Abstract
The crystal structure of the high affinity Escherichia coli MetNI methionine uptake transporter, a member of the ATP Binding Cassette (ABC) family, has been solved to 3.7 Å resolution. The overall architecture of MetNI reveals two copies of the ATPase MetN in complex with two copies of the transmembrane domain MetI, with the transporter adopting an inward-facing conformation exhibiting widely separated nucleotide binding domains. Each MetI subunit is organized around a core of five transmembrane helices that correspond to a subset of the helices observed in the larger membrane spanning subunits of the molybdate ModBC and maltose MalFGK ABC transporters. In addition to the conserved nucleotide binding domain of the ABC family, MetN contains a C-terminal extension belonging to the ACT-domain family previously proposed to represent a conserved regulatory binding fold. These domains separate the nucleotide binding domains and would interfere with their association required for ATP binding and hydrolysis. Methionine binds to the dimerized C-terminal domain and is shown to inhibit ATPase activity. These observations are consistent with an allosteric regulatory mechanism operating at the level of transport activity, where increased intracellular levels of the transported ligand stabilize an inward-facing, ATPase-inactive state of MetNI to inhibit further ligand translocation into the cell.
The high affinity uptake of methionine by Escherichia coli is mediated by the MetD system (1, 2), a member of the methionine uptake transporter family (3) of ATP Binding Cassette (ABC) transporters (4-8). ABC transporters such as MetD translocate substrates across the membrane, a process that is coupled to the hydrolysis of ATP by a cytoplasmic nucleotide binding domain (NBD). Architecturally, an ABC transporter contains four domains, two NBDs responsible for the interactions with ATP and two transmembrane domains (TMDs) that form the translocation pathway; the NBD and TMD components of the E. coli MetD system have been identified as MetN and MetI, respectively (9, 10). Substrate translocation by ABC transporters may be described in terms of an alternating access model that involves the interconversion of outward- and inward-facing conformations driven by the binding and hydrolysis of ATP by the NBDs. Additional domains of diverse structure may be fused to the TMDs or NBDs to regulate transport activity (11); examples include ionic strength sensing by OpuA (12), the inducer exclusion phenomenon mediated by MalK (13), and post-translational modifications of the cystic fibrosis transmembrane conductance regulator (14). Structural studies of the intact ABC importers for vitamin B12 (BtuCD, (15, 16)) and the homologous HI1470/71 (17), molybdate (ModBC, (18)) and maltose (MalFGK, (19)), and the exporters Sav1866 (20) and MsbA (21) have established the basic molecular architecture of ABC transporters and have provided structural models for some of the states of the transport cycle.
The MetD system was originally identified as an importer of both L- and D-methionine, either of which may be used as a source of methionine by E. coli (2, 22). MetD is additionally capable of transporting L-selenomethionine and other methionine derivatives. Intriguingly, early studies by Kadner (23) established the phenomenon of trans-inhibition in this system, whereby uptake of external methionine is inhibited by intracellular methionine levels in a fashion consistent with the direct action of methionine on the transporter. To further investigate this system, we have determined the crystal structure at 3.7 Å resolution of the E. coli methionine ABC transporter MetNI solubilized in the detergent dodecylmaltoside (24). The crystallographic analyses, together with kinetic studies, indicate that the binding of methionine to the C-terminal domain of the ABC subunits stabilizes an inward-facing, ATPase-inactive conformation of the transporter, and provide a structural basis for the regulatory properties discovered by Kadner.
