (ii) Nearly all of the ASPP2 binding residues in Bcl-2 are surface-exposed in the full-length protein, further supporting their importance for the interaction with ASPP2. (iii) The ASPP2 binding sites on all Bcl proteins tested were homologous and known to be involved in PPI.
Molecular Basis of the ASPP2–Bcl Interactions.
The SPR studies showed selectivity in the binding of ASPP2Ank-SH3 to the Bcl-derived peptides. The peptide array screening results, although semiquantitative, are in good agreement with the SPR results (compare peak intensities in Fig. 1A). The docking studies, sequence alignment, NMR studies, and binding studies of mutated peptides all suggest that the extra positive charge of the Bcl-derived peptides is the reason for the selectivity in binding. The moderate ionic strength dependence of the interaction and the specific residues found by NMR to mediate the binding of the Bcl-XL peptide suggest that the binding is mediated mainly by specific local interactions between amino acids in both interaction sites in the Bcl proteins. Most of these residues are positively charged, and they bind negative amino acids in ASPP2. Binding does not seem to be governed by a global electrostatic effect. The tight binding of ASPP2 to Bcl-2 peptides may suggest that Bcl-2 is the major target protein from this family for apoptosis stimulation by ASPP2. The two sites identified in our study are in close spatial proximity, and it is likely that ASPP2 binds both of them in a cooperative manner: binding to the high-affinity BH4 site is followed by binding to the low-affinity proapoptotic site. Thus, apoptosis may be regulated by the ASPP2-Bcl interaction simultaneously via both binding sites (Fig. 5). The affinity of ASPP2 to the Bcl-2 BH4 peptide is at the low-micromolar range, while the affinity of Bcl-2 to the ASPP2-derived peptides is submicromolar. The order of magnitude difference is probably due to the use of peptides, which form only part of the binding site.
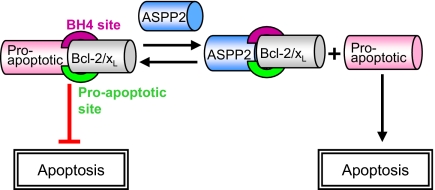
Fig. 5.
Proposed mechanism for induction of apoptosis after binding of ASPP2 to Bcl-2 family proteins. ASPP2 binds the antiapoptotic Bcl-2 family members at two functional sites and mediates apoptosis by inducing the release of other proapoptotic proteins that also bind Bcl-2 at those sites, which may enable their apoptotic activity, as specified in the text.
In our recent studies we discovered an intramolecular interaction between the Ank-SH3 domains and the proline-rich region in ASPP2 (14). The site in the Ank-SH3 domain that binds the proline-rich domain (residues 931–961) overlaps the binding site we found for Bcl-2. This supports our model that the intramolecular interaction in ASPP2 regulates the intermolecular interactions of the protein (14).
Implications for Apoptosis Regulation by ASPP2: A Proposed Mechanism.
Binding between the proapoptotic ASPP2 and antiapoptotic Bcl-2 family members is likely to maintain the balance between the pro- and antiapoptotic activity of both proteins. Based on our in vitro results, we propose a mechanism by which ASPP2 regulates apoptosis by preventing the antiapoptotic function of Bcl-2 (Fig. 5). It may do so by (i) binding the BH4 domain, which is essential for inhibition of apoptosis, and blocking its activity; or (ii) binding at the known binding site for proapoptotic proteins of the Bcl-2 family, inhibiting their binding to Bcl-2 and allowing their apoptotic activity. For example, the BH4 domains of Bcl-XL and Bcl-2 are known to bind and inhibit the proapoptotic mitochondrial protein VDAC (28). Binding of ASPP2 to Bcl-2 via the same site may release VDAC and allow its activity of releasing cytochrome C and inducing apoptosis. ASPP2 may also activate apoptosis by binding the known binding site for proapoptotic regulators from the Bcl-2 family such as Bak/Bak (8, 9), inducing their release from the complex, resulting in proapoptotic protein homooligomerization, mitochondrial membrane permeabilization, and apoptosis.
