Abstract
The expression of interleukin-4 (IL-4) is viewed as the hallmark of a Th2 lymphocyte, whereas the subsequent action of IL-4 and IL-13, mediated through the STAT6 signaling pathway, is seen as a prerequisite for the full development of Th2 immune responses to parasites and allergens. G4 mice, whose IL-4 gene locus contains the fluorescent reporter eGFP, were used to quantify the number of Th2 cells that develop during Nippostrongylus brasiliensis- or allergen-induced immune responses under conditions where IL-4 or STAT6 was absent. Here, we show that deletion of IL-4 or STAT6 had little impact on the number or timing of appearance of IL-4-producing Th2 cells. These data indicate that in vivo differentiation of naïve CD4 T cells to Th2 status often occurs independently of IL-4 and STAT6 and that recently described pathways of Th2 cell differentiation may explain how allergens and parasites selectively induce Th2-mediated immunity.
Keywords: allergen, cytokine, asthma, Nippostrongylus
An exquisite feature of the adaptive immune system is that the specific properties of different infectious agents or their toxins are effectively countered by the selective development of appropriate effector immune responses. The key to this process is the antigen-driven differentiation of naïve CD4 lymphocytes into distinct T helper (Th) cell subsets, each of which is able to direct its own type of effector immune response through the production of specific cytokines (1). One of most striking examples of this selective process is the differentiation of Th2 cells, which mediate allergic diseases and control metazoan parasites. A central paradigm of the Th2 immune response is that Th2 differentiation is regulated by the cytokine IL-4 and its signaling pathway STAT6 (2–4). However, the issue of how relevant IL-4/STAT6 is to the development of in situ T cell immune responses during natural infections or disease-relevant immune responses is not fully resolved (5–8). The initial source of IL-4 in this process has remained elusive, and although IL-4 and STAT6 have been firmly implicated in the production of serum IgE and IL5-mediated eosinophilia (3, 4, 9–11), the difficulties of measuring IL-4 in situ have hampered more direct studies on the role of IL-4 in the development of IL-4-producing Th2 cells. Finkelman et al. (12) have used an antibody capture assay to measure serum IL-4 and showed that production of IL-4 is not entirely STAT6-dependent. However, the use of indirect cytokine capture techniques could not exclude the contribution of eosinophil- (13), NKT cell- (9), or basophil- (14) derived IL-4 to this response. In studies of Schistosoma mansoni-infected IL-4 receptor (IL-4R)- or STAT6-deficient mice, it was shown that although IL-4-producing Th2 cells could be generated in the absence of IL-4 or the STAT6 signaling pathway, the numbers of IL-4-producing CD4 T cells were significantly reduced after ex vivo culture and restimulation (15). Similarly, in studies using the 4get reporter mouse strain, it was reported that Th2 cells could be generated in STAT6-deficient mice (16).
The development of G4 mice, which express GFP as a surrogate for IL-4 expression, has made it possible to follow directly the development of IL-4-producing immune cells in situ (14, 17). The replacement of the first exon and first intron of the Il4 gene by the gene encoding enhanced GFP creates mice that are IL-4-deficient when bred in the homozygous form and IL-4-sufficient when maintained as a heterozygous strain. By comparing immune responses in the two strains of mice, we took the opportunity to study the role of IL-4 in the in vivo development of Th2 cells. By using the greater sensitivity and longer half-life of the eGFP reporter system, those CD4+ Th2 lymphocytes that had expressed the Il4 gene locus in vivo could be detected directly, avoiding the use of in vitro reactivation culture systems, which only provide an indirect measure of the reactivation potential of Th2 cells.
Infection of mice with the nematode Nippostrongylus brasiliensis (Nb) is a well characterized experimental system for the study of Th2 immune responses. Specific features of the Th2 response to Nb infection include the rapid (5–10 days) development of highly polarized Th2 cells producing high levels of IL-4, IL-5, IL-13, and the sequestration of these Th2 cells to lung and gut tissues leading to the clearance of the worms from the gut (18). A convenient feature of the Nb infection model is that the infection is rapid and transient with no replication of worms. Here, we used the G4 reporter system to follow the development and fate of IL-4-producing Th2 cells after infection with Nb in IL-4- and STAT6-deficient animals. Cells were harvested from key lymphoid and nonlymphoid tissue sites, and the percentage and number of GFP-positive CD4 T cells were determined. Having established the in vivo pattern of induction of GFP/IL-4-producing Th2 cells, we were able to address the question of how the magnitude, timing, and character of the Th2 immune response was affected by the absence of IL-4 or of the STAT6 signaling pathway. Our results show that deletion of IL-4 or STAT6 had very little effect on the timing or magnitude of the IL-4/GFP response. Furthermore, analysis of sorted GFP-positive Th2 cells from either IL-4+/G4, IL-4G4/G4, or STAT6−/− IL-4G4/G4 Nb-infected mice revealed that all had similar highly enriched levels of Th2 cytokines and GATA-3. Our results clearly indicate that, contrary to the paradigm that has been developed from previous in vitro and indirect in vivo studies, Th2 differentiation in vivo in response to Nb infection is independent of IL-4 and STAT6.
