Abstract
Biological responses to mechanical stress require strain-sensing molecules, whose mechanically induced conformational changes are relayed to signaling cascades mediating changes in cell and tissue properties. In vertebrate muscle, the giant elastic protein titin is involved in strain sensing via its C-terminal kinase domain (TK) at the sarcomeric M-band and contributes to the adaptation of muscle in response to changes in mechanical strain. TK is regulated in a unique dual autoinhibition mechanism by a C-terminal regulatory tail, blocking the ATP binding site, and tyrosine autoinhibition of the catalytic base. For access to the ATP binding site and phosphorylation of the autoinhibitory tyrosine, the C-terminal autoinhibitory tail needs to be removed. Here, we use AFM-based single-molecule force spectroscopy, molecular dynamics simulations, and enzymatics to study the conformational changes during strain-induced activation of human TK. We show that mechanical strain activates ATP binding before unfolding of the structural titin domains, and that TK can thus act as a biological force sensor. Furthermore, we identify the steps in which the autoinhibition of TK is mechanically relieved at low forces, leading to binding of the cosubstrate ATP and priming the enzyme for subsequent autophosphorylation and substrate turnover.
Keywords: atomic force microscopy, force–probe molecular dynamics simulation, muscle signaling, protein kinase regulation, single-molecular force spectroscopy
Mechanical activity and adaptive responses to changes in work load in muscle are tightly linked, but the mechanosensors triggering the sweeping adaptive changes seen in vivo are as yet poorly understood on the molecular level. In the vertebrate muscle sarcomere, titin serves as a molecular ruler for sarcomere assembly and is responsible for resting elasticity of muscle (1, 2) (Fig. 1A). At the M-band, titin contains a serine/threonine protein kinase domain (TK) (Fig. 1B) (3, 4). TK is regulated in a dual autoinhibition mechanism by a C-terminal regulatory tail, blocking the ATP binding site, and tyrosine autoinhibition of the catalytic base by tyrosine-170 (5). For access to the ATP binding site and the autoinhibitory tyrosine, the C-terminal autoinhibitory tail must be removed.
Fig. 1.
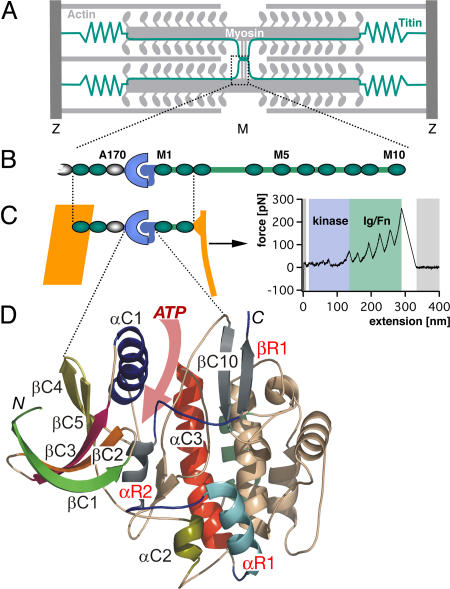
Sarcomeric location and structure of the investigated TK protein construct. (A) Schematic diagram of the sarcomere showing the transverse Z- and M-bands and actin and myosin filaments, linked by the elastic titin filament. M-bands cross-link myosin filaments by a complex of titin (green), obscurin, and myomesin (15). (B) Domain structure of M-band titin showing the array of structural Ig (green), Fn3 domains (white), and unique sequences (green lines) surrounding titin kinase domain (TK) (blue). (C) The titin construct A168-M2 contains the kinase domain surrounded by Fn3 and Ig domains. ATP binding requires relief of the C-terminal autoinhibitory tail (blue) from the active site, which can be achieved by external force. In mechanical single-molecule experiments, A168-M2 is pulled off the gold support (yellow line) by a cantilever, resulting in a unique force spectrum when the protein is stretched and domains sequentially unfold. Analysis of the unfolding force spectrum (see SI Text) identifies the peaks shaded in blue as kinase unfolding peaks; the five unfolding peaks shaded in green correspond to sequential Ig and Fn domain unfolding. (D) Kinase domain structure, with the ATP binding site highlighted by the pink arrow and individual secondary structure elements color-coded. Numbering is from N to C terminus, where C1 to C10 refer to catalytic core structures, and R1 to R3 (in red) refer to the regulatory tail (5). The N and C termini are marked.