Our structural model for the ASPP2–Bcl-2 interaction shows that the interacting peptides from both proteins do not bind in cavities but rather bind extended protein–protein interaction interfaces. These interfaces should be the target for further development of inhibitory peptides and small molecules. ASPP2 controls cell fate and hence is an important potential target for the development of anticancer drugs that will stimulate apoptosis by interfering with its regulation. Understanding the ASPP2–Bcl-2 interaction at the molecular and cellular levels could serve as a basis for such novel anticancer drug leads.
Materials and Methods
Expression and Purification of ASPP2Ank-SH3.
pRSET HLT ASPP2 893-1128 vector (kind gift from A. R. Fersht, University of Cambridge) was expressed, and the protein was purified as described in ref. 14.
Expression and Purification of Bcl-2 1–217.
pHISparallel BCl-2 1–217 vector (provided by V. Shoshan-Barmatz, Ben-Gurion University of the Negev) was transformed into E. coli BL21 cells. Cultures were grown at 37°C in 2× YT medium to an optical density at 600 nm of ≈0.8 and induced with 1 mM isopropyl-1-thio-β-d-galactoside (IPTG). Cells were harvested after 16 h of incubation at 22°C. The protein was purified on a fast-flow, 7.8-ml Ni Sepharose HP (100 × 10 mm) column (Amersham Biosciences), using an FPLC system (ÁKTA Explorer; Amersham Biosciences). The sample was further purified by filtration on a Sephacryl S100 column (950 × 26 mm; Amersham Biosciences). Ten percent glycerol was used throughout the purification to reduce aggregation.
Peptide Array Screening.
The Bcl peptide array was synthesized by JPT. The peptides were acetylated at their N terminus (3). Binding of human ASPP2Ank-SH3 to the cellulose-bound peptides was screened according to the protocol by Rüdiger et al. (5) with slight modifications as described in ref. 14. The binding was detected by using rabbit anti-ASPP2Ank-SH3 antibody that was produced in house.
A second peptide array, derived from the ASPP2 sequence, was synthesized by INTAVIS Bioanalytical Instruments (29). The peptide array was immersed for 4 h in blocking solution (BS) [50 mM Tris·HCl (pH 7.5), 0.15 M NaCl, 0.05% Tween 20, nonfat dry milk 3.5%] and prewashed three times in TBST. Bcl-2 in final concentration of 2 μM was diluted with BS and incubated with the array overnight at 4°C. Washing steps included two times for 5 min in BS and three times for 5 min TBST. The binding was detected with Bcl-2-specific polyclonal rabbit antiserum, using a chemiluminescence blotting substrate Super Signal reagent (Beit Haemek) according to the manufacturer's instructions.
Peptide Synthesis and Purification.
Peptides were synthesized on Applied Biosystems 433A peptide synthesizer, using standard Fmoc chemistry as described in ref. 27. Peptides were biotin- or fluorescein-labeled at the N terminus as described in refs. 27 and 29.
Enzyme-Linked Immunosorbent Assay (ELISA).
ELISA detecting of the binding between biotinylated peptides (derived from Bcl-2 proteins) to ASPP2Ank-SH3 adsorbed to MaxiSorp plates (Nunc) was performed as described in ref. 30.
Fluorescence Spectroscopy.
Measurements were performed as described in refs. 14 and 26. Briefly, the fluorescein-labeled peptide [0.1 μM in 20 mM Hepes buffer (pH 7.3), 42 mM NaCl, 5% glycerol, and 5 mM DTT] was placed in a cuvette, and the nonlabeled protein was added at 1-min intervals and stirred for 10 sec. Dissociation constants (Kd) were calculated by fitting the fluorescence titration curves (corrected for dilution) to 1:1 binding model (14, 26).
Kinetic Measurements.
All experiments were performed by using a Bio-Rad ProteOn XPR36 array biosensor that provides high throughput and a parallel processing approach with an SPR-based detector. The system was equilibrated with PBS-T buffer [20 mM Na-phosphate, 5 mM DTT, 105 mM NaCl, 0.005% Tween 20 (pH 7.4)]. Each SPR experiment used multichannel detection. At each channel, a different biotinylated peptide was captured to a ProteOn NLC sensor chip at 25°C, using a flow rate of 30 μl/min. One clear channel served as reference. This resulted in peptides coupled at response levels of 100–500 RU. For binding measures, ASPP2Ank-SH3 was injected simultaneously (a single injection) at six different concentrations at a flow rate of 50 μl/min. The data for each analyte concentration series collected over the same target density surface were fit to the Langmuir 1:1 interaction model by using the integrated ProteOn Manager software. The average standard deviation was approximately ±5%. The equilibrium dissociation constant (Kd) was derived from the kdiss/kass ratio (kinetics). Data from the binding equilibria of the peptides were also used to calculate an affinity value independent of the binding kinetics (equilibrium).
