Abstract
Tolerance to abiotic stress is an important agronomic trait in crops and is controlled by many genes, which are called quantitative trait loci (QTLs). Identification of these QTLs will contribute not only to the understanding of plant biology but also for plant breeding, to achieve stable crop production around the world. Previously, we mapped three QTLs controlling low-temperature tolerance at the germination stage (called low-temperature germinability). To understand the molecular basis of one of these QTLs, qLTG3–1 (quantitative trait locus for low-temperature germinability on chromosome 3), map-based cloning was performed, and this QTL was shown to be encoded by a protein of unknown function. The QTL qLTG3–1 is strongly expressed in the embryo during seed germination. Before and during seed germination, specific localization of beta-glucuronidase staining in the tissues covering the embryo, which involved the epiblast covering the coleoptile and the aleurone layer of the seed coat, was observed. Expression of qLTG3–1 was tightly associated with vacuolation of the tissues covering the embryo. This may cause tissue weakening, resulting in reduction of the mechanical resistance to the growth potential of the coleoptile. These phenomena are quite similar to the model system of seed germination presented by dicot plants, suggesting that this model may be conserved in both dicot and monocot plants.
Keywords: natural variation, QTLs, seed germination
The control of seed dormancy and germination is important for the adaptability of plants, and germination under favorable environmental conditions is needed for their survival. Seed dormancy and germination is a complex trait influenced by many genes and environmental conditions (1). In addition, the plant hormones gibberellin and abscisic acid (ABA) play important roles in the expression of seed dormancy and germination (2, 3).
Strong seedling vigor under low temperatures is an important objective of rice breeding programs in direct-seeding cultivation methods in temperate rice-growing areas, at high altitudes in tropical and subtropical areas, and in areas with a cold irrigation water supply where low temperature induces retardation of early seedling growth. Germinability (germination rate) and early seedling growth are major components of seedling vigor. A wide range of phenotypic variation of low-temperature germinability was found in rice cultivars. Quantitative trait loci (QTL) analysis for low temperature germinability revealed that many genes control this trait (4–6). However, the molecular functions of the genes for these QTLs have not been identified.
A QTL mapping approach is effective to detect genes controlling traits involved in seed germination. Under several kinds of stress, such as temperature, NaCl, and osmotic pressure, germination is delayed or inhibited. QTLs for the speed of germination may be important for germination in general and may not be affected by any kind of stress (7, 8). QTLs for the speed of germination collocated with those of ABA sensitivity and salt tolerance (9). The molecular identification of the genes responsible for these QTLs provides not only new insights into seed biology, but also understanding of their adaptability in natural variation (10).
Previously, we mapped three QTLs controlling low-temperature germinability using backcross inbred lines (BILs) derived from a cross between Italica Livorno and Hayamasari (vigorous and weak low-temperature germinability, respectively) (6). A major QTL for low temperature germinability on chromosome 3, qLTG3–1, explained >30% of the total phenotypic variation in the mapping population. This study sought to clone qLTG3–1 by a map-based strategy and revealed that qLTG3–1 encodes a protein of unknown function. Histological analyses indicated that the expression of qLTG3–1 was tightly associated with vacuolation of cells in the tissues covering the embryo. Based on the results, it was considered that qLTG3–1 might be involved in tissue weakening, which is known as one of the key regulators in seed germination observed in dicot plants.
Results
Genetic Effect of qLTG3–1.
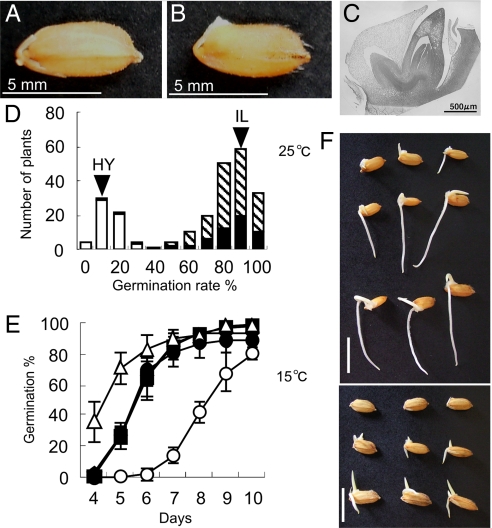
In rice seed germination, the coleoptile and coreorhiza are elongated (Fig. 1 A–C). At first, the coleoptile emerges from the seed, which is the definition for seed germination in rice (see Fig. 1 B). To determine the genetic effect of qLTG3–1, we performed a segregation analysis of low-temperature germinability using advanced backcross progeny (see Methods). Frequency distribution of low temperature germinability in the population showed a clear bimodal pattern, validating a large phenotypic effect of qLTG3–1 as a single Mendelian factor (Fig. 1 D), and indicating that qLTG3–1 is a dominant gene with a large effect. To clarify the effect on low-temperature germinability of qLTG3–1, we developed a near-isogenic line (NIL) containing a small chromosomal segment around qLTG3–1 (≈360 kb) (see Methods) from Italica Livorno in a Hayamasari genetic background. This NIL clearly showed more vigorous low-temperature germinability than Hayamasari (Fig. 1 E and F). These results clearly suggested that qLTG3–1 was required to express the high-level germination rate under low temperature conditions.
