Abstract
Mutations affecting the seemingly unrelated gene products, SepN1, a selenoprotein of unknown function, and RyR1, the major component of the ryanodine receptor intracellular calcium release channel, result in an overlapping spectrum of congenital myopathies. To identify the immediate developmental and molecular roles of SepN and RyR in vivo, loss-of-function effects were analyzed in the zebrafish embryo. These studies demonstrate the two proteins are required for the same cellular differentiation events and are needed for normal calcium fluxes in the embryo. SepN is physically associated with RyRs and functions as a modifier of the RyR channel. In the absence of SepN, ryanodine receptors from zebrafish embryos or human diseased muscle have altered biochemical properties and have lost their normal sensitivity to redox conditions, which likely accounts for why mutations affecting either factor lead to similar diseases.
Keywords: congenital myopathy, disease model, intracellular calcium release
Congenital myopathies comprise a genetically and clinically heterogeneous group of muscle disorders typically associated with weakness early in life and often exhibiting only mild progression (1). Many of these diseases demonstrate abnormal sarcomere structure and disruption of the aligned organization of neighboring myofibrils, leading to ultrastructurally recognized irregularities such as “Z-band streaming” (2). Mutations affecting sarcomere components underlie some of the diseases, but other genes not clearly related to myofibril assembly also have been linked to them. Whether these genes regulate few or many pathways that contribute to the disease state has been difficult to ascertain and holds relevance for the development of treatments.
Several factors, including the long interlude between disease onset and diagnosis, make it difficult to recognize the initial defects leading to the disease state and may magnify the appearance of phenotypic heterogeneity among related diseases (3). Genetic heterogeneity may also contribute to phenotypic variability, because almost all identified mutations associated with the myopathies are specialized missense alleles (4). To detect primary cellular pathways regulated by disease-associated genes and to determine whether loss of function of seemingly unrelated genes might affect common processes, we analyzed gene function in the zebrafish embryo. Although the loss-of-function phenotype in the zebrafish embryo may not precisely recapitulate the prevalent human disease phenotype, modeling muscle disorders in the zebrafish can uncover the primary cellular functions of disease genes (5).
We first analyzed Selenoprotein N (SepN), a member of the selenocysteine-containing protein family, because complete loss of SEPN1 gene function results in classical congenital myopathy phenotypes (6–10). Whereas no biochemical activity has been linked to SepN, its function appears conserved, because reduced expression in zebrafish causes muscle abnormalities resembling those observed in diseased tissue (11). Because loss of SepN is a viable condition in humans, we postulated it might modulate a molecular pathway required for myofibril formation. Here, we show SepN and the ryanodine receptor (RyR) intracellular calcium release channel are both required for normal muscle development and differentiation and for some calcium mobilization events in the embryo. The two proteins are physically associated in vivo, where SepN is required for full activity of the RyR channel.
Results
SepN Is Required Cell-Autonomously for Muscle Formation.
The zebrafish sepN orthologue was identified by the multiple distinguishing primary sequence and gene organization features it shares with its human counterpart, SEPN1 (11, 12) [see supporting information (SI) Fig. S1]. In the embryo, sepN transcripts are expressed at high levels in the notochord, the tailbud, the presomitic mesoderm, and the emerging somites (Fig. 1A, B, D, and E; refs. 11 and 12), precursors of tissues affected in individuals with SEPN1-associated disease. As the notochord and somitic muscle differentiate overtly, sepN expression in these tissues is greatly reduced (Fig. 1C), paralleling its down-regulation in differentiating mammalian muscle cells (13).
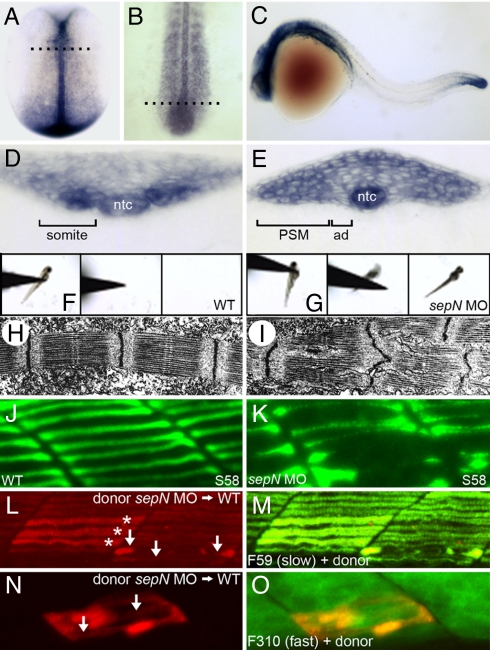
Fig. 1.
