Abstract
Many bacterial species are capable of biofilm growth, in which cells live and replicate within multicellular community groups. Recent work shows that biofilm growth by a wide variety of bacterial species can generate genetic diversity in microbial populations. This finding is significant because the presence of diverse subpopulations can extend the range of conditions in which communities can thrive. Here, we used biofilms formed by the pathogen Pseudomonas aeruginosa to investigate how this population diversity is produced. We found that some cells within biofilms incur double-stranded DNA breaks caused by endogenous oxidative stress. Genetic variants then result when breaks are repaired by a mutagenic mechanism involving recombinatorial DNA repair genes. We hypothesized that the mutations produced could promote the adaptation of biofilm communities to changing conditions in addition to generating diversity. To test this idea, we exposed biofilms to an antibiotic and found that the oxidative stress-break repair mechanism increased the emergence of antibiotic-resistant bacteria. The diversity and adaptability produced by this mechanism could help biofilm communities survive in harsh environments.
Keywords: antibiotic resistance, DNA breaks, population diversity, recombinatorial repair
In many natural environments and chronic human infections bacteria live in matrix-encased groups known as biofilms, rather than as free-living (planktonic) cells (1). Biofilm growth increases the ability of cells to persist in adverse conditions by inducing bacterial phenotypes that are not apparent in planktonically grown cells. For example, biofilm bacteria are more resistant to killing by antibiotics, biocides, and host defenses than planktonic cells and may be less vulnerable to predators (1). Biofilm growth also enhances resistance to desiccation and radiation and may increase metabolic cooperation between cells (1, 2).
Recent work from a number of groups has uncovered another interesting characteristic of biofilms. Several labs have found that biofilm growth can produce extensive genetic diversity in bacterial populations even though no external stress or mutagen is applied. Diverse genetic variants (called variants below) are generated by biofilms of Pseudomonas aeruginosa, Pseudomonas fluorescence, Vibro cholera, Streptococcus pneumoniae, and Staphylococcus aureus (3–9).
In work with P. aeruginosa, we found that the diversity affects many bacterial traits. Biofilm growth produced variants in colony morphology; swimming and surface motility; pigment, surfactant, and exopolysaccharide production; nutritional requirements; and other phenotypes (8). These variants were not seen in batch or chemostat planktonic cultures grown for a comparable number of generations in the same medium (8). Furthermore, the variant phenotypes are generally heritable, indicating that genetic changes have occurred (8). Of note, diverse variants are also commonly isolated from patients with biofilm infections such as bronchiectasis (10–12), but are not seen in infections associated with planktonic growth (12, 13). Thus, extensive genetic variation appears to be generated by biofilms in vitro and in vivo.
The presence of phenotypically distinct variants could have a major effect on the functioning of biofilm communities. Population diversity can lessen the impact of environmental upsets because the presence of diverse subpopulations extends the range of conditions in which the community will thrive (14, 15). Diversity can also enhance the productivity and sustainability of populations because of positive interactions between community members and because individuals with different (rather than superimposed) resource requirements use available habitats more effectively (14–17). These effects have been observed in plant and animal communities (15, 16) and are also likely to occur in bacterial populations (8). Given the potential impact of diversity in biofilms, we investigated how it arises.
Results
Variants Do Not Preexist in the Biofilm Inoculum.
Diversity is a consequence of mutation and the amplifying effects of selection. Two points suggest that selection could play a large role in the diversity that we observe. First, the structured organization and metabolic activity of biofilm bacteria generate many different selective environments (see Discussion and refs. 1 and 18–21). A large diversity of selective pressures could explain the great variety of variant phenotypes that emerge. Second, the sheer numbers of variants generated suggests strong selective amplification. In the biofilms we studied, 20–30% of the population formed colonies that were morphologically distinct from the inoculum after only 5 days of growth (8). Increased mutation frequency alone would not be expected to produce this outcome unless rates were extremely high. Thus, at least some selective amplification probably occurs as the variants replicate.
The likelihood that selection is important in biofilm-mediated diversity led us to explore the possibility that the diversity we observe is entirely caused by the selective amplification of variants that exist undetected in the inoculum. Indeed, previous work shows that variants can arise in planktonic cultures after prolonged growth (22).
To test this idea, we markedly reduced the inoculum (to ≈1,000 cells). Half the bacteria were directly plated so that colonies made by each cell could be examined for the variant phenotypes, and the other half used to establish biofilms. As shown in Fig. 1, no colony morphology, motility, pigment production, or auxotrophic variants were found in the small inoculum. Despite this, biofilms grown from these cells produced the same degree of diversity as the larger inocula previously used (Fig. 1). Thus, variants do not preexist in the inoculum and must be generated during biofilm growth.