In most autoinhibited kinases, the relief of intramolecular autoinhibition is essentially a partial unfolding event of the autoinhibited conformation, driven by ligand binding or posttranslational modification. Although TK activity can be moderately stimulated by calmodulin when tyrosine phosphorylation is mimicked, calmodulin or other calcium binding proteins are unable on their own to activate it (5). Because titin is firmly embedded in the contractile machinery (Fig. 1A), its conformation and function can readily be affected by mechanical forces (1, 6). The M-band, being much more compliant than the Z-disk (7, 8), is ideally placed as a strain sensor (9, 10). Because the M-band lattice is deformed only during active contraction, it is optimal for detecting the actual workload on the myofibril (10). Force–probe molecular dynamics simulations of the mechanical properties of TK suggested that kinase activation might be possible by mechanical forces (11). Indeed, a mechanosensitive signaling complex (signalosome) was identified that interacts with an open conformation of TK, and by controlling protein turnover and muscle gene transcription (12) seems to contribute to the adaptation of muscle in response to changes in mechanical strain. The importance of TK in maintaining the turnover of muscle proteins is highlighted by a point mutation in the human kinase domain that causes a myopathy with failure of load-dependent protein turnover (12).
Two recent reports on single-molecule force spectroscopy of titin kinase and C. elegans giant muscle protein kinase (13, 14) showed that these giant muscle protein kinases can unfold in a stepwise fashion, as predicted (11). To be strain regulated, association of the cytoskeletal lattice with the N- and C-terminal ends of the kinase domain is required. Titin is firmly integrated into the M-band lattice by interactions with obscurin, obscurin-like 1, and myomesin, which form a ternary complex at its C terminus (15). Because of their I-band (16) or broad A-band localization (17), the nematode giant muscle protein kinases have been implicated in contraction regulation. It is therefore as yet unclear whether the invertebrate giant protein kinases are similarly integrated into the cytoskeleton, follow the same activation pathways, and serve analogous functions as the M-band-associated TK.
However, experimental proof of direct mechanical activation, rather than simply partial unfolding, is lacking not only for titin kinase, but for all biological force sensors in muscle (2). Can mechanical force really induce a catalytically competent kinase conformation that will be able to bind substrates? The complex protein composition of the sarcomere M-band precludes an unequivocal experimental answer. Studying single molecules in isolation, however, can unravel their intrinsic properties in molecular detail and allow these to be compared with the known properties of intact sarcomeres and measurable enzymatic properties. We therefore used atomic force microscope (AFM)-based single-molecule force spectroscopy, molecular dynamics simulations, and enzymatics to investigate the molecular details of mechanical TK activation.
Results
Sequential Unfolding of TK at Low Forces.
We expressed a TK construct A168-M2, encompassing the human kinase domain flanked by its naturally surrounding Ig/Fn domains [Fig. 1 B and C; see details in supporting information (SI) SI Text (section 1), Fig. S1, and Table S1]. The TK construct was attached to an AFM cantilever [see SI Text (sections 4 and 5) and Fig. S2] and stretched with nanometer accuracy. The resulting force, recorded with piconewton precision (Fig. 1D), showed a characteristic saw-tooth appearance as TK was gradually stretched and unfolded, mimicking the mechanical stress in muscle. (In a very simple comparison, the slowest experimental pulling speed per folded protein length amounts to 300 nm/s/25 nm = 12/s and is close to physiological rates. A rabbit sarcomere of 2 μm length can contract with 6 μm/s, yielding a contraction rate of 3/s.) Typically, a series of five initial low-force peaks below 50 pN was followed by up to five distinct saw-tooth-shaped high-force peaks that correlated exactly with the number and contour lengths of the flanking Ig/Fn-domains (18, 19) [Fig. 1; and see SI Text (sections 6–8) and Figs. S3–S5, and Table S2]. Therefore, the low-force peaks, occurring before Ig/Fn unfolding, derive from unfolding events within the kinase domain (see Fig. S2b for a schematic). These low-force unfolding events are strictly ordered, although their height is similar. In contrast to the independently unfolding Ig/Fn domains, their fixed sequence is not determined by mechanical stability but rather by topology. The forces required even for complete unfolding of the kinase do not exceed 50 pN at 23°C, or 30 pN at 37°C [see SI Text (section 9) and Fig. S6] and at pulling speeds of 1 μm/s. Such low forces were also predicted from force probe simulations (11). The fact that the mechanically more stable Ig/Fn domains always unfold after the kinase domain shows that the force acts on all domains in series, and that the protein construct is therefore completely stretched in the beginning of a retraction cycle.
Mechanically Activated ATP Binding Detected by AFM.