Loss of sepN function disrupts muscle differentiation in the zebrafish embryo. (A–E) sepN is expressed transiently in newly formed somites, adaxial cells (ad), presomitic mesoderm (PSM), and the notochord (ntc). sepN expression in (A and D) 10.5 hpf (two-somite stage), (B and E) 16 hpf, and (C) 24 hpf WT embryos. (A and B), Dorsal views with rostral up; (C) lateral view with rostral left. (D and E) Transverse sections through embryos at the levels indicated in B and C, respectively. (F and G) Three sequential frames illustrate that morphants, unlike WT embryos, rarely move >0.5 cm in response to touch. (H and I) Transmission electron micrographs of fast muscle fibers of 24 hpf (H) WT and (I) sepN morphant embryos indicate disruption of sarcomere organization in morphants. (J and K) Superficial slow muscle fibers of 24-hpf embryos stained with S58 antibody. (L–O) Development of sepN-depleted muscle fibers in WT hosts. sepN-depleted slow (L and M) or fast (N and O) dye-labeled muscle cells (red) were analyzed in mosaic embryos at 24 hpf stained with F59 or F310 antibody (green), respectively. (L and N) Dye-labeled donor cells; (M and O) merged views. *, normal-appearing fibers; arrows, dysmorphic donor fibers.
Formation of mature sepN transcripts was completely inhibited by injecting embryos with combinations of two splice-blocking morpholino oligonucleotides (sbMOs) complementary to exon–intron junctions present in nascent sepN transcripts (Fig. S1). SepN-depleted embryos invariably exhibited diminished spontaneous and touch-induced movements normally displayed by 24- to 72-hour postfertilization (hpf) embryos (Fig. 1 F and G). Patterning and differentiation of many neural cell types were unaltered by lack of SepN (Fig. S2). In contrast, newly formed somitic muscle of SepN-depleted 24-hpf embryos exhibited distinct structural abnormalities (11). Ultrastructural analysis of the deeper and more prevalent fast fibers of the somite revealed that SepN-depleted muscle exhibited myofibril disorganization with prominent Z-band anomalies not seen in WT muscle (Fig. 1 H and I).
Overall muscle cell morphology was also perturbed in SepN-depleted embryos. Individual slow muscle fibers at the superficial surface of the somite can be readily visualized in the 24-hpf embryo (14). In control embryos, the slow muscle fibers extend continuously from the anterior to the posterior border of the somite, with nearly uniform thickness along their entire length (Fig. 1J). In contrast, slow fibers of SepN-depleted embryos were often distended or broken (Fig. 1K). Loss of SepN1 function in humans has also been associated with hypotrophic slow twitch (type 1) fibers (6).
To determine which fibers directly require sepN function, we analyzed development of SepN-depleted muscle fibers in WT hosts. SepN-depleted gastrula-stage precursors of the slow (Fig. 1 L and M) or fast (Fig. 1 N and O) muscle cells were transplanted homotopically (15) into WT host gastrulae, and mosaic embryos were analyzed at 24 hpf. Individual SepN-depleted slow or fast muscle fibers exhibited discontinuities and morphological anomalies even when differentiating in a generally WT somitic environment. For example, among mosaic embryos in which slow muscle cell differentiation was analyzed, none of the 10 slow fibers analyzed in five WT→WT transplants were dysmorphic, whereas 74% (n = 23) of the slow fibers analyzed in 15 sepN MO→WT transplants were dysmorphic. The muscle fiber abnormalities can be detected very shortly after cellular differentiation, indicating they likely do not result from aberrant muscle cell function.
SepN Is Required for Development of the Slow Muscle Fiber Lineage.
SepN-depleted embryos were defective also in generating embryonic slow muscle cells, a finding that differs from the conclusions of a study in which SepN expression was only partially inhibited (11). SepN-depleted embryos had a significant reduction in the number of all slow muscle fibers, identified by expression of the S58 antigen, and the 4D9+ subclass of “pioneer” slow muscle fibers that reside at the horizontal myoseptum (14) (Fig. 2 A–D; Fig. S3). Consistent with the loss of these fibers, somites of embryos lacking SepN exhibited the “u-shaped” phenotype (Fig. 2 E and F), a tell-tale indicator of slow muscle cell deficiencies.
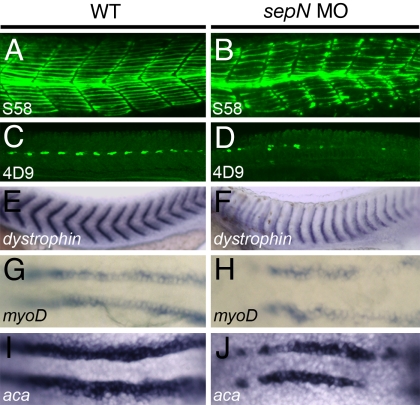
Fig. 2.
Slow muscle fiber formation is reduced in sepN morphants. (A–F) Somitic muscle in midtrunk region of 24-hpf WT and sepN morphant embryos. (A and B) S58+ slow muscle fibers and (C and D) 4D9+ slow muscle pioneer fiber nuclei are present in reduced numbers in sepN morphants. Staining for dystrophin expression (E and F) reveals “u-shaped” somites in sepN morphant embryos. (G–J) Expression of myogenic lineage genes in adaxial cells of three-somite stage embryos. myoD (G and H) and α-cardiac actin (aca; I and J) are expressed discontinuously and at reduced levels in the adaxial cell population of sepN morphants. (A–F) Lateral views, rostral left. (G–J) Dorsal views, rostral left.