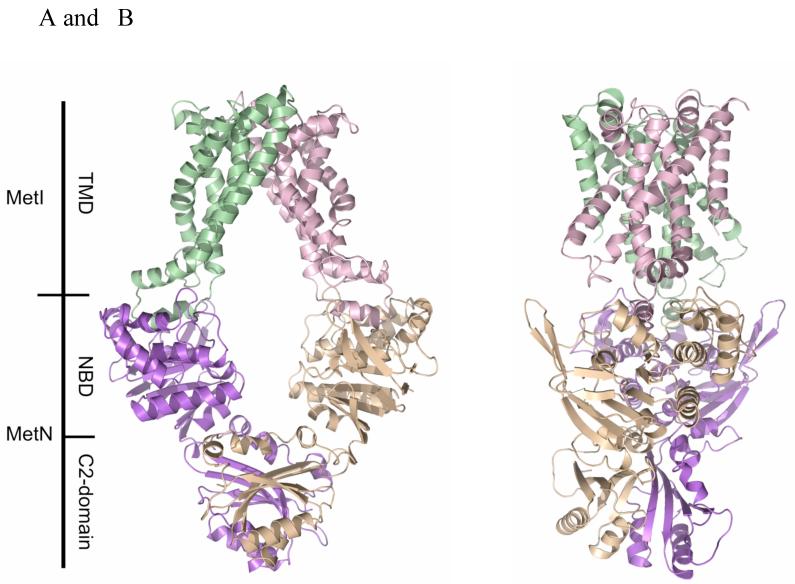
The overall architecture of MetNI consists of two copies of the ATPase MetN in complex with two copies of the transmembrane domain MetI, with the transporter adopting an inward-facing conformation having the TMDs open to the cytoplasm and the NBDs widely separated (Figure 1). The folds of MetN and MetI resemble those previously observed in the ModBC (18) and MalFGK (19) transporters, with three principal differences: (i) the transmembrane domain of the methionine transporter is smaller, as each MetI subunit contains only five transmembrane helices per monomer, (ii) the structures of the methionine, molybdate and maltose transporters represent a progression of inward-facing conformations of the translocation pathway that vary from wide open to closed, respectively, and (iii) the MetN ABC subunits contain a C-terminal extension that is distinct from those present in the maltose and molybdate transporters, and that separate the NBDs in this inward-facing conformation.
Figure 1.
(A) The ABC transporter MetNI consists of four subunits: two membrane-spanning MetI subunits (green and pink) and two MetN ABC subunits (purple and tan). The molecular rotation axis relating the MetI subunits is vertical, with the cytoplasmic (inward) facing surface of the transporter towards the bottom. The C2-domains forming the dimer interface between MetN subunits are at the bottom. (B) A view of MetNI rotated ∼90° about the vertical axis from that of (A), illustrating the asymmetrical orientation of the C2 domains relative to the MetI subunits and the MetN nucleotide binding domains. This figure was prepared and rendered with PyMOL (38).
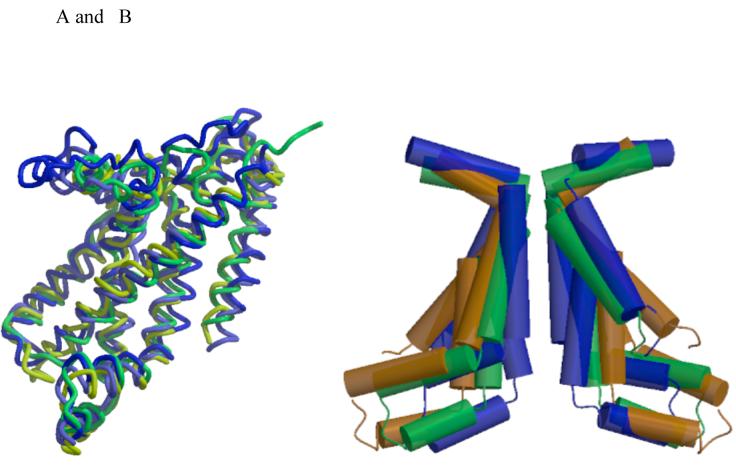
Each MetI subunit is organized around a core of five transmembrane helices that correspond to a subset of the helices observed in the larger membrane spanning subunits of the molybdate and maltose ABC transporters; the membrane spanning ModB, MalF and MalG subunits in those transporters contain 6, 8 and 6 helices, respectively, with the common set corresponding to the five most C-terminal helices in these structures (Figure 2A). The root mean square deviations (rmsd) between equivalent Cα positions in MetN and these other subunits are ∼2.5 Å. The region of highest sequence and structural similarity between these structures coincides with the “coupling helix” and the two transmembrane helices on either side (TM3 and TM4 in MetI). This region forms the core of the distinct conformations of the translocation pathway observed in the methionine, molybdate and maltose transporters (Figure 2B), with the coupling helix (including the consensus “EAA” motif (25, 26)) contributing significantly to the interface between the TMD and NBD subunits. The sequence and