Results
In Vitro Requirements for Th2 Differentiation.
As a first step, we sought to confirm that in vitro T cell responses using cells from G4 mice replicated the well established observations seen in wild-type (WT) mice. As reported, IL-4 and STAT6 were essential for in vitro αCD3/αCD28 culture-induced Th2 differentiation using the G4 reporter assay (2, 4). After restimulation, CD4 T cells derived from both IL-4+/G4 STAT6+/+ (Fig. 1A) and IL-4+/G4 STAT6−/− (Fig. 1B) mice failed to express IL-4 or GFP when cultured in Th neutral or Th1 conditions (Fig. 1A). However, CD4 T cells derived from IL-4+/G4 STAT6+/+ mice and cultured under Th2 conditions expressed IL-4 (6%), GFP (8%), and IL-4+ GFP+ (1%) as described in ref. 17. In comparison, IL-4+/G4 STAT6−/− CD4+ cells failed to express IL-4 or GFP after activation under Th2 conditions and instead produced IFNγ (8%) (Fig. 1B). Therefore, using GFP as a surrogate marker for IL-4 expression by CD4 T cells, we confirmed that IL-4 signaling through STAT6 is required for expression of both IL-4 and GFP. The expression of GFP mRNA by activated CD4 T cells was also found to correlate with the previously identified “Th2 genes” IL-13, IL-5, IL-10, and GATA-3 (Fig. 1C), extending the evidence that GFP expression in the G4 model system is a faithful indicator of differentiation to Th2 status.
Fig. 1.
Th2 differentiation after αCD3/αCD28 stimulation requires IL-4 and STAT6 signaling. (A and B) FACS analysis of CD4+ T cells from IL-4+/G4 (A) and IL-4+/G4 STAT6−/− (B) mice after in vitro Th neutral (TH0), 1, and 2 differentiation and restimulation. Results shown are representative of three separate experiments. (C) Real-time PCR analysis of IL-4, GFP, IL-13, and GATA-3 in sorted CD4+GFP− (open bars) and CD4+GFP+ lymphocytes (solid bars) after in vitro TH2 differentiation with 6-h restimulation. N.D., not determined.
In Vivo Requirements for Th2 Differentiation.
To assess the role of IL-4 and STAT6 signaling in an in vivo context, WT, IL-4+/G4 STAT6+/+, IL-4G4/G4 STAT6+/+, IL-4+/G4 STAT6−/−, and IL-4G4/G4 STAT6−/− were infected with the nematode parasite Nb. The numbers of lymphocytes, CD4 T cells, and CD4+ GFP+ Th2 cells appearing in the lung-draining mediastinal lymph node (Md) (Fig. 2), mesenteric lymph node (Ms) [supporting infor-mation (SI) Figs. S1–S5], and skin draining lymph nodes (d.Ln) (Figs. S1–S5) were measured during the course of both the primary and secondary infections. The CD4+GFP+ Th2 response peaked at day 9–12 after primary infection and day 5–7 after secondary infection. The deletion of IL-4 or STAT6 did not affect the number of total lymphocytes, CD4+ T cells, or CD4+GFP+ T cells appearing at the peak of response, nor were the kinetics of the expansion or contraction phases significantly altered (Fig. 2 and Figs. S1–S4). Although previous reports have indicated that STAT6-specific sites are integral for the establishment of a memory conformation at the Th2 locus (19), we found that IL-4- and STAT6-deficient animals did not display any evidence of a failure to induce a Nb-specific memory Th2 response (12). Furthermore, although it has been observed that STAT6-deficient animals develop Th2 cells in response to Nb (16), here we quantify the numbers of CD4+GFP+ cells that evolve during response to Nb, and we demonstrate that neither IL-4 nor STAT6 is required for a full Th2 response.
Fig. 2.
In vivo Th2 differentiation after Nb infection proceeds independently of IL-4/STAT6 signaling. Flow cytometric analysis of mediastinal lymph node responses to primary and secondary intradermal Nb infection in IL-4+/+ (●), IL-4+/G4 (■), and IL-4G4/G4 (▲), IL-4+/+ STAT6−/− (○), IL-4+/G4 STAT6−/− (□), and IL-4G4/G4 STAT6−/− (△) mice is shown. (A) Kinetics of total live cell counts. (B) Live CD4+ cell counts. (C) Percentage of CD4+ cells expressing GFP. (D) Total number of CD4+GFP+ cells. Data points shown indicate mean ± SE from three individual animals. Live infection was performed simultaneously on each strain with data shown representing an independent experiment. (E) At day 9 after infection, mesenteric lymph node CD4+GFP− and CD4+GFP+ cells were isolated and sorted by flow cytometry. Total RNA was isolated from sorted cells and analyzed by real time-PCR for the presence of IL-4, GFP, IL-13, IL-5, and GATA-3 transcripts, as indicated. For each real-time PCR analysis, data were normalized to 18S RNA, and total mRNA present was calculated by comparison with a serially diluted standard. Results consistent with the data shown have been obtained in one repeat independent experiment.