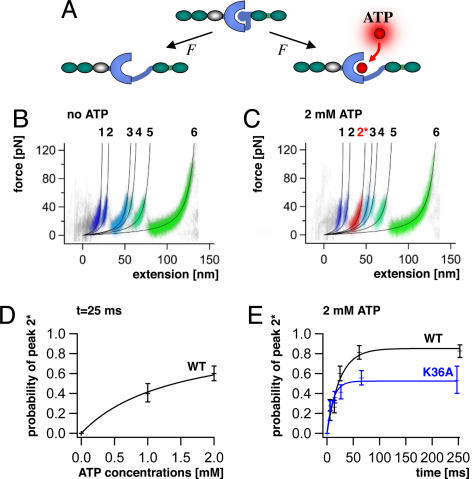
Mechanical activation of TK must at an early stage uncover its ATP binding site while leaving the active site intact. Subsequent ATP binding alters the stability of the enzyme (Fig. 2A) and should therefore give rise to ATP-dependent changes in the TK unfolding profile (20). In the absence of ATP, five energy barriers separated by 9.1, 28.6, 7.3, 18.0, and 57.9 nm in contour length are observed (Fig. 2B). In the presence of Mg2+-ATP at physiological concentrations (2 mM), a certain fraction of the traces shows an additional well pronounced peak (peak 2*) at 51.6 nm (Fig. 2C). This peak denotes an early interaction with ATP during the sequential unfolding of the kinase and thus demonstrates the initial opening of the active site.
Fig. 2.
Unfolding profile of TK and kinetics of mechanically induced ATP binding. (A) External force can open the ATP binding site of TK by unfolding of the autoinhibitory domain (blue ball). (B) Superimposed traces of 66 single-molecule unfolding events in TK show a fixed sequence of local unfolding events, numbered 1–5. (C) Mechanically induced ATP binding leads to a distinctly altered force profile with the appearance of an extra force peak, 2*, absent in unfolding events in the absence of ATP (44 traces). (D) ATP binding probability (i.e., the occurrence of peak 2*) depends on ATP concentrations, giving access to virtual direct kinetics though the system is not in equilibrium (ATP peaks detected at pulling rates of 720 nm/s, or after 25 ms). (E) Probability of ATP binding depends on pulling rates, decreasing at faster speeds because of reduced opening time of the ATP binding site. ATP binding is strongly reduced by mutation of lysine-36 to alanine (K36A).
The probability of observing the ATP-dependent peak should depend on the likelihood for ATP binding during the time span between the opening of the binding site and the moment when the ATP barrier (peak 2*) is probed and, therefore, on the ATP concentration (Fig. 2D). More interestingly, because this time span is controlled by the pulling speed, we gain direct experimental access to the ATP binding kinetics (see SI Text, section 10). This experiment can be seen as a mechanical pump–probe experiment: first the binding pocket of the TK is “pumped” open, and after a certain time ATP binding is probed. Variation of this time window provides the kinetic constants. The calibration of the time axis was estimated by the ratio of the MD-determined extension, during which the binding pocked is open but not deformed, and by the pulling speed. This mechanical pump–probe experiment showed saturation after ≈100 ms (Fig. 2E). Toward higher pulling rates, the probability of ATP binding decreased strongly and approached zero (Fig. 2E). This time dependence demonstrates that ATP binding is mechanically induced and perfectly agrees with the absence of catalytic activity of autoinhibited TK in solution (Table 1). Furthermore, this experiment allows estimates of the apparent on and off rates and resulting dissociation constant, which compare with affinity values observed for titin (Table 1) and other kinases in solution (21).
Table 1.
Kinetic parameters for ATP of autoinhibited titin kinase (WT and K36A) and the constitutively activated WT-kin3 measured by AFM or in solution
| Construct | Kon, 1/M (AFM) | Koff, 1/s (AFM) | Kd, μM (AFM) | KM, μM (solution) | Autophosphorylation |
|---|---|---|---|---|---|
| WT | 1.8 ± 0.3 × 104 | 6 ± 3 | 347 | No activity | − |
| K36A | 2.3 ± 0.5 × 104 | 41 ± 11 | 1,810 | No activity | − |
| WT-kin3 | 270 | + |
Following a suggestion from our MD simulations, we mutated lysine-36 to alanine (K36A), a highly conserved residue equivalent to lysine-72 interacting with the α/β phosphates of ATP in cAMP-dependent protein kinase (22, 23). This mutation abolishes kinase activity in TK (5). Now the ATP affinity of TK was dramatically reduced, with a >6-fold increase of koff and a concomitant increase of Kd to millimolar values (Fig. 2D, SI Text, section 10 and Table 1). These results localize mechanically induced ATP binding to the canonical site in TK and confirm that the conserved lysine residue, known to play a crucial part in ATP binding of homologous protein kinases, is also a key residue in the TK binding pocket.
