Abstract
In the yeast Saccharomyces cerevisiae, the Apg12p–Apg5p conjugating system is essential for autophagy. Apg7p is required for the conjugation reaction, because Apg12p is unable to form a conjugate with Apg5p in the apg7/cvt2 mutant. Apg7p shows a significant similarity to a ubiquitin-activating enzyme, Uba1p. In this article, we investigated the function of Apg7p as an Apg12p-activating enzyme. Hemagglutinin-tagged Apg12p was coimmunoprecipitated with c-myc–tagged Apg7p. A two-hybrid experiment confirmed the interaction. The coimmunoprecipitation was sensitive to a thiol-reducing reagent. Furthermore, a thioester conjugate of Apg7p was detected in a lysate of cells overexpressing both Apg7p and Apg12p. These results indicated that Apg12p interacts with Apg7p via a thioester bond. Mutational analyses of Apg7p suggested that Cys507 of Apg7p is an active site cysteine and that both the ATP-binding domain and the cysteine residue are essential for the conjugation of Apg7p with Apg12p to form the Apg12p–Apg5p conjugate. Cells expressing mutant Apg7ps, Apg7pG333A, or Apg7pC507A showed defects in autophagy and cytoplasm-to-vacuole targeting of aminopeptidase I. These results indicated that Apg7p functions as a novel protein-activating enzyme necessary for Apg12p–Apg5p conjugation.
INTRODUCTION
Autophagy is the process of bulk degradation of cytoplasmic components by the lysosomal/vacuolar system (Seglen and Bohley, 1992; Dunn, 1994). The phenomenon is dramatically enhanced under nutrient starvation conditions. In the initial step of the macroautophagy, a cup-shaped membrane sac surrounds cytosolic components to form an autophagosome (Baba et al., 1994). The outer membrane of the autophagosome fuses with a lysosome/vacuole (Baba et al., 1995). A transient single-membrane structure, the autophagic body, is released into the lumen and subsequently degraded in the lysosome/vacuole. Although biochemical and cell–biological approaches have revealed several aspects of autophagy in mammalian cells, the molecular mechanism of autophagy remains unclear.
In the yeast Saccharomyces cerevisiae, macroautophagy was first described by Takeshige et al. (1992), and the process is similar to that in higher eukaryotes (Baba et al., 1994, 1995). Taking advantage of yeast genetics, autophagy-defective mutants (14 apg mutants and 9 aut mutants) have been isolated in two different laboratories (Tsukada and Ohsumi, 1993; Thumm et al., 1994; Harding et al., 1996). Surprisingly, most of the apg and aut mutants overlap genetically and phenotypically with some cvt mutants, which have defects in the cytoplasm-to-vacuole targeting of aminopeptidase I (API)1. These results suggest that the APG and AUT gene products function even in a vegetative growth (APG1/CVT10/AUT3, APG7/CVT2, APG8/CVT5/AUT7, APG9/CVT7/AUT9, APG14/CVT12, APG15/CVT11, and CVT17/AUT5) (Harding et al., 1996; Scott et al., 1996). Several APG and AUT genes have been identified, and most encode novel proteins (APG1/CVT10/AUT3, APG5, APG6, APG7/CVT2, APG12, APG13, APG14, AUT1, AUT2, and AUT7) (Kametaka et al., 1996; Funakoshi et al., 1997; Matsuura et al., 1997; Schlumpberger et al., 1997; Straub et al., 1997; Kametaka et al., 1998; Lang et al., 1998; Mizushima et al., 1998a). Biochemical characterization of the gene products has revealed some molecular aspects of autophagy: Apg1p/Aut3p is a novel protein kinase (Matsuura et al., 1997; Straub et al., 1997). Apg6p/Vps30p and Apg14p form a protein complex (Kametaka et al., 1998). Aut2p interacts with Aut7p/Apg8p, a homologue of rat microtubule-associated protein light chain 3 (Lang et al., 1998).
Recently, we found that the Apg12p–Apg5p conjugation system is essential for autophagy (Mizushima et al., 1998a). Apg12p has no significant homology to ubiquitin or ubiquitin-related modifiers; however, Apg12p is conjugated to Apg5p through an isopeptide bond between the C-terminal Gly residue of Apg12p and the Lys149 residue of Apg5p. Ubiquitination is a posttranslational modification to present the degradation signal for proteolytic attack by 26S proteasomes (reviewed by Finley and Chau, 1991; Hershko and Ciechanover, 1992; Hershko, 1996; Hochstrasser, 1996a,b; Haas and Siepmann, 1997; Varshavsky, 1997). Ubiquitin is activated by a ubiquitin-activating enzyme (E1) in an ATP-dependent manner in which a thioester bond is formed between the C terminus of ubiquitin and a Cys residue within the E1 enzyme (Ciechanover et al., 1982; Haas et al., 1982). The ubiquitin is transferred from the E1 enzyme to a Cys residue within a ubiquitin-conjugating enzyme (E2). Finally, ubiquitin is covalently attached to a target protein by an isopeptide linkage directly from E2 or by a ubiquitin-protein ligase (E3) (Hershko et al., 1983; Bartel et al., 1990; Scheffner et al., 1993, 1995; Peters et al., 1996; Zachariae et al., 1996).
Recent discoveries have revealed that the ubiquitin-related modifiers other than ubiquitin play essential roles in eukaryotes (reviewed by Johnson and Hochstrasser, 1997; Saitoh et al., 1997; Dolan, 1998; Hochstrasser, 1998). A mammalian ubiquitin-related protein, SUMO-1 [small ubiquitin-related modifier; also called GMP1, PIC1, UBL1, or sentrin (Boddy et al., 1996; Matunis et al., 1996; Okura et al., 1996; Shen et al., 1996)] is covalently attached to the RanGAP1 and PML proteins (Matunis et al., 1996; Mahajan et al., 1997; Muller et al., 1998). This posttranslational modification affects the subcellular localization of these proteins (Mahajan et al., 1998; Matunis and Blobel, 1998; Muller et al., 1998). A yeast SUMO-1 homologue, Smt3p, is activated by an E1-like heterodimer Aos1p/Uba2p (Dohmen et al., 1995; Johnson et al., 1997). The E2 enzyme for Smt3p and SUMO-1 are Ubc9p and its mammalian homologue (Gong et al., 1997; Johnson and Blobel, 1997; Lee et al., 1998; Schwarz et al., 1998). Another family of ubiquitin-related proteins is RUB1 and NEDD8 (Kumar et al., 1993; Callis et al., 1995; Hochstrasser, 1996; Kamitani et al., 1997; Lammer et al., 1998; Liakopoulos et al., 1998). A major substrate of RUB1/NEDD8 is Cdc53p/Cullin in yeast and mammalian cells, which play an essential role in regulating the cell cycle (Lammer et al., 1998; Liakopoulos et al., 1998; Osaka et al., 1998). In Arabidopsis thaliana, the auxin response depends on the RUB1 modification of nuclear proteins (del Pozo et al., 1998). RUB1 and NEDD8 are activated by E1-like heterodimers: Ula1p (Enr2p)/Uba3p in yeast, an APP-BP1 (a 59-kDa β-amyloid-protein-precursors-binding protein)/human UBA3 homologue in humans, and AXR1/ECR1 in A. thaliana (Leyser et al., 1993; Chow et al., 1996; del Pozo et al., 1998; Liakopoulos et al., 1998; Osaka et al., 1998). The E2 enzyme for RUB1 and NEDD8 are Ubc12p and its mammalian homologue (del Pozo et al., 1998; Liakopoulos et al., 1998; Osaka et al., 1998).
These findings strongly suggest that there must be E1- and E2-like enzymes for the Apg12p–Apg5p conjugation system in yeast. Candidates in the Apg12p conjugation system are Apg7p/Cvt2p and Apg10p. In apg7 and apg10 mutants, no Apg12p–Apg5p conjugate is observed, indicating that Apg7p and Apg10p play indispensable roles in the Apg12p conjugation system (Mizushima et al., 1998a). A region of Apg7p (residues 322 to 407 out of 633 amino acids) shows significant homology to the corresponding region of a ubiquitin-activating enzyme, Uba1p, although the other regions show no homology (McGrath et al., 1991; Mizushima et al., 1998a). We took particular interest in Apg7p/Cvt2p and investigated functions of Apg7p through both biochemical and molecular biological techniques. In this study, we provide several lines of evidence showing that Apg7p is an Apg12p-activating enzyme.