structural homologies extend to TM2 in these structures, including the relatively short length of this membrane spanning helix (∼14 residues) and the kinking at Pro 67 near the periplasmic interface; these irregular features are not uncommon near the translocation pathways of various transporters (17). One distinctive aspect of MetI is that it contains an odd number of transmembrane helices, so that the N- and C-termini are on opposite sides of the membrane (the periplasm and cytoplasm, respectively); this arrangement contrasts with all other structurally characterized ABC transporters which possess an even-number of transmembrane helices per TMD and have both termini in the cytoplasm.
Figure 2.
(A) Superposition of the five membrane spanning helices forming the common core of the transmembrane domains of MetI (yellow), ModB (green), MalF (blue) and MalG (light blue). TM1 (using the MetI number) is oriented diagonally across the front, while remaining helices are arranged in the sequence TM2 to TM5, from left to right. The coupling helices between TM3 and TM4 are the helical elements at the bottom of the figure. (B) Comparison of the TM2-TM3-TM4 helices lining the translocation pathways of MetI (brown), ModB (green) and MalFG (blue), illustrating the progressive narrowing of cytoplasmic opening of the translocation pathway in the sequence from the methionine to molybdate to maltose transporters. The views in this figure are from the membrane, with the cytoplasmic surface oriented down. Figures 2 and 3 were prepared with MOLSCRIPT and RASTER3D (39, 40).
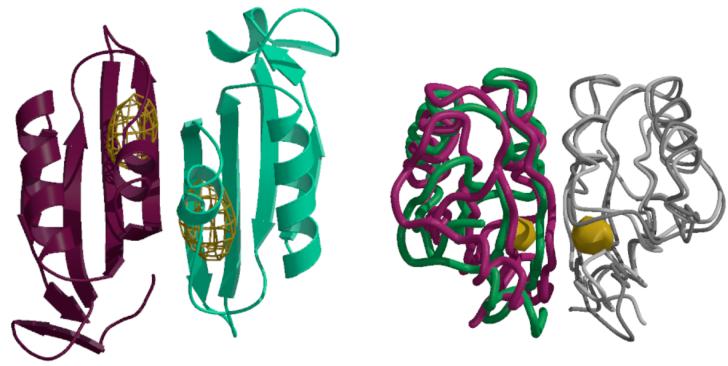
The MetN ABC subunit consists of two domains, the conserved NBD (residues 1-230), homologous to other ABC subunits (rmsds to ModC and MalK ∼1 Å), and the C-terminal C2-domain (residues 265-343) that are connected by a linker of ∼35 residues. While the pair of NBDs are approximately related by a two-fold axis, as are the pair of C2-domains, these axes differ in orientation by ∼20°, which generates a pronounced asymmetry in the transporter structure (Figure 1B). The distinct orientations of the rotation axes relating the NBD pairs and the C2-domain pairs within a transporter are accommodated through different conformations of the linker separating the NBD and C2-domains in the two MetN subunits. The C2-domain exhibits a ferredoxin-like fold with a 4 stranded antiparallel β-sheet (Figure 3A) that belongs to the ACT-domain family, proposed to represent a conserved regulatory ligand binding fold (27-29). As with other members of this family, the C2-domains from each MetN subunit in a transporter dimerize to form an 8-stranded β-sheet; a similar arrangement is also observed in the structure of the isolated C2-domain determined in the course of this project (24).