Molecular Mechanism of TK Activation by Force.
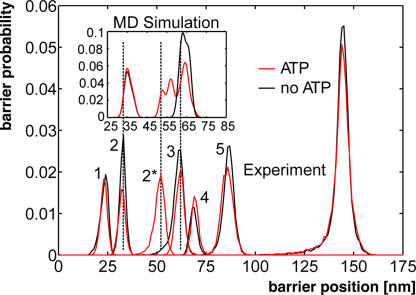
We used force–probe MD simulations (24, 25) to characterize the force-induced unfolding of TK at the atomic level and to correlate the structural states with the energy barriers observed by the single-molecule force spectroscopy experiments. Force–probe molecular dynamics simulations (24, 25) used the TK x-ray structure [Protein Data Bank entry 1TKI (5)] as the starting structure, with the autoinhibitory tail partly removed [see SI Text (sections 11–16) and Tables S3 and S4]. Two sets of simulations (five each) were carried out for this truncated TK: one set with an empty binding pocket, and one set with an ATP molecule and magnesium ions inserted into the (closed) binding pocket. As a control, the autoinhibited complete TK was also subjected to force–probe MD simulations (see SI Text, section 17). As in the experiment, the two force profiles obtained from the simulations of the truncated TK (Fig. 3B Top and Middle) are largely similar. A notable exception is the more pronounced force peak seen in the presence of ATP (see Fig. 3B Inset) at the position of the measured force peak 2*. To allow direct comparison of the unfolding pathways between experiment and simulation, we transformed the force extension traces of Fig. 2 into barrier position histograms (14) and derived the same from our simulations (see SI Text, section 18). The two histograms agree well both in the presence and absence of ATP [Fig. 4 (dashed lines) and Fig. S7], allowing the conclusion that the main unfolding events are correctly described by the simulations.
Fig. 3.
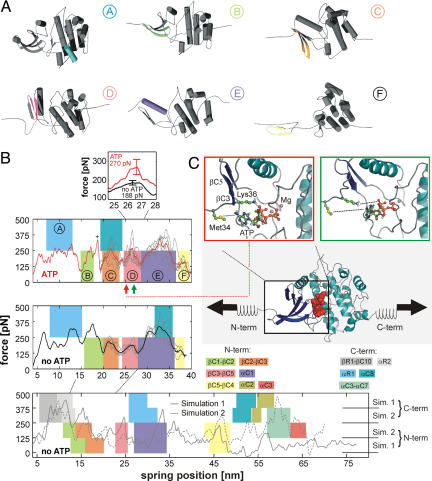
Molecular dynamics (MD) simulations of the force-induced unfolding of titin kinase (TK). (A) Representative unfolding intermediates with the unfolding secondary structure elements colored according to the scheme in Fig. 1D; the β-strands unfold pairwise, and colors refer to the respective N-terminal strand of each pair. (B) Unfolding forces of truncated TK with ATP (Top), without ATP (Middle), and of the complete TK (Bottom). For the complete TK, two independent 90-ns simulations were carried out (solid and dashed lines). Starting from a partially unfolded structure at ≈19 nm, five 26-ns trajectories (thin gray lines) were averaged for both sets of simulations (thick lines in Top and Middle). Color-shaded areas indicate main unfolding events, which correspond to the colors used in A and in Fig. 1D. An additional force peak in the presence of ATP is predicted (plus sign and pink-shaded area in Top). This force peak (Inset) is higher for bound ATP (270 pN) than for an empty binding pocket (188 pN). Because of the necessarily much faster pulling rates of 0.8 m/s used for the simulations, larger unfolding forces are seen, which can be related to the experimental loading rates (11). (C) In the force–probe MD simulations, harmonic springs were attached to the protein and retracted with constant velocity (lower schematic, ATP shown as red spheres). (C Insets) Representative structures shortly before (Left) and after (Right) the ATP force peak. ATP and the two key residues methionine-34 and lysine-36 are shown in ball-and-stick representation, and the rupture of molecular interactions is indicated by dotted lines.
Fig. 4.
Contour length histograms obtained from single-molecule force spectroscopy experiments (transformation with QM-WLC and P = 0.8 nm) and from MD simulations (Inset). The folded kinase construct has a length of 25 nm. The peak positions with (red) and without (black) ATP are similar in both histograms (dashed lines), except for one additional peak in the presence of ATP (red peak at ≈51.6 nm). The experimentally determined contour length increments are, in the absence of ATP, 9.1, 28.6, 7.3, 18.0, 57.9 nm; and, in the presence of ATP, 9.1, 19.4, 10.1, 7.5, 16.4, 58.3 nm—with an estimated error of ±2%. The position of the initial peak (24 nm) reflects the mean length of the TK construct with completely folded domains.