MATERIALS AND METHODS
Strains, Media, Materials, and Genetic Techniques
Escherichia coli strain DH5α as the host for plasmids and protein expression was grown in Luria Broth medium in the presence of antibiotics as required (Ausubel et al., 1995). The S. cerevisiae strains and plasmids used in this study are listed in Table 1. All yeast strains were cultured in rich medium (YPD, pH 5.0: 1% yeast extract, 2% polypeptone, 2% glucose, 20 mg/l adenine, 20 mg/l tryptophan, 20 mg/l uracil, and 50 mM succinate/NaOH, pH 5.0), MVD medium (0.67% yeast nitrogen base without amino acids, 0.5% casamino acids, and 2% glucose), or SD medium (0.67% yeast nitrogen base without amino acids, 2% glucose, and appropriate amino acids as described by Kaiser et al. [1994]). Nitrogen-starvation medium contained 0.17% yeast nitrogen base without amino acids and ammonium sulfate and 2% glucose. Solid medium contained 2% Bacto agar. Standard genetic and molecular biological techniques were performed as described by Kaiser et al. (1994) and Ausubel et al. (1995). The PCR was performed with a program temperature control system PC-701 (ASTEC, Fukuoka, Japan). The DNA sequence was determined by an ABI 373A DNA sequencer (PE Applied Biosystems, Foster City, CA). Restriction enzymes were purchased from TOYOBO (Osaka, Japan) and New England Biolabs (Beverly, MA). Oligonucleotides were synthesized by Sawady Technology (Tokyo, Japan) or ESPEC oligo-service (Ibaraki, Japan). pYO324 was a kind gift from Y. Ohya, pRS series vectors were kind gifts from P. Hieter, and pGAD-C1 vector, pGBD-C1 vector, and PJ69-4A strain were kind gifts from P. James (Sikorski and Hieter, 1989; James et al., 1996; Homma et al. 1998). pGEX-3X and cyanogen bromide-activated Sepharose beads were purchased from Amersham Pharmacia Biotech (Uppsala, Sweden); pGEM-T was from Promega (Madison, WI).
Table 1.
Yeast strains and plasmids
| Genotype | Source | |
|---|---|---|
| Yeast strains | ||
| YW5-1B | MATa leu2-3, 112 trp1 ura3-52 | Tsukada and Ohsumi (1993) |
| YIT701 | MATa leu2-3, 112 trp1 ura3-52 apg7Δ::LEU2 | This study |
| YIT702 | MATa leu2-3, 112 trp1 ura3-52 apg7Δ::LEU2 | This study |
| [pAPG7myc-314, pAPG12HA-316] | ||
| YIT703 | MATa leu2-3, 112 trp1 ura3-52 apg7Δ::LEU2 | This study |
| [pAPG7myc-324, pAPG12HA-426] | ||
| YIT704 | MATa leu2-3, 112 trp1 ura3-52 apg7Δ::LEU2 | This study |
| [pYO324, pAPG12HA-426] | ||
| YIT7G333A | MATa leu2-3, 112 trp1 ura3-52 apg7Δ::LEU2 | This study |
| [pAPG7G333Amyc-314, pAPG12HA-316] | ||
| YIT7C507A | MATa leu2-3, 112 trp1 ura3-52 apg7Δ::LEU2 | This study |
| [pAPG7C507 Amyc-314, pAPG12HA-316] | ||
| YTS2 | MATa ura3-52 lys2-801 ade2-101 trp1 his3 | This study |
| leu2 pho8::pho8Δ60 apg7::HIS3 | ||
| PJ69-4A | MATa trp1-901 leu2-3, 112 ura3-52 his3-200 | James et al. (1996) |
| gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 | ||
| met2::GAL7-lacZ | ||
| Plasmids | ||
| pRS314 | CEN6 TRP1 | Sikorski and Hieter (1989) |
| pRS316 | CEN6 URA3 | Sikorski and Hieter (1989) |
| pYO324 | 2μ TRP1 | Homma et al. (1998) |
| pAPG7myc-314 | CEN6 TRP1 APG7-3 × cmyc epitope | This study |
| pAPG7myc-324 | 2μ TRP1 APG7-3 × cmyc epitope | This study |
| pAPG7G333Amyc-314 | CEN6 TRP1 APG7G333A-3 × cmyc epitope | This study |
| pAPG7C507Amyc-314 | CEN6 TRP1 APG7C507A-3 × cmyc epitope | This study |
| pAPG12HA-316 | CEN6 URA3 3 × HA epitope-APG12 | Mizushima et al. (1998) |
| pAPG12HA-426 | 2μ URA3 3 × HA epitope-APG12 | Mizushima et al. (1998) |
| pGAD-C1 | 2μ LEU2 PADH1::GAL4 AD | James et al. (1996) |
| pGAD-APG12 | 2μ LEU2 PADH1::GAL4 AD-APG12 | This study |
| pGBD-C1 | 2μ TRP1 PADH1::GAL4 BD | James et al. (1996) |
| pGBD-APG7 | 2μ TRP1 PADH1::GAL4 BD-APG7-3 × cmyc epitope | This study |
| pGBD-APG7G333A | 2μ TRP1 PADH1::GAL4 BD-APG7G333A-3 × cmyc epitope | This study |
| pGBD-APG7C507A | 2μ TRP1 PADH1::GAL4 BD-APG7C507A-3 × cmyc epitope | This study |
Gene Disruption of the APG7 Gene
Gene disruption with the PCR product was performed as described by Lorenz et al. (1995). Briefly, PCR was performed with APG7-pRSF primer (5′-GTCGTCAGAAAGGGTCTTAAGTTATGCACCAGCTTTTAAATCATTTCTGGTATCACGAGGCCCTTTCGTC-3′), APG7pRSR primer (5′-GCAATCTCATCAGATTCATCATCTTCCCATTCAAAAACATCGTTGCCTAGGTGCGGTATTTCACACCGC-3′), and pRS30X plasmids as a template. The amplified PCR product was transformed into a yeast strain. APG7 gene disruption was confirmed by PCR with APG7Bgl2ATG primer (5′-AGATCTATGTCGTCAGAAAGGGTCTTAAG-3′) and APG7SalISTOP primer (5′-GTCGACTATTAAGCAATCTCATCAGATTCATC-3′).
Plasmid Construction and Site-directed Mutagenesis
A BamHI fragment (3.9 kb) containing the APG7 gene (ORF YHR171w) was introduced into the BamHI site of pRS314 and pYO324 (pAPG7-314 and pAPG7-324, respectively). To construct an expression plasmid for c-myc–tagged Apg7p, SacI and ApaI sites were introduced just before the termination codon of the APG7 gene by nested PCR with APG7SAF primer (5′-GAATCTGATGAGATTGC-TGAGCTCGGGCCCTAATATTTTGCATATAATAGC-3′), APG7SAR primer (5′-GCTATTATATGCAAAATATTAGGGCCCGAGCTCAG-CAATCTCATCAGATTC-3′), M13-47 primer (5′-CGCCAGGGTTT-TCCCAGTCACGAC-3′), and M13-RV primer (5′-GAGCGGATAA-CAATTTCACACAGG-3′). The amplified DNA fragment was cloned into pGEM-T Vector (pAPG7SA-GEMT). A StuI–SacI fragment (0.12 kb) of pAPG7-GEMT, a SacI–ApaI fragment (0.16 kb) of pMPY-3xMYC, and an ApaI–SalI fragment (1.0 kb) of pAPG7-GEMT were replaced with the StuI–SalI region of pAPG7-314 and pAPG7-324 (pAPG7myc-314 and pAPG7myc-324). The junction and PCR-amplified region were confirmed by DNA sequencing. The plasmid complemented the apg7Δ mutation.
Site-directed mutagenesis of the APG7 gene (Gly333 and Cys507 changed to Ala) was performed by nested PCR with APG7E1928F primer (5′-CTTTAAAAATTGCTGACCAATCCGTGG-3′), APG7X2298F primer (5′-GAGCATTAATAAAAGAGCATG-3′), APG7G333AR primer (5′-CAACCTAGTGTAGCAGCACCTAGTAGTAG-3′), APG7C507AR primer (5′-CTAGTTACTGTGGCCATTTGATCC-3′), APG7S2855R primer (5′-CCTGCTTTATGACTGACAAACCGC-3′), and pAPG7-314 as a template. The amplified EcoRI–StuI fragment (0.8 kb) was replaced with the same region of pAPG7myc-314. The mutation site and PCR-amplified region were confirmed by DNA sequencing. The resultant plasmids were designated pAPG7G333Amyc-314 and pAPG7C507Amyc-314.
For the two-hybrid experiment, a BglII site was introduced just before the start codon of the APG7 gene by PCR with APG7Bgl2ATG primer, APG7SalISTOP primer, and pAPG7-314 as a template. The PCR product was cloned into pGEM-T. The AflII–SalI fragment (∼1.9 kb) of the clone was replaced with the AflII–SalI fragment (∼3.8 kb) of pAPG7myc-314 (pAPG7BS-GEMT). The BglII–SalI fragment of pAPG7BS-GEMT was subcloned into the BamHI–SalI sites of the pGBD-C1 vector (pGBD-APG7). The AflII–SalI fragment of pGBD-APG7 was replaced with the AflII–SalI fragment from pAPG7G333Amyc-314 or pAPG7C507Amyc-314 to construct pGBD-APG7G333A and pGBD-APG7C507A, respectively. The whole APG12 coding region was PCR-amplified and then introduced into the PstI site of pGAD-C1 (pGAD-APG12).