Fig. 1.
Seed germination and the effect of qLTG3–1 on low temperature germinability. (A and B) Rice seed germination. (C) Microscopic observation of the embryo. (D) Frequency distribution of low temperature germinability in the advanced backcrossed progeny. Arrowheads indicate the means of Italica Livorno (IL) and Hayamasari (HY). Three classified genotypes, homozygous for the Italica Livorno allele (black), heterozygous (hatched), and homozygous for the Hayamasari allele (white), assessed using the marker GBR3001 (6) are indicated. (E) Germination behavior of Hayamasari (open circle), the NIL (closed), and Italica Livorno (open triangle) under low temperature at 15°C. The means and SD of triplicates are shown. (F) Phenotype of germination of Hayamasari (Top), the NIL (Middle), and Italica Livorno (Bottom) incubated for 3 days at 25°C and 7 days at 15°C. The bars are 1 cm.
Germinability Under Several Kinds of Stresses.
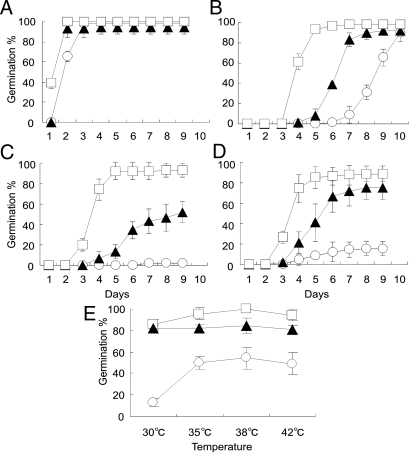
To determine whether qLTG3–1 shows tolerance to different stresses, germination tests were performed using Italica Livorno, Hayamasari, and the NIL under low and high temperature, high salinity, and high osmotic conditions (Fig. 2). Under the optimum temperature conditions for seed germination of rice, 25°C, there was no difference among these lines (see Fig. 2A). The NIL clearly showed increased germinability under low temperature 13°C, 300-mM NaCl, and 500-mM mannitol conditions compared with Hayamasari (see Fig. 2 B–D). Below 150-mM NaCl and 250-mM mannitol, all varieties showed delayed germination with relatively small varietal differences (data not shown). Under high temperature conditions, varietal differences were detected in hourly observations. Although all varieties could not germinate at 45°C, the NIL showed more vigorous germinability than Hayamasari under all high temperature conditions, 30–42°C (see Fig. 2E). These results revealed that qLTG3–1 showed tolerance for all of these stress conditions.
Fig. 2.
Germination behavior of qLTG3–1 under different stress conditions. (A) The optimum temperature, 25°C. (B) Low temperature, 13°C. (C) High salt, 300-mM NaCl. (D) High osmolarity, 500-mM mannitol. (E) Germination rates at 28 h after incubation under high temperature conditions. The means and SD of triplicates are shown for Hayamasari (open circle), the NIL (closed triangle), and Italica Livorno (open square).
Map-Based Cloning of qLTG3–1.
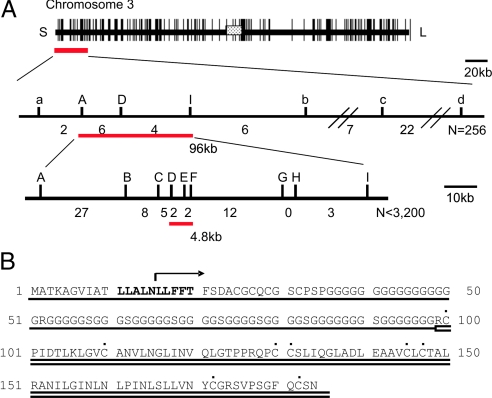
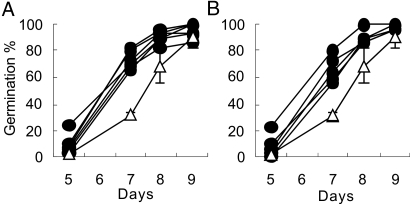
To determine the molecular function of qLTG3–1, the gene was cloned. High-resolution mapping placed qLTG3–1 in a 4.8-kb region between the markers SSR118673–13.1 (D) and S107 (F) on chromosome 3 (Fig. 3A) (see Methods). In this region, only one gene, Os03g0103300, was predicted in the Rice Annotation Project Database (RAP-DB) (http://rapdb.lab.nig.ac.jp/index.html). Sequence analyses of Os03g0103300 and its flanking regions of both 5′ and 3′ sides in Italica Livorno and Hayamasari identified a 71-bp deletion at position 47, causing a frame-shift in Hayamasari (Fig. 3B). To prove that Os03g0103300 functions in vigorous low temperature germinability, a 3-kb fragment containing the coding and both the 5′ and 3′ regions from Italica Livorno was cloned and transformed into Hayamasari using Agrobacterium tumefaciens (11, 12). Five T3 lines from three independent T0 transformants, which were homozygous for the transgene, clearly showed vigorous low temperature germinability (Fig. 4A).
