Abstract
Background
Leukemia evolves through a multistep process from premalignancy to malignancy. Epigenetic alterations, including histone modifications, have been proposed to play an important role in tumorigenesis. The involvement of two chromatin remodeling genes, retinoblastoma-binding protein 1 (Rbbp1/Arid4a) and Rbbp1-like 1 (Rbbp1l1/Arid4b), in leukemogenesis was not characterized.
Methods
The leukemic phenotype of mice deficient for Arid4a with or without haploinsufficiency for Arid4b was investigated by serially monitoring complete blood counts together with microscopic histologic analysis and flow cytometric analysis of bone marrow and spleen from the Arid4a−/− mice or Arid4a−/−Arid4b+/− mice. Regulation in bone marrow cells of downstream genes important for normal hematopoiesis was analyzed by reverse transcription–polymerase chain reaction. Genotypic effects on histone modifications were examined by western blotting and immunofluorescence analysis. All statistical tests were two-sided.
Results
Young (2–5 months old) Arid4a-deficient mice had ineffective blood cell production in all hematopoietic lineages. Beyond 5 months of age, the Arid4a−/− mice manifested monocytosis, accompanied by severe anemia and thrombocytopenia. These sick Arid4a−/− mice showed bone marrow failure with myelofibrosis associated with splenomegaly and hepatomegaly. Five of 42 Arid4a−/− mice and 10 of 12 Arid4a−/−Arid4b+/− mice progressed to acute myeloid leukemia (AML) and had rapid further increases of leukocyte counts. Expression of Hox genes (Hoxb3, Hoxb5, Hoxb6, and Hoxb8) was decreased in Arid4a-deficient bone marrow cells with or without Arid4b haploinsufficiency, and FoxP3 expression was reduced in Arid4a−/−Arid4b+/− bone marrow. Increases of histone trimethylation of H3K4, H3K9, and H4K20 (fold increases in trimethylation = 32, 95% confidence interval [CI] = 27 to 32; 45, 95% CI = 41 to 49; and 2.2, 95% CI = 1.7 to 2.7, respectively) were observed in the bone marrow of Arid4a-deficient mice.
Conclusions
Arid4a-deficient mice initially display ineffective hematopoiesis, followed by transition to chronic myelomonocytic leukemia (CMML)–like myelodysplastic/myeloproliferative disorder, and then transformation to AML. The disease processes in the Arid4a-deficient mice are very similar to the course of events in humans with CMML and AML. This mouse model has the potential to furnish additional insights into the role of epigenetic alterations in leukemogenesis, and it may be useful in developing novel pharmacological approaches to treatment of preleukemic and leukemic states.
CONTEXT AND CAVEATS
Prior knowledge
Epigenetic modifications of DNA have been proposed to play an important role in carcinogenesis. The involvement of two chromatin remodeling genes, Arid4a and Arid4b, in the etiology of leukemia was unknown.
Study design
Mice deleted for Arid4a or mice deleted for Arid4a and haploinsufficient for Arid4b were characterized in terms of blood counts, bone marrow abnormalities as revealed by histologic analysis, gene transcription, and epigenetic modification in their bone marrow cells.
Contribution
The fact that the genetically modified mice progressed to acute myeloid leukemia suggests a critical role for epigenetic modifications mediated by Arid4a and Arid4a in suppression of leukemogenesis.
Implications
The mouse model described here may be useful for studying the etiology of leukemia and the role that epigenetic modification plays in this process.
Limitations
The suitability of the mouse strains described here as a model for human leukemia and the precise roles of Arid4a and Arid4b in chromatin remodeling remain to be determined.
From the Editors
The human ARID4A and ARID4B genes, previously known as retinoblastoma-binding protein 1 (RBBP1, RBP1) (1,2) and RBBP1–like protein 1 (RBBP1L1) (3), respectively, are homologous members of the AT-rich interaction domain (ARID) gene family (4). The ARID4A amino acid sequence, but not that of ARID4B, contains an LVCHE sequence as a conserved LXCXE motif known to interact with the pocket of retinoblastoma protein (RB) (1,2). We reported that ARID4A interacts with ARID4B (5). Both ARID4A and ARID4B contain a Tudor domain, an ARID domain, and a chromodomain (4), as well as two repression domains, R1 and R2, with part of the R1 domain overlapping the ARID domain (6). The ARID domain contains a helix-turn-helix structure with DNA binding activity (7). Chromodomains are found in a variety of proteins that play roles in the functional organization of chromosome structure through chromatin remodeling (8–10). Both chromodomains and Tudor domains mediate binding to methylated lysines of histones H3 and H4 (11–14). ARID4A recruited by RB has been defined as a repressor of E2F-dependent transcription (15). Through their R2 regions, ARID4A and ARID4B serve as adapters to recruit the mSin3A–histone deacetylase (HDAC) histone-modifying complex to E2F-dependent promoters (6,16,17). ARID4A and ARID4B also provide a repressive function in an HDAC-independent manner through their R1 regions. Transcriptional repression activity via the R1 region is controlled by SUMOylation, a posttranslational modification by small ubiquitin-related modifier proteins (18).