Antibodies
To express the GST-Apg7p fusion protein in E. coli, an EcoRV fragment (0.6 kb) and an EcoRV–EcoRI fragment (1.6 kb) of pAPG7-316 were subcloned into the SmaI and SmaI–EcoRI sites of pGEX-3X, respectively (pGEX-APG7N and pGEX-APG7C). GST-Apg7p fusion proteins were expressed in DH5α cells carrying pGEX-APG7N and pGEX-APG7C according to manufacturer’s protocol (Amersham Pharmacia Biotech). The GST–C-terminal region of Apg7p was expressed in cells carrying pGEX-APG7C and purified on glutathione Sepharose 4B. The GST–N-terminal region of Apg7p was expressed in cells carrying pGEX-APG7N but was included in the inclusion bodies. The protein was extracted from the inclusion bodies with extraction buffer (8 M urea, 50 mM Tris-Cl, pH 9.5), subjected to SDS-PAGE, and excised from the gel. Polyclonal antibodies against the carboxyl and amino terminal fragments of Apg7p were raised in Japanese white rabbits using purified proteins as antigens (αApg7C and αApg7N, respectively). The IgG fraction was precipitated with 50% saturated ammonium sulfate and dissolved in TBS buffer (150 mM NaCl, 20 mM Tris-Cl, pH 7.5). Anti-Apg7p antibodies were purified on a GST-Apg7p Sepharose column. A polyclonal anti-API antibody was a kind gift from D. J. Klionsky. An anti-hemagglutinin (HA) mouse mAb (16B12) was purchased from Berkeley Antibody Company (Berkeley, CA), an anti–c-myc mouse mAb (9E10)-Agarose conjugate was from Santa Cruz Biotechnology (Santa Cruz, CA), and anti-yeast 3-phosphoglycerate kinase and anti-yeast Dol-P-Man synthase mouse mAbs were from Molecular Probes (Eugene, OR).
Immunoprecipitation
Cells (OD600 = 10) grown to early logarithmic phase in MVD or YPD pH 5.0 medium were harvested and converted to spheroplasts in spheroplasting solution (1% yeast extract, 2% polypeptone, 0.5% glucose, 1 mg/ml Zymolyase 100T). The spheroplasts were harvested in 1.3 M sorbitol as a cushion, lysed with lysis buffer (1% SDS, 150 mM NaCl, 20 mM sodium phosphate, pH 7.5), vortexed, boiled for 5 min, and chilled on ice. When indicated, 1 mM DTT was added to the lysate before boiling. Ten volumes of IP buffer (2% Triton X-100, 150 mM NaCl, 20 mM sodium phosphate, pH 7.5) were added to the cell lysate, and the mixture was centrifuged at 10,000 × g for 5 min at 4°C to remove debris. The supernatant was precleared with 50 μl Protein A-Agarose (20% slurry, Santa Cruz Biotechnology). Fifty microliters of Agarose beads conjugated with anti–c-myc antibody (9E10) (20% slurry, Santa Cruz Biotechnology) were added to the lysate, and the mixture was incubated for 2 h at 4°C. The immunoprecipitate–bead complex was washed six times with ice-cold RIPA buffer (10 mM Tris-Cl, pH 7.4, 1% Nonidet P40, 0.1% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 1 mM EDTA). Proteins were eluted from the beads with 0.1N glycine-HCl, pH 2.5, and precipitated by incubation with 10% TCA on ice for 1 h. The sediment was washed twice in cold acetone, subjected to reducing SDS-PAGE, transferred to a polyvinylidene difluoride membrane (Millipore, Bedford, MA), and analyzed by Western blotting. All solutions contained a protease-inhibitor mixture for use with fungal and yeast extracts (Sigma, St. Louis, MO).
Other Techniques
Two-hybrid analysis was performed as described by James et al. (1996). A biochemical assay monitoring autophagy in yeast by alkaline phosphatase processing was performed as described by Noda et al. (1998). Subcellular fractionation of yeast cells was performed as described by Huang and Chiang (1997). Advanced BLAST search was performed on the National Center for Biotechnology Information web site (http://www.ncbi.nlm.nih.gov/BLAST/). Multiple alignment of amino acids sequences was performed using a CLUSTAL W program by the computer laboratory in the National Institute for Basic Biology.
RESULTS
Apg12p Interacts with Apg7p via a Thioester Bond
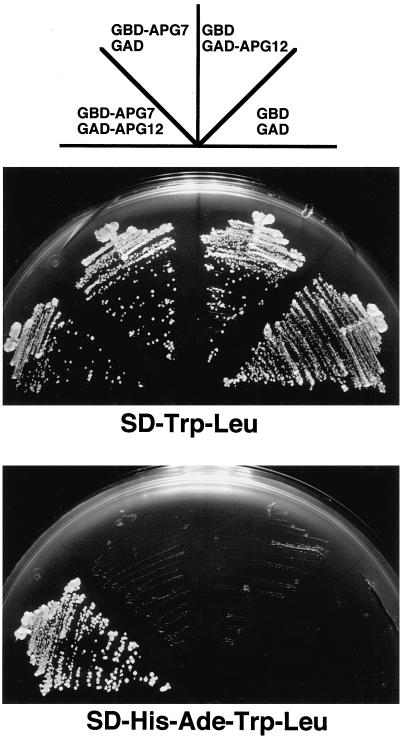
A significant homology between Apg7p and Uba1p suggests that Apg7p functions as an Apg12p-activating enzyme. In this case, Apg12p should first form a transient enzyme–substrate complex with Apg7p as demonstrated with Uba1p and ubiquitin. To examine this possibility, we constructed a strain of yeast expressing both c-myc–tagged Apg7p and HA-tagged Apg12p (YIT702 strain), and immunoprecipitated the c-myc–tagged Apg7p with anti–c-myc antibody. Western analysis using αApg7N antibody recognized c-myc–tagged Apg7p (∼78 kDa) in the immunoprecipitates (Figure 1A, WB αApg7N). HA-tagged Apg12p was also detected in the same immunoprecipitate with anti-HA antibody (Figure 1A, WB αHA). The results correspond to those of the two-hybrid experiment (Figure 2). PJ69-4A strain (trp1-901 leu2-3112 gal4Δ gal80Δ GAL2-ADE2 GAL1-HIS3) expressing both GAL4AD-Apg12p and GAL4BD-Apg7p grew well on SD-Ade-His-Trp-Leu plate, whereas strains expressing GAL4AD and GAL4BD, GAL4AD-Apg12p and GAL4BD, or GAL4AD and GAL4BD-Apg7p did not. These results suggest that Apg12p interacts with Apg7p.
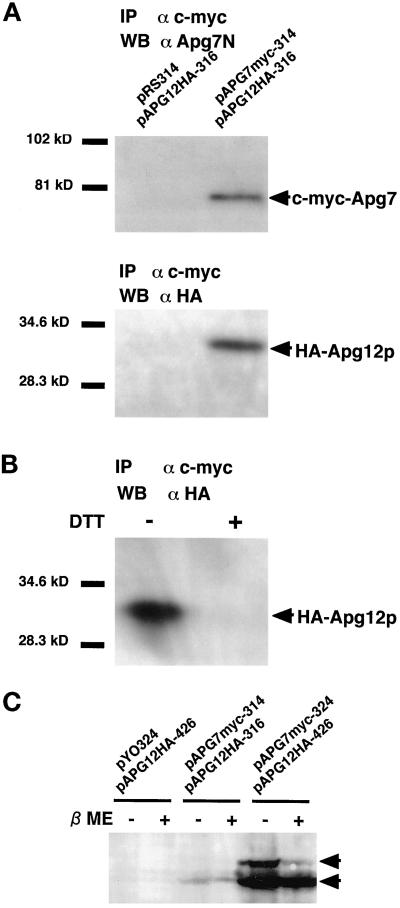
Figure 1.
Apg12p conjugates with Apg7p via a thioester bond. (A) Apg12p was coimmunoprecipitated with Apg7p. pAPG7myc-314 plasmid (CEN) was transformed into apg7Δ cells producing HA-tagged Apg12p to express c-myc–tagged Apg7p. A transformant was designated as the YIT702 strain. The pRS314 vector was used as a control. Cell lysates were prepared as described in MATERIALS AND METHODS. A c-myc–tagged Apg7p was immunoprecipitated with Agarose beads conjugated with anti–c-myc mAb (9E10). The immunoprecipitates were subjected to SDS-PAGE on a 10% gel and transferred to a polyvinylidene difluoride membrane. Proteins were detected by Western blotting with αApg7N antibody (for Apg7p) and anti-HA mouse mAb (16B12; for HA-tagged Apg12p). (B) The coimmunoprecipitation of Apg12p with Apg7p is sensitive to DTT. A cell lysate of the YIT702 strain was prepared as described above. The lysate was boiled with (DTT+) or without 1 mM DTT (DTT−). Immunoprecipitation and Western blotting were performed as described above. (C) The higher molecular weight band of Apg7p in cells overexpressing Apg7p and Apg12p is sensitive to β-mercaptoethanol. Cells grown to early logarithmic phase in MVD medium were harvested and converted to spheroplasts in spheroplasting solution. The spheroplasts were harvested in 1.3 M sorbitol as a cushion, lysed with a 4× SDS sample buffer (Ausubel et al., 1995) with a protease-inhibitor mixture (Sigma). The lysate was boiled for 5 min in the presence (βME+) or absence (βME−) of 3% β-mercaptoethanol. SDS-PAGE on a 7% gel and Western blotting wereperformed as described above. pYO324/pAPG12HA-426: strain YIT704 cells; pAPG7myc-314/pAPG12HA-316: strain YIT702 cells; pAPG7myc-324/pAPG12HA-426: strain YIT703 cells.