Next, we investigated which molecular interactions determine the observed force peaks. For the ATP peak 2*, two strong interactions are seen, a salt bridge from lysine-36 to the α-phosphate group of ATP, and a contact between methionine-34 and the adenine moiety of ATP (Fig. 3C). Both interactions break irreversibly upon βC3-βC4 rupture, giving rise to the significantly larger force peak of 270 ± 39 pN in the simulations with bound ATP as compared with 188 ± 13 pN without ATP (Fig. 3B Inset and Movie S1). Notably, in the AFM experiment, the contour length of 51.6 nm for the ATP peak position (Fig. 4, peak 2*) also points to a residue close to lysine-36. An additional peak is seen at 18 nm for the simulation with ATP present (plus sign in Fig. 3B Top). Here, a force-induced deformation of the N-terminal domain triggers the transient rupture and reformation of the methionine-34–ATP and lysine-36–ATP interactions.
Closer structural analysis of our simulations suggests the following sequence of events (colors in Fig. 3 A and B, and Fig. 4). Peak 1 (Fig. 4) is caused by unfolding of the 23-residue linker at the N terminus of TK, which is not present in the simulations (see SI Text for details). At peak 2, the autoinhibitory tail is unfolded and removed, rendering the ATP binding site accessible (region shaded in gray in Fig. 3B Bottom). Subsequently, N-terminal β-sheets βC1-βC2 and βC2-βC3 rupture (regions B and C). For these events, no force peak is seen in the experiment, because it would fall into the lag time after force peak 2. Peak 2* described above is dominated by interactions of ATP with the binding pocket. The truncated construct necessarily lacks part of the autoinhibitory tail stabilizing the adjacent C-terminal α-helix αR1 in the full-length TK. Accordingly, αR1 unfolds first in the truncated kinase (Fig. 3B Top and Middle, region A) but after βR1 and αR2 in the autoinhibited kinase (Fig. 3B Bottom). Hence, and in agreement with the complete TK unfolding simulations (Fig. 3B Bottom), peaks 3 and 4 are assigned to unfolding of αC1 and βC4-βC5, respectively (regions D and E). Finally, peak 5 arises from the combined effect of αC2 and αC8 rupture (Fig. 3B Bottom). At peak 6, the complete TK is unfolded and stretched. Taking the diameter of the folded TK into account (5.5 nm), the contour length increment to peak 1 (121 nm) corresponds to (5.5 + 121 ± 2) nm/0.365 nm = 346 ± 5 residues, in agreement with the 344 aa of TK including its N-terminal linker (see SI Text, section 7).
Autophosphorylation of TK.
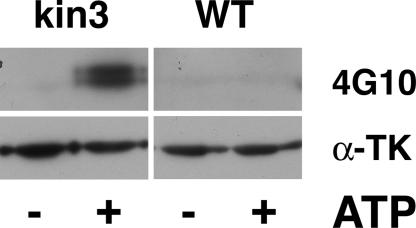
Our simulations show that the open ATP binding site does not relieve autoinhibition of the catalytic base aspartate-127 by tyrosine-170. However, our model of the ATP-bound state of TK suggests that this semi-opened state might autophosphorylate, in agreement with previous predictions of the open apo-enzyme (11). We tested this notion by assaying recombinant TK with its ATP binding site released (TK-kin3, mimicking the mechanically induced open state after peak 2), and found that the release of ATP binding not only activates kinase activity toward substrates like telethonin, but also allows tyrosine autophosphorylation (see SI Text, sections 1 and 2, for experimental details). As shown in Fig. 5, although low levels of phosphotyrosine are detected by the 4G10 antibody before incubation with ATP, tyrosine phosphorylation is strongly stimulated by ATP, with a preference toward Mn2+, similar to other enzymes (26, 27).
Fig. 5.
Autophosphorylation of TK on tyrosine. Incubation of the highly purified TK-kin3 enzyme in the absence (−) and presence (+) of ATP and Mn2+ ions leads to tyrosine phosphorylation detected by Western blot, using the phosphotyrosine antibody 4G10. The autoinhibited kinase construct A168-M2 (WT) shows no appreciable phosphotyrosine incorporation under any tested condition. Lower blot: loading control, detection with anti-titin kinase antibody (α-TK).