In Vivo-Derived CD4+GFP+ Cells Have a Th2 Gene Expression Pattern.
CD4+GFP+ T cells were purified from the lymph nodes of IL-4+/G4, IL-4G4/G4, IL-4+/G4 STAT6−/−, and IL-4G4/G4 STAT6−/− mouse strains at the day 9 peak of their response to Nb infection and analyzed for expression of the Th2-associated genes IL-4, GFP, IL-13, and GATA-3 (Fig. 2E). GFP-expressing CD4+ cells sorted from IL-4+/G4, IL-4G4/G4, IL-4+/G4 STAT6−/−, and IL-4G4/G4 STAT6−/− mouse strains expressed comparable levels of GFP, IL-13, and GATA-3 mRNA (Fig. 2E). Most importantly, CD4+GFP+ cells from IL-4+/G4 and IL-4+/G4 STAT6−/− mice expressed comparable levels of IL-4, IL-13, and GATA-3 mRNA, confirming that the CD4+GFP+ T cells derived from STAT6−/− mice were “bona fide” Th2 cells. In particular, this finding implies that GATA-3, known to be essential for the differentiation in response to Nb infection (20), is induced in the absence of the engagement of the IL-4/STAT6 pathway.
A comparison of the surface marker phenotype of CD4+GFP− and CD4+GFP+ cells derived from Md lymph nodes at the day 9 peak of infection found significant differences in CD62L (Fig. 3A), CD44 (Fig. 3A), and CD69 (Fig. 3B) expression consistent with the GFP+ cells possessing an activation phenotype. Although the CD4+GFP+ T cells represent a relatively minor (3–5%) subset of cells present in the lymph node at the peak of response, they exhibit a highly activated phenotype compared with the CD4+GFP− population. In all three G4 strains analyzed, the CD4+GFP+ populations recovered from the lymph nodes comprised >80% CD44Hi cells and >50% CD44Hi CD62llo cells. These data indicate that acquisition of the capacity to express GFP is coincident with a highly activated surface marker phenotype. Importantly, a comparison of the activation profiles for both CD4+GFP− and CD4+GFP+ cells revealed no major difference between each of the mouse strains analyzed. Taken together, CD4+GFP+ cells that develop in Nb-infected mice exhibit a highly activated phenotype according to expression levels of CD62L, CD44, and CD69. The finding that the activation phenotype was similar irrespective of whether CD4 T cells were isolated from either IL-4+/G4, IL-4G4/G4, or IL-4G4/G4 STAT6−/− mice indicates that neither IL-4 nor its key signaling intermediate STAT6 was needed for generation of fully activated CD4+GFP+ Th2 cells.
Fig. 3.
Phenotype of in vivo generated CD4+GFP+ cells and CD4+GFP− cells. (A) Naïve IL-4+/+, IL-4+/G4, IL-4G4/G4, IL-4+/+ STAT6−/−, and IL-4G4/G4 STAT6−/− mice were infected with Nb. At day 9 after infection mediastinal lymph nodes were harvested, and CD4+GFP+ and CD4+GFP− cells were analyzed for the expression of CD44 and CD62Ll by flow cytometry. (B) CD69 expression in CD4+GFP− (shaded) and CD4+GFP+ (dashed line) populations was analyzed. FACS plots are representative of three animals per group. Data shown represent one of multiple independent experiments.
Effect of IL-4/STAT6 Deficiency on Th2 Effector Responses.
The effect of IL-4/STAT6 deficiency on Nb infection-induced airway eosinophilia and B cell IgE class switching was found to depend on IL-4 and STAT6 signaling, as reported (3, 4) (Fig. 4 A and B). Furthermore, the recruitment of eosinophils to the airways was decreased in the absence of IL-4 and almost abrogated in animals lacking STAT6 (Fig. 4B). Interestingly the absence of IL-4 did not significantly alter the kinetics of the diminished eosinophil infiltration, with peak airway eosinophilia occurring at day 11 after primary and day 9 after secondary infection in both models. Serum IgE was followed in the Nb-infected mice by fluorometric bead assay (FBA) and was only detectable in samples taken from IL-4-sufficient mice (Fig. 4C). Also, analysis of the day 15 peak serum IgE response in the indicated mouse strains confirmed that STAT6 signaling was required for IgE production (Fig. 4D). These results demonstrate the absolute requirement for IL-4/STAT6 signaling in the induction of an IgE response (3, 4) and confirm that the IL-4G4/G4 strain represents a selective and complete inactivation of in vivo IL-4 activity.