Figure 2.
Apg7p interacts with Apg12p in vivo. pGBD-APG7 (TRP1) and pGAD-APG12 (LEU2) were transformed into PJ69-4A strain (gal4Δ gal80Δ trp1 leu2 GAL2-ADE2 GAL1-HIS3) to express GAL4BD-Apg7p and GAL4AD-Apg12p, respectively. pGAD-C1 and pGBD-C1 were used as controls. Cells were plated on SC-Trp-Leu plate (positive control) and SC-Ade-His-Trp-Leu plate, and incubated at 30°C for 3 d. PJ69-4A strain counterclockwise from the bottom carried pGBD-C1 and pGAD-C1 (GBD GAD), pGBD-C1 and pGAD-APG12 (GBD GAD-APG12), pGBD-APG7 and pGAD-C1 (GBD-APG7 GAD), and pGBD-APG7 and pGAD-APG12 (GBD-APG7 GAD-APG12). A strain expressing both GAL4BD-Apg7p and GAL4AD-Apg12p grew well on SC-Ade-Trp-Leu plate, indicating that Apg7p interacts with Apg12p.
We next determined whether this interaction is mediated by a thioester bond. When the lysate was treated with 1 mM DTT before immunoprecipitation, no Apg12p coimmunoprecipitated with the c-myc–tagged Apg7p (Figure 1B, DTT+). In contrast, Apg12p coimmunoprecipitated with c-myc–tagged Apg7p when the lysate was not treated with DTT (Figure 1B, DTT−). Furthermore, when a lysate of cells overexpressing both Apg7p and Apg12p (YIT703 strain) was analyzed on a nonreducing gel, two bands of Apg7p were detected by Western analysis using αApg7N antibody (Figure 1C, pAPG7myc-324/pAPG12HA-426, βME−). When a sample was treated with a thiol-reducing reagent, β-mercaptoethanol, the upper band disappeared, indicating that the upper band is a thioester conjugate of Apg7p. The conjugates were hardly detected in a lysate of cells expressing Apg7p and Apg12p on centromere-type plasmids (Figure 1C, pAPG7myc-314/pAPG12HA-316, YIT702 strain). This is probably due to the low amount of the conjugate form of Apg7p in the strain. These results indicate that Apg12p binds with Apg7p via a thioester bond.
The ATP-binding Domain of Apg7p Is Essential for the Interaction with Apg12p to Form the Apg5p–Apg12p Conjugate
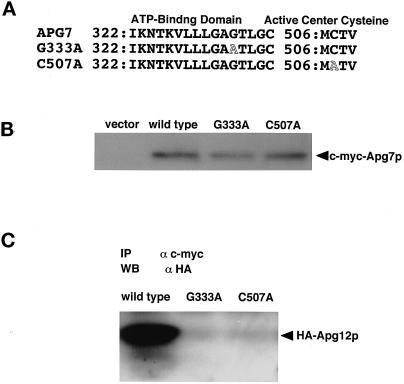
Apg12p–Apg5p conjugation was reconstituted in an ATP-dependent manner in vitro (Mizushima et al. 1998a). A Uba1p-homologous region in Apg7p contains a putative ATP-binding domain (GXGXXG; residues 331 to 336) (Figure 3A) (Wierenga and Hol, 1983; McGrath et al., 1991). According to Wierenga and Hol (1983), the second Gly of the ATP-binding domain will be essential for function. To investigate whether the ATP-binding domain is essential for Apg7p function, we used the site-directed mutagenesis to change the Gly333 of Apg7p to Ala (Figure 3A). The mutant protein was expressed in an amount quite similar to wild-type Apg7p (Figure 3B); however, it could not precipitate HA-tagged Apg12p (Figure 3C). This result indicates that the ATP-binding domain of Apg7p is essential for the conjugation of Apg12p with Apg7p.
Figure 3.
The ATP-binding domain and active site cysteine of Apg7p are essential for conjugation with Apg12p via a thioester bond. (A) Point mutation sites are schematically represented. Gly333 in a predicted ATP-binding domain of Apg7p was changed to Ala by site-directed mutagenesis, and Cys507 in Apg7p was also changed to Ala. (B) Apg7pG333A and Apg7pC507A are expressed in yeast cells at similar levels to Apg7p. The apg7Δ strain carrying pRS314 (vector), strain YIT702 (wild type), strain YIT7G333A (G333A), and strain YIT7C507A (C507A) cells were grown in MVD medium and lysed. c-myc–tagged Apg7p proteins were immunoprecipitated as described in Figure 1A. (C) No Apg12p is coimmunoprecipitated with Apg7pG333A and Apg7pC507A. c-myc–tagged Apg7p in the cell lysate of YIT702 (wild type), YIT7G333A (G333A), and YIT7C507A (C507A) strains were immunoprecipitated, and the coimmunoprecipitates were detected by Western blotting with anti-HA antibody as described above.
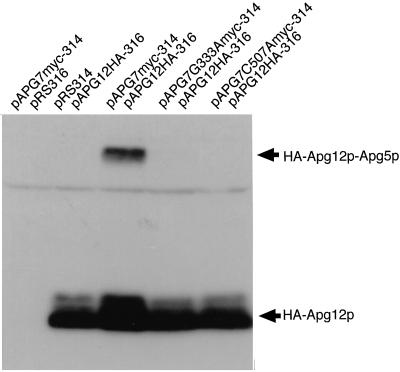
If Apg7p functions as an activating enzyme for Apg12p–Apg5p conjugation, the loss of conjugation of Apg7pG333A with Apg12p may result in a defect in the formation of the Apg12p–Apg5p conjugate. In wild-type cells expressing HA-tagged Apg12p, the HA-tagged Apg12p–Apg5p conjugate was recognized, whereas no conjugate was recognized in apg7Δ cells expressing HA-tagged Apg12p, as described previously (Figure 4, pAPG7myc-314/pAPG12HA-316 and pRS314/pAPG12HA-316) (Mizushima et al., 1998a). In cells expressing Apg7pG333A, very little conjugate was recognized (Figure 4, pAPG7G333Amyc-314/pAPG12HA-316). These results suggest the ATP-binding domain of Apg7p is essential for the formation of the Apg12p–Apg5p conjugate.
Figure 4.
Apg7pG333A and Apg7pC507A show significant defects in the formation of the Apg12p–Apg5p conjugate. The conjugate in cell lysates expressing HA-tagged Apg12p was detected by Western blotting using αHA antibody as described previously (Mizushima et al. 1998a). pAPG7myc-314/pRS316: apg7Δ cells expressing c-myc–tagged wild-type Apg7p only; pRS314/pAPG12HA-316: apg7Δ cells expressing HA-tagged Apg12p only; pAPG7myc-314/pAPG12HA-316: strain YIT702 cells; pAPG7G333Amyc-314/pAPG12HA-316: strain YIT7G333A cells; pAPG7C507Amyc-314/pAPG12HA-316: strain YIT7C507A cells.
Cys507 of Apg7p Is an Active Center Cysteine Essential for Its E1 Function
The active center cysteine (Cys600) of yeast Uba1p is essential for the function of a ubiquitin-activating enzyme. If Apg7p is a protein-activating enzyme for Apg12p, a Cys residue of Apg7p will be essential for Apg12p–Apg7p conjugation. The active center cysteine of Apg7p is strongly suggested to be Cys507, because the amino acid sequence of a neighboring region of Apg7p (MCTV) is identical to the corresponding regions in wheat UBA1 (MCTV; GenBank accession number [Ac.No.] P20973) and A. thaliana UBA1 (MCTV; GenBank Ac.No. U80808), and homologous to the corresponding regions in yeast Uba1p (LCTL; GenBank Ac.No. X55386), human UBA1 (ICTL; GenBank Ac.No. M58028), and mouse UBA1 (ICTL; GenBank Ac.No. D10576) (Hatfield et al., 1990, 1997; Hatfield and Vierstra, 1992; Handley et al., 1991; McGrath et al., 1991; Imai et al., 1992).