All embryonic slow muscle cells derive from an identifiable precursor population, the adaxial cells, which arise as continuous rows of cells that border the notochord (14). In response to signals from the notochord, adaxial cells up-regulate expression of muscle lineage genes (Fig. 2 G and I) and subsequently migrate radially away from the notochord to generate the slow muscle fibers at the surfaces of the somites. In SepN-depleted embryos, myogenic gene expression in the adaxial region was reduced and discontinuous, interrupted by patches of cells that expressed the genes at low or undetectable levels (Fig. 2 H and J). Loss of SepN did not affect viability or organization of cells surrounding the notochord (Fig. S4). In sum, complete loss of SepN results in a diminution of the expression of myogenic lineage genes in slow muscle cell precursors and a reduction in the generation and development of the slow muscle cell population.
RyRs and SepN Have Similar Developmental Functions.
Because altered function of the RyR calcium release channel causes defects that resemble the effects of loss of sepN function (4, 16–19), we investigated whether SepN and RyRs contribute to similar cell events in the zebrafish embryo. Two ryr homologues are expressed in the zebrafish embryo throughout somitogenesis in patterns that overlap sepN expression (18, 20). ryr1a is expressed at high levels in the adaxial cells during somitogenesis; later, at 24 hpf, ryr1a transcripts are abundant in the pioneer slow muscle fibers and present at low levels throughout the somite (Fig. 3 A and C). In contrast, ryr3 is expressed in many tissues at somitogenesis stages and continues to be expressed broadly in the somites of 24 hpf embryos (Fig. 3 B and D). Depletion of RyR1a or RyR3 with sbMOs (Fig. S5) caused slow muscle defects similar to those seen after loss of SepN function. RyR-depleted embryos had “u-shaped” somites (data not shown), severely reduced expression of muscle lineage genes in adaxial cells (Fig. 3 E–H) and reduced numbers of 4D9+ pioneer (Fig. 3 I–L) and F59+ nonpioneer (Fig. 3 M–P) slow muscle cells. The activity of each RyR contributes to development of the slow muscle lineage (Fig. 3 K, L, O, and P; Fig. S5).
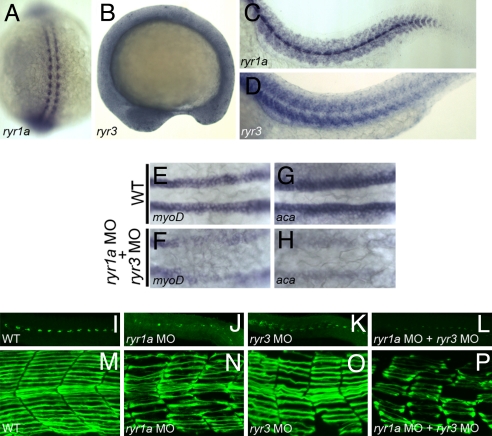
Fig. 3.
Ryanodine receptors and SepN have similar functions in muscle formation. (A–D) Expression of ryr1a and ryr3 transcripts in 15-somite (A and B) and 24 hpf (C and D) embryos. (E–H) Expression of the myogenic lineage genes, myoD and α-cardiac actin, is greatly reduced in the adaxial cell population of ryr1a; ryr3 double morphants. (I–L) 4D9+ slow muscle pioneer fiber nuclei and (M–P) F59+ slow muscle fibers are reduced in number and irregularly shaped in 24-hpf embryos depleted for RyR1a, RyR3, or both RyRs. (A) Dorsal view, rostral up. B–D and I–P, lateral views, rostral left. (E–H) Dorsal views, rostral left.
RyR1a and RyR3 activities are also required for the formation of normal-appearing slow muscle cells. Reduction of RyR1a or RyR3 resulted in disrupted, nonuniform slow muscle fibers (Fig. 3 N and O) producing a phenotype closely resembling that seen after complete depletion of SepN (Fig. 2F). In addition, muscle differentiation appeared to be sensitive to the combined levels of SepN and RyRs (Fig. S6 and Table S1), consistent with the possibility that they function in a common molecular pathway.
SepN and RyRs Are Required for Calcium Flux Activity.
To determine whether SepN, like the RyRs, contributes to calcium mobilization in the embryo, we examined free calcium levels around the Kupffer's Vesicle (KV), a site at which calcium fluxes have been measured in the zebrafish embryo and where sepN and ryr3 (but not ryr1a) are expressed (Fig. 4 A and B). The KV is a small structure at the base of the elongating notochord with similarities to the mammalian node (21). Basal levels of calcium flux are detectable all around the KV. However, at midsomitogenesis, in a process that may be linked to the patterning of left–right asymmetries, prolonged elevated levels of free calcium can be detected on the left side of the KV (22). We asked whether sepN and ryr3 gene functions were required for calcium signaling at the KV.
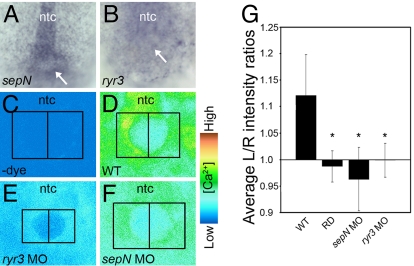
Fig. 4.