Discussion
Our results show that mechanical stress is able to activate titin kinase by releasing the active site for ATP binding, and they unravel the first step of this mechanical signaling pathway. We also show that mechanical release of the ATP binding site allows a second step in TK activation by triggering autophosphorylation on the inhibitory tyrosine. That TK can thus indeed act as a biological force sensor is supported by the fact that the forces activating ATP binding are within the physiological range and, importantly, lower than the ones unfolding the surrounding structural titin domains. Small force imbalances of four to eight myosin motor domains, equivalent to ≈3% of the 147 myosin molecules (1) between adjacent thick filaments could thus translate into a physiologically significant signal by activation of the TK mechanosensor. Similarly, the “gating distance” between the open and closed state of the TK active site, as the distance between peaks 1 and 2, is 9 nm. The reversible increase in the width of the 14.5-nm x-ray reflection during contraction of striated muscle indicates axial displacement between adjacent myosin filaments on the order of 10 nm (28, 29), which would translate into shear strain on the M-band. Both the forces and displacements required for mechanical TK activation are therefore within the ranges observed in muscle.
Once the ATP binding site is opened by mechanical force, not only does the enzyme bind ATP, but it actually undergoes the next necessary step for full activation, the phosphorylation of the autoinhibitory tyrosine-170. Rather than leading to a dead end, mechanical activation of ATP binding thus activates the full catalytic activity of TK. This mechanism may be particularly relevant when the sarcomere is extended while generating active tension due to an opposing force greater than that generated by the muscle, also called eccentric exercise (9). Under such conditions, large changes in M-band structure are observed (30). Eccentric exercise is a strong stimulator for muscle growth and repair (31–34), and the interaction of titin kinase with ubiquitin-associated scaffold proteins with links to multiple signaling pathways controlling muscle gene expression and protein turnover (12) supports plausibly that titin kinase can act as a force sensor in the activated sarcomere. Unlike the homologous nematode kinases, which retain catalytic activity in their inhibited form (13), we show that TK is completely inactive in its autoinhibited form. Mechanical switching of its ATP-binding site thus confers a significant signal between active and inactive kinase, as expected for a signal that modulates energy-costly processes like protein breakdown and transcriptional activity (12).
Our surprising observation that a protein kinase can be activated by local protein unfolding induced by mechanical force may find analogies in the small GTPase Rab8, whose activation by the nucleotide exchange factor MSS4 also involves local protein unfolding (35). The mechanoenzymatic sensor found in titin kinase may therefore be paradigmatic also for other members of the family of cytoskeletal autoregulated protein kinases, a branch of the calcium-calmodulin-regulated enzymes of the human kinome (36), containing myosin light-chain kinase and obscurin kinases. These enzymes share with titin the N- and C-terminal cytoskeletal association (37) or specific residues involved in autoinhibition (38) and may thus bear features of mechanical modulation. Furthermore, other autoregulated cytoskeletal signaling domains, like GDP-GTP exchange factor domains, may be similarly activated. Our single-molecule approach will therefore be useful for investigating the mechanochemistry of many cellular systems that may share similar mechanosensitive regulation mechanisms.
Materials and Methods
Titin kinase expression was carried out in sf9 insect cells by using a recombinant baculovirus system essentially as described in ref. 5. Purification and enzymatic assays were performed essentially as described in refs. 5 and 12 (for details, see SI Text). Atomic force microscopy using a custom-built instrument, and analysis of the data were carried out essentially as described (14, 19); for details see the SI. Force–probe molecular dynamics simulations (24, 25) used the TK x-ray structure [Protein Data Bank entry 1TKI (5)] as the starting structure, with the autoinhibitory tail partly removed. Two sets of simulations (five each) were carried out for this truncated TK, one set with an empty binding pocket, and one with an ATP molecule and magnesium ions inserted into the (closed) binding pocket. As a control, the autoinhibited complete TK was also subjected to force–probe MD simulations. For further details, see SI Text.
Supplementary Material
Acknowledgments.
We thank Thorsten Kampmann for help with the ATP force field and Carsten Kutzner for help with the GROMACS force probe code. U.H. and L.V.S. were supported by the Deutsche Forschungsgemeinschaft (research training group 782 and SFB Grant 755). L.V.S. was supported by the Boehringer Ingelheim Fonds and by the European Union. This work was supported by the Center for Integrated Protein Science Munich and the Medical Research Council of the United Kingdom.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805034105/DCSupplemental.