To investigate whether the Cys residue is essential for Apg7p function, we changed the Cys507 of Apg7p to Ala by site-directed mutagenesis and expressed both c-myc–tagged Apg7pC507A and HA-tagged Apg12p in the apg7Δ mutant with centromere-type plasmids (YIT7C507A strain) (Figure 3A). Immunoprecipitation using anti–c-myc antibody showed that no HA-tagged Apg12p was coimmunoprecipitated with c-myc–tagged Apg7pC507A (Figure 3C), although Apg7pC507A was stably expressed in the YIT7C507A strain (Figure 3B). We further examined the formation of the Apg12p–Apg5p conjugate. No HA-tagged Apg12p–Apg5p conjugate was recognized in the lysate of the YIT7C507A strain (Figure 4, pAPG7C507Amyc-314/pAPG12HA-316). These results indicate that Cys507 of Apg7p is an active site cysteine essential for the Apg12p-activating enzyme to form the Apg12p–Apg5p conjugate.
The ATP-binding Domain and Active Site Cysteine of Apg7p Are Essential for Autophagy
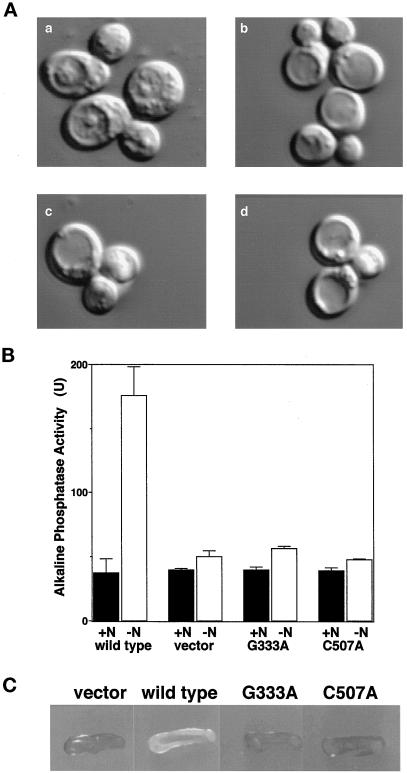
If the formation of the Apg12p–Apg5p conjugate via Apg7p is essential for autophagy in yeast, cells expressing Apg7pG333A and Apg7pC507A will show defects in autophagy. We first examined the accumulation of autophagic bodies in cells under starvation conditions in the presence of PMSF. Nomarski optics showed that no autophagic bodies accumulated in YIT7G333A and YIT7C507A cells cultured in nitrogen-starvation medium in the presence of PMSF similar to the result seen with apg7Δ cells (Figure 5A, b–d). In contrast, many autophagic bodies were seen in apg7Δ cells expressing wild-type Apg7p under the same conditions (Figure 5A, a).
Figure 5.
The ATP-binding domain and active site cysteine of Apg7p are essential for autophagy. (A) Gly333 and Cys507 in Apg7p are essential for the accumulation of autophagic bodies under nitrogen-starvation conditions. Cells grown to early-logarithmic phase in MVD+Ura medium were transferred to nitrogen-starvation medium in the presence of PMSF and incubated for 8 h at 30°C. Representative Nomarski images of the cells are shown. Autophagic bodies accumulated in vacuoles of apg7Δ cells carrying pAPG7myc-314 (a). Few autophagic bodies accumulated in vacuoles of apg7Δ cells carrying pRS314 (b), pAPG7G333Amyc-314 (c), and pAPG7C507Amyc-314 (d). (B) Autophagy in apg7Δ cells expressing Apg7pG333A and Apg7pC507A was monitored by an alkaline phosphatase processing assay. We constructed strain YTS2 (pho8::pho8Δ60 apg7::HIS3 trp1) and transformed it with plasmids pRS314 (vector), pAPG7myc-314 (wild type), pAPG7G333Amyc-314 (G333A), and pAPG7C507Amyc-314 (C507A). Cells growing logarithmically in MVD+Ade+Ura medium (N+) were transferred to nitrogen-starvation medium and incubated for 4 h at30°C (N−). Alkaline phosphatase activity in cells under rich or nitrogen-starvation conditions was measured as described by Noda et al. (1998). (C) Gly333 and Cys507 in Apg7p are essential for cell viability under nitrogen-starvation conditions. Cells were plated on nitrogen-starvation medium containing 10 μg/ml phloxine B and incubated at 30°C for 3 d. Nonviable cells were stained red (gray in monochrome), whereas viable cells were not stained (white in monochrome).
We next investigated Apg7pG333A and Apg7pC507A for defects in autophagy using a biochemical assay monitoring autophagy-dependent alkaline phosphatase processing (Noda and Ohsumi, 1998). The principle of this assay is as follows. A modified version of vacuolar alkaline phosphatase, Pho8Δ60p, remains in the cytosol as a proform (inactive form). When autophagy is enhanced, Pho8Δ60p in the cytosol is transferred to the vacuole and processed to the active form. This processing is monitored by assaying the activity of alkaline phosphatase in the cell lysate.
pAPG7G333Amyc-314 and pAPG7C507Amyc-314 were introduced into an apg7Δ tester strain (YTS2; apg7Δ pho8::pho8Δ60). Alkaline phosphatase activity was measured in the transformants under rich or nitrogen-starvation conditions. Under rich conditions, the alkaline phosphatase activities in cells expressing Apg7pG333A and Apg7pC507A were as low as that of wild-type Apg7p, indicating that autophagy was suppressed (Figure 5B, +N). When apg7Δ cells expressing wild-type Apg7p were incubated in nitrogen-starvation medium for 4 h at 30°C, the activity increased drastically (Figure 5B, −N), indicating that autophagy is induced by nitrogen starvation; however, the activities in apg7Δ cells expressing Apg7pG333A and Apg7pC507A were not enhanced, indicating that autophagy is suppressed in the mutant cells.
A defect in autophagy results in a loss of cell viability under nutrient starvation conditions (Tsukada and Ohsumi, 1993). We further examined the loss of viability of YIT7G333A and YIT7C507A strains using phloxine B. Phloxine B specifically stains dead cells. The colonies of YIT7G333A and YIT7C507A cells turned red (gray in monochrome) on nitrogen-starvation plates containing 10 μg/ml phloxine B, whereas the color of apg7Δ mutant colonies expressing wild-type Apg7p was pink (white in monochrome) (Figure 5C). The results indicate that the viability of YIT7G333A and YIT7C507A cells was markedly decreased under nitrogen-starvation conditions. From these results, we conclude that the ATP-binding domain and Cys507 are essential for the function of Apg7p in autophagy.
The ATP-binding Domain and Active Site Cysteine of Apg7p Are Essential for Cytoplasm-to-Vacuole Targeting of API
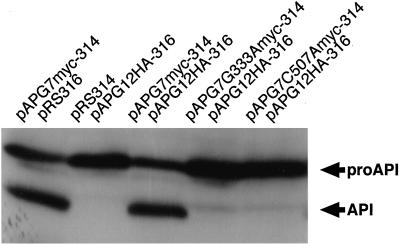
The apg7 mutant also has a defect in the cytoplasm-to-vacuole targeting of API. API is synthesized in the cytoplasm as a precursor form (Klionsky et al., 1992). After targeting to the vacuole, the precursor is processed to the mature form in the vacuole (Klionsky et al., 1992). We examined whether Gly333 and Cys507 in Apg7p are also essential for the targeting. We prepared lysates of cells in logarithmic phase and detected the precursor and mature forms of API by Western blotting using αAPI antibody. In wild-type cells, the mature form of API was detected in addition to the precursor, indicating that the precursor transferred from the cytoplasm to the vacuole to be processed into the mature form (Figure 6, pAPG7myc-314/pRS316 and pAPG7myc-314/pAPG12HA-316). In contrast, the processing of API was markedly decreased in cells expressing Apg7pG333A and completely inhibited in cells expressing Apg7pC507A, as in the case of apg7Δ cells (Figure 6, pAPG7G333Amyc-314/pAPG12HA-316, pAPG7C507Amyc-314/pAPG12HA-316, and pRS314/pAPG12HA-316). These results indicate that the ATP-binding domain and Cys507 of Apg7p are essential for the cytoplasm-to-vacuole targeting of API under rich conditions.
Figure 6.
The ATP-binding domain and active center cysteine of Apg7p are essential for cytoplasm-to-vacuole targeting of API. The processing of API in cells growing logarithmically was measured as described by Mizushima et al. (1998a). Lanes correspond to those in Figure 4.
Apg7p Is Present in Cytosol
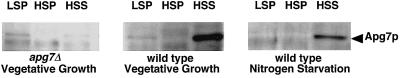
It has been shown that most free Apg5p, Apg5p–Apg12p conjugate, and more than half of Apg12p are present in the 100,000 × g pellet (Mizushima et al., 1998a). To determine the intracellular localization of Apg7p, a lysate of wild-type cells was fractionated by centrifugation as described previously (Huang and Chiang, 1997). Apg7p in each fraction was detected by Western blotting using αApg7C antibody. Apg7p in vegetatively growing cells was present mainly in the 100,000 × g supernatant (Figure 7, wild type, Vegetative Growth). Under nitrogen-starvation conditions, the amount and localization of Apg7p remained unchanged (Figure 7, wild type, Nitrogen Starvation). Similar results were obtained with the immunoprecipitation of fractions prepared from cells expressing c-myc–tagged Apg7p. These results indicate that Apg7p is mainly present in the cytoplasm.