Docking Analysis.
For the docking studies, we used PatchDock (21), which is based on shape complementarity of surface patches, and RosettaDock (22), which is based on simultaneous optimization of side-chain conformation and rigid body position of the two docking partners.
Supplementary Material
Acknowledgments.
We thank Prof. Gideon Schreiber, Prof. Atan Gross, Prof. Varda Shoshan-Barmaz, Prof. Moshe Kotler, Dr. Ora Furman, and Prof. Abraham Loyter for their invaluable help. This study was supported by a career development award from the Human Frontier Science Program Organization (to A.F.) and by a grant from the Israeli Cancer Association (to A.F.). H.B. is supported by a postdoctoral fellowship from the Lady Davis Fellowship Trust. S. Rüdiger was supported by a Marie Curie Excellence Grant of the European Union, a VIDI grant of the Netherlands Science Organization, and a High Potential grant of Utrecht University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711269105/DCSupplemental.
References
- 1.Lee LC, Hunter JJ, Mujeeb A, Turck C, Parslow TG. Evidence for α-helical conformation of an essential N-terminal region in the human Bcl2 protein. J Biol Chem. 1996;271:23284–23288. doi: 10.1074/jbc.271.38.23284. [DOI] [PubMed] [Google Scholar]
- 2.Sattler M, et al. Structure of Bcl-xL-Bak peptide complex: Recognition between regulators of apoptosis. Science. 1997;275:983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 3.Frank R. The SPOT-synthesis technique. Synthetic peptide arrays on membrane supports—Principles and applications. J Immunol Methods. 2002;267:13–26. doi: 10.1016/s0022-1759(02)00137-0. [DOI] [PubMed] [Google Scholar]
- 4.Reimer U, Reineke U, Schneider-Mergener J. Peptide arrays: From macro to micro. Curr Opin Biotechnol. 2002;13:315–320. doi: 10.1016/s0958-1669(02)00339-7. [DOI] [PubMed] [Google Scholar]
- 5.Rüdiger S, Germeroth L, Schneider-Mergener J, Bukau B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 1997;16:1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu GW, et al. The central region of HDM2 provides a second binding site for p53. Proc Natl Acad Sci USA. 2006;103:1227–1232. doi: 10.1073/pnas.0510343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansson LO, Friedler A, Freund S, Rüdiger S, Fersht AR. Two sequence motifs from HIF-1α bind to the DNA-binding site of p53. Proc Natl Acad Sci USA. 2002;99:10305–10309. doi: 10.1073/pnas.122347199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26:1324–1337. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris MH, Thompson CB. The role of the Bcl-2 family in the regulation of outer mitochondrial membrane permeability. Cell Death Differ. 2000;7:1182–1191. doi: 10.1038/sj.cdd.4400781. [DOI] [PubMed] [Google Scholar]
- 10.Bruncko M, et al. Studies leading to potent, dual inhibitors of Bcl-2 and Bcl-xL. J Med Chem. 2007;50:641–662. doi: 10.1021/jm061152t. [DOI] [PubMed] [Google Scholar]
- 11.Samuels-Lev Y, et al. ASPP proteins specifically stimulate the apoptotic function of p53. Mol Cell. 2001;8:781–794. doi: 10.1016/s1097-2765(01)00367-7. [DOI] [PubMed] [Google Scholar]
- 12.Slee EA, Lu X. The ASPP family: Deciding between life and death after DNA damage. Toxicol Lett. 2003;139:81–87. doi: 10.1016/s0378-4274(02)00421-6. [DOI] [PubMed] [Google Scholar]
- 13.Rotem S, Katz C, Friedler A. Insights into the structure and protein-protein interactions of the pro-apoptotic protein ASPP2. Biochem Soc Trans. 2007;35:966–969. doi: 10.1042/BST0350966. [DOI] [PubMed] [Google Scholar]
- 14.Rotem S, et al. The structure and interactions of the proline-rich domain of ASPP2. J Biol Chem. 2008;283:18990–18999. doi: 10.1074/jbc.M708717200. [DOI] [PubMed] [Google Scholar]