References
- 1.Tskhovrebova L, Trinick J. Titin: Properties and family relationships. Nat Rev Mol Cell Biol. 2003;4:679–689. doi: 10.1038/nrm1198. [DOI] [PubMed] [Google Scholar]
- 2.Hoshijima M. Mechanical stress-strain sensors embedded in cardiac cytoskeleton: Z disk, titin, and associated structures. Am J Physiol. 2006;290:H1313–H1325. doi: 10.1152/ajpheart.00816.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Labeit S, Gautel M, Lakey A, Trinick J. Towards a molecular understanding of titin. EMBO J. 1992;11:1711–1716. doi: 10.1002/j.1460-2075.1992.tb05222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Obermann WMJ, et al. The structure of the sarcomeric M band: Localization of defined domains of myomesin, M-protein and the 250 kD carboxy-terminal region of titin by immunoelectron microscopy. J Cell Biol. 1996;134:1441–1453. doi: 10.1083/jcb.134.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayans O, et al. Structural basis of the activation of the titin kinase domain during myofibrillogenesis. Nature. 1998;395:863–869. doi: 10.1038/27603. [DOI] [PubMed] [Google Scholar]
- 6.Vogel V. Mechanotransduction involving multimodular proteins: Converting force into biochemical signals. Annu Rev Biophys Biomol Struct. 2006;35:459–488. doi: 10.1146/annurev.biophys.35.040405.102013. [DOI] [PubMed] [Google Scholar]
- 7.Horowits R, Podolsky RJ. The positional stability of thick filaments in activated skeletal muscle depends on sarcomere length: Evidence for the role of titin filaments. J Cell Biol. 1987;105:2217–2223. doi: 10.1083/jcb.105.5.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akiyama N, Ohnuki Y, Kunioka Y, Saeki Y, Yamada T. Transverse stiffness of myofibrils of skeletal and cardiac muscles studied by atomic force microscopy. J Physiol Sci. 2006;56:145–151. doi: 10.2170/physiolsci.RP003205. [DOI] [PubMed] [Google Scholar]
- 9.Agarkova I, Ehler E, Lange S, Schoenauer R, Perriard JC. M-band: A safeguard for sarcomere stability? J Muscle Res Cell Motil. 2003;24:191–203. doi: 10.1023/a:1026094924677. [DOI] [PubMed] [Google Scholar]
- 10.Agarkova I, Perriard JC. The M-band: An elastic web that crosslinks thick filaments in the center of the sarcomere. Trends Cell Biol. 2005;15:477–485. doi: 10.1016/j.tcb.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Grater F, Shen J, Jiang H, Gautel M, Grubmuller H. Mechanically induced titin kinase activation studied by force-probe molecular dynamics simulations. Biophys J. 2005;88:790–804. doi: 10.1529/biophysj.104.052423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lange S, et al. The kinase domain of titin controls muscle gene Expression and protein turnover. Science. 2005;308:1599–1603. doi: 10.1126/science.1110463. [DOI] [PubMed] [Google Scholar]
- 13.Greene D, et al. Single-molecule force spectroscopy reveals a stepwise unfolding of Caenorhabditis elegans giant protein kinase domains. Biophys J. 2008;95:1360–1370. doi: 10.1529/biophysj.108.130237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puchner E, Franzen G, Gautel M, Gaub H. Comparing proteins by their unfolding pattern. Biophys J. 2008;95:426–434. doi: 10.1529/biophysj.108.129999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukuzawa A, et al. Interactions with titin and myomesin target obscurin and its small homologue, obscurin-like 1, to the sarcomeric M-band: Implications for hereditary myopathies. J Cell Sci. 2008;121:1841–1851. doi: 10.1242/jcs.028019. [DOI] [PubMed] [Google Scholar]
- 16.Flaherty D, et al. Titins in C. elegans with unusual features: Coiled-coil domains, novel regulation of kinase activity and two new possible elastic regions. J Mol Biol. 2002;323:533–549. doi: 10.1016/s0022-2836(02)00970-1. [DOI] [PubMed] [Google Scholar]
- 17.Moerman DG, Benian GM, Barstead RJ, Schreifer LA, Waterston RH. Identification and intracellular localization of the unc-22 gene product of Caenorhabditis elegans. Genes Dev. 1988;2:93–105. doi: 10.1101/gad.2.1.93. [DOI] [PubMed] [Google Scholar]