Figure 7.
Apg7p is present in the cytosol. A wild-type strain (YW5–1B) was cultured in MVD+Ura+Trp medium, transferred to nitrogen-starvation medium, and incubated for 30 min. Cells growing vegetatively were converted to spheroplasts in rich buffer (1% yeast extract, 2% polypeptone, 0.5% glucose, and 0.7 M sorbitol), whereas nitrogen-starved cells were converted to spheroplasts in nitrogen-starvation buffer (0.15% yeast nitrogen base without amino acids and ammonium sulfate, 2% glucose, and 0.9 M sorbitol). Cells were lysed and fractionated by centrifugation as described by Huang and Chiang (1997). LSP, 15,000 × g pellet; HSP, 100,000 × g pellet; HSS, 100,000 × g supernatant.
DISCUSSION
In this article, we have characterized Apg7p as an Apg12p-activating enzyme and demonstrated its indispensable role in the yeast autophagy. Apg12p, a novel modifier protein, binds to Apg7p via a thioester bond. This binding requires both the active site cysteine (Cys507) and the ATP-binding domain. Accordingly, both the cysteine residue and the ATP-binding domain are essential for Apg12p–Apg5p conjugation and for autophagy as well. From these results, we conclude that Apg7p is a novel E1-like enzyme that activates Apg12p and is essential for autophagy.
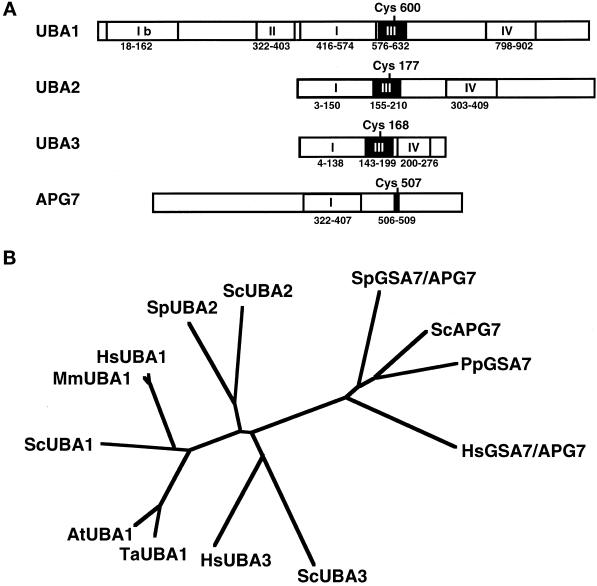
Although we clearly showed the E1-like function of Apg7p, only the C-terminal region of Apg7p (residues 322 to 407) shows homology to Uba1p. According to the similarity boxes within other E1-like enzymes as described by Johnson et al. (1997) and Liakopoulos et al. (1998), Apg7p has an ATP-binding domain within box I and an active site cysteine within box III that are essential for E1 function (Figure 8A); however, box III of Apg7p is not similar to those of Uba1p, Uba2p, or Uba3p. Also, neither similarity box II nor IV is present within Apg7p. Phylogenetic analysis of the regions containing box I and box III in the E1 enzymes also suggested that the relationship of Apg7p to the corresponding regions in UBA1, UBA2, and UBA3 family proteins is distant (Figure 8B) (Hatfield et al., 1990, 1997; Handley et al., 1991; McGrath et al., 1991; Imai et al., 1992; Dohmen et al., 1995; Liakopoulos et al., 1998; Mizushima et al., 1998a; Osaka et al., 1998; Yuan et al., 1998). The comparison of the similarity boxes and phylogenetic analysis showed that Apg7p is distinct from other E1-like enzymes. It is of interest to know whether Apg7p functions as a heterodimer, because the Aos1p/Uba2p and Ula1p/Uba3p complexes function as heterodimers (del Olmo et al., 1997; Johnson et al., 1997; Liakopoulos et al., 1998). At present, we have no answer.
Figure 8.
Apg7p is distantly related to Uba1p and its relatives. (A) Schematic representation of the similarity domains among UBA1, UBA2, UBA3, and APG7. The putative active site cysteines are located within similarity box III (UBA domain), as indicated. The essential ATP-binding domains are located within similarity box I. UBA1 contains another ATP-binding motif within similarity box Ib, which is nonessential for function. The similarity boxes correspond to those of Johnson et al. (1997) and Liakopoulos et al. (1998), except box Ib. (B) The phylogenetic tree of E1 enzymes. The homologous regions of E1-like enzymes containing the ATP-binding domain and active site cysteine were multi-aligned and analyzed by a CLUSTAL W multiple alignment program (Thompson et al., 1994). Shown are ScAPG7 (residues 321 to 520 of S. cerevisiae Apg7p; PID: g731742), PpAPG7 (residues 322 to 521 of P. pastoris Gsa7p), HsAPG7/GSA7 (residues 235 to 434 of Homo sapiens APG′7/GSA7), SpAPG7/GSA7 (residues 332 to 531 of Schizosaccharomyces pombe Apg7p/Gsa7p; PID g2894280), HsUBA1 (residues 465 to 664 of H. sapiens UBA1; PID: g340072), MmUBA1 (residues 465 to 664 of Mus musculus UBA1; PID: g220629), ScUBA1 (residues 431 to 630 of S. cerevisiae Uba1p; PID: g4715), AtUBA1 (residues 489 to 688 of A. thaliana UBA1; PID: g1750376), TaUBA1 (residues 459 to 658 of Triticum aestivum UBA1; PID g136632), ScUBA2 (residues 18 to 219 of S. cerevisiae Uba2p; PID: g1717852), SpUBA2 (residues 22 to 221 of S. pombe Uba2p; PID: g2956755), HsUBA3 (residues 45 to 244 of H. sapiens UBA3; PID: g3342564), and ScUBA3 (residues 1 to 200 of S. cerevisiae Uba3p; PID: g2980755) (Hatfield et al., 1990; Handley et al., 1991; McGrath et al., 1991; Imai et al., 1992; Dohmen et al., 1995; Hatfield et al., 1997; Liakopoulos et al., 1998; Mizushima et al., 1998a; Osaka et al., 1998, Yuan et al., 1999).
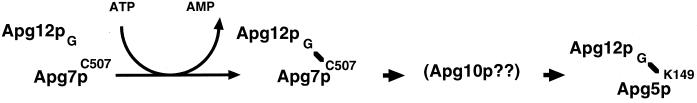
A working hypothesis of the Apg12p conjugation system consistent with these results is proposed in Figure 9. The C-terminal Gly of Apg12p is activated by Apg7p in an ATP-dependent manner. The Apg12p–Apg7p conjugate forms via the C-terminal Gly of Apg12p and Cys507 of Apg7p. The reaction probably occurs in the cytosol or the cytoplasmic side of an Apg12p-associated compartment(s), because Apg7p is mainly present in the cytoplasm (Figure 7) (Kim et al., 1999). By analogy to ubiquitination and related modifications, there may be novel E2 and E3 enzymes for the conjugation system. One such candidate for E2 is Apg10p, because the Apg12p–Apg5p conjugate is not observed in an apg10 mutant. Cloning of the APG10 gene and biochemical analysis will reveal the function of Apg10p. Screening other candidates will be performed using an Apg12p-affinity column, an Apg7p-affinity column, or two-hybrid screening.
Figure 9.
Working hypothesis for the function of Apg7p in the Apg12p conjugation system. Apg12p associates with Apg7p. The C-terminal Gly of Apg12p conjugates with Cys507 in Apg7p via a thioester bond in an ATP-dependent manner. The reaction occurs in the cytoplasm or on the cytoplasmic side of an Apg12p-associated compartment(s). Although the E2 enzyme has not been identified, one candidate is Apg10p.
It remains to be determined at which step in autophagy the Apg12p conjugation system works. Apg7p is mainly present in the cytosol, whereas more than half of Apg12p is localized in membrane compartment(s) or a large complex. The Apg12p–Apg7p binding probably occurs in the cytosol or the cytoplasmic side of an Apg12p-associated structure(s). Recently, Yuan et al. (1999) suggested that a quite similar conjugation system is required for microautophagy in Pichia pastoris. Microautophagy is the sequestration of cytoplasmic components (peroxisomes in this case) by an invagination of the vacuolar membrane. In a conjugation-deficient mutant (gsa7), the sequestration is not accomplished completely, probably because of defects in the membrane fusion step. Gsa7p, a P. pastoris homologue of Apg7p, was shown to be conjugated to a small protein through a thioester bond. Although a modifier and substrate(s) have not been identified, it strongly suggests that the Apg12p conjugation system is essential for microautophagy. Because the process of microautophagy is quite different from that of macroautophagy, it would be interesting if similar machinery is involved in both processes.