SepN and RyR3 are required for calcium signaling at the KV. (A and B) sepN and ryr3 transcripts are present in and around the KV (indicated by arrow) at the base of the notochord (ntc). (C–F) Visualization of free calcium in the region of the KV in 5- to 8-somite embryos. Images shown represent a single confocal section, taken from a dorsal view at the plane of the maximal apparent diameter of the KV. (C) Without calcium-sensitive dye, fluorescence is not detected. (D) Elevated levels of free calcium are detected on the left compared with the right side of the KV in WT embryos. (E) Loss of ryr3 function results in a dramatic overall reduction of free calcium. (F) Loss of sepN function results in complete loss of asymmetric distribution of free calcium. (G) For quantitative analyses, left- and right-side intensity values were determined for each embryo by integrating pixel values within symmetric boxed areas abutting the KV midline as indicated in C-F. Left/right intensity ratios were determined for each embryo [WT controls, n = 17; rhodamine dextran-injected (RD), n = 11; sepN MO-injected, n = 22; ryr3 MO-injected, n = 19] and average ratios are graphed. *, significant difference from WT ratio (P < 0.001 using paired Student's t test). Error bars are ± SD.
Embryos were injected at the one-cell stage with the calcium-sensitive fluorescent dye indicator Oregon green 488 BAPTA-1 dextran, fluorescence levels surrounding the KV of 5- to 8-somite stage embryos were analyzed by confocal microscopy, and left–right relative fluorescence was determined for each embryo. Every WT embryo (n = 17) exhibited higher levels of fluorescence on the left side of the KV; in addition, elevated levels were always seen at the base of the notochord (Fig. 4 D and G). The asymmetric levels of fluorescence reflected the true distribution of calcium as no signal was detected in the absence of dye (Fig. 4C), and dextran-linked dyes were delivered uniformly around the KV (Fig. 4G). Upon loss of RyR3, little calcium-dependent signal was observed (Fig. 4 E and G). Significantly, upon depletion of SepN, the left-side-specific elevation of calcium appeared blocked (Fig. 4F), and levels of left- and right-side calcium-dependent signal were consistently similar in each SepN MO-treated embryo (Fig. 4G). Both SepN and RyR3 are needed for calcium mobilization around the KV.
SepN Is Stably Associated with RyRs.
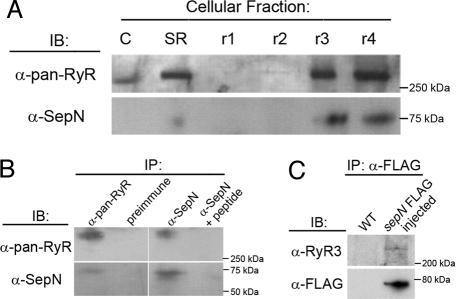
Previous studies indicated SepN is a transmembrane protein of undetermined topology, likely to be “part of an integral protein complex” associated with the SR (13). Several lines of evidence indicated RyR and SepN are physically associated in vivo. Stratification of the SR from adult rabbit skeletal muscle by differential centrifugation (23) revealed the two proteins were colocalized and highly enriched in terminal cisternae (Fig. 5A). SepN was immunoprecipitated from rabbit muscle homogenate with an antiserum recognizing all isoforms of RyR (Fig. 5B). Similarly, RyR proteins were immunoprecipitated from the homogenate with an anti-SepN antibody generated against a peptide derived from human SepN1 protein (Fig. 5B). Inclusion of the SepN1 peptide as a competitor in the reaction blocked recovery of the RyR proteins. Moreover, RyR proteins were coimmunoprecipitated with C terminus-FLAG-tagged zebrafish SepN that had been expressed in embryos (Fig. 5C and data not shown). Although these experiments support association of SepN and RyRs in vivo, they do not indicate the stoichiometry of the association nor whether the proteins are always complexed.
Fig. 5.
SepN and RyRs are physically associated. (A) Immunoblotting (IB) of fractionated SR indicates SepN and RyRs are colocalized in terminal cisternae. C, crude membranes; SR, crude SR; r1, longitudinal SR/t-tubules/plasma membrane; r2, longitudinal SR; r3, longitudinal SR/terminal cisternae; r4, terminal cisternae. (B) Coimmunoprecipitation (IP) of RyR and SepN from rabbit skeletal muscle SR protein homogenates. (C) Coimmunoprecipitation of RyR and SepN-FLAG from homogenates of zebrafish embryos expressing SepN-FLAG.
SepN Is Required for Normal RyR Activity.
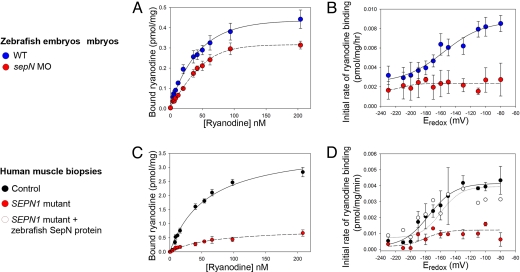
Because neither the expression patterns nor the overall levels of RyR proteins in zebrafish embryos were altered detectably by loss of SepN (Fig. S7), we examined the activity of RyRs in SepN-depleted embryos. High affinity binding of the plant alkaloid ryanodine strongly correlates with the activity of the Ca2+ release channel, and an increase in ryanodine binding is an indicator of enhanced channel activity (24). Protein homogenates were prepared from independently generated paired groups of control or SepN-depleted 14–18-somite stage embryos and concentration-dependent equilibrium binding of [3H]ryanodine to protein was determined. SepN-depleted tissue had decreased ryanodine binding capacity (control tissue Bmax = 0.52 ± 0.01 pmol/mg vs. SepN-depleted tissue Bmax = 0.42 ± 0.02 pmol/mg) and decreased binding affinity for ryanodine (control tissue Kd = 37.2 ± 1.9 nM vs. SepN-depleted tissue Kd = 49.9 ± 6.2 nM) (Fig. 6A). Although a decrease in Bmax in the SepN-depleted samples could reflect a loss of RyR or some additional RyR-complexed protein, the observed increase in Kd indicates a fundamental functional change in the RyRs after loss of SepN. Moreover, because the SepN-depleted homogenates were derived from thousands of MO-injected embryos, they likely contained some residual WT SepN protein, resulting in underestimation of the true effect of complete loss of SepN.