Several lines of evidence suggest that ARID4A and ARID4B may be involved in the pathogenesis of breast and other cancers. RB (with which ARID4A interacts and which is inactivated in many tumors) has been shown to be involved in many cellular processes, including control of the cell cycle, cell differentiation, DNA-damage responses, DNA replication, and protection against apoptosis (19). Repression of E2F-dependent transcription by ARID4A and ARID4B in conjunction with RB leads to cell arrest reminiscent of senescence (15,18). Screening a cDNA library from the MCF7 breast cancer cell line with IgG purified from the serum of a breast cancer patient identified ARID4A and ARID4B as tumor-associated antigens, and breast cancer patients have high titers of antibodies against both proteins (3,20,21). Furthermore, human cytotoxic T cells stimulated with ARID4A peptides kill breast cancer cells (22). ARID4A also interacts with the breast cancer metastasis suppressor 1 (BRMS1) and the BRMS1 homologue p40 in the mSin3–HDAC complex (23,24). BRMS1 reduces the metastatic activity of cancer cells without affecting tumorigenicity (25). Despite these reports, the role of ARID4A and ARID4B in cancer development is unclear.
Preleukemic conditions are relatively common in aging populations, and little is known about their pathogenesis or how they predispose to leukemia. Many physicians and patients face this distressing clinical situation on a chronic basis, and there is little that the physician can offer to prevent the potential transformation to malignancy because the mechanisms by which cancer progresses from premalignancy to malignancy are not fully understood.
Epigenetic alterations such as histone modifications of chromatin structure have been suggested to be involved in the development of leukemia and other cancers (26). Although an increasing number of chromatin remodeling proteins have been defined biochemically (27), their biological functions, particularly in the context of an animal model, are poorly characterized, and the molecular events governing chromatin reorganization in cancer cells remain relatively unexplored. Recently, we developed mouse models for Arid4a and Arid4b deficiency that demonstrated the function of Arid4a and Arid4b in the regulation of genomic imprinting through control of epigenetic modifications (5). Here, we use these mouse models deficient for Arid4a, alone or in combination with haploinsufficiency for Arid4b, to analyze a premalignant hematopoietic disorder and eventual leukemia.
Methods
Animal Care
The mice (100 wild-type mice, 150 Arid4a−/− mice, 40 Arid4a+/−Arid4b+/− mice, and 40 Arid4a−/−Arid4b+/− mice) were all bred and maintained according to a protocol approved by the Baylor College of Medicine Animal Care and Use Committee at the institution's specific pathogen–free mouse facility, which is approved by the American Association for Accreditation of Laboratory Animal Care and is operated in accordance with current regulations and standards of the US Departments of Agriculture and of Health and Human Services.
Beginning at 5 months of age, the Arid4a−/− mice and Arid4a−/−Arid4b+/− mice were monitored weekly for signs of morbidity. Blood was obtained weekly for complete blood cell counts. Mice were sacrificed when they met any of the following criteria: 1) obvious morbidity, in which case mice proved to have either severe anemia or greatly increased white blood cell (WBC) counts; 2) severe anemia or high WBC counts with milder or impending morbidity; or 3) marked splenomegaly or hepatomegaly. Mice were anesthetized with isoflurane and then killed by cervical dislocation. The mice were dissected, and organs were examined for the presence or absence of tumor enlargement. The lung, spleen, and liver were removed, and bone marrow cells from both femurs were flushed with syringes.
Hematologic Analysis
Mice (52 wild-type mice, 50 Arid4a−/− mice, 40 Arid4a+/−Arid4b+/− mice, and 10 Arid4a−/−Arid4b+/− mice) were anesthetized with isoflurane, and blood from each mouse was obtained from the retro-orbital venous plexus. Complete blood cell counts were determined with an analyzer (ADVIA 120 Hematology System; Bayer Diagnostics).