Figure 3.
(A) Anomalous difference Fourier map calculated at 5.2 Å resolution illustrating the binding of selenomethionine to the C2-domains of MetNI following a 1 mM soak of transporter crystals. The electron density is contoured at 6 times the standard deviation of the map. (B) Comparison of the relative orientations of C2-domains observed in the MetNI and free MetN-C2 structures, following superposition of one subunit in each dimer. The green and dark gray traces correspond to the C2-domain dimer in MetNI, while the magenta and light gray traces represent the isolated C2-domain structure. The binding site for selenomethionine is denoted by the gold surface. The view is roughly perpendicular about the horizontal axis from that in (A).
Since ATP binds to conserved sequence motifs at the interface between two closely juxtaposed NBDs (30-33), the crystallographically observed inward-facing conformation of the MetNI transporter corresponds to an ATPase-inactive state. The extent of separation of the NBDs in this structure relative to other ABC importers (and the correlation between NBD separation and the conformation of the translocation pathway) may be assessed from the intersubunit distances between the P-loops and signature motifs of different ABC subunits. Using the Cα residues of MetN residues Gly 43 and 143 to define the positions of the P-loop and signature motifs, respectively, the intersubunit distances between these two motifs are found to be 26-30 Å (the range reflects the asymmetry of this complex). For comparison, the corresponding distances in the maltose and molybdate transporter structures are ∼11 Å and ∼16 Å, respectively (based on the intersubunit distances between residues Gly 36 and 129 of ModB, and Gly 41 and 136 of MalK, respectively), and were previously reported as 14 Å and 16 Å in the structures of BtuCD and HI1470/71 (17). The short 11 Å intersubunit distance observed in the maltose transporter between the P-loop and the signature motif reflects both the closed interface between the NBDs and that the translocation pathway is closed to the cytoplasm. While BtuCD has a somewhat larger separation (14 Å) between NBDs than observed in the maltose transporter, the translocation pathway is also closed to the cytoplasm. Although the intersubunit separation between NBDs is only 2 Å longer in HI1470/71 and the molybdate transporter, the translocation pathways in both structures are open to the cytoplasm, suggesting that this represents a critical parameter relating the relative positioning of the NBDs to the conformation of the translocation pathway. The ∼28 Å distance observed in the methionine structure is significantly larger than the previously reported NBD separations in ABC importers and suggests that the positioning of the C2-domain dimer is crucial for stabilizing this arrangement.
To identify potential methionine binding sites, crystals of the wild-type methionine transporter were soaked in 1 mM L-selenomethionine for two hours and diffraction data were collected to 5.2 Å resolution at the selenium edge. Anomalous difference Fourier maps revealed two binding sites for selenomethionine located near the dimer interface between C2-domains (Figure 3A). The binding sites are located near the side chains of methionines 301 and 312 (Figure S2), although the resolution was insufficient to permit a detailed modeling of the bound selenomethionine. The presence of ligand binding sites near the dimer interface parallels observations on other ACT domain containing proteins (29). Significantly, binding of serine to allosteric sites at the interface between ACT domains of phosphoglycerate dehydrogenase results in a sequence of conformational changes that are propagated to distinct catalytic domains (34). Evidence for the flexibility of the corresponding interface in MetNI is provided by a ∼15° twist in the relative orientations between C2-domains present in the transporter and in free form (Figure 3B), along with rearrangements of several loops, that could plausibly be influenced by the binding of methionine.