We have identified a human Apg12p homologue and have observed that the Apg12p homologue also conjugates with a human Apg5p homologue, which has been identified as an apoptosis-specific protein (Hammond et al. 1998; Mizushima et al. 1998b). A BLAST search of Apg7p in the Expressed Sequence Tag database showed potential mammalian homologues of Apg7p. Recently, an Apg7p homologue in P. pastoris, Gsa7p, and a human Gsa7p/Apg7p homologue were identified by Yuan et al. (1999). The human Gsa7p/Apg7p homologue is expressed in various tissues as revealed by Northern analysis (Tanida and Kominami, unpublished observations). These findings suggest that the Apg12p conjugation system generally functions in eukaryotes. By analogy to the Apg5p homologue, an Apg7p homologue may play a significant role in the autophagic and apoptotic pathways in mammalian cells. Cloning and biochemical analyses of Apg7p homologues will reveal the function of the novel protein conjugation system in mammalian cells.
Apg7p is distantly related to UBA1, UBA2, and UBA3 family proteins (Figure 8A). Nevertheless, Apg7p functions as an E1 enzyme for Apg12p–Apg5p conjugation. These findings suggest that the ubiquitin-like protein modification system will form a ubiquitous regulatory system in eukaryotes, not specific to ubiquitin and ubiquitin-like proteins. Further analyses of potential E1 enzymes may reveal a novel regulatory system by posttranslational modification.
ACKNOWLEDGMENTS
We thank D.J. Klionsky (University of California Davis) for providing information, helpful discussion, and antibody, W.A. Dunn, Jr. (University of Florida) for providing information, A. Ogiwara (National Institute for Basic Biology) for analyzing sequence data, Y. Ohya (University of Tokyo), P. James (University of Wisconsin), and P. Hieter (Johns Hopkins University) for plasmids and strains, K. Ishidoh, J. Ezaki, D. Muno (Juntendo University), and members of Y. Ohsumi’s laboratories for helpful discussions. This work was supported in part by grants-in-aid 09680629 (to T.U.) for Scientific Research, grants-in-aid 08278103 (to E.K.) for Scientific Research on Priority areas from the Ministry of Education, Science, Sports, and Culture of Japan, and The Science Research Promotion Fund from the Japan Private School Promotion Foundation (to E.K.).
Abbreviations used:
- Ac.No.
accession number
- API
aminopeptidase I
- GAL4AD
GAL4 activation domain
- GAL4BD
GAL4 DNA binding domain
- HA
hemagglutinin
REFERENCES
- Ausubel F, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Short Protocols in Molecular Biology. 3rd ed. New York: John Wiley & Sons; 1995. [Google Scholar]
- Baba M, Ohsumi M, Ohsumi Y. Analysis of the membrane structures involved in autophagy in yeast by freeze-replica method. Cell Struct Funct. 1995;20:465–471. doi: 10.1247/csf.20.465. [DOI] [PubMed] [Google Scholar]
- Baba M, Takeshige K, Baba N, Ohsumi Y. Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J Cell Biol. 1994;124:903–913. doi: 10.1083/jcb.124.6.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel B, Wunning I, Varshavsky A. The recognition component of the N-end rule pathway. EMBO J. 1990;9:3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy MN, Howe K, Etkin LD, Solomon E, Freemont PS. PIC1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukemia. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- Callis J, Carpenter T, Sun C-W, Vierstra RD. Structure and evolution of genes encoding polyubiquitin and ubiquitin-like proteins in Arabidopsis thaliana ecotype Columbia. Genetics. 1995;139:921–939. doi: 10.1093/genetics/139.2.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow N, Korenberg JR, Chen XN, Neve RL. APP-BP1, a novel protein that binds to the carboxyl-terminal region of the amyloid precursor protein. J Biol Chem. 1996;271:11339–11346. doi: 10.1074/jbc.271.19.11339. [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Elias S, Heller H, Hershko A. “Covalent affinity” purification of ubiquitin-activating enzyme. J Biol Chem. 1982;257:2537–2542. [PubMed] [Google Scholar]
- del Olmo M, Mizrahi N, Gross S, Moore CL. The Uba2 and Ufd1 proteins of Saccharomyces cerevisiae interact with poly(A) polymerase and affect the polyadenylation activity of cell extracts. Mol Gen Genet. 1997;255:209–218. doi: 10.1007/s004380050491. [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Timpte C, Tan S, Callis J, Estelle M. The ubiquitin-related protein RUB1 and auxin response in Alabidopsis. Science. 1998;280:1760–1763. doi: 10.1126/science.280.5370.1760. [DOI] [PubMed] [Google Scholar]
- Dohmen RJ, Stappen R, McGrath JP, Forrova H, Kolarov J, Goffeau A, Varshavsky A. An essential yeast gene encoding a homolog of ubiquitin-activating enzyme. J Biol Chem. 1995;270:18099–18109. doi: 10.1074/jbc.270.30.18099. [DOI] [PubMed] [Google Scholar]
- Dolan L. Pointing roots in the right direction: the role of auxin transport in response to gravity. Genes Dev. 1998;12:2091–2095. doi: 10.1101/gad.12.14.2091. [DOI] [PubMed] [Google Scholar]
- Dunn WA., Jr Autophagy and related mechanism of lysosome-mediated protein degradation. Trends Cell Biol. 1994;4:139–143. doi: 10.1016/0962-8924(94)90069-8. [DOI] [PubMed] [Google Scholar]
- Finley D, Chau V. Ubiquitination. Annu Rev Cell Biol. 1991;7:25–69. doi: 10.1146/annurev.cb.07.110191.000325. [DOI] [PubMed] [Google Scholar]
- Funakoshi T, Matsuura A, Noda T, Ohsumi Y. Analyses of APG13 gene involved in autophagy in yeast: Saccharomyces cerevisiae. Gene. 1997;192:207–213. doi: 10.1016/s0378-1119(97)00031-0. [DOI] [PubMed] [Google Scholar]
- Gong L, Kamitani T, Fujise K, Caskey LS, Yeh TH. Preferential interaction of sentrin with a ubiquitin-conjugating enzyme, Ubc9. J Biol Chem. 1997;272:28198–28201. doi: 10.1074/jbc.272.45.28198. [DOI] [PubMed] [Google Scholar]
- Haas AL, Siepmann TJ. Pathways of ubiquitin conjugation. FASEB J. 1997;11:1257–1268. doi: 10.1096/fasebj.11.14.9409544. [DOI] [PubMed] [Google Scholar]
- Haas AL, Warms JVB, Hershko A, Rose I. Ubiquitin-activating enzyme. J Biol Chem. 1982;257:2543–2548. [PubMed] [Google Scholar]
- Hammond EM, Brunet CL, Johnson GD, Parkhill J, Milner AE, Brady G, Gregory CD, Grand RJ. Homology between a human apoptosis specific protein and the product of APG5, a gene involved in autophagy in yeast. FEBS Lett. 1998;425:391–395. doi: 10.1016/s0014-5793(98)00266-x. [DOI] [PubMed] [Google Scholar]
- Handley PM, Mueckler M, Siegel NR, Ciechanover A, Schwartz AL. Molecular cloning, sequence, and tissue distribution of the human ubiquitin-activating enzyme E1. Proc Natl Acad Sci USA. 1991;88:258–262. doi: 10.1073/pnas.88.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding TM, Hefner-Gravink A, Thumm M, Klionsky DJ. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein targeting pathway. J Biol Chem. 1996;271:17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- Hatfield PM, Callis J, Vierstra RD. Cloning of ubiquitin activating enzyme from wheat and expression of a functional protein in Escherichia coli. J Biol Chem. 1990;265:15813–15817. [PubMed] [Google Scholar]
- Hatfield PM, Gosink MM, Carpenter TB, Vierstra RD. The ubiquitin-activating enzyme (E1) gene family in Arabidopsis thaliana. Plant J. 1997;11:213–226. doi: 10.1046/j.1365-313x.1997.11020213.x. [DOI] [PubMed] [Google Scholar]
- Hatfield PM, Vierstra RD. Multiple forms of ubiquitin-activating enzyme E1 from wheat. Identification of an essential cysteine by in vitro mutagenesis. J Biol Chem. 1992;267:14799–14803. [PubMed] [Google Scholar]
- Hershko A. Lessons from the discovery of the ubiquitin system. Trends Biochem Sci. 1996;21:445–449. doi: 10.1016/s0968-0004(96)10054-2. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983;258:8206–8214. [PubMed] [Google Scholar]
- Hochstrasser M. Protein degradation or regulation: Ub the judge. Cell. 1996a;84:813–815. doi: 10.1016/s0092-8674(00)81058-2. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996b;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. There’s the Rub: a novel ubiquitin-like modification linked to cell cycle regulation. Genes Dev. 1998;12:901–907. doi: 10.1101/gad.12.7.901. [DOI] [PubMed] [Google Scholar]