Peripheral blood and bone marrow smears were stained with Wright–Giemsa stain. On the peripheral blood smears (from 10 wild-type mice, more than 20 chronic myelomonocytic leukemia [CMML]–like Arid4a−/− mice, three acute myeloid leukemia [AML] Arid4a−/− mice, and five AML Arid4a−/−Arid4b+/− mice) and bone marrow smears (from five wild-type mice, five CMML-like Arid4a−/− mice, two AML Arid4a−/− mice, and three AML Arid4a−/−Arid4b+/− mice), we analyzed blasts, immature precursors, WBCs (including lymphocytes, neutrophils, monocytes, eosinophils, and basophils), red blood cells (RBCs), and platelets.
Histologic Analysis
For bone marrow examination, femurs from three wild-type mice and three Arid4a−/− mice were fixed and decalcified by immersion in Cal-EXII solution (Fisher Scientific, Pittsburgh, PA). Spleen and liver from five wild-type mice, five CMML-like mice, and five AML mice were fixed in 10% formalin (Fisher Scientific). Histology was performed on 5-μm paraffin-embedded tissue sections that were stained either with reticulin to show myelofibrosis for femur bone sections (three slides from three wild-type mice and three slides from three Arid4a−/− mice) or with hematoxylin and eosin to show extramedullary hematopoiesis in spleen and liver (one slide each from five wild-type mice, five CMML-like Arid4a−/− mice, and five AML Arid4a−/−Arid4b+/− mice).
Flow Cytometry
Spleens were removed from mice and dissociated into single cells by sliding tissue between two superfrost microscope slides. RBCs were lysed by hypotonic buffer (NH4Cl, 0.14 M; Tris, 0.017 M, pH 7.2). Cells from spleen or bone marrow were gently filtered through 70-μm cell strainers (BD Falcon, VWR, Batavia, IL) and were stained with directly fluorescein-5-isothiocyanate (FitC)-conjugated antibodies to c-kit (2B8, eBioscience, San Diego, CA), Sca1 (D7, eBioscience), Mac1 (M1/70, BD Biosciences Pharmingen, San Jose, CA), Gr1 (RB6-8C5, BD Biosciences Pharmingen), B220 (RA3-6B2, BD Biosciences Pharmingen), CD19 (1D3, BD Biosciences Pharmingen), CD3 (145-2C11, BD Biosciences Pharmingen), or Ter119 (Ter119, eBioscience). A lineage antibody cocktail (Lin) included antibodies against Mac1 (M1/70), Gr1 (RB6-8C5), CD4 (L3T4, BD Biosciences Pharmingen), CD8 (53-6.7, BD Biosciences Pharmingen), CD19 (1D3), and B220 (RA3-6B2) to stain bone marrow cells. All antibodies were diluted (1:100) in phosphate-buffered saline (PBS). Hematopoietic stem cells (HSCs) (Lin−Sca1+c-Kit+), common myeloid progenitors (CMPs) (Lin−Sca1−c-Kit+), granulocytes and monocytes (Gr1+Mac1+), erythroid cells (Ter119+), T cells (CD3+), and B cells (B220+CD19+) were analyzed. For apoptosis analysis, bone marrow cells were stained with annexin V–FITC conjugates (BD Biosciences Pharmingen). Cells were then washed and analyzed by flow cytometry (Beckman-Coulter EPICS XL-MCL).
Immunofluorescence
Bone marrow cells flushed from femur bones were fixed in 4% paraformaldehyde, spread on glass slides, and permeabilized in cold acetone. Subsequently, cells were blocked with 5% bovine serum albumin in PBS, followed by incubation with primary antibodies against trimethylated H3K9, H3K4, or H4K20 (07-442 for H3K9me3, 05-745 for H3K4me3, and 07-463 for H4K20me3, Upstate, Charlottesville, VA) at a dilution of 1:200 in blocking solution. Then, cells were washed with blocking solution and incubated with Alexa 488–conjugated goat anti–rabbit secondary antibody (Molecular Probes, Invitrogen, Carlsbad, CA). Cells were mounted with Vectashield containing 4’,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA) and analyzed on a deconvolution fluorescence microscope (DeltaVision Restoration Microscope, Zeiss, Jena, Germany).