Fig. 3.
Cloning of qLTG3–1. (A) Positional cloning of the qLTG3–1 gene. Fine mapping delimited the 96-kb region of qLTG3–1 on chromosome 3 (Middle). High-resolution mapping (Bottom) revealed that qLTG3–1 was located in the 4.8-kb region between the markers D and F and cosegregated with the marker E. The number indicates the number of recombinants between the markers. (B) Sequence alignment of qLTG3–1. A deletion identified in the Hayamasari allele is shown above the aligned sequences by arrows. The two domains as defined by Pfam, GRP, and LTP, are underlined by single and double lines, respectively. The conserved eight Cys residues in the LTP motif are indicated by dots. The 10 conserved amino acids at the N terminus are in bold.
Fig. 4.
Introductions of the functional qLTG3–1 gene from Italica Livorno into Hayamasari. The germination of homozygous transformants obtained by Agrobacterium-mediated transformation of Hayamasari with the genomic fragment of qLTG3–1 (A) and the overexpression of qLTG3–1 by the CaMV 35S promoter (B) are shown. The germination behavior (n = 30) of Hayamasari (open triangle) and T3 lines (closed symbols) at 15°C is shown. The mean and SD of triplicats in Hayamasari are shown.
To determine the effects of qLTG3–1 overexpression, we generated transgenic rice plants of six T3 lines from three independent T0 transformants that expressed qLTG3–1 under the control of the Cauliflower mosaic virus (CaMV) 35S promoter. These transgenic plants showed higher expression levels of the functional qLTG3–1 than the NIL in the leaves, panicles, and embryos during seed germination [supporting information (SI) Text and Fig. S1]. The higher expression level in the transgenic plants was detected before the start of the expression of qLTG3–1 in the NIL during seed germination. The phenotype of low temperature germinability in the overexpression plants was very similar to the transgenic plants that express qLTG3–1 under their own promoter (Fig. 4B). These findings confirmed that the Italica Livorno allele of Os03g0103300 is responsible for the vigorous low temperature germinability of qLTG3–1.
qLTG3–1 Is a Plant-Specific Gene.
qLTG3–1 consisted of one exon of 555-bp length, which encodes a novel protein of 184 aa with unknown function (see Fig. 3B). Based on domain searches, the qLTG3–1 protein has two conserved domains, glycine-rich cell wall protein (GRP) of the glycine rich protein family from amino acid 1 to 100, and Tryp_alpha_amyl of the protease inhibitor/seed storage/LTP family from amino acid 100 to 182 according to Pfam analysis (http://motif.genome.jp). In addition, 10 conserved amino acids, LLALNLLFFT, were identified at the N termini as the results of the comparison of qLTG3–1-like proteins in plants (see Fig. 3B and Fig. S2).
A database search found 21 genes with significant homology to qLTG3–1 only in plant species (Fig. S3). Homologues of the qLTG3–1-like proteins appeared to be expressed in a small range of higher plants, Poaceae, Solanaceae, Cucurbitaceae, and Leguminosae.
Expression of qLTG3–1 Is Tissue-Specific.
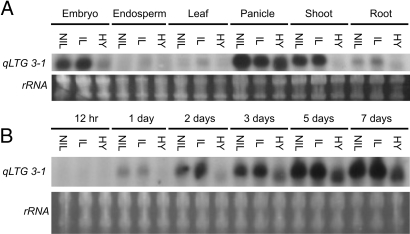
We analyzed qLTG3–1 expression in different tissues: embryo and endosperm from germinating seed, leaf from 2-month-old plants, young panicle before heading, shoot and root of seedlings. The expression of qLTG3–1 was tissue-specific to the embryo, panicle, and shoot (Fig. 5A). During seed germination at 15°C, qLTG3–1 expression was detected at 1 day from the start of incubation before germination, then the expression levels of qLTG3–1 increased during germination in the NIL and Italica Livorno (Fig. 5 B). The expression patterns in the NIL were the same as those of Italica Livorno. The expression of qLTG3–1 was also detected under optimum conditions at 30°C (data not shown). The patterns of increasing expression levels during seed germination under both conditions were the same. This finding indicated that qLTG3–1 was not induced by low temperature stress. In the process of seed development after pollination, the expression of qLTG3–1 was not detected (data not shown). The expression levels and patterns in the three varieties corresponded well with the phenotype of low-temperature germinability. These results indicated that the timing and level of expression of qLTG3–1 were strongly associated with the promotion of seed germination.