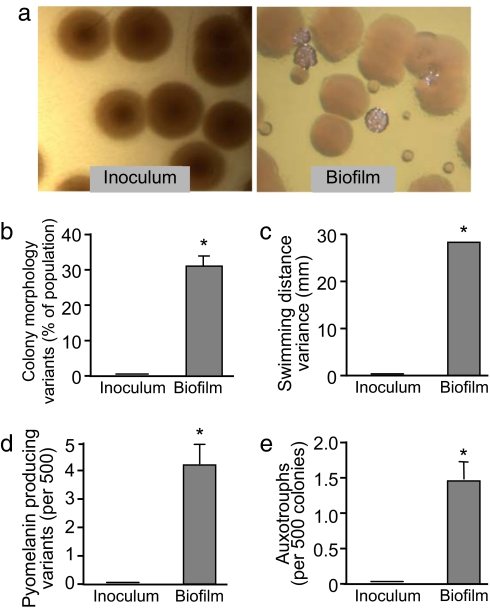
Fig. 1.
Diversity does not preexist in the inoculum used to establish biofilms. Diversity in the phenotypes shown is heritable, indicating that genetic changes have occurred. (a) Photographs of bacterial colonies on agar plates grown from the inoculum or from cells harvested from 5-day-old biofilms. Whereas the inoculum was uniform, biofilm growth produced diversity in colony morphology. (b) The proportion of the inoculum and biofilm population that formed colonies with variant morphologies. A total of 500 colonies were examined per experiment; data are the mean of four experiments. Error bars show SEM. *, P < 0.0004 versus the inoculum. (c) Variance in swimming induced by biofilm growth. The swimming ability of colonies from the inoculum was compared with those from biofilms. Only colonies with the typical appearance were tested so swimming variance and colony morphology variation are independent measures of diversity. Data are the variance of 50 randomly picked colonies from both conditions. *, P < 0.0001 versus the inoculum. (d) Abundance of bacteria overproducing pyomelanin in the inoculum and biofilms. Graph shows the mean of four experiments. Error bars show SEM. *, P < 0.0008 versus the inoculum. (e) Abundance of amino acid auxotrophs in the inoculum and biofilms. Graph shows the mean of four experiments. Error bars show SEM. *, P < 0.01 versus the inoculum.
Variants Are Not Generated by Spontaneous Mutation or by the Emergence of Mutator Strains.
How then are the variants produced? The simplest explanation is that spontaneous, growth-related mutations produced by DNA replication errors are responsible. However, our previous finding that inactivation of the recA gene markedly reduced diversity in P. aeruginosa biofilms argues against this possibility (8). RecA is necessary for homologous recombination and recombinatorial DNA repair (23) but it is not thought to be required for spontaneous, growth-related mutations (24). Consistent with this fact, we found no significant difference in the frequency of spontaneously arising rifampicin-resistant mutants in WT and recA P. aeruginosa cultures [supporting information (SI) Table S1]. Thus, inactivation of recA prevents the emergence of large numbers of variants in biofilms, but does not significantly change the spontaneous mutation frequency.
We also explored the possibility that variants are produced by hypermutable strains that might arise in biofilms. To accomplish this aim, we measured the frequency at which spontaneous mutations arise in 10 different colony morphology variants from independent biofilm experiments. The mutation frequency of the variants did not differ significantly from that of the parental strain. These data suggest that the variants in biofilms arise through some mechanism other than spontaneous mutation or mutator strains.
Endogenous Oxidative Stress Generates Diversity in Biofilms.
A clue to the mechanism emerged when we investigated whether biofilm-derived variants would themselves generate the full range of variant types after biofilm growth. Whereas most variants did, one variant type repeatedly failed to do so (Fig. 2a). These variants overproduced pyomelanin (Fig. 2b), a pigment that protects against oxidative stress in other organisms (25). We confirmed that the P. aeruginosa pyomelanin-expressing variant was resistant to oxidative stress by measuring bacterial viability after exposure to lethal concentrations of peroxide (Fig. 2c).
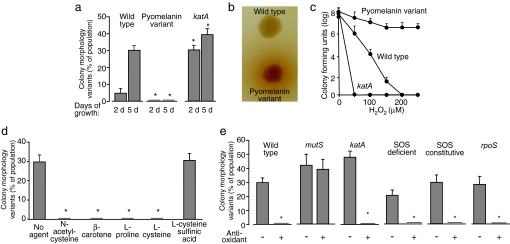
Fig. 2.