Fig. 6.
Ryanodine receptor activity depends on SepN. Concentration-dependent (A and C) and redox potential-dependent (B and D) binding of [3H]ryanodine to homogenates prepared from control (A) or sepN MO-injected (B) 14- to 18-somite stage zebrafish embryos, and from muscle biopsies from control (C) or SEPN1 mutant (D) individuals. Concentration-dependent equilibrium binding was performed in quadruplicate by using three independent matched sets embryo homogenates or one matched set of biopsies; error bars are ± SEM. The redox potential-dependent initial rate of ryanodine binding to protein from human control or SEPN1-diseased muscle tissue was determined in two independent experiments. In one set of experiments, diseased homogenate was supplemented with in vitro-synthesized zebrafish SepN protein.
The homotetrameric RyR channel contains ≈400 cysteines and its activity is exquisitely sensitive to redox regulation (25–27). Because several selenoproteins mediate redox reactions (28) and selenium-containing compounds stimulate the calcium release channel by oxidizing endogenous thiols on the RyR (29), we tested whether loss of SepN affected the responsiveness of RyRs to the redox state of the environment.
The initial rate of ryanodine binding to WT zebrafish embryo homogenates was found to be strongly dependent on the redox potential of the aqueous environment, which was manipulated by the GSSG/GSH2 ratio in the binding buffer (Fig. 6B). As critical thiols on the channel were oxidized in controls, binding of ryanodine was enhanced. The redox potential of the zebrafish RyR (the midpoint of the redox titration) was approximately −150 mV, similar to that observed with preparations from rabbit skeletal muscle SR (26). In contrast, the RyRs of SepN-depleted embryos exhibited greatly diminished responsiveness to the redox potential of the environment (Fig. 6B). These results indicate that a fundamental physiological mechanism by which RyR activity is regulated is blocked upon loss of SepN.
As an independent test of the role of SepN, we examined the properties of RyRs isolated from human muscle tissue lacking SepN1. Homogenates were prepared from muscle biopsies obtained from an individual with congenital myopathy who was homozygous for a SepN1 mutation (R466Q) and from age- and sex-matched controls. The R466Q allele results in absence of significant expression of the SepN1 protein and therefore apparently is a functional null allele (M.T.H. and K.M.F., unpublished work). Equilibrium binding analyses (Fig. 6C) indicated protein from SepN1-deficient tissue had dramatically diminished capacity for ryanodine binding (control tissue Bmax = 3.77 ± 0.19 pmol/mg vs. SEPN1-diseased tissue Bmax = 0.84 ± 0.07 pmol/mg). Although the reduced binding of ryanodine to RyRs of diseased tissue made it difficult to measure Kd with certainty, SepN-depleted tissue displayed a small decrease in affinity for ryanodine (data not shown). These results are consistent with a requirement for SepN1 in the maintenance of normal levels and/or activity of RyR in human muscle.
In contrast to control homogenates, positive (oxidizing) redox potentials failed to stimulate binding of human RyRs from SEPN1-diseased muscle (Fig. 6D). We tested whether the RyR activity from the SEPN1-diseased patient could be restored by addition of WT SepN protein. Full-length zebrafish SepN protein was synthesized in vitro, purified, and added to muscle homogenates prepared from the SEPN1-diseased patient. Addition of exogenous SepN partially restored the binding capacity of the human RyRs from SEPN1-diseased tissue (Bmax = 1.43 ± 0.19 pmol/mg). Most strikingly, addition of zebrafish SepN restored the ability of human RyRs from diseased tissue to respond to changes in the redox potential of the binding environment (Fig. 6D), indicating that RyR channel behavior is modulated by the presence of SepN protein. Further studies are required to determine the biochemical mechanism by which SepN affects RyR activity.
Discussion
We show here that two previously unrelated gene products, Selenoprotein N and ryanodine receptor, each of which is associated with an overlapping spectrum of congenital muscle disorders, are physically associated in vivo where they contribute to a single biochemical pathway regulating intracellular calcium release. The phenotypic analyses presented here revealed that SepN and RyRs contribute to common developmental events and provided a rationale for examining how they might share biochemical function. Loss of function analysis in the zebrafish embryo served to simplfy conceptualization of diseases that appeared to have heterogeneous etiologies.
We note that the zebrafish embryonic phenotypes do not precisely recapitulate the human disease phenotypes. Two factors likely contribute to the differences. First, whereas a limited set of RYR1 mutant alleles are associated with disease in humans (4), we analyzed the effects of combined loss of ryr1a and ryr3, with the purpose of generally identifying RyR-dependent processes in the zebrafish embryo. Second, the human and zebrafish orthologues may not be used in precisely the same developmental contexts (30). Thus, although sepN is required for calcium mobilization at the KV, another selenoprotein gene may provide an equivalent role at the mammalian node.