Fig. 5.
Expression of qLTG3–1. Northern blot analysis was used to measure the qLTG3–1 expression levels in total RNA extracted from different tissues of Hayamasari (HY), the NIL (NIL), and Italica Livorno (IL). Ethidium bromide stained rRNA was used as a loading control. (A) Tissue specificity. (B) Germination at 15°C.
Promoter and Subcellular Localization Analyses.
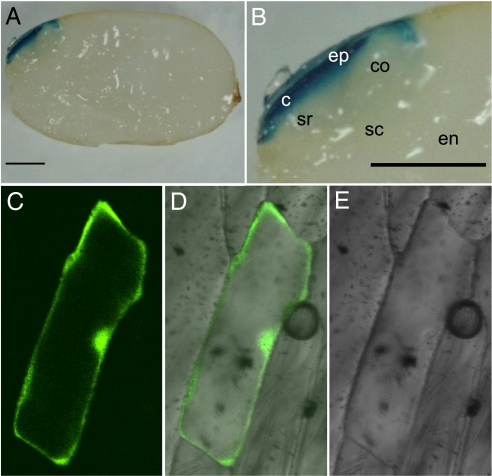
We analyzed qLTG3–1 promoter activity in transgenic rice plants carrying an qLTG3–1::GUS (beta-glucuronidase) reporter gene fusion. The expression of GUS in germinating seeds incubated for 1 day at 30°C was detected only in the tissues covering the embryo (Fig. 6 A and B). These tissues involved the seed coat and the epiblast covering the coleoptile. No signal was detected in the seed before incubation (data not shown). To define the tissues with GUS expression, the seed coat covering the embryo was removed from the seed, then the seed coat and the seed without it were stained individually. GUS expression was detected in both the inner cell layer of the seed coat and the tissues covering the embryo in the seed without the seed coat (data not shown). The seed covering tissues consist of nonliving tissues, pericarp and testa, and a single layer of living aleurone cells (Fig. 7 A and C). These indicated that GUS was expressed in both the epiblast covering the embryo and the aleurone cells.
Fig. 6.
GUS expression and subcellular localization. (A) GUS expression under the control of the qLTG3–1 gene promoter. Transgenic plants expressing GUS under the control of the 2-kb upstream region of qLTG3–1 were stained for GUS activity. The seeds were incubated for 1 day. The bars are 1 mm. (B) Close up view of the embryo in (A). c, coleorhiza; co, coleoptile; en, endosperm; ep, epiblast; sc, scutellum; sr, seminal root. (C–E) Subcellular localization of GFP in onion epidermal cells expressing a qLTG3–1-GFP fusion as shown by confocal microscopy.
Fig. 7.
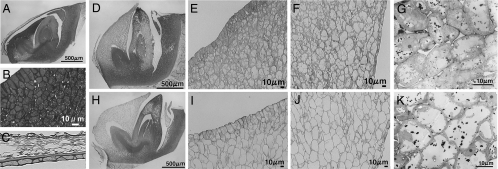
Characterization of the morphological changes in seed germination: Hayamasari (D–G) and the NIL (H–K). (A) Embryo before incubation. (B) Close up view of an area on the epidermal-side of the epiblast. (C) Close up view of the seed covering tissue. (D and H) Embryo incubated for 1 day at 30°C. (E and I) Close up view of an area of the epidermal-side of the epiblast. (F and J) Close up view of an area on the inner-side of the epiblast. (G and K) Transmission electron microscopy (TEM) sections of epidermis cells of the epiblast.
To determine the subcellular localization of the qLTG3–1 protein, the coding region of qLTG3–1 fused to modified green fluorescent protein (sGFP) at the N-terminal end (13) was expressed under the control of the CaMV 35S promoter. Fluorescence was detectable in both the cytoplasm and the nucleus in onion epidermal cells following particle bombardment (Fig. 6 C–E).
Histological Characterization.