- Homma K, Terui S, Minemura M, Qadota H, Anraku Y, Kanaho Y, Ohya Y. Phosphatidylinositol-4-phosphate 5-kinase localized on the plasma membrane is essential for yeast cell morphogenesis. J Biol Chem. 1998;273:15779–15786. doi: 10.1074/jbc.273.25.15779. [DOI] [PubMed] [Google Scholar]
- Huang P-H, Chiang H-L. Identification of novel vesicles in the cytosol to vacuole protein degradation pathway. J Cell Biol. 1997;136:803–810. doi: 10.1083/jcb.136.4.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai N, Kaneda S, Nagai Y, Seno T, Ayusawa D, Hanaoka F, Yamao F. Cloning and sequence of a functionally active cDNA encoding the mouse ubiquitin-activating enzyme E1. Gene. 1992;118:279–282. doi: 10.1016/0378-1119(92)90200-9. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designated for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES, Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Schwienhorst I, Dohmen RJ, Blobel G. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 1997;16:5509–5519. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PR, Hochstrasser M. SUMO-1: ubiquitin gains weight. Trends Cell Biol. 1997;7:408–413. doi: 10.1016/S0962-8924(97)01132-X. [DOI] [PubMed] [Google Scholar]
- Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1994. [Google Scholar]
- Kametaka S, Matsuura A, Wada Y, Ohsumi Y. Structural and functional analyses of APG5, a gene involved in autophagy in yeast. Gene. 1996;178:139–143. doi: 10.1016/0378-1119(96)00354-x. [DOI] [PubMed] [Google Scholar]
- Kametaka S, Okano T, Ohsumi M, Ohsumi Y. Apg14p and Apg6p/Vps30p form a protein complex essential for autophagy in the yeast Saccharomyces cerevisiae. J Biol Chem. 1998;273:22284–22291. doi: 10.1074/jbc.273.35.22284. [DOI] [PubMed] [Google Scholar]
- Kamitani T, Kito K, Nguyen HP, Yeh ET. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem. 1997;272:28552–28562. doi: 10.1074/jbc.272.45.28557. [DOI] [PubMed] [Google Scholar]
- Kim J, Dalton VM, Eggerton KP, Scott SV, Klionsky DJ. Mol. Biol. Cell. 1337–1351. 1999. Apg7p, a ubiquitin E1-like activating enzyme, is an essential, shared component of Cvt transport, autophagy, and the peroxisomal degradation pathway. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Cueva R, Yaver DS. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Yoshida Y, Noda M. Cloning of a cDNA which encodes a novel ubiquitin-like protein. Biochem Biophys Res Commun. 1993;195:393–399. doi: 10.1006/bbrc.1993.2056. [DOI] [PubMed] [Google Scholar]
- Lammer D, Mathias N, Laplaza JM, Jiang W, Liu Y, Callis J, Goebl M, Estelle M. Modification of yeast Cdc53p by the ubiquitin-related protein Rub1p affects function of the SCFCdc4 complex. Genes Dev. 1998;7:914–926. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang T, Schaeffeler E, Bernreuther D, Bredschneider M, Wolf DH, Thumm M. Aut2p and Aut7p, two novel microtubule-associated proteins are essential for delivery of autophagic vesicles to vacuole. EMBO J. 1998;17:3597–3607. doi: 10.1093/emboj/17.13.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GW, Melchior F, Matunis MJ, Mahajan R, Tian Q, Anderson P. Modification of Ran GTPase-activating protein by small ubiquitin-related modifier SUMO-1 requires Ubc9, an E2-type ubiquitin-conjugating enzyme homologue. J Biol Chem. 1998;273:6503–6507. doi: 10.1074/jbc.273.11.6503. [DOI] [PubMed] [Google Scholar]
- Leyser D, Lincoln CA, Timpte C, Lammer D, Turner J, Estelle M. Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature. 1993;364:161–164. doi: 10.1038/364161a0. [DOI] [PubMed] [Google Scholar]
- Liakopoulos D, Doenges G, Matuschewski K, Jentsch S. A novel protein modification pathway related to the ubiquitin system. EMBO J. 1998;17:2208–2214. doi: 10.1093/emboj/17.8.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Muir RS, Lim E, McElver J, Weber SC, Heitman J. Gene disruption with PCR products in Saccharomyces cerevisiae. Gene. 1995;158:113–117. doi: 10.1016/0378-1119(95)00144-u. [DOI] [PubMed] [Google Scholar]
- Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- Mahajan R, Gerace L, Melchior F. Molecular characterization of the SUMO-1 modification of RanGAP1 and its role in nuclear envelope association. J Cell Biol. 1998;140:259–270. doi: 10.1083/jcb.140.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- Matunis MJ, Blobel G. SUMO-1 modification and its role in targeting the RanGTPase-activating protein, RanGAP1, to the nuclear pore complex. J Cell Biol. 1998;140:499–509. doi: 10.1083/jcb.140.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath JP, Jentsch S, Varshavsky A. UBA1: an essential yeast gene encoding ubiquitin-activating enzyme. EMBO J. 1991;10:227–236. doi: 10.1002/j.1460-2075.1991.tb07940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A novel protein conjugation system essential for autophagy. Nature. 1998a;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J Biol Chem. 1998b;273:33889–33892. doi: 10.1074/jbc.273.51.33889. [DOI] [PubMed] [Google Scholar]
- Muller S, Matunis MJ, Dejean A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- Okura T, Gong L, Kamitani T, Wada T, Okura I, Wei CF, Chang HM, Yeh ET. Protection against Fas/Apo-1- and tumor necrosis factor mediated cell death by a novel protein, sentrin. J Immunol. 1996;157:4277–4281. [PubMed] [Google Scholar]
- Osaka F, Kawasaki H, Aida N, Saeki M, Chiba T, Kawashima M, Tanaka K, Kato S. A new NEDD8-ligating system for cullin-4A. Genes Dev. 1998;12:2263–2268. doi: 10.1101/gad.12.15.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, King RW, Hoog C, Kirschner MW. Identification of BIME as a subunit of the anaphase-promoting complex. Science. 1996;274:1199–1201. doi: 10.1126/science.274.5290.1199. [DOI] [PubMed] [Google Scholar]
- Saitoh H, Pu RT, Dasso M. SUMO-1: wrestling with a new ubiquitin-related modifier. Trends Biochem Sci. 1997;22:374–376. doi: 10.1016/s0968-0004(97)01102-x. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Nuber U, Huibregtse JM. Protein ubiquitination involving an E1–E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- Schlumpberger M, Schaeffeler E, Straub M, Bredschneider M, Wolf DH, Thumm M. AUT1, a gene essential for autophagocytosis in the yeast Saccharomyces cerevisiae. J Bacteriol. 1997;179:1068–1076. doi: 10.1128/jb.179.4.1068-1076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz SE, Matuschewski K, Liakopoulos D, Scheffner M, Jentsch S. The ubiquitin-like proteins SMT3 and SUMO-1 are conjugated by the UBC9 E2 enzyme. Proc Natl Acad Sci USA. 1998;95:560–564. doi: 10.1073/pnas.95.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Hefner-Gravink A, Morano KA, Noda T, Ohsumi Y, Klionsky DJ. Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc Natl Acad Sci USA. 1996;93:12304–12308. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seglen PO, Bohley P. Autophagy and other vacuolar protein degradation mechanism. Experientia. 1992;48:158–172. doi: 10.1007/BF01923509. [DOI] [PubMed] [Google Scholar]
- Shen Z, Pardington-Purtymun PE, Comeaux JC, Moyzis RK, Chen DJ. Associations of UBE21 with RAD52, UBL1, p53 and RAD51 proteins in a yeast two-hybrid system. Genomics. 1996;37:183–186. doi: 10.1006/geno.1996.0540. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub M, Bredschneider M, Thumm M. AUT3, a serine/threonine kinase gene, is essential for autophagocytosis in Saccharomyces cerevisiae. J Bacteriol. 1997;197:3875–3883. doi: 10.1128/jb.179.12.3875-3883.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119:301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, Wolf DH. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. The ubiquitin system. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- Wierenga RK, Hol WGJ. Predicted nucleotide-binding properties of p21 protein and its cancer-associated variant. Nature. 1983;302:842–844. doi: 10.1038/302842a0. [DOI] [PubMed] [Google Scholar]
- Yuan W, Stromhaug PE, Dunn WA., Jr Glucose-induced microautophagy of peroxisomes in Pichia pastoris requires a unique E1-like protein. Mol Biol Cell. 1999;10:000–000. doi: 10.1091/mbc.10.5.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae W, Shin TH, Galova M, Obermaier B, Nasmyth K. Identification of subunits of the anaphase-promoting complex of Saccharomyces cerevisiae. Science. 1996;274:1201–1204. doi: 10.1126/science.274.5290.1201. [DOI] [PubMed] [Google Scholar]