Oxidative stress mediates diversity in biofilms. (a) An oxidant-resistant strain produces less diversity, and an oxidant-sensitive strain produces more diversity than WT P. aeruginosa. Graph shows the proportion of the population in WT, pyomenanin-overproducing, and katA biofilms that form colonies with variant morphologies after 2 and 5 days of growth. Data are the mean of three experiments. Error bars show SEM. *, P < 0.03 versus corresponding WT control. Bacteria from biofilms formed by the pyomenanin-overproducing strain also showed less diversity in swimming motility and pyomelanin production than WT biofilm bacteria (Fig. S3). Bacteria from katA biofilms showed more diversity in these assays (Fig. S3). (b) Photograph of colonies of the WT and pyomelanin-overproducing strain grown on indicator agar plates. The dark pigment surrounding the variant colony is pyomelanin. (c) Susceptibility of planktonically grown WT, katA, and pyomelanin-overproducing P. aeruginosa to killing by H2O2. Data are the mean of four replicates from one experiment and are representative of three others. Error bars show SEM. (d) Effect of various antioxidants on diversity in biofilms after 5 days of growth. Indicated antioxidants were added to the biofilm medium at the beginning of growth, and the proportion of bacteria with variant colony morphologies was determined. Cysteine sulfinic acid is an oxidized form of cysteine lacking antioxidant activity. Data are means of four experiments. Error bars show SEM. *, P = 0.002 versus no agent control. Addition of antioxidants also reduced diversity in swimming motility and pyomelanin production (Fig. S3). (e) The effect of hypermutability, bacterial catalase, the SOS response, and rpoS on diversity in biofilms. P. aeruginosa strains in which mutS, katA, and rpoS was inactivated, and in which the SOS response was deficient and constitutively active (see Fig. S2) were grown in biofilms for 5 days. Antioxidants decrease diversity in biofilms formed by catalase deficient P. aeruginosa, but not in biofilms formed by a hypermutable (mutS) strain. Neither the SOS response nor rpoS was required to generate colony morphology diversity in biofilms. Antioxidants eliminated the observed diversity in the SOS constitutive strain even though its SOS activity was unaffected (see Fig. S2). Similar results were seen in assays measuring diversity in swimming motility and pyomelanin production (Fig. S3). Data shown are for l-proline. In other experiments the antioxidants N-acetyl-cysteine, β-carotene, and l-cysteine had a similar effect on diversity (Fig. 2d and Fig. S3). Data are means of three experiments. Error bars show SEM. *, P < 0.002 versus the untreated control.
The failure of an oxidant-resistant variant to diversify led us to hypothesize that oxidative stress produces the mutations that generate the variants. If this hypothesis is correct, increasing the sensitivity of bacteria to oxidants should increase diversity in biofilms and reducing oxidative stress should have the opposite effect. We first tested this by using an oxidant-sensitive catalase mutant (katA; see Fig. 2c) and found that it produced variants earlier and to a greater extent than the WT parent (Fig. 2a). We also added the antioxidants N-acetyl-cysteine, β-carotene, l- cysteine, or l-proline (26) to the biofilm medium and found that they all eliminated the observed diversity in both WT (Fig. 2d) and katA biofilms (Fig. 2e).
Given that starvation stress can increase mutation frequency (22, 27), we considered the possibility that these antioxidant compounds act by increasing carbon and energy in the medium. However, the addition of an oxidized form of l-cysteine, cysteine sulfinic acid, to the biofilm medium had no affect on diversity (Fig. 2d). We also tested other carbon and energy sources not known to have antioxidant activity. Adding glycerol, methionine, alanine, or sucrose, or using a 10-fold higher concentration of tryptic soy broth, did not significantly change the degree of diversity present (Fig. S1).
Finally, if the hypothesis linking oxidant stress to genetic variation in biofilms is correct, supplying mutations by another method should overcome the diversity-inhibiting effect of the antioxidants. We tested this by using a mutS mutant that constitutively generates mutations at ≈100 times the frequency of WT P. aeruginosa due to defective mismatch repair functions (see Table S1). As shown in Fig. 2e, mutS biofilms generated extensive diversity even if grown in the presence of antioxidants. Thus, an oxidant-sensitive strain produced more diversity, an oxidant-resistant strain and antioxidants block diversity, and increasing mutations (via mutS inactivation) overcomes the diversity-inhibiting effects of antioxidants. Given that no exogenous oxidants were added in these experiments, these data link the genetic variation in biofilms with oxidative stress that is endogenous to the bacteria.
Neither the SOS Response Nor rpoS Is Required to Generate Diversity in Biofilms.
The above experiments and our previous work indicate that both oxidative stress and recA gene function are required to generate the diversity we observe in biofilms. One mechanism that could tie these findings together involves a bacterial stress response known as SOS. The SOS response can induce mutations by activating error-prone DNA polymerases (28), and SOS is required in Esherichia coli stress-induced mutagenesis systems (27, 29). Moreover, reactive oxygen species are known triggers of SOS, and RecA function is required for SOS activation.
To investigate the role of SOS, we used a recA+ P. aeruginosa strain that was SOS-deficient (because of a lexA point mutation), and a strain constitutively expressing SOS genes (Fig. S2). As shown in Fig. 2e, both strains produced a similar degree of population diversity as the WT. Furthermore, the addition of antioxidants eliminated the observed diversity in both strains, even though the SOS activity of the constitutive strain was unaffected (Fig. 2e and Fig. S2). Thus, whereas RecA function is required to generate diversity in biofilms, the SOS response is not. We also found that inactivation of the rpoS stress and stationary-phase regulator [required in stress-induced mutagenesis systems (29, 30)] had no significant effect on diversity in biofilms (Fig. 2e).
dsDNA Break (DSB) Repair Is Required for Biofilm-Mediated Diversity.
Another mechanism potentially linking oxidative stress and recA gene function to genetic variation involves direct DNA damage. Although oxidants can produce a variety of DNA lesions (31, 32), we hypothesized that DSBs mediated diversity in biofilms for two reasons. Recent work shows DSB repair can be mutagenic under some conditions (29), which could explain the generation of diverse variants. In addition, DSBs are lethal unless corrected, and repair requires recA gene function (29). Thus, the absence of observable diversity in recA biofilms could be explained by the death of cells that incur breaks. We tested this hypothesis in several ways.