To determine why qLTG3–1 causes vigorous low-temperature germinability, the cell morphology of the tissue where qLTG3–1 is expressed was compared between the NIL and Hayamasari. A clear varietal difference was detectable at 30°C in the hourly observations. Although germination rates were different among the varieties after 1 day at 30°C, we selected seeds in the same stage, which was just before germination (Fig. 7 D and H), and compared the cell morphology in the embryo. The cell morphology was drastically different from each other (Fig. 7 D–K). The cells in the tissues covering the embryo were filled with protein storage vacuoles (PSV) and lipid-bodies in mature seeds (Fig. 7 A and B). The coalescence of smaller PSV into one large central vacuole was observed during the incubation of seeds for germination. Vacuolation of the cytoplasm in the epiblast was observed in most cells in Hayamasari, but cytoplasm organelles still remained (see Fig. 7 D–G), whereas most cells in the NIL were fully vacuolated (Fig. 7 H–K). The same morphological changes of the cells were observed in germinating seeds at 15°C (Fig. S4). Vacuolation was likely to begin from the inner cells to the epidermal cells in the epiblast. It is known that genes involved in cell wall modification or tissue weakening are expressed in the endosperm cap before radicle emergence in dicot plants (14). Elevated vacuolation observed in the NIL suggested that the function of qLTG3–1 may be to accelerate vacuolation and to be involved in weakening of the tissues covering the embryo during the seed germination process.
Discussion
Despite the importance of the control of seed dormancy and germination in cereal crops, knowledge of the physiological mechanisms and genetic basis of these traits is very limited in monocots. This study elucidated the molecular basis of qLTG3–1, which has been identified as the most effective locus controlling low-temperature germinability in the rice variety Italica Livorno (6). In the present work, the gene responsible for this QTL was identified by a map-based cloning method.
qLTG3–1 represents a gene with unknown function, containing two known conserved domains, GRP and LTP, and a different conserved amino acid motif found in this study (see Fig. 3B). GRP-containing proteins in plants are divided into two known classes: RNA-binding proteins and structural proteins in the cell wall (15, 16). The former has a putative RNA-binding domain, but qLTG3–1 does not show any homology to this class of proteins. Because many genes with diverse functions involve each domain (17), the function of qLTG3–1 carrying both domains could not be estimated from domain information alone. Because of the unique features of the combination of the two domains, expression analyses with Tfm5 in tomato (18) and NT16 in tobacco (19) have been carried out. Tfm5 is specifically expressed in immature green fruit, whereas NT16 expression is developmentally regulated and induced by wound-stress conditions. However, their functions are still not clear.
qLTG3–1 was expressed in the tissues covering the embryo in the seed. Similar to the tissue specific expression of qLTG3–1, it is known that genes for chitinase, class I β-1,3-glucanase, expansin, endo-β-d-mannanase, and xyloglucan endotransglycosylase are involved in cell wall modification or tissue weakening in the endosperm cap before radicle emergence in tomato (20–24). Vacuolation was observed in the region of the aleurone layer that ruptures first during germination (14, 25). These cellular changes are associated with physical weakening of the endosperm cap before radicle emergence. The expression of qLTG3–1 was tightly associated with vacuolation, and the elevated vacuolation observed in the NIL suggests that the function of qLTG3–1 may be to accelerate vacuolation and to be involved in the weakening of tissues covering the embryo during the seed germination process.
A model system of seed germination and dormancy was proposed from biochemical and molecular information using dicot plants. Germination is determined by the balance between the growth potential of the embryo and the mechanical resistance of the tissues covering the embryo (26–29). However, the mechanistic basis of the effect of the genes encoding cell wall proteins and hydrolases on seed germination is not well understood. Furthermore, little information is known about the analogous process in monocot plants. This study provides new insights into seed germination in monocot plants. qLTG3–1 was genetically and molecularly identified as a gene involved in the regulation of germination. The tissue specificity of gene expression, the estimated function, and the vacuolation of the cells are quite similar to the model system of seed germination presented by dicot plants. This indicates that the model of seed germination may be conserved in both dicot and monocot plants.
The results of this study indicate that qLTG3–1 may not be involved in the response to low temperature but involved in seed germination itself. The NIL also showed vigorous germinability under several kinds of stress (see Fig. 2). This suggested that the vigor of seed germination as regulated by qLTG3–1 may show tolerance to several kinds of stress. Recently, QTL mapping has been successfully applied to dissecting the complexity of abiotic stress tolerance in rice leading to the isolation of SKC1 for salt tolerance (30) and Sub1A for submergence tolerance (31), and these genes could enhance abiotic stress tolerance in rice (32). qLTG3–1 has a large genetic effect on the increase in low-temperature germinability in rice. Because of the complex inheritance of this trait and many undesirable associated traits in the commercial cultivars, it has not been possible to introduce it into such cultivars as a single QTL. The gene, the molecular markers, and the NIL identified and developed in this study are useful to improve low-temperature germinability in rice breeding programs. In addition, the molecular identification of this QTL controlling low-temperature germinability is important for the elucidation of the complex genetic regulation of low-temperature germinability. This study clearly indicates that the combination of qLTG3–1 with the genes encoded that exhibits high growth potential of the coleoptile could be the most effective to establish rice cultivars with vigorous low-temperature germinability.
Methods
Plant Materials.