Fig. 4.
Induction of type 2 effector responses in the absence of IL-4/STAT6 signaling. (A and B) Kinetics of blood (A) and BAL eosinophilia (B) after primary and secondary s.c. Nb infection were determined by flow cytometry in IL-4+/+ (●), IL-4G4/G4 (▲), IL-4+/+STAT6−/− (○), and IL-4G4/G4STAT6−/− (△) mice. (C) Serum IgE levels were determined by FBA in IL-4+/+ (●), IL-4+/G4 (■), and IL-4G4/G4 (▲) mice infected with Nb as above. (D) Naïve IL-4+/+, IL-4+/G4, IL-4G4/G4, IL-4+/+ STAT6−/−, IL-4G4/G4 STAT6−/−, and IL-4G4/G4 STAT6−/− mice were infected with Nb via s.c. injection. At day 15 after infection, peak serum IgE concentration was determined by FBA. Data points shown indicate mean ± SE from three individual animals. Data shown represent one independent experiment. *, IgE concentration below the limit of detection.
Effect of IL-4 Deletion on Other Models of in Vivo TH2 Induction.
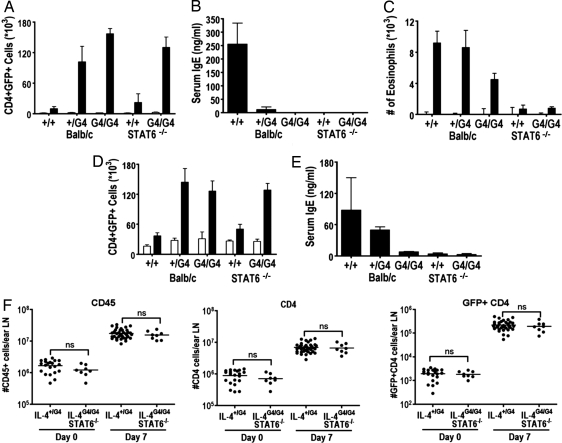
The Th2-priming models seen in allergic airway immunity using keyhole limpet hemocyanin (KLH) sensitization (21), the αIgD injection model (22), and the intradermal ear injection model using dead Nb were also assessed in the G4 assay system (Fig. 5). Flow cytometric analysis of the number of CD4+GFP+ T cells in the Md lymph node draining the airways of KLH-challenged mice revealed that similar numbers of CD4+GFP+ cells were induced in both the presence and absence of IL-4/STAT6 signaling (Fig. 5A). Similar results were obtained in mice that were challenged with OVA by the intranasal route (data not shown), although in all groups the numbers of CD4+GFP+ cells were lower than those induced by KLH, as would be expected on the basis of the lower antigenic complexity of OVA compared with KLH. However, production of IgE was abrogated in the IL-4G4/G4, IL-4+/+STAT6−/−, and IL-4G4/G4 STAT6−/− mice (Fig. 5B), and levels of BAL eosinophilia were diminished in the absence of IL-4 and almost fully inhibited in mice lacking STAT6 (Fig. 5C). Similarly, the numbers of CD4+GFP+ T cells induced by injection of αIgD mAbs (250 μg of Hδa/1 and 250 μg of FF1-4D5) were found to be similar in IL-4+/G4, IL-4G4/G4, IL-4+/+STAT6−/−, and IL-4G4/G4 STAT6−/− mice (Fig. 5D). However, IgE responses were abrogated in the absence of IL-4 or STAT6 as observed (Fig. 5E). Further, when dead Nb were injected intradermally into the ears of IL-4G4/G4 IL-4+/+ STAT6G4/G4 mice (Fig. 5F), no detectable difference could be detected in the numbers of CD45+, CD4+, or CD4+GFP+ cells present at the peak (day 7) of the ear-draining lymph node response. Taken together, these data indicate that the IL-4/STAT6-independent Th2 differentiation observed using the G4 reporter mice represents a genuinely distinct pathway of Th2 induction from that observed when using in vitro culture conditions.
Fig. 5.
Effects of IL-4/STAT6 deficiency on CD4+GFP+ induction and Th2 effector responses after KLH-induced airway inflammation, anti-IgD injection, or ear model of Th2 induction. (A) Cytometric analysis of mediastinal lymph node CD4+GFP+ cell numbers. (B) FBA of serum IgE levels after KLH challenge. (C) BAL eosinophilia after administration of KLH challenge. (D) Cytometric analysis of mediastinal lymph node CD4+GFP+ cell numbers. (E) FBA of serum IgE levels at day 7 after administration of αIgD. Data points shown indicate mean ± SE from three individual animals. (F) Cytometric analysis of ear-draining lymph node cell numbers after injection of 600 dead L3 Nb. Naive mice (open bars), challenged mice (solid bars). Data were compiled from multiple independent experiments. Statistical analysis was performed by one-way ANOVA with Turkey's post hoc testing, P < 0.001 except where indicated by ns (no significant difference, P > 0.95).