- 18.Rief M, Gautel M, Schemmel A, Gaub HE. The mechanical stability of immunoglobulin and fibronectin III domains in the muscle protein titin measured by atomic force microscopy. Biophys J. 1998;75:3008–3014. doi: 10.1016/S0006-3495(98)77741-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin Ig-domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 20.Kedrov A, Krieg M, Ziegler C, Kuhlbrandt W, Muller DJ. Locating ligand binding and activation of a single antiporter. EMBO Rep. 2005;6:668–674. doi: 10.1038/sj.embor.7400455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lew J, Taylor SS, Adams JA. Identification of a partially rate-determining step in the catalytic mechanism of cAMP-dependent protein kinase: A transient kinetic study using stopped-flow fluorescence spectroscopy. Biochemistry. 1997;36:6717–6724. doi: 10.1021/bi963164u. [DOI] [PubMed] [Google Scholar]
- 22.Knighton DR, et al. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:407–420. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- 23.Bossemeyer D, Engh R, Kinzel V, Ponstingl H, Huber R. Phosphotransferase and substrate binding mechanism of the cAMP-dependent protein kinase catalytic subunit from porcine heart as deduced from the 2.0 Å structure of the complex with Mn2+ adenylyl imidodiphosphate and inhibitor peptide PKI(5–24) EMBO J. 1993;12:849–859. doi: 10.1002/j.1460-2075.1993.tb05725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grubmuller H, Heymann B, Tavan P. Ligand binding: Molecular mechanics calculation of the streptavidin-biotin rupture force. Science. 1996;271:997–999. doi: 10.1126/science.271.5251.997. [DOI] [PubMed] [Google Scholar]
- 25.Izrailev S, Stepaniants S, Balsera M, Oono Y, Schulten K. Molecular dynamics study of unbinding of the avidin-biotin complex. Biophys J. 1997;72:1568–1581. doi: 10.1016/S0006-3495(97)78804-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cobb MH, Sang BC, Gonzalez R, Goldsmith E, Ellis L. Autophosphorylation activates the soluble cytoplasmic domain of the insulin receptor in an intermolecular reaction. J Biol Chem. 1989;264:18701–18706. [PubMed] [Google Scholar]
- 27.Tennagels N, Hube-Magg C, Wirth A, Noelle V, Klein HW. Expression, purification, and characterization of the cytoplasmic domain of the human IGF-1 receptor using a baculovirus expression system. Biochem Biophys Res Commun. 1999;260:724–728. doi: 10.1006/bbrc.1999.0968. [DOI] [PubMed] [Google Scholar]
- 28.Huxley HE, Faruqi AR, Kress M, Bordas J, Koch MH. Time-resolved x-ray diffraction studies of the myosin layer-line reflections during muscle contraction. J Mol Biol. 1982;158:637–684. doi: 10.1016/0022-2836(82)90253-4. [DOI] [PubMed] [Google Scholar]
- 29.Linari M, et al. Interference fine structure and sarcomere length dependence of the axial x-ray pattern from active single muscle fibers. Proc Natl Acad Sci USA. 2000;97:7226–7231. doi: 10.1073/pnas.97.13.7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki S, Sugi H. Extensibility of the myofilaments in vertebrate skeletal muscle as revealed by stretching rigor muscle fibers. J Gen Physiol. 1983;81:531–546. doi: 10.1085/jgp.81.4.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldspink G, Williams P, Simpson H. Gene expression in response to muscle stretch. Clin Orthop Relat Res. 2002:S146–152. doi: 10.1097/00003086-200210001-00017. [DOI] [PubMed] [Google Scholar]
- 32.Barash IA, Mathew L, Ryan AF, Chen J, Lieber RL. Rapid muscle-specific gene expression changes after a single bout of eccentric contractions in the mouse. Am J Physiol. 2004;286:C355–C364. doi: 10.1152/ajpcell.00211.2003. [DOI] [PubMed] [Google Scholar]
- 33.Carlsson L, Yu J-G, Moza M, Carpen O, Thornell L-E. Myotilin—A prominent marker of myofibrillar remodelling. Neuromuscular Disorders. 2007;17:61–68. doi: 10.1016/j.nmd.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 34.Eliasson J, et al. Maximal lengthening contractions increase p70 S6 kinase phosphorylation in human skeletal muscle in the absence of nutritional supply. Am J Physiol. 2006;291:E1197–E1205. doi: 10.1152/ajpendo.00141.2006. [DOI] [PubMed] [Google Scholar]
- 35.Itzen A, Pylypenko O, Goody RS, Alexandrov K, Rak A. Nucleotide exchange via local protein unfolding—Structure of Rab8 in complex with MSS4. EMBO J. 2006;25:1445–1455. doi: 10.1038/sj.emboj.7601044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 37.Kamm KE, Stull JT. Dedicated myosin light chain kinases with diverse cellular functions. J Biol Chem. 2001;276:4527–4530. doi: 10.1074/jbc.R000028200. [DOI] [PubMed] [Google Scholar]
- 38.Fukuzawa A, Idowu S, Gautel M. Complete human gene structure of obscurin: Implications for isoform generation by differential splicing. J Muscle Res Cell Motil. 2005;26:427–434. doi: 10.1007/s10974-005-9025-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.