Hayamasari from Japan and Italica Livorno from Italy, which are temperate japonica rice varieties, were used. Based on the phenotype of low-temperature germinability and the genotype of qLTG3–1, one line, BIL116, was selected from the BILs derived from the cross between Hayamasari and Italica Livorno (6). BIL116 was backcrossed with Hayamasari using marker-assisted selection for the development of NIL and mapping populations. The NIL has at most a 360-kb chromosomal region introgressed from Italica Livorno around qLTG3–1 in Hayamasari (Fig. S5). Germination tests were as described by Fujino et al. (6). For low- and high-temperature stress experiments, 30 seeds in a Petri dish were placed in an incubator. For NaCl and mannitol stress tests, different concentration solutions were added to the Petri dish and placed in an incubator at 25°C.
Map-Based Cloning of qLTG3–1.
The F2 population consisting of 256 plants derived from the cross between BIL116 and Hayamasari was used for fine mapping. The genotype of F2 plants at the qLTG3–1 locus was determined by germination tests with their F3 progeny. As a result, qLTG3–1 was located on a 96-kb region between the markers SSR125411–4.1 (A) and STS73–28 (I) (see Fig. 3A). In addition, a BC1F2 population consisting of approximately 3,200 plants was used for the high-resolution mapping. There are no simple sequence repeat (SSR) markers showing polymorphisms available in the target region because the two parents are genetically closely related. We sequenced the target regions of both Hayamasari and Italica Livorno by direct sequencing of both strands of PCR products using a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems) (33). Fourteen polymorphisms including SSR, single nucleotide polymorphisms, and indels were detected and six of these were used for high-resolution mapping.
DNA Analysis.
Genomic DNA was extracted according to the procedure described by Fujino et al. (6). Genomic DNA was used for PCR analysis to score cosegregation of the phenotype with the PCR-based molecular markers. The molecular markers for mapping are listed in Table S1.
Plasmid Constructs and Transformation.
A genomic DNA fragment of 3-kb of qLTG3–1 from Italica Livorno was amplified by PCR using primers Ano13-LA5U (CGCGGATCCCTTCGTAATTCAGCAGGGCCGGGCAAATAA) and Ano13-LA5L (AATGAGCTCGTGTTGTGAAAACAAACAGCTAGTATGTATGTGTG). To make the qLTG3–1 promoter-GUS gene fusion construct, a 2-kb genomic DNA fragment of the 5′ upstream region of qLTG3–1 from Italica Livorno was amplified by PCR using primers Ano13–10U (GTTAAGCTTCTTCGTAATTCAGCAGGGCCGGG) and Ano13–10L (CGAGGATCCGCCCACCCACCGCACTGCACCTG). Because the length of the regulatory sequence sufficient for true qLTG3–1 gene expression was unclear, a 2-kb 5′ upstream region from the start codon of qLTG3–1 was used as the promoter. These fragments and the GUS gene were cloned into the pPZP2H-lac Ti-plasmid vector (11). To make the overexpression plants, a construct with a CaMV 35S promoter driving qLTG3–1 expression was developed. The PCR product derived from the primers Ano13-LA5U and Ano13-LA5L was digested with PstI and SacI. This fragment was cloned into the pPZP2Ha3 Ti-plasmid vector (11). Agrobacterium-mediated transformation was used for the transformation of Hayamasari (12). Plants regenerated from hygromycin-resistant calli (T0 plants) were grown, and then self-pollinated plants of each T0 plant were grown. T1 and T2 transformants were selected on the basis of PCR for the transgene and cultivated to set T3 seeds that were collected to perform germination experiments.
GUS Staining.
For the detection of GUS expression, seeds of transgenic plants with a qLTG3–1::GUS transgene were incubated at 15°C and 30°C and sampled at the indicated times. Whole seeds and longitudinally cut seeds of transgenic rice were vacuum-infiltrated with 50-mM NaH2PO4 (pH 7.0) containing 0.5-mM X-Gluc, 0.5-mM K3[Fe(CN)6], 0.5-mM K4[Fe(CN)6], and 0.5% (vol/vol) Triton X-100 and incubated for 6 h at 37°C. Then, 70% EtOH was added to terminate the enzyme reaction at room temperature.
Expression Analysis.
Total RNA was extracted from various organs (embryo and endosperm from germinating seed, leaves from 2-month-old plants, young panicle before heading, shoot and root of 4- to 7-day-old seedlings) according to the procedure described by Fujino et al. (34). For Northern blot analysis, total RNA (4 μg per sample) was separated on agarose-denaturing formaldehyde gels. Hybridization and signal detection were performed using a digoxygenin system and CDP-star (Roche Diagnostics), respectively. A PCR fragment derived from the primers 13–5U (TGCTGAATGGGCTGATAAAC) and 13–5L (ATGCAGAAAAGACGAGATGCAG) was used as a probe for Northern blot analysis.