Discussion
The reigning dogma of Th2 biology remains that IL-4/STAT6 signaling is required for full Th2 differentiation in vivo, after which Th2 cells orchestrate effector responses to parasitic infections and mediate allergic disease (1, 19, 23–26). To quantify more accurately the in vivo function of the IL-4/STAT6 signaling pathway in Th2 immune responses, we used G4 mice to address the question of how the magnitude, timing, and character of the Th2 response were affected by the absence of IL-4 and/or STAT6 signaling. In using the G4 murine model system backcrossed to STAT6−/− mice, we found that in direct contrast to the observation that IL-4/STAT6 is absolutely required for in vitro Th2 differentiation, deletion of the Il4 gene or its STAT6-signaling pathway had little impact on the magnitude or timing of Th2 differentiation in several in vivo settings. Furthermore, the absence of IL-4 and/or STAT6 did not noticeably alter the expansion or kinetics of CD4+ cells during the memory response, and in particular there was no discernable decrease in the total numbers of Th2 cells evoked during the memory response, further indicating the redundancy of the IL-4/STAT6 signaling pathway in vivo.
Although IL-4/STAT6 signaling was demonstrated to be dispensable for Th2 differentiation in the four models examined, IL-4/STAT6 signaling was still found to be essential for key effector arms of the type 2 response, such as the optimal induction and migration of eosinophils and for B cell class switching and production of IgE as shown in previous studies (3, 4, 10, 12, 27). Therefore, a major role for IL-4 is in promoting and directing type 2 effector responses, whereas it is dispensable for directing the fate of responding CD4+ T cells in the experimental models we have tested. Thus, IL-4 was not required for robust Th2 induction, including up-regulation of GATA-3, in responses to living or killed Nb, to induction of airway inflammation to KLH, and in responses to anti-IgD injection. Whether other in vivo Th2 differentiation processes require IL-4 is not resolved. For example, Medzhitov and colleagues (28) have argued that responses to the cysteine protease papain are at least partially IL-4-dependent. In sum, our data, taken together with previous results, indicate the existence of a robust pathway of Th2 differentiation. Among the mechanisms proposed to underlie this “IL-4-independent” Th2 differentiation are responses driven by T cell costimulatory receptors including OX40 (29, 30), ICOS (31), TIM-1 (32), and Notch Delta/Jagged (33). How these molecules actually cause Th2 differentiation remains to be established, and whether additional in vivo pathways exist needs to be determined.
Methods
Mice.
The generation of GFP/IL-4 knockin (G4) mice has been reported in ref. 17. Briefly, G4 mice were backcrossed to BALB/c mice for 10 generations; they were obtained from the National Institute of Allergy and Infectious Diseases (NIAID)/National Institutes of Health contract facility at Taconic Farms. BALB/c and BALB/c STAT6−/− were also obtained from a NIAID/NIH facility. BALB/c IL-4+/G4 mice were obtained by crossing IL-4G4/G4 mice with BALB/c WT mice. BALB/c IL-4G4/G4 STAT6−/− mice were obtained by crossing G4 mice with BALB/c STAT6−/−. BALB/c IL-4G4/G4 STAT6−/− mice were obtained by crossing BALB/c IL-4G4/G4 STAT6−/− mice with BALB/c STAT6−/−. All experimental procedures involving animals were approved by the Victoria University Animal Ethics Committee and carried out in accordance with the guidelines set forth by the Monash Institute of Medical Research and Victoria University. Mice aged 6–12 weeks were used for experimental work, and mice were age-matched for each individual experiment.
Nb Infection.
Nb life cycle was maintained as described. Mice were inoculated with 600 third-stage larvae by either s.c. or intradermal injection (ear) as specified. Secondary infection was initiated at day 30 after primary infection. In experiments that examined responses to a secondary infection in STAT6−/− mice, primary infections were cured by treating mice by oral gavage with 1 mg of pyrantel pamoate (Strongid T; Pfizer) before secondary infection (34).
αIgD Injection Model.
Mice were injected s.c. with 250 μg of Hδa/1 and 250 μg of FF1-4D5 Abs in 0.2 ml of PBS. At day 8 after infection animals were euthanized, lymph nodes were removed, and CD4+GFP+ expression was analyzed by flow cytometry, as described in ref. 22.
KLH Airway Inflammation Model.
Mice were primed by injection (i.p.) with 4 μg of KLH (Calbiochem) in 200 μl of alum (20 μg/ml; Serva). Day 7 after prime infection, animals were challenged by intranasal application of 100 μg of KLH in 50 μl of PBS (2 mg/ml). Animals were euthanized, and lymph nodes were removed and analyzed for CD4+ GFP expression by flow cytometry, as described in ref. 21.
Ear Model of Th2 Induction.
Mice were injected with 600 dead Nb L3 larvae (killed by three freeze/thaw cycles) in the outer dermis of the ear. At indicated time points, mice were euthanized. The ear draining lymph node was taken, and single cell suspensions were prepared and analyzed by flow cytometry.