- 15.Gorina S, Pavletich NP. Structure of the p53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2. Science. 1996;274:1001–1005. doi: 10.1126/science.274.5289.1001. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi S, et al. 53BP2 induces apoptosis through the mitochondrial death pathway. Genes Cells. 2005;10:253–260. doi: 10.1111/j.1365-2443.2005.00835.x. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi N, et al. Inhibition of the 53BP2S-mediated apoptosis by nuclear factor κB and Bcl-2 family proteins. Genes Cells. 2005;10:803–811. doi: 10.1111/j.1365-2443.2005.00878.x. [DOI] [PubMed] [Google Scholar]
- 18.Espanel X, Sudol M. Yes-associated protein and p53-binding protein-2 interact through their WW and SH3 domains. J Biol Chem. 2001;276:14514–14523. doi: 10.1074/jbc.M008568200. [DOI] [PubMed] [Google Scholar]
- 19.Sugioka R, et al. BH4-domain peptide from Bcl-xL exerts anti-apoptotic activity in vivo. Oncogene. 2003;22:8432–8440. doi: 10.1038/sj.onc.1207180. [DOI] [PubMed] [Google Scholar]
- 20.Petros AM, et al. Rationale for Bcl-xL/Bad peptide complex formation from structure, mutagenesis, and biophysical studies. Protein Sci. 2000;9:2528–2534. doi: 10.1110/ps.9.12.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneidman-Duhovny D, et al. Taking geometry to its edge: Fast unbound rigid (and hinge-bent) docking. Proteins. 2003;52:107–112. doi: 10.1002/prot.10397. [DOI] [PubMed] [Google Scholar]
- 22.Gray JJ, et al. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J Mol Biol. 2003;331:281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 23.Oltersdorf T, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 24.Petros AM, et al. Solution structure of the antiapoptotic protein bcl-2. Proc Natl Acad Sci USA. 2001;98:3012–3017. doi: 10.1073/pnas.041619798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manion MK, et al. Bcl-XL mutations suppress cellular sensitivity to antimycin A. J Biol Chem. 2004;279:2159–2165. doi: 10.1074/jbc.M306021200. [DOI] [PubMed] [Google Scholar]
- 26.Friedler A, et al. A peptide that binds and stabilizes p53 core domain: Chaperone strategy for rescue of oncogenic mutants. Proc Natl Acad Sci USA. 2002;99:937–942. doi: 10.1073/pnas.241629998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayouka Z, et al. Inhibiting HIV-1 integrase by shifting its oligomerization equilibrium. Proc Natl Acad Sci USA. 2007;104:8316–8321. doi: 10.1073/pnas.0700781104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimizu S, Konishi A, Kodama T, Tsujimoto Y. BH4 domain of antiapoptotic Bcl-2 family members closes voltage-dependent anion channel and inhibits apoptotic mitochondrial changes and cell death. Proc Natl Acad Sci USA. 2000;97:3100–3105. doi: 10.1073/pnas.97.7.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coster G, et al. The DNA damage response mediator MDC1 directly interacts with the anaphase promoting complex/cyclosome. J Biol Chem. 2007;282:32053–32064. doi: 10.1074/jbc.M705890200. [DOI] [PubMed] [Google Scholar]
- 30.Rosenbluh J, et al. Positively charged peptides can interact with each other, as revealed by solid phase binding assays. Anal Biochem. 2006;352:157–168. doi: 10.1016/j.ab.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 31.Hinds MG, et al. The structure of Bcl-w reveals a role for the C-terminal residues in modulating biological activity. EMBO J. 2003;22:1497–1507. doi: 10.1093/emboj/cdg144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delano WL. San Carlos, CA: DeLano Scientific; 2002. The PyMOL Molecular Graphics System. www.pymol.org. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.