First, this hypothesis predicts that the abundance of DSBs should be increased in biofilms and should decrease with the addition of antioxidants. As shown in Fig. 3a, biofilm-grown cells contained more breaks than planktonic bacteria, and the addition of antioxidants reduced the numbers of breaks in biofilms. As an additional test, we measured breaks in biofilms grown from the pyomelanin-overproducing P. aeruginosa that show reduced biofilm-mediated diversity and are protected from oxidant stress (Fig. 2 a and c). As expected, we found fewer breaks in biofilms grown from this strain.
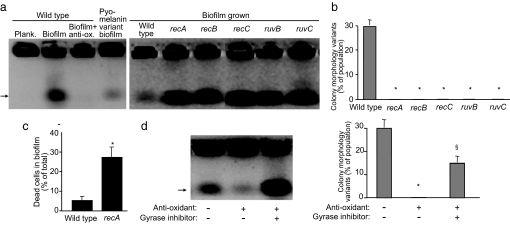
Fig. 3.
DSBs mediate diversity in biofilms. (a) The abundance of DSBs in biofilms is related to oxidative stress. Planktonic and biofilm grown bacteria of indicated strains were treated with DNase to remove extracellular DNA (see Fig. S4 for control experiments). The abundance of DSBs was measured by using a pulse field gel electrophoresis technique in which chromosomes with DSBs migrate into the gel (arrow), whereas unbroken chromosomes cannot (38). (Left) WT biofilms contained more chromosomes with breaks than WT planktonic bacteria, WT biofilms grown in antioxidant-containing medium, and biofilms formed by the oxidant-resistant pyomelanin-producing variant. (Right) Inactivation of genes involved in DSB repair increases the abundance of DSBs in biofilms. Gels shown are representative of three experiments. (b) Inactivation of genes involved in DSB repair reduces biofilm-mediated diversity in colony morphology, swimming motility, and pyomelanin production (Fig. S3). Data are the mean of three experiments. Error bars show SEM. *, P < 0.001 versus the WT. (c) Inactivation of a gene involved in DSB repair increases the abundance of dead cells in biofilms. WT and recA biofilms were mechanically dispersed and stained with a viability strain (BacLight; Molecular Probes). A total of 3,000 bacteria were scored by fluorescent microscopy in each experiment. Data are the mean of three experiments. Error bars show SEM. *, P < 0.003 versus the WT. (d) Addition of the DNA gyrase inhibitor ciprofloxacin to antioxidant-treated biofilms restored DSBs (Left), diversity in colony morphology (Right), and swimming motility (Fig. S3a). Gel shown is representative of three experiments. Data in graph are the mean of three experiments. Error bars show SEM. *, P < 0.0001 versus the untreated control; §, P < 0.01 versus the antioxidant treated, without gyrase inhibitor condition.
A second prediction is that inactivating genes involved in DSB repair will increase the abundance of breaks and eliminate the diversity we observe in biofilms. This prediction is based on reasoning that DSBs will accumulate without repair, and cells incuring breaks cannot become variants because unrepaired DSBs are lethal. As shown in Fig. 3, inactivation of recA, recB, recC, ruvB, and ruvC [all involved in DSB repair in E. coli (33, 34)] increased the abundance of DSBs (Fig. 3a) and eliminated biofilm-mediated diversity (Fig. 3b). We also found that inactivation of break repair increased the number of dead cells in biofilms by 5-fold (Fig. 3c). This finding is consistent with the explanation that variants fail to arise in repair mutants because bacteria incurring breaks die.
A third prediction made by this hypothesis is that inducing DSBs should overcome the inhibitory effect of antioxidants on diversity. To test this we grew biofilms in the presence of the DNA gyrase inhibitor ciprofloxacin, which is known to generate breaks. Addition of ciprofloxacin restored DSBs and diversity to WT biofilms grown in the presence of antioxidants (Fig. 3d). Furthermore, the gyrase inhibitor had no effect on population diversity in biofilms grown from the recA, recB, recC, ruvB, or ruvC repair mutants even though breaks were abundant (data not shown). These data are all consistent with the hypothesis that DSBs mediate the genetic diversity that we observe in biofilms.
Endogenous Oxidative Stress in Biofilms Can Promote Antibiotic Resistance.
Although some chromosomal regions may be more susceptible than others, oxidative stress will likely cause breaks throughout the genome. If this is true, variants with an enormous diversity of fitness characteristics should be produced in biofilms, including variants resistant to antibiotics and other stresses. However, many of these will not be evident in unstressed laboratory biofilms as they will not be amplified in typical growth conditions.
To test this idea, we added low concentrations of the antibiotic gentamicin to the biofilm medium. Gentamicin is a protein synthesis inhibitor that inhibits ribosome function. This antibiotic was chosen because the selective pressures it imposes are likely to be very different from those inherent to biofilms, and because it is used clinically. As Fig. 4 shows, biofilms grown in gentamicin generated large numbers of resistant bacteria. Furthermore, both the addition of antioxidants and the inactivation of break repair genes markedly reduced the number of resistant bacteria (by ≈1,000 fold). These data suggest that the oxidative stress-break repair mechanism can generate genetic variants with a wide range of fitness characteristics. Some can be amplified by selective pressures inherent to biofilms, and others can be amplified by extrinsic stresses like antibiotics.