Tissue Preparation.
At the time points indicated, mice were euthanized, and cells were prepared from indicated tissues. Lymphocytes from the BAL were collected by lavage of the airways (three times with 1 ml of cold PBS). Lymph nodes draining the s.c. infection site (d.LN), mediastinal lymph nodes (Md), and mesenteric lymph nodes (Ms) were harvested as indicated, and single-cell suspensions were prepared by passing the organs through a nylon cell strainer. After centrifugation at 200 × g for 4 min, cells were collected, counted, and analyzed further by flow cytometry. Serum was isolated from blood, which was removed from the inferior vena cava and incubated at 4°C for 12 h before centrifugation at 3,000 × g and removal of serum supernatant.
Flow Cytometry.
Flow cytometry reagents were purchased from BD Biosciences. All cell suspensions were first incubated with 2.4G2 to block nonspecific binding of antibodies to Fc receptors. The following mAbs were used: CD45-PE, B220-biotin, CD4-APC, CD4-PerCp, IL-4APC, and IFNγ-PE. Flow cytometry data were acquired on a FACSCalibur (Becton Dickinson) flow cytometer. Data were analyzed by using FlowJo (TreeStar) software. Cell sorting was conducted by using a FACSVantage SE (Becton Dickinson).
Cell Culture.
For Th2 culture Ms LN cells were isolated as previously outlined and stimulated (3 × 106/ml) in tissue culture plates coated with αCD3 (5 μg/ml) in Iscoves modified Dulbecco's medium, 5% FCS with αCD28 (37:51, 1:50) in the presence of IL-2 (100 units/ml), IL-4 (1,000 units/ml), and IL-6 (2 ng/ml) for 5 days. Cells were then rested in Iscoves modified Dulbecco's medium, 5% FCS and IL-2 (100 units/ml) for 2 days. For restimulation, purified cells were cultured on tissue culture plates coated with 10 μg/ml αCD3 cells for 6 h in the presence of αCD28 (37:51, 1:50) and IL-2 (100 units/ml). For intracellular staining, Monensin was added for the last 2 h; quantitative PCR cells were taken at 6 h.
Quantitative RT-PCR.
Total RNA was isolated using TRIzol (Invitrogen), and cDNA was made using the SuperScript III first-strand synthesis systems for RT-PCR (Invitrogen), according to the manufacturer's directions. Quantitative real-time PCR analysis was performed on a 7500 Real Time PCR system (Applied Biosystems). Primers and MGB and FAM MGB probe sets for the murine cytokines IL-4, IL-5, IL-10, IL-13, GATA-3, and 18S ribosomal RNA (VIC MGB probe) were purchased from Applied Biosystems.
Primers and MGB probes for eGFP were custom-made by Applied Biosystems. Sequences for GFP primers and MGB probes are 5′-GCTACCCCGACCACATGAAG-3′, 5′-CGGGCATGGCGGACTT-3′, and 5′-FAM-CAGCACGACTTCTTC-3′. Relative and absolute quantification were conducted as described (35, 36). Briefly, cytokine-specific DNA was generated using the probe sets outlined. The DNA generated were run on 2% agarose gel with low DNA mass ladder (Invitrogen) to determine the concentration of DNA present. Serial dilutions of specific DNA were run in parallel with samples being tested to allow construction of a standard curve and calculation of specific cDNA concentration in each sample.
FBA.
A polylink coupling kit (Bangslab) was used to bind IgE (R1E4) to carboxylated polystyrene beads (Bangslab) according to product protocol. Briefly, beads were washed [10 mM Tris (pH 8.0), 0.05% BSA, 0.05% ProClin 300] and resuspended in coupling buffer [50 mM Mes (pH 5.2), 0.05% ProClin 300) before the addition of 200 mg/ml EDAC solution (carbodiimide 750) and 70 μg of Ab per 5 × 107 beads. Beads were then incubated at room temperature with mixing for 1 h. Beads were then washed again in wash buffer and stored at 4°C until required. Samples for analysis were prepared for analysis in Costar V-bottomed 96-well plates (Sigma–Aldrich). Ab-coated beads (1,000 per well) were added to wells prewetted with FBA buffer (0.5% BSA, 0.01% NaAz, 0.05% Tween 20), and beads were washed with PBS. Samples to be tested were diluted in FBA buffer and added to wells. Purified IgE was serially diluted in FBA buffer and added to the corresponding wells. Biotinylated detector reagents IgE (12.3) at 10 μg/ml in FBA buffer were added to corresponding wells and incubated at room temperature in the dark for 10 min. Wells were then washed twice with FBA buffer. Streptavidin-FITC (BD PharMingen) was diluted 1:1,000 in FBA buffer and added to wells, followed by incubation at room temperature in the dark for 10 min. Beads were then washed as above and resuspended in FBA buffer for analysis. Sample acquisition was conducted in a fluorescence-activated cell sorter (FACS) array (Becton Dickinson), and data were analyzed using FlowJo; at least 50 beads per sample were analyzed. Standard curves and regression analysis were calculated with Prism (GraphPad).