Subcellular Localization of qLTG3–1.
The qLTG3–1 coding sequence amplified by PCR using the primers GFP-qLTGU (TAAGGTACCGGTGGAGGTGGAGGGATGGCGACGAAAGCTGGGGTGAT) and GFP-qLTGL (TTAGGTACCTTAGTTGCTGCACTGGAAGCC) was cloned into the down-stream of the CaMV 35S promoter and in frame with GFP in the binary vector pTH-2 (13). Transient GFP assays were conducted by introducing the plasmid DNA into onion epidermal cells by particle bombardment using IDERA (Tanaka products). The epidermis samples were incubated on 0.5 × MS media for 16 h at 26°C, then mounted on slides and examined by confocal laser scanning microscopy, FLUOVIEW (Fuji-film).
Microscopy Analysis.
The samples were prefixed in a mixture of 2% paraformaldehyde and 2.5% glutaraldehyde with 50-mM cacodylate buffer (pH 7.2) and then postfixed in 2% osmium tetroxide solution. The samples were further dehydrated through a graded ethanol series, substituted with qy-1, and embedded in epon812 resin. Then, semithin sections (1-μm thick) were obtained with an ultra-microtome and stained with toluidine blue. Ultra-thin sections were cut with a diamond knife, mounted on copper grids, and stained with 2% uranyl acetate (20 min) and lead citrate (3 min). After staining, the sections were observed using a TEM (Hitachi H-800) at an accelerating voltage of 75 kV.
Supplementary Material
Acknowledgments.
This work was supported in part by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Integrated research project for plant, insect and animal using genome technology QT-3007) (to K.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AB369214 and AB369215).
This article contains supporting information online at www.pnas.org/cgi/content/full/0805303105/DCSupplemental.
References
- 1.Koornneef M, Bentsink L, Hilhorst H. Seed dormancy and germination. Curr Opin Plant Biol. 2002;5:33–36. doi: 10.1016/s1369-5266(01)00219-9. [DOI] [PubMed] [Google Scholar]
- 2.Peng J, Harberd NP. The role of GA-mediated signalling in the control of seed germination. Curr Opin Plant Biol. 2002;5:376–381. doi: 10.1016/s1369-5266(02)00279-0. [DOI] [PubMed] [Google Scholar]
- 3.Kucera B, Cohn MA, Leubner-Metzger G. Plant hormone interactions during seed dormancy release and germination. Seed Sci Res. 2007;15:281–307. [Google Scholar]
- 4.Miura K, Lin SY, Yano M, Nagamine T. Mapping quantitative trait loci controlling low temperature germinability in rice (Oryza sativa L.) Breeding Sci. 2001;51:293–299. [Google Scholar]
- 5.Cui H, et al. Molecular dissection of seedling-vigor and associated physiological traits in rice. Theor Appl Genet. 2002;105:745–753. doi: 10.1007/s00122-002-0908-2. [DOI] [PubMed] [Google Scholar]
- 6.Fujino K, et al. Mapping of quantitative trait loci controlling low-temperature germinability in rice (Oryza sativa L.) Theor Appl Genet. 2004;108:794–799. doi: 10.1007/s00122-003-1509-4. [DOI] [PubMed] [Google Scholar]
- 7.Foolad R, Zhang P, Khan AA, Niño-Liu D, Lin Y. Identification of QTLs for early blight (Alternaria solani) resistance in tomato using backcross populations of a Lycopersicon esculentum × L hirsutum cross. Theor Appl Genet. 2002;104:945–958. doi: 10.1007/s00122-002-0870-z. [DOI] [PubMed] [Google Scholar]
- 8.Bettey M, Finch-Savage WE, King GJ, Lynn JR. Quantitative genetic analysis of seed vigour and pre-emergence seedling growth traits in Brassica oleracea. New Phytol. 2000;148:277–286. [Google Scholar]
- 9.Clerkx EJ, et al. Analysis of natural allelic variation of Arabidopsis seed germination and seed longevity traits between the accessions Landsberg erecta and Shakdara, using a new recombinant inbred line population. Plant Physiol. 2004;135:432–443. doi: 10.1104/pp.103.036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bentsink L, Jowett J, Hanhart CJ, Koornneef M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc Natl Acad Sci USA. 2006;103:17042–17047. doi: 10.1073/pnas.0607877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuse T, Sasaki T, Yano M. Ti-plasmid vectors useful for functional analysis of rice genes. Plant Biotechnol. 2001;18:219–222. [Google Scholar]
- 12.Toki S, et al. Early infection of scutellum tissue with agrobacterium allows high-speed transformation of rice. Plant J. 2006;47:969–976. doi: 10.1111/j.1365-313X.2006.02836.x. [DOI] [PubMed] [Google Scholar]
- 13.Niwa Y. A synthetic green fluorescent protein gene for plant biotechnology. Plant Biotechnol. 2003;20:1–11. [Google Scholar]