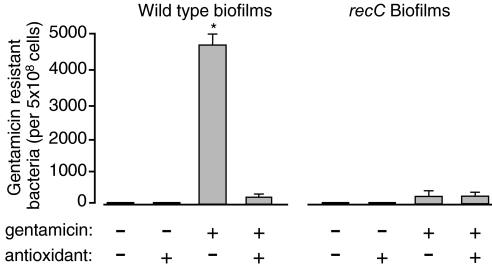
Fig. 4.
The oxidative stress break-repair mechanism increases the frequency at which resistant bacteria arise in antibiotic-exposed biofilms. WT and recC biofilms were grown for 5 days in medium with and without low concentrations of gentamicin (0.5 μg/ml) and antioxidants. In the presence of the antibiotic, biofilms generated large numbers of resistant bacteria, and the gentamicin-resistant phenotype was retained upon serial passaging (data not shown). Blocking the oxidative stress-break repair mechanism by the addition of antioxidants or the inactivation of break repair genes (using the recC strain) markedly reduced the number of antibiotic-resistant bacteria. Data shown are for l-proline. In other experiments the antioxidants N-acetyl-cysteine, β-carotene, and l- cysteine had a similar effect on the emergence of variants (Fig. 2d and Fig. S3). Data are means of three experiments. Error bars show SEM. *, P < 0.0001 versus all other conditions tested; other values are not significantly different.
Discussion
Diverse genetic variants are generated by biofilm growth in a wide range of bacterial species (3–9); however, the mechanism producing this variation was unknown. A fundamental question is whether the genetic variation that underlies the diversity is a consequence of spontaneous growth-related mutations or some other process. Our data with P. aeruginosa suggest that it is largely caused by the mutagenic repair of DSBs caused by oxidative stress. This conclusion is supported by experiments showing that increasing the sensitivity of bacteria to oxidants increases both breaks and diversity, reducing oxidant stress has the opposite effect, and an agent producing breaks overcomes the inhibitory effects of antioxidants on diversity. Furthermore, inactivation of DSB repair genes eliminates the observed diversity without appreciably changing the frequency of spontaneous mutations (see model; Fig. 5).
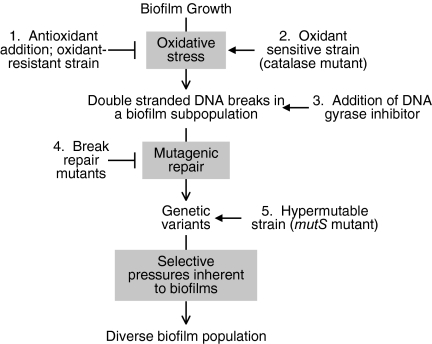
Fig. 5.
Proposed model for the generation of genetic diversity in biofilms. Experimental interventions from this study that support this model are numbered; ┤ indicates an inhibitory effect, and → indicates an augmenting or overriding effect. Endogenous oxidative stress produces DSBs in some biofilm bacteria. Mutagenic repair of breaks generates genetic variants. Different variant types are then amplified by selective pressures inherent to biofilms, producing diversity in the population. The model is consistent with the following findings from this study. (1) Adding antioxidants to the medium or using an oxidant-resistant strain reduces DSB's in biofilm bacteria (Fig. 3a) and reduces diversity (Fig. 2 a and d). (2) Inactivation of catalase biosynthesis increases sensitivity to oxidative stress (Fig. 2c) and diversity in biofilms (Fig. 2a). (3) Addition of a gyrase inhibitor produces DSBs in antioxidant-treated biofilms (Fig. 3d) and overrides the inhibitory effect of antioxidants on diversity (Fig. 3d). (4) Inactivation of break repair genes eliminates the observed diversity (Fig. 3b and Fig. S3), despite the presence of breaks (Fig. 3a), likely because mutagenic repair is required to generate variants. Consistent with this, repair mutant biofilms contain more dead cells than WT (Fig. 3c). (5) The mutS mutant constitutively produces mutants at a high frequency (Table S1) and generates diversity even when grown in antioxidant-containing medium (Fig. 2e and Fig. S3).
These experiments link the emergence of extensive diversity in biofilms to oxidative stress. However, we do not yet know the identity of the oxygen species involved or how biofilm growth changes cell physiology to produce this stress. One possibility is that DSBs are produced by oxidants generated as a consequence of aerobic bacterial respiration. This explanation may seem paradoxical given the low oxygen tensions detected deep within biofilm structures (35). However, recent work shows that respiration can produce enough oxidative stress to produce DNA damage (36), and some biofilm bacteria may express lower levels of antioxidants such as catalase (37). Thus, an imbalance between oxidant burden and antioxidant defenses could exist in some biofilm regions. Oxidative stress could also be produced if electron transport is somehow altered or by increased production of redox-active metabolic products such as phenazines. Whatever the source of the oxidants, our experiments suggest that genetic variation is caused by physiological changes in some biofilm bacteria leading to direct DNA damage, rather than by some “programmed response” to biofilm growth.