Statistical Analysis.
Statistical analysis for Fig. 5 was performed using one-way ANOVA with Turkey's post hoc testing to compare column variances. P > 0.001 except where indicated by ns (no significant difference detected). Day 0 IL-4+/G4 n = 15 and IL-4G4/G4 STAT6−/− n = 8, day 8 IL-4+/G4 n = 31 and IL-4G4/G4 STAT6−/− n = 8. Data were compiled from multiple experiments (Fig. S5). Statistical analysis was performed with Prism, using two-tailed Mann–Whitney U testing (*, P < 0.95, n = 3), with 2 degrees of freedom.
Supplementary Material
Acknowledgments.
We thank J. Zhu for helpful discussions and K. Price for expertise with cell sorting. This work was supported by Research Program funding from the Health Research Council of New Zealand, the Marjorie Barclay Trust, the Marsden Fund, AMI Insurance Ltd, and the Intramural Research Program of the National Institute of Allergy and Infectious Diseases/National Institutes of Health through Project Z01 AI000493.22.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806372105/DCSupplemental.
References
- 1.Murphy KM, et al. Signaling and transcription in T helper development. Annu Rev Immunol. 2000;18:451–494. doi: 10.1146/annurev.immunol.18.1.451. [DOI] [PubMed] [Google Scholar]
- 2.Le Gros G, Ben-Sasson SZ, Seder R, Finkelman FD, Paul WE. Generation of interleukin 4 (IL-4)-producing cells in vivo and in vitro: IL-2 and IL-4 are required for in vitro generation of IL-4-producing cells. J Exp Med. 1990;172:921–929. doi: 10.1084/jem.172.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kopf M, et al. Disruption of the murine IL-4 gene blocks Th2 cytokine responses. Nature. 1993;362:245–248. doi: 10.1038/362245a0. [DOI] [PubMed] [Google Scholar]
- 4.Kaplan MH, Schindler U, Smiley ST, Grusby MJ. STAT6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4:313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 5.Cote-Sierra J, et al. Interleukin 2 plays a central role in Th2 differentiation. Proc Natl Acad Sci USA. 2004;101:3880–3885. doi: 10.1073/pnas.0400339101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noben-Trauth N, Hu-Li J, Paul WE. IL-4 secreted from individual naïve CD4+ T cells acts in an autocrine manner to induce Th2 differentiation. Eur J Immunol. 2002;32:1428–1433. doi: 10.1002/1521-4141(200205)32:5<1428::AID-IMMU1428>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 7.Sun J, Pearce EJ. Suppression of Early IL-4 production underlies the failure of CD4 T cells activated by TLR-stimulated dendritic cells to differentiate into Th2 cells. J Immunol. 2007;178:1635–1644. doi: 10.4049/jimmunol.178.3.1635. [DOI] [PubMed] [Google Scholar]
- 8.Zhu J, Guo L, Watson CJ, Hu-Li J, Paul WE. STAT6 is necessary and sufficient for IL-4's role in Th2 differentiation and cell expansion. J Immunol. 2001;166:7276–7281. doi: 10.4049/jimmunol.166.12.7276. [DOI] [PubMed] [Google Scholar]
- 9.Noben-Trauth N, et al. An interleukin 4 (IL-4)-independent pathway for CD4+ T cell IL-4 production is revealed in IL-4 receptor-deficient mice. Proc Natl Acad Sci USA. 1997;94:10838–10843. doi: 10.1073/pnas.94.20.10838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimoda K, et al. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature. 1996;380:630–633. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- 11.Finkelman FD, et al. IL-4 is required to generate and sustain in vivo IgE responses. J Immunol. 1988;141:2335–2341. [PubMed] [Google Scholar]
- 12.Finkelman FD, et al. STAT6 regulation of in vivo IL-4 responses. J Immunol. 2000;164:2303–2310. doi: 10.4049/jimmunol.164.5.2303. [DOI] [PubMed] [Google Scholar]
- 13.Voehringer D, Shinkai K, Locksley RM. Type 2 immunity reflects orchestrated recruitment of cells committed to IL-4 production. Immunity. 2004;20:267–277. doi: 10.1016/s1074-7613(04)00026-3. [DOI] [PubMed] [Google Scholar]
- 14.Min B, et al. Basophils produce IL-4 and accumulate in tissues after infection with a Th2-inducing parasite. J Exp Med. 2004;200:507–517. doi: 10.1084/jem.20040590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jankovic D, et al. Single cell analysis reveals that IL-4 receptor/STAT6 signaling is not required for the in vivo or in vitro development of CD4+ lymphocytes with a Th2 cytokine profile. J Immunol. 2000;164:3047–3055. doi: 10.4049/jimmunol.164.6.3047. [DOI] [PubMed] [Google Scholar]