Whereas diseases associated with SEPN1 or RYR1 mutations are generally viewed as affecting subcellular aspects of muscle fiber formation, our analyses indicate the possibility that SepN and RyRs also have roles in fiber specification. Other studies have implicated SepN and RyRs in muscle fiber generation. A previous study of sepN function in zebrafish also appeared to indicate deficits of muscle fibers in embryos, although this was not quantified (11). In addition, mice homozygous for the human disease-associated RYR1I4895T allele have reduced muscle production (19).
RyRs are best known for their roles in the excitation–contraction (EC) coupling pathway that mediates muscle function (31). However, EC coupling does not appear critical for the normal assembly of muscle fibers in the zebrafish somite (32). Thus, RyR channels are likely to be used in additional contexts. The defects caused by loss of SepN or RyR in the present study, reduction in the generation of slow muscle fibers, and aberrant myofibrillogenesis, may in fact represent two distinct requirements for calcium mobilization during muscle development.
Increasing evidence supports a model in which regulation of the redox state of critical thiol groups of the ryanodine receptor is a major physiological pathway by which calcium flow is regulated (27, 33, 34). Oxidation of selected “hyperreactive” cysteines can occur through formation of disulfides, by S-nitrosylation, or by S-glutathionylation, and the redox state of these cysteines correlates closely with the open state of the calcium release channel (35, 36). Redox modification of specific cysteine residues also affects the ability of calcium release channel components to maintain their association with the RyR core protein (37). Moreover, well characterized modulators of calcium release channel activity affect the redox potential of the channel, whether through direct or indirect means (26, 38). Thus, the RyR channel is a “redox sensor” regulated by a number of associated proteins including SepN.
Only some selenoproteins have proven biochemical activity, but among these, several are associated with protein folding, disulfide bond shuffling, or other redox activities in biosynthetic pathways (3, 39). Here, we show that SepN is a modulator of RyR activity, affecting the ability of the calcium release channel to function as a redox sensor. The finding that SepN is required for normal RyR activity is consistent with the presence in SepN orthologues of a CxxS domain that has been linked to redox activity (40). Given the viability of apparent null SEPN1 mutations in humans, it is likely that the central function of Selenoprotein N is as a facilitator of RyR function. The finding that the RyR activity of one SEPN1-mutant muscle can be partially rescued by addition of exogenous SepN in vitro to muscle homogenates suggests the interaction between SepN and RyR is highly specific and efficient. Furthermore, it demonstrates that RyR activity may not be irreversibly damaged in patients with SEPN1-associated disease and might provide a target for therapeutic intervention. Analysis of additional biopsy samples is required to establish whether RyR activity is generally depressed in SEPN1-diseased muscle.
Materials and Methods
Preparation of Embryo and Muscle Homogenates.
Live 14–18-somite stage embryos were rinsed twice and dounce-homogenized (15 strokes) in muscle buffer (see SI Methods), flash-frozen in liquid N2, and stored at −80°C. Each homogenate was prepared from 1,000–1,500 pooled injected or control embryos (600–700 embryos/ml homogenate; 10–12 mg of protein/ml homogenate). Approximately 100 mg (wet weight) of diseased or age-/sex-matched control muscle biopsy (MB) material, snap-frozen in liquid nitrogen-cooled isopentane (2) and stored at −80°C, was processed similarly in MB buffer. Diseased tissue was from a 12-year-old girl with a rigid spine syndrome phenotype who was homozygous for a mutation in exon 11 of the SEPN1 gene (c.1397G→A; R466Q).
Ryanodine-Binding Analyses.
Equilibrium and redox-sensitive ryanodine binding was measured as in ref. 26 with slight modifications (see SI Methods). For the SepN protein add-back experiment, 8X His-tagged zebrafish SepN was synthesized in vitro (see SI Methods) and SepN protein (0.2 μg/ml) was added to SEPN1 mutant human muscle homogenates before binding studies.
Additional Materials and Methods.
Additional materials described in the SI Methods include: Animals and embryo manipulations; sequences of antisense MOs and primers for RT-PCR; sources of antibodies and gene probes; procedures for in situ hybridization, immunohistochemistry, immunoprecipitation, and protein synthesis; and details of ryanodine-binding conditions.
Supplementary Material
Acknowledgments.
We thank Steve DeVoto, Fred Clayton, and our Utah colleagues for insights and Katrina Lister and Christine Anderson for technical assistance. We also thank T. V. McCarthy (University College Cork) and V. Sorrentino (University of Siena) for generously sharing anti-RyR antibodies. M.J.J. was supported by a National Research Service Award Predoctoral National Institutes of Health fellowship and a postdoctoral National Institutes of Health fellowship (National Institute of Neurological Disorders and Stroke Grant T32 NS07493). Research was supported by funding from the Muscular Dystrophy Association (to D.J.G. and M.H.), the University of Utah Catalyst Grant Research Program (to D.J.G., M.H., and K.M.F.), the Science Foundation of Ireland (Grant RFP2007/BCIF165, to J.J.M.), and the National Institute of Arthritis and Musculoskeletal and Skin Diseases (RO1-AR-48911 to J.J.A.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession no. DQ160295).