Although our data point to DSBs as the proximate lesion and show that break repair genes are required, further work will be needed to understand how mutagenic repair occurs. In E. coli, break repair becomes mutagenic during stationary phase and requires the rpoS sigma factor, the SOS response, and the error-prone DNA polymerase pol IV (29). Our experiments suggest that none of these factors is required in P. aeruginosa (this study and ref. 8), perhaps because DSBs are inherently mutagenic in P. aeruginosa or mutagenic repair involves different genetic mechanisms.
In addition to experiments relating to the mechanism of genetic variation, our studies underscore the importance of selection in biofilm-mediated diversity. The fact that large numbers of different variant types emerge indicate that strong and varied selective pressures exist in biofilms, and that these can amplify many different phenotypes. Our experiments also indicate that the selective pressures responsible are in large part independent of the oxidant-mediated mechanism generating variation. This conclusion is supported by experiments in which diversity was generated even if oxidative stress was blocked, provided mutations are supplied by other means (see experiments with the mutS strain and gyrase inhibitor; Figs. 2e and 3d).
How does biofilm growth produce such strong diversifying selection? In our view, two biofilm characteristics may be particularly important. First, the group structure of biofilms will, on its own, produce many microenvironments with differing conditions, because of chemical gradients generated by the metabolism of resident bacteria and transport limitation (19, 20). For example, the consumption of oxygen will generate oxygen gradients that vary with physical location within the biofilm (in the X, Y, and Z spatial dimensions) and as a function of time (35). Gradients of other nutrients, metabolites, and wastes will be similarly produced, even when biofilms are grown in unstressed conditions. The superimposed effect of these gradients will produce a tremendous number of different microenvironments, all of which could theoretically select for different variant phenotypes.
A second diversity-promoting effect relates to the fact that biofilm growth can lead to the physical separation of cells in different biofilm regions. Geographic isolation makes it more likely that different bacterial lineages will have sustained exposure to different selective pressures, which in turn increases the probability that lineages will take different paths of evolution. The impact would be even greater if the separated cells are different variant types, as distinct selective pressures would then be operating on cells with different initial fitness characteristics. Together, geographic separation and the many distinct microenvironments in biofilms could produce a pronounced diversifying effect.
A key question for the future is whether the oxidative stress-break repair mechanism operates in biofilms found in natural environments and human infections. If it does, this process could significantly affect the functioning of these bacterial communities, carrying both costs and benefits. Because this mechanism will likely produce mutations throughout the genome, an extensive variety of variants will be generated. Many variants will not survive, because most random mutations are deleterious. However, our data show that some variants will be amplified by the heterogeneous selective pressures inherent to biofilms, leading to population diversity. Diversity in biofilms could mitigate the effect of environmental upsets, just as it does for other biological communities like forests. Other variants that survive, but are not amplified under prevailing conditions, are likely to remain scarce. These rare variants could increase the capacity of biofilms to adapt to adversity by being “on-call” and ready to multiply should extrinsic stresses like antibiotics arise.
Methods
Bacterial Strains, Plasmids, and Growth Conditions.
Bacteria were grown at 37°C in tryptic soy broth or agar as the growth medium. Colony phenotypes, motility, pyomelanin, and auxotrophy was analyzed as described (8). P. aeruginosa mutants (transposon insertions) were obtained from the University of Washington Genome Center or constructed as described below. Strain ID numbers from the University of Washington are: WT PA01, katA ptl5765, recB ptl47550, recC ptl39347, ruvB ptl48439, ruvC ptl17611, lexA ptl32620, mutS ptl2006, and rpoS ptl48855; recA has been described (8). A SOS noninducible mutant was constructed by using overlap extension PCR to change lexA glycine at position 86 to valine by mutating the corresponding codon from GTG to GTT. This was cloned into miniCTX1 and inserted on the chromosome of a lexA null mutant. A recA::gfp reporter was made by cloning a 136-bp promoter region of recA in front of a promoterless gfp in pUCP18. Expression of this reporter was monitored by using a fluormeter (Tecan) with λex 435 nm and λem 535 nm. Antibiotic concentrations (per ml) were: 400 μg nalixidic acid, 60 μg tetracycline, 300 μg rifampicin, 50 or 0.5 μg gentamycin, and 1 μg ciprofloxacin. Drip-flow biofilm reactors were used as in ref. 8. Biofilm medium was 1% tryptic soy broth supplemented as indicated. Antioxidants used were: N-acetyl cysteine, 0.55 mM; l- proline, 2 mM; β-carotene, 0.35 mM, l- cysteine, 0.55 mM.
Detection of DSBs.
To detect DSBs, 108 bacteria (grown planktonically or in a biofilm for 3 days) were suspended in 100 μl of 10 mM Tris, pH 7.5 plus 5 units of Dnase and incubated at 30°C for 3 h. Cells were washed in Tris buffer, resuspended in 100 μl of 10 mM Tris pH 7.5/20 mM NaCl/50 mM EDTA and mixed with 100 μl of molten 2% InCert agarose to create a plug containing intact bacteria. Plug treatment, pulse field gel electrophoresis, and gel straining have been described (34). In some cases, biofilms were grown for 24 h, then 1 μg/ml ciprofloxacin was added.
Viability and Diversity Assays.