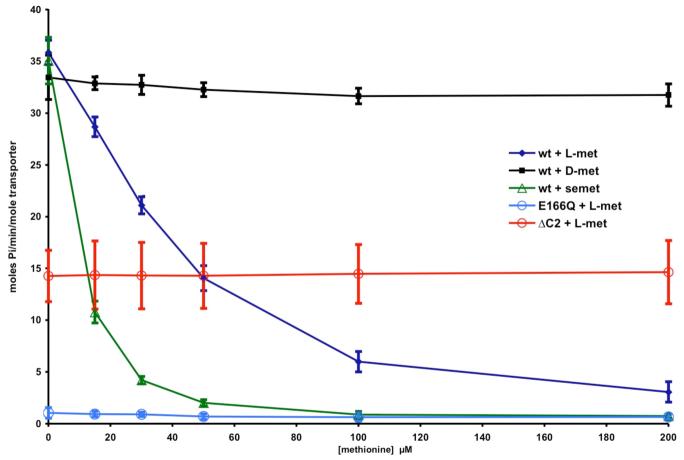
The juxtaposition of the methionine binding C2-domains between the NBDs in the present structure suggests they may be relevant to the transinhibition mechanism established by Kadner (23). To test this proposal, the effect of methionine and methionine derivatives on the ATPase activity of detergent solubilized transporter was characterized. Although this assay does not directly monitor the influence of methionine on transport activity, it is nevertheless appropriate to assess the ability of methionine to stabilize an ATPase-inactive conformation of the transporter. As detailed in Figure 4, L-methionine does inhibit the ATPase activity, with half-maximal inhibition occurring at ∼30 μM. L-Selenomethionine is an even more potent inhibitor, with half-maximal inhibition occurring at ∼10 μM, while D-methionine has little effect. As controls, truncation of the C2-domain, although reducing ATPase activity under the assay conditions by ∼60%, eliminates the inhibitory effects of methionine, while the E166Q mutant in the NBD (corresponding to a mutation shown to prevent ATP hydrolysis in other systems (32)) exhibits little ATPase activity. These results are consistent with a model where an equilibrium exists between conformational states of MetNI differing in the extent of NBD displacement. Despite their separation in the crystal structure of detergent solubilized MetNI, activity assays demonstrate that these domains can associate to form an ATPase competent state. As methionine binds to the C2-domains, the equilibrium shifts towards transporter conformations with separated NBDs, and as a consequence, the rate of ATP hydrolysis decreases.
Figure 4.
Dependence of the ATPase activity of various MetNI constructs on the concentration of methionine analogues. The ATPase activity was measured from the rate of phosphate production as assayed by the method of Webb (41), and converted to moles phosphate/minute/mole transporter. An initial ATP concentration of 1 mM was used, which is ∼3 times the apparent Km (Figure S3). The blue diamonds, black squares and green triangles correspond to L-methionine, D-methionine and L-selenomethionine, while the open blue and red circles represent the effect of L-methionine on the E166Q (ATPase-inactive mutant) and the ΔC2-MetNI truncation mutant, respectively. The error bars represent the standard deviations calculated from 8 measurements.
The structure of MetNI described here provides the first crystallographic analysis of a member of the methionine uptake transporter family (3) of the superfamily of ABC transporters. Beyond the relevance for the mechanistic understanding of this family, the MetNI structure provides two general insights into the structural organization of ABC importers - (i) a minimal core of 5 transmembrane helices is identified that may be elaborated by additional helices in other transporters and (ii) a mechanistic basis is proposed for the regulation of transport activity by domains fused to the core transporter. The binding of methionine by the C2-domain at the C-terminus of the MetN subunit effectively stabilizes the inward-facing, ATPase-inactive conformation of the transporter. By sterically interfering with NBD association, the ATP-driven engine that powers transport will be disrupted, thereby providing a molecular mechanism for Kadner’s observation that increasing levels of internal methionine inhibit transport. The ability of methionine, and particularly selenomethionine, to inhibit transport further suggests that the deletion of the C2-domain from the endogenous methionine transporter of E. coli strains used for selenomethionine labeling may increase the incorporation efficiency by allowing uptake to a higher intracellular concentration. Of a more general consideration, transporter activity can represent a major component of the ATP requirement of bacteria (35, 36). Consequently, it is not surprising that transporters can be regulated at the level of protein function to more efficiently control the allocation of cellular energy resources. The structure of the MetNI ABC transporter provides a foundation for the characterization of regulatory mechanisms relevant to the uptake of methionine, and how these processes are more generally integrated with cellular metabolism.