This article contains supporting information online at www.pnas.org/cgi/content/full/0806015105/DCSupplemental.
References
- 1.Tubridy N, Fontaine B, Eymard B. Congenital myopathies and congenital muscular dystrophies. Curr Opin Neurol. 2001;14:575–582. doi: 10.1097/00019052-200110000-00005. [DOI] [PubMed] [Google Scholar]
- 2.Dubowitz V. Muscle Biopsy: A Practical Approach. 2nd Ed. London: Balliere Tindall; 1985. [Google Scholar]
- 3.Rederstorff M, Krol A, Lescure A. Understanding the importance of selenium and selenoproteins in muscle function. Cell Mol Life Sci. 2006;63:52–59. doi: 10.1007/s00018-005-5313-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou H, et al. Molecular mechanisms and phenotypic variation in RYR1-related congenital myopathies. Brain. 2007;130:2024–2036. doi: 10.1093/brain/awm096. [DOI] [PubMed] [Google Scholar]
- 5.Lieschke GJ, Currie PD. Animal models of human disease: Zebrafish swim into view. Nat Rev Genet. 2007;8:353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- 6.Clarke NF, et al. SEPN1: Associated with congenital fiber-type disproportion and insulin resistance. Ann Neurol. 2006;59:546–552. doi: 10.1002/ana.20761. [DOI] [PubMed] [Google Scholar]
- 7.D'Amico A, et al. Two patients with ‘Dropped head syndrome’ due to mutations in LMNA or SEPN1 genes. Neuromuscul Disord. 2005;15:521–524. doi: 10.1016/j.nmd.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 8.Ferreiro A, et al. Mutations of the selenoprotein N gene, which is implicated in rigid spine muscular dystrophy, cause the classical phenotype of multiminicore disease: Reassessing the nosology of early-onset myopathies. Am J Hum Genet. 2002;71:739–749. doi: 10.1086/342719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moghadaszadeh B, et al. Mutations in SEPN1 cause congenital muscular dystrophy with spinal rigidity and restrictive respiratory syndrome. Nat Genet. 2001;29:17–18. doi: 10.1038/ng713. [DOI] [PubMed] [Google Scholar]
- 10.Flanigan KM, et al. Congenital muscular dystrophy with rigid spine syndrome: A clinical, pathological, radiological, and genetic study. Ann Neurol. 2000;47:152–161. [PubMed] [Google Scholar]
- 11.Deniziak M, et al. Loss of selenoprotein N function causes disruption of muscle architecture in the zebrafish embryo. Exp Cell Res. 2007;313:156–167. doi: 10.1016/j.yexcr.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Thisse C, et al. Spatial and temporal expression patterns of selenoprotein genes during embryogenesis in zebrafish. Gene Expr Patterns. 2003;3:525–532. doi: 10.1016/s1567-133x(03)00054-1. [DOI] [PubMed] [Google Scholar]
- 13.Petit N, et al. Selenoprotein N: An endoplasmic reticulum glycoprotein with an early developmental expression pattern. Hum Mol Genet. 2003;12:1045–1053. doi: 10.1093/hmg/ddg115. [DOI] [PubMed] [Google Scholar]
- 14.Devoto SH, Melancon E, Eisen JS, Westerfield M. Identification of separate slow and fast muscle precursor cells in vivo, prior to somite formation. Development. 1996;122:3371–3380. doi: 10.1242/dev.122.11.3371. [DOI] [PubMed] [Google Scholar]
- 15.Hirsinger E, Stellabotte F, Devoto SH, Westerfield M. Hedgehog signaling is required for commitment but not initial induction of slow muscle precursors. Dev Biol. 2004;275:143–157. doi: 10.1016/j.ydbio.2004.07.030. [DOI] [PubMed] [Google Scholar]
- 16.Pietrini V, Marbini A, Galli L, Sorrentino V. Adult onset multi/minicore myopathy associated with a mutation in the RYR1 gene. J Neurol. 2004;251:102–104. doi: 10.1007/s00415-004-0220-y. [DOI] [PubMed] [Google Scholar]
- 17.Herasse M, et al. Abnormal distribution of calcium-handling proteins: A novel distinctive marker in core myopathies. J Neuropathol Exp Neurol. 2007;66:57–65. doi: 10.1097/NEN.0b013e31802d47ce. [DOI] [PubMed] [Google Scholar]
- 18.Brennan C, Mangoli M, Dyer CE, Ashworth R. Acetylcholine and calcium signalling regulates muscle fibre formation in the zebrafish embryo. J Cell Sci. 2005;118:5181–5190. doi: 10.1242/jcs.02625. [DOI] [PubMed] [Google Scholar]
- 19.Zvaritch E, et al. An Ryr1I4895T mutation abolishes Ca2+ release channel function and delays development in homozygous offspring of a mutant mouse line. Proc Natl Acad Sci USA. 2007;104:18537–18542. doi: 10.1073/pnas.0709312104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirata H, et al. Zebrafish relatively relaxed mutants have a ryanodine receptor defect, show slow swimming and provide a model of multi-minicore disease. Development. 2007;134:2771–2781. doi: 10.1242/dev.004531. [DOI] [PubMed] [Google Scholar]