- 16.Mohrs M, Shinkai K, Mohrs K, Locksley RM. Analysis of type 2 immunity in vivo with a bicistronic IL-4 reporter. Immunity. 2001;15:303–311. doi: 10.1016/s1074-7613(01)00186-8. [DOI] [PubMed] [Google Scholar]
- 17.Hu-Li J, et al. Regulation of expression of IL-4 alleles: Analysis using a chimeric GFP/IL-4 gene. Immunity. 2001;14:1–11. doi: 10.1016/s1074-7613(01)00084-x. [DOI] [PubMed] [Google Scholar]
- 18.Urban JF, Jr, et al. IL-13, IL-4Rα, and STAT6 are required for the expulsion of the gastrointestinal nematode parasite Nippostrongylus brasiliensis. Immunity. 1998;8:255–264. doi: 10.1016/s1074-7613(00)80477-x. [DOI] [PubMed] [Google Scholar]
- 19.Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: Regulation by cis elements and epigenetics. Immunity. 2006;24:369–379. doi: 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Zhu J, et al. Conditional deletion of Gata3 shows its essential function in T(H)1–T(H)2 responses. Nat Immunol. 2004;5:1157–1165. doi: 10.1038/ni1128. [DOI] [PubMed] [Google Scholar]
- 21.Finkelman FD, Katona IM, Urban JF, Jr, Paul WE. Control of in vivo IgE production in the mouse by interleukin 4. Ciba Found Symp. 1989;147:3–22. doi: 10.1002/9780470513866.ch2. [DOI] [PubMed] [Google Scholar]
- 22.Finkelman FD, et al. Suppression of in vivo polyclonal IgE responses by monoclonal antibody to the lymphokine B-cell stimulatory factor 1. Proc Natl Acad Sci USA. 1986;83:9675–9678. doi: 10.1073/pnas.83.24.9675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paul WE, Seder RA. Lymphocyte responses and cytokines. Cell. 1994;76:241–251. doi: 10.1016/0092-8674(94)90332-8. [DOI] [PubMed] [Google Scholar]
- 24.Sprent J, Surh CD. T cell memory. Annu Rev Immunol. 2002;20:551–579. doi: 10.1146/annurev.immunol.20.100101.151926. [DOI] [PubMed] [Google Scholar]
- 25.Dooms H, Abbas AK. Control of CD4+ T-cell memory by cytokines and costimulators. Immunol Rev. 2006;211:23–38. doi: 10.1111/j.0105-2896.2006.00387.x. [DOI] [PubMed] [Google Scholar]
- 26.Mowen KA, Glimcher LH. Signaling pathways in Th2 development. Immunol Rev. 2004;202:203–222. doi: 10.1111/j.0105-2896.2004.00209.x. [DOI] [PubMed] [Google Scholar]
- 27.Takeda K, et al. Essential role of STAT6 in IL-4 signalling. Nature. 1996;380:627–630. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 28.Sokol CL, Barton GM, Farr AG, Medzhitov R. A mechanism for the initiation of allergen-induced T helper type 2 responses. Nat Immunol. 2008;9:310–318. doi: 10.1038/ni1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Croft M. Costimulation of T cells by OX40, 4–1BB, and CD27. Cytokine Growth Factor Rev. 2003;14:265–273. doi: 10.1016/s1359-6101(03)00025-x. [DOI] [PubMed] [Google Scholar]
- 30.Salek-Ardakani S, et al. OX40 (CD134) controls memory T helper 2 cells that drive lung inflammation. J Exp Med. 2003;198:315–324. doi: 10.1084/jem.20021937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shilling RA, et al. Cutting edge: Polymorphisms in the ICOS promoter region are associated with allergic sensitization and Th2 cytokine production. J Immunol. 2005;175:2061–2065. doi: 10.4049/jimmunol.175.4.2061. [DOI] [PubMed] [Google Scholar]
- 32.Umetsu SE, et al. TIM-1 induces T cell activation and inhibits the development of peripheral tolerance. Nat Immunol. 2005;6:447–454. doi: 10.1038/ni1186. [DOI] [PubMed] [Google Scholar]
- 33.Amsen D, et al. Direct regulation of GATA-3 expression determines the T helper differentiation potential of Notch. Immunity. 2007;27:89–99. doi: 10.1016/j.immuni.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urban JF, Jr, Katona IM, Paul WE, Finkelman FD. Interleukin 4 is important in protective immunity to a gastrointestinal nematode infection in mice. Proc Natl Acad Sci USA. 1991;88:5513–5517. doi: 10.1073/pnas.88.13.5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giulietti A, et al. An overview of real-time quantitative PCR: Applications to quantify cytokine gene expression. Methods. 2001;25:386–401. doi: 10.1006/meth.2001.1261. [DOI] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.