The recent report by Gerber et al. (published online Science DOI: 10.1126/science.1156213) details the structural basis of trans-inhibition of the ModBC molybdate ABC transporter from Methanosarcina acetivorans; although the molybdate/tungstate binding C-terminal regulatory domain of ModC has a distinct fold from the methionine binding C2-domain of MetN, the consequences for transporter conformation are quite similar.
Supplementary Material
Acknowledgments
We thank Heather Pinkett, Zhenfeng Liu, Oded Lewinson, Lynmarie Thompson, David Tirrell and James B. Howard for helpful discussions, and the staff of the Stanford Synchrotron Radiation Laboratory (SSRL) and the Advanced Light Source (ALS) for their assistance during crystal screening and data collection. This work was supported in part by NIH grant GM45162. We would like to acknowledge the Gordon and Betty Moore Foundation for support of the Molecular Observatory at Caltech. Operations at SSRL and ALS are supported by the US DOE and NIH. Coordinates and structure factors for MetNI and the MetN-C2 domain will be deposited in the Protein Data Bank (www.rcsb.org/pdb; (37)) for release upon publication.
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. Their manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
References
- 1.Kadner RJ. J. Bact. 1974;117:232. doi: 10.1128/jb.117.1.232-241.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kadner RJ, Watson WJ. J. Bact. 1974;119:401. doi: 10.1128/jb.119.2.401-409.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Z, et al. Arch. Microbiol. 2003;180:88. doi: 10.1007/s00203-003-0561-4. [DOI] [PubMed] [Google Scholar]
- 4.Higgins CF. Annu. Rev. Cell Biol. 1992;8:67. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 5.Holland IB, Cole SPC, Kuchler K, Higgins CF, editors. ABC proteins: From Bacteria to Man. Academic; London: 2003. p. 647. [Google Scholar]
- 6.Davidson AL, Chen J. Annu. Rev. Biochem. 2004;73:241. doi: 10.1146/annurev.biochem.73.011303.073626. [DOI] [PubMed] [Google Scholar]
- 7.Higgins CF, Linton KJ. Nat. Struct. Mol. Biol. 2004;11:918. doi: 10.1038/nsmb836. [DOI] [PubMed] [Google Scholar]
- 8.Hollenstein K, Dawson RJP, Locher KP. Curr. Opin. Struct. Biol. 2007;17:412. doi: 10.1016/j.sbi.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 9.Gál J, Szvetnik A, Schnell R, Kálmán M. J. Bact. 2002;184:4930. doi: 10.1128/JB.184.17.4930-4932.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merlin C, Gardiner G, Durand S, Masters M. J. Bact. 2002;184:5513. doi: 10.1128/JB.184.19.5513-5517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biemans-Oldehinkel E, Deoven MK, Poolman B. FEBS Lett. 2006;580:1023. doi: 10.1016/j.febslet.2005.11.079. [DOI] [PubMed] [Google Scholar]
- 12.Biemans-Oldehinkel E, Mahmood NABN, Poolman B. Proc. Natl. Acad. Sci. USA. 2006;103:10624. doi: 10.1073/pnas.0603871103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kühnau S, Reyes M, Sievertsen A, Shuman HA, Boos W. J. Bacteriol. 1991;173:2180. doi: 10.1128/jb.173.7.2180-2186.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rich DP, et al. Science. 1991;253:205. doi: 10.1126/science.1712985. [DOI] [PubMed] [Google Scholar]