- 21.Essner JJ, Amack JD, Nyholm MK, Harris EB, Yost HJ. Kupffer's vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development. 2005;132:1247–1260. doi: 10.1242/dev.01663. [DOI] [PubMed] [Google Scholar]
- 22.Sarmah B, Latimer AJ, Appel B, Wente SR. Inositol polyphosphates regulate zebrafish left-right asymmetry. Dev Cell. 2005;9:133–145. doi: 10.1016/j.devcel.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 23.Saito A, Seiler S, Chu A, Fleischer S. Preparation and morphology of sarcoplasmic reticulum terminal cisternae from rabbit skeletal muscle. J Cell Biol. 1984;99:875–885. doi: 10.1083/jcb.99.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pessah IN, Stambuk RA, Casida JE. Ca2+-activated ryanodine binding: mechanisms of sensitivity and intensity modulation by Mg2+, caffeine, and adenine nucleotides. Mol Pharmacol. 1987;31:232–238. [PubMed] [Google Scholar]
- 25.Feng W, Liu G, Allen PD, Pessah IN. Transmembrane redox sensor of ryanodine receptor complex. J Biol Chem. 2000;275:35902–35907. doi: 10.1074/jbc.C000523200. [DOI] [PubMed] [Google Scholar]
- 26.Xia R, Stangler T, Abramson JJ. Skeletal muscle ryanodine receptor is a redox sensor with a well defined redox potential that is sensitive to channel modulators. J Biol Chem. 2000;275:36556–36561. doi: 10.1074/jbc.M007613200. [DOI] [PubMed] [Google Scholar]
- 27.Zissimopoulos S, Lai FA. Redox regulation of the ryanodine receptor/calcium release channel. Biochem Soc Trans. 2006;34:919–921. doi: 10.1042/BST0340919. [DOI] [PubMed] [Google Scholar]
- 28.Hatfield DL, Gladyshev VN. How selenium has altered our understanding of the genetic code. Mol Cell Biol. 2002;22:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia R, Ganther HE, Egge A, Abramson JJ. Selenium compounds modulate the calcium release channel/ryanodine receptor of rabbit skeletal muscle by oxidizing functional thiols. Biochem Pharmacol. 2004;67:2071–2079. doi: 10.1016/j.bcp.2004.02.018. [DOI] [PubMed] [Google Scholar]
- 30.Postlethwait JH. The zebrafish genome in context: Ohnologs gone missing. J Exp Zoolog B. 2007;308:563–577. doi: 10.1002/jez.b.21137. [DOI] [PubMed] [Google Scholar]
- 31.Hamilton SL. Ryanodine receptors. Cell Calcium. 2005;38:253–260. doi: 10.1016/j.ceca.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 32.Schredelseker J, et al. The beta 1a subunit is essential for the assembly of dihydropyridine-receptor arrays in skeletal muscle. Proc Natl Acad Sci USA. 2005;102:17219–17224. doi: 10.1073/pnas.0508710102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abramson JJ, Salama G. Sulfhydryl oxidation and Ca2+ release from sarcoplasmic reticulum. Mol Cell Biochem. 1988;82:81–84. doi: 10.1007/BF00242520. [DOI] [PubMed] [Google Scholar]
- 34.Meissner G. Regulation of mammalian ryanodine receptors. Front Biosci. 2002;7:d2072–2080. doi: 10.2741/A899. [DOI] [PubMed] [Google Scholar]
- 35.Aracena-Parks P, et al. Identification of cysteines involved in S-nitrosylation, S-glutathionylation, and oxidation to disulfides in ryanodine receptor type 1. J Biol Chem. 2006;281:40354–40368. doi: 10.1074/jbc.M600876200. [DOI] [PubMed] [Google Scholar]
- 36.Liu G, Abramson JJ, Zable AC, Pessah IN. Direct evidence for the existence and functional role of hyperreactive sulfhydryls on the ryanodine receptor-triadin complex selectively labeled by the coumarin maleimide 7-diethylamino-3-(4′-maleimidylphenyl)-4-methylcoumarin. Mol Pharmacol. 1994;45:189–200. [PubMed] [Google Scholar]
- 37.Zissimopoulos S, Docrat N, Lai FA. Redox sensitivity of the ryanodine receptor interaction with FK506-binding protein. J Biol Chem. 2007;282:6976–6983. doi: 10.1074/jbc.M607590200. [DOI] [PubMed] [Google Scholar]
- 38.Marinov BS, Olojo RO, Xia R, Abramson JJ. Nonthiol reagents regulate ryanodine receptor function by redox interactions that modify reactive thiols. Antioxid Redox Signal. 2007;9:609–621. doi: 10.1089/ars.2006.1426. [DOI] [PubMed] [Google Scholar]
- 39.Moghadaszadeh B, Beggs AH. Selenoproteins and their impact on human health through diverse physiological pathways. Physiology (Bethesda) 2006;21:307–315. doi: 10.1152/physiol.00021.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fomenko DE, Gladyshev VN. CxxS: Fold-independent redox motif revealed by genome-wide searches for thiol/disulfide oxidoreductase function. Protein Sci. 2002;11:2285–2296. doi: 10.1110/ps.0218302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.