To measure oxidative stress sensitivity, bacteria were grown in TSB medium overnight, diluted to 107 cfu/ml, and exposed to hydrogen peroxide for 4 h. Cells were then serial-diluted and plated. To assess the viability of WT and recA biofilm bacteria, biofilms were grown for 3 days and then stained with 0.01 μM SYTO9 and 0.03 μM propidium iodide for 20 min and then examined microscopically. Mutation frequencies were determined by plating 5 × 109 planktonic bacteria onto rifampicin plates and counting colonies after 48 h.
To determine the effect of extrinsic selection, 0.5 μg/ml gentamycin was added to biofilm medium at 1 day and growth continued for 5 days. Biofilms were dispersed by using a tissue homogenizer and plated on nutrient agar with and without gentamycin (50 μg/ml) to determine total and gentamycin-resistant counts. Unpaired Student's t test was used to generate P values, except for Fig. 1c in which the F test was used.
Supplementary Material
Acknowledgments.
We thank G. Doern for assistance with the pulse field gel electrophoresis studies, C. Manoil (University of Washington, Seattle, WA) for P. aeruginosa mutants, and M. Parsek, T. Yahr, C. Manoil, J Penterman, and F. Fang for helpful discussions. Funding for this study was provided by the National Heart, Lung, and Blood Institute of the National Institutes of Health, the Cystic Fibrosis Foundation, and the Burroughs Wellcome Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801499105/DCSupplemental.
References
- 1.Stoodley P, Sauer K, Davies DG, Costerton JW. Biofilms as complex differentiated communities. Annu Rev Microbiol. 2002;56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 2.Hansen SK, Rainey PB, Haagensen JA, Molin S. Evolution of species interactions in a biofilm community. Nature. 2007;445:533–536. doi: 10.1038/nature05514. [DOI] [PubMed] [Google Scholar]
- 3.Waite RD, Struthers JK, Dowson CG. Spontaneous sequence duplication within an open reading frame of the pneumococcal type 3 capsule locus causes high-frequency phase variation. Mol Microbiol. 2001;42:1223–1232. doi: 10.1046/j.1365-2958.2001.02674.x. [DOI] [PubMed] [Google Scholar]
- 4.Allegrucci M, Sauer K. Characterization of colony morphology variants isolated from Streptococcus pneumoniae biofilms. J Bacteriol. 2007;189:2030–2038. doi: 10.1128/JB.01369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deziel E, Comeau Y, Villemur R. Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J Bacteriol. 2001;183:1195–1204. doi: 10.1128/JB.183.4.1195-1204.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirisits MJ, Prost L, Starkey M, Parsek MR. Characterization of colony morphology variants isolated from Pseudomonas aeruginosa biofilms. Appl Environ Microbiol. 2005;71:4809–4821. doi: 10.1128/AEM.71.8.4809-4821.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palmer RJ, Jr, Stoodley P. Biofilms 2007: Broadened horizons and new emphases. J Bacteriol. 2007;189:7948–7960. doi: 10.1128/JB.00787-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boles BR, Thoendel M, Singh PK. Self-generated diversity produces “insurance effects” in biofilm communities. Proc Natl Acad Sci USA. 2004;101:16630–16635. doi: 10.1073/pnas.0407460101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yarwood JM, Paquette KM, Tikh IB, Volper EM, Greenberg EP. Generation of virulence factor variants in Staphylococcus aureus biofilms. J Bacteriol. 2007;189:7961–7967. doi: 10.1128/JB.00789-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haussler S, Tummler B, Weissbrodt H, Rohde M, Steinmetz I. Small-colony variants of Pseudomonas aeruginosa in cystic fibrosis. Clin Infect Dis. 1999;29:621–625. doi: 10.1086/598644. [DOI] [PubMed] [Google Scholar]
- 11.Haussler S, et al. Highly adherent small-colony variants of Pseudomonas aeruginosa in cystic fibrosis lung infection. J Med Microbiol. 2003;52:295–301. doi: 10.1099/jmm.0.05069-0. [DOI] [PubMed] [Google Scholar]
- 12.Sheehan DJ, Janda JM, Bottone EJ. Pseudomonas aeruginosa: Changes in antibiotic susceptibility, enzymatic activity, and antigenicity among colonial morphotypes. J Clin Microbiol. 1982;15:926–930. doi: 10.1128/jcm.15.5.926-930.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oliver A, Canton R, Campo P, Baquero F, Blazquez J. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science. 2000;288:1251–1254. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- 14.McCann KS. The diversity-stability debate. Nature. 2000;405:228–233. doi: 10.1038/35012234. [DOI] [PubMed] [Google Scholar]
- 15.Carr MH, Anderson TW, Hixon MA. Biodiversity, population regulation, and the stability of coral-reef fish communities. Proc Natl Acad Sci USA. 2002;99:11241–11245. doi: 10.1073/pnas.162653499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tilman D, Wedin D, Knops J. Productivity and sustainability influenced by biodiversity in grassland ecosystems. Nature. 1996;379:718–720. [Google Scholar]