- 15.Locher KP, Lee AT, Rees DC. Science. 2002;296:1091. doi: 10.1126/science.1071142. [DOI] [PubMed] [Google Scholar]
- 16.Hvorup RN, et al. Science. 2007;317:1387. doi: 10.1126/science.1145950. [DOI] [PubMed] [Google Scholar]
- 17.Pinkett HW, Lee AT, Lum P, Locher KP, Rees DC. Science. 2007;315:373. doi: 10.1126/science.1133488. [DOI] [PubMed] [Google Scholar]
- 18.Hollenstein K, Frei DC, Locher KP. Nature. 2007;446:213. doi: 10.1038/nature05626. [DOI] [PubMed] [Google Scholar]
- 19.Oldham ML, Khare D, Quiocho FA, Davidson AL, Chen J. Nature. 2007;450:515. doi: 10.1038/nature06264. [DOI] [PubMed] [Google Scholar]
- 20.Dawson RJP, Locher KP. Nature. 2006;443:180. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 21.Ward A, Reyes CL, Yu J, Roth CB, Chang G. Proc. Natl. Acad. Sci. U.S.A. 2007;104:19005. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kadner RJ. J. Bact. 1977;129:207. doi: 10.1128/jb.129.1.207-216.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kadner RJ. J. Bact. 1975;122:110. doi: 10.1128/jb.122.1.110-119.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Material and methods are available as supporting material on Science online.
- 25.Saurin W, Koster W, Dassa E. Mol. Micro. 1994;12:993. doi: 10.1111/j.1365-2958.1994.tb01087.x. [DOI] [PubMed] [Google Scholar]
- 26.Mourez M, Hofnung N, Dassa E. EMBO J. 1997;16:3066. doi: 10.1093/emboj/16.11.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aravind L, Koonin EV. J. Mol. Biol. 1999;287:1023. doi: 10.1006/jmbi.1999.2653. [DOI] [PubMed] [Google Scholar]
- 28.Chipman DM, Shaanan B. Curr. Opin. Struct. Biol. 2001;11:694. doi: 10.1016/s0959-440x(01)00272-x. [DOI] [PubMed] [Google Scholar]
- 29.Grant GA. J. Biol. Chem. 2006;281:33825. doi: 10.1074/jbc.R600024200. [DOI] [PubMed] [Google Scholar]
- 30.Jones PM, George AM. FEMS Microbiol. Lett. 1999;179:187. doi: 10.1111/j.1574-6968.1999.tb08727.x. [DOI] [PubMed] [Google Scholar]
- 31.Hopfner K-P, et al. Cell. 2000;101:789. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 32.Smith PC, et al. Mol. Cell. 2002;10:139. doi: 10.1016/s1097-2765(02)00576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J, Lu G, Lin J, Davidson AL, Quiocho FA. Mol. Cell. 2003;12:651. doi: 10.1016/j.molcel.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 34.Thompson JR, Bell JK, Bratt J, Grant GA, Banaszak LJ. Biochemistry. 2005;44:5763. doi: 10.1021/bi047944b. [DOI] [PubMed] [Google Scholar]
- 35.Stouthamer AH. Int. Rev. Biochem. 1979;21:1. [Google Scholar]
- 36.Neijsel OM, Teixeira de Mattos MJ, Tempest DW. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt FC, editor. ASM Press; Washington, D.C.: 1996. [Google Scholar]
- 37.Berman HM, et al. Nuc. Acids Res. 2000;28:235. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; Palo Alto, CA: 2002. http://www.pymol.org. [Google Scholar]
- 39.Kraulis PJ. J. Appl. Crystallogr. 1991;24:946. [Google Scholar]
- 40.Merritt EA, Murphy MEP. Acta crystallogr. 1994;D50:869. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 41.Webb MR. Proc. Natl. Acad. Sci. U.S.A. 1992;89:4884. doi: 10.1073/pnas.89.11.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.