- 14.Nonogaki H, Chen F, Bradford KJ. Mechanisms and genes involved in germination sensu stricto. In: Bradford K, Nonogaki H, editors. Seed Development, Dormancy and Germination. Oxford: Blackwell; 2007. pp. 264–304. [Google Scholar]
- 15.Sachetto-Martins G, Franco LO, Oliveira DED. Plant glycine-rich proteins: a family or just proteins with a common motif? Biochimica et Biophysica Acta. 2000;1492:1–14. doi: 10.1016/s0167-4781(00)00064-6. [DOI] [PubMed] [Google Scholar]
- 16.Mousavi A, Hotta Y. Glycine-rich proteins: a class of novel proteins. Appl Biochem Biotechnol. 2005;120:169–174. doi: 10.1385/abab:120:3:169. [DOI] [PubMed] [Google Scholar]
- 17.Kader JC. Lipid-transfer proteins: a puzzling family of plant proteins. Trends Plants Sci. 1997;2:66–70. [Google Scholar]
- 18.Santino CG, Stanford GL, Conner TW. Developmental and transgenic analysis of two tomato fruit enhanced genes. Plant Mol Biol. 1997;33:405–416. doi: 10.1023/a:1005738910743. [DOI] [PubMed] [Google Scholar]
- 19.Yasuda E, Ebinuma H, Wabiko H. A novel glycine-rich/hydrophobic 16 kDa polypeptide gene from tobacco: similarity to proline-rich protein genes and its wound-inducible and developmentally regulated expression. Plant Mol Biol. 1997;33:667–678. doi: 10.1023/a:1005714119561. [DOI] [PubMed] [Google Scholar]
- 20.Nonogaki H, Gee OH, Bradford KJ. A germination-specific endo-beta-mannanase gene is expressed in the micropylar endosperm cap of tomato seeds. Plant Physiol. 2000;123:1235–1246. doi: 10.1104/pp.123.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen F, Bradford KJ. Expression of an expansin is associated with endosperm weakening during tomato seed germination. Plant Physiol. 2000;124:1265–1274. doi: 10.1104/pp.124.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu CT, Leubner-Metzger G, Meins F, Jr, Bradford KJ. Class I {beta}-1,3-glucanase and chitinase are expressed in the micropylar endosperm of tomato seeds prior to radicle emergence. Plant Physiol. 2001;126:1299–1313. doi: 10.1104/pp.126.3.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen F, Nonogaki H, Bradford KJ. A gibberellin-regulated xyloglucan endotransglycosylase gene is expressed in the endosperm cap during tomato seed germination. J Exp Bot. 2002;53:215–223. doi: 10.1093/jexbot/53.367.215. [DOI] [PubMed] [Google Scholar]
- 24.Ogawa M, et al. Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell. 2003;15:1591–1604. doi: 10.1105/tpc.011650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bethke PC, et al. The Arabidopsis aleurone layer responds to nitric oxide, gibberellin, and abscisic acid and is sufficient and necessary for seed dormancy. Plant Physiol. 2007;143:1173–1188. doi: 10.1104/pp.106.093435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bewley JD, Black M. New York: Plenum Press; 1994. Seeds: Physiology of development and germination. [Google Scholar]
- 27.Debeaujon I, Koornneef M. Gibberellin requirement for Arabidopsis seed germination is determined both by testa characteristics and embryonic abscisic acid. Plant Physiol. 2000;122:415–424. doi: 10.1104/pp.122.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bentsink L, Koornneef M. Rockville, MD: American Society of Plant Biologists; 2002. Seed dormancy and germination. [Google Scholar]
- 29.Nonogaki H. Seed germination:The biochemical and molecular mechanisms. Breeding Sci. 2006;56:93–105. [Google Scholar]
- 30.Ren ZH, et al. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet. 2005;37:1141–1146. doi: 10.1038/ng1643. [DOI] [PubMed] [Google Scholar]
- 31.Xu K, et al. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature. 2006;442:705–708. doi: 10.1038/nature04920. [DOI] [PubMed] [Google Scholar]
- 32.Neeraja CN, et al. A marker-assisted backcross approach for developing submergence-tolerant rice cultivars. Theor Appl Genet. 2007;115:767–776. doi: 10.1007/s00122-007-0607-0. [DOI] [PubMed] [Google Scholar]
- 33.Fujino K, Sekiguchi H, Kiguchi T. Identification of an active transposon in intact rice plants. Mol Genet Genomics. 2005;273:150–157. doi: 10.1007/s00438-005-1131-z. [DOI] [PubMed] [Google Scholar]
- 34.Fujino K, et al. NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol Genet Genomics. 2008;279:499–507. doi: 10.1007/s00438-008-0328-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.