- 17.Tilman D, et al. Diversity and productivity in a long-term grassland experiment. Science. 2001;294:843–845. doi: 10.1126/science.1060391. [DOI] [PubMed] [Google Scholar]
- 18.Haagensen JA, et al. Differentiation and distribution of colistin- and sodium dodecyl sulfate-tolerant cells in Pseudomonas aeruginosa biofilms. J Bacteriol. 2007;189:28–37. doi: 10.1128/JB.00720-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasmussen K, Lewandowski Z. Microelectrode measurements of local mass transport rates in heterogeneous biofilms. Biotechnol Bioeng. 1998;59:302–309. doi: 10.1002/(sici)1097-0290(19980805)59:3<302::aid-bit6>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 20.Teal TK, Lies DP, Wold BJ, Newman DK. Spatiometabolic stratification of Shewanella oneidensis biofilms. Appl Environ Microbiol. 2006;72:7324–7330. doi: 10.1128/AEM.01163-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boles BR, Thoendel M, Singh PK. Genetic variation in biofilms and the insurance effects of diversity. Microbiology. 2005;151:2816–2818. doi: 10.1099/mic.0.28224-0. [DOI] [PubMed] [Google Scholar]
- 22.Finkel SE, Kolter R. Evolution of microbial diversity during prolonged starvation. Proc Natl Acad Sci USA. 1999;96:4023–4027. doi: 10.1073/pnas.96.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuzminov A. Unraveling the late stages of recombinational repair: Metabolism of DNA junctions in Escherichia coli. BioEssays. 1996;18:757–765. doi: 10.1002/bies.950180911. [DOI] [PubMed] [Google Scholar]
- 24.Rosenberg SM, Hastings PJ. Adaptive point mutation and adaptive amplification pathways in the Escherichia coli Lac system: Stress responses producing genetic change. J Bacteriol. 2004;186:4838–4843. doi: 10.1128/JB.186.15.4838-4843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nosanchuk JD, Casadevall A. The contribution of melanin to microbial pathogenesis. Cell Microbiol. 2003;5:203–223. doi: 10.1046/j.1462-5814.2003.00268.x. [DOI] [PubMed] [Google Scholar]
- 26.Chen C, Dickman MB. Proline suppresses apoptosis in the fungal pathogen Colletotrichum trifolii. Proc Natl Acad Sci USA. 2005;102:3459–3464. doi: 10.1073/pnas.0407960102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bjedov I, et al. Stress-induced mutagenesis in bacteria. Science. 2003;300:1404–1409. doi: 10.1126/science.1082240. [DOI] [PubMed] [Google Scholar]
- 28.Sanders LH, Rockel A, Lu H, Wozniak DJ, Sutton MD. Role of Pseudomonas aeruginosa dinB-encoded DNA polymerase IV in mutagenesis. J Bacteriol. 2006;188:8573–8585. doi: 10.1128/JB.01481-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ponder RG, Fonville NC, Rosenberg SM. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol Cell. 2005;19:791–804. doi: 10.1016/j.molcel.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 30.Lombardo MJ, Aponyi I, Rosenberg SM. General stress response regulator RpoS in adaptive mutation and amplification in Escherichia coli. Genetics. 2004;166:669–680. doi: 10.1534/genetics.166.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bridges BA, Sekiguchi M, Tajiri T. Effect of mutY and mutM/fpg-1 mutations on starvation-associated mutation in Escherichia coli: Implications for the role of 7,8-dihydro-8-oxoguanine. Mol Gen Genet. 1996;251:352–357. doi: 10.1007/BF02172526. [DOI] [PubMed] [Google Scholar]
- 32.Saumaa S, Tover A, Tark M, Tegova R, Kivisaar M. Oxidative DNA damage defense systems in avoidance of stationary-phase mutagenesis in Pseudomonas putida. J Bacteriol. 2007;189:5504–5514. doi: 10.1128/JB.00518-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benson FE, Illing GT, Sharples GJ, Lloyd RG. Nucleotide sequencing of the ruv region of Escherichia coli K-12 reveals a LexA regulated operon encoding two genes. Nucleic Acids Res. 1988;16:1541–1549. doi: 10.1093/nar/16.4.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He AS, Rohatgi PR, Hersh MN, Rosenberg SM. Roles of E. coli double-strand-break-repair proteins in stress-induced mutation. DNA Repair. 2006;5:258–273. doi: 10.1016/j.dnarep.2005.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu KD, Stewart PS, Xia F, Huang CT, McFeters GA. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl Environ Microbiol. 1998;64:4035–4039. doi: 10.1128/aem.64.10.4035-4039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park S, You X, Imlay JA. Substantial DNA damage from submicromolar intracellular hydrogen peroxide detected in Hpx− mutants of Escherichia coli. Proc Natl Acad Sci USA. 2005;102:9317–9322. doi: 10.1073/pnas.0502051102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hassett DJ, et al. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol Microbiol. 1999;34:1082–1093. doi: 10.1046/j.1365-2958.1999.01672.x. [DOI] [PubMed] [Google Scholar]
- 38.Schapiro JM, Libby SJ, Fang FC. Inhibition of bacterial DNA replication by zinc mobilization during nitrosative stress. Proc Natl Acad Sci USA. 2003;100:8496–8501. doi: 10.1073/pnas.1033133100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.