Western Blotting
Bone marrow cells flushed from femurs were resuspended in lysis buffer (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, and protease inhibitors [Complete protease inhibitor cocktail tablets, Roche, Indianapolis, IN]). Histones were acid extracted by 0.2 N HCl and precipitated with 20% trichloroacetic acid. Proteins were electrophoresed on 7.5% Tris–Cl ready gels (Bio-Rad, Hercules, CA) and then transferred to nitrocellulose membranes (Bio-Rad). The incubations with the appropriate primary antibodies were performed as follows: rabbit anti-H3 (1:1000 dilution, ab1791, Abcam, Cambridge, MA), goat anti-H4 (1:100 dilution, sc-8658, Santa Cruz Biotechology, Santa Cruz, CA), rabbit anti-H2AX (1:5000 dilution, BL179, Bethyl, TX), rabbit anti-H3K4me3 (1:4000 dilution, 05-745, Upstate), rabbit anti-H3K9me3 (1:500 dilution, 07-523, Upstate), or rabbit anti-H4K20me3 (1:200 dilution, 07-749, Upstate). The membranes were then incubated with either goat anti–rabbit IgG horseradish peroxidase (HRP) (1:5000 dilution, ac-2004, Santa Cruz Biotechnology) or donkey anti–goat HRP (1:5,000 dilution, ac-2020, Santa Cruz Biotechnology). Antibody binding was detected by enhanced chemiluminesence (ECL, Amersham, Piscataway, NJ).
Reverse Transcription–Polymerase Chain Reaction
Total RNA was purified from bone marrow cells using an RNeasy plus kit (Qiagen, Hilden, Germany). Total RNA (5 μg) was used for reverse transcription to synthesize the first-strand cDNA (Superscript III First-strand synthesis system, Invitrogen). cDNA (5 μg) was used for polymerase chain reaction (PCR). PCR conditions and primer sequences have been described (28) and/or are listed in Supplementary Table 1, available online. Hprt transcripts were amplified as a control for gene expression.
Statistical Analysis
Means and the accompanying 95% confidence intervals were calculated from at least three independent experiments. Group means were compared using a two-sided Student t test. P values less than .05 were considered to be statistically significant.
Results
A CMML-like Myelodysplastic/Myeloproliferative Disorder in Arid4a-Deficient Mice
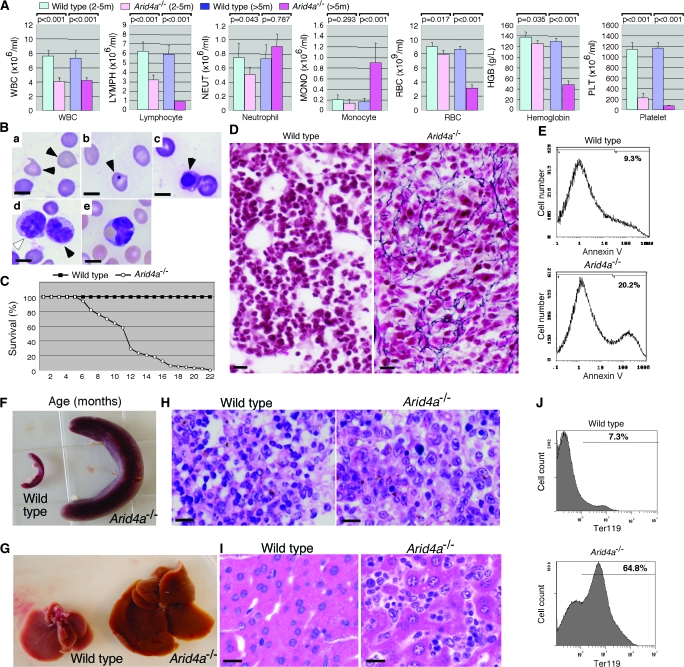
To investigate the function of Arid4a and Arid4b, we bred Arid4a null (Arid4a−/−) mice and mice null for Arid4a and heterozygous for the Arid4b deletion (Arid4a−/−Arid4b+/−); homozygous null mutants for Arid4b (both Arid4a+/+Arid4b−/− and Arid4a−/−Arid4b−/−) were not viable and died before E7.5 (5). Young (2–5 months old) adult Arid4a-deficient mice initially had ineffective blood cell production in all hematopoietic lineages, with mild leukopenia (WBC: mean wild-type count = 7.64 × 106/mL, mean Arid4a−/− count = 4.09 × 106/mL, difference = 3.55 × 106/mL, 95% confidence interval [CI] = 2.72 × 106/mL to 4.38 × 106/mL, P < .001. Lymphocyte: mean wild-type count = 6.29 × 106/mL, mean Arid4a−/− count = 3.21 × 106/mL, difference = 3.08 × 106/mL, 95% CI = 2.22 × 106/mL to 3.94 × 106/mL, P < .001. Neutrophil: mean wild-type count = 0.84 × 106/mL, mean Arid4a−/− count = 0.51 × 106/mL, difference = 0.33 × 106/mL, 95% CI = 0.16 × 106/mL to 0.51 × 106/mL, P = .043) and mild anemia (RBC: mean wild-type count = 8.96 × 109/ml, mean Arid4a−/− count = 7.88 × 109/mL, difference = 1.08 × 109/mL, 95% CI = 0.5 × 109/mL to 1.66 × 109/mL, P = .017; hemoglobin: mean wild-type count = 137.8 g/L, mean Arid4a−/− count = 125.0 g/L, difference = 12.8 g/L, 95% CI = 5.0 g/L to 20.7 g/L, P = .035) with statistically significant thrombocytopenia (platelet: mean wild-type count = 1144.3 × 106/mL, mean Arid4a−/− count = 225.8 × 106/mL, difference = 918.5 × 106/mL, 95% CI = 802.3 × 106/mL to 1034.7 × 106/mL, P < .001) (Figure 1, A). Beyond 5 months of age, the Arid4a−/− mice manifested monocytosis (monocyte: mean wild-type count = 0.2 × 106/mL, mean Arid4a−/− count = 0.9 × 106/mL, difference = 0.7 × 106/mL, 95% CI = 0.55 × 106/mL to 0.85 × 106/mL, P < .001), accompanied with severe anemia (RBC: mean wild-type count = 8.6 × 109/mL, mean Arid4a−/− count = 3.1 × 109/mL, difference = 5.5 × 109/mL, 95% CI = 5.0 × 109/mL to 6.0 × 109/mL, P < .001; hemoglobin: mean wild-type count = 130 g/L, mean Arid4a−/− count = 48 g/L, difference = 82 g/L, 95% CI = 76 g/L to 88 g/L, P < .001) and severe thrombocytopenia (platelet: mean wild-type count = 1169 × 106/mL, mean Arid4a−/− count = 76 × 106/mL, difference = 1093 × 106/mL, 95% CI = 1036 × 106/mL to 1150 × 106/mL, P < .001) (Figure 1, A). Peripheral blood smears from older (more than 5 months old) sickly Arid4a−/− mice showed the presence of teardrop poikilocytes (Figure 1, B, a) and increased numbers of immature erythroid cells (Figure 1, B, b and c). Immature and maturing mononuclear cells morphologically consistent with the monocyte lineage were also noted in the peripheral blood (Figure 1, B, d). Some monocytoid cells that contained phagocytosed RBCs were also present (Figure 1, B, e). The Arid4a−/− mice showed signs of morbidity (eg, ruffled hair, decreased activity, and rapid respiration). Mortality of the Arid4a−/− mice increased sharply from 6 months of age onward, with no Arid4a−/− mice surviving past 22 months of age (Figure 1, C). Increased mortality appeared to be associated with increasing severity of the hematologic abnormalities, especially severe anemia, in Arid4a−/− mice.
Figure 1.
A chronic myelomonocytic leukemia (CMML)–like myelodysplastic/myeloproliferative disorder in Arid4a-deficient mice. A) Complete blood counts (white blood cell, lymphocyte, neutrophil, monocyte, red blood cell [RBC], hemoglobin, platelet) in wild-type mice at 2–5 months of age (n = 35), in Arid4a−/− mice at 2–5 months of age (n = 30), in wild-type mice more than 5 months old (n = 25), and in Arid4a−/− mice more than 5 months old with symptoms of CMML (n = 25). Means (and 95% confidence intervals) for cell concentrations are shown, and P values were calculated using Student t test. B) Wright–Giemsa staining of peripheral blood from an Arid4a−/− mouse with symptoms of CMML, showing teardrop poikilocytes (a, black arrowheads), red cells with Howell–Jolly bodies (b, black arrowhead), and nucleated red cells (c, black arrowhead). Immature (d, white arrowhead) and maturing (d, black arrowhead) mononuclear cells were also observed, together with phagocytosis of RBC by a monocyte (e, arrowhead). Ten separate analyses were performed. Scale bars = 5 μm. C) Survival of Arid4a−/− (n = 25) mice and wild-type (n = 25) mice. D) Reticulin staining of paraffin sections of bone marrow from a wild-type mouse and a sick Arid4a−/− mouse. The Arid4a−/− sample shows fibrous tissue stained with black color. Scale bars = 20 μm. E) Flow cytometric analysis of apoptotic cells in bone marrow from a wild-type and a sick Arid4a−/− mouse. The percentages of cells positive for annexin V are indicated. Five separate cytometric analyses were performed. F) Splenomegaly and G) hepatomegaly in a sick Arid4a−/− mouse. Hematoxylin and eosin–stained sections of H) spleen and I) liver from a wild-type mouse and a sick Arid4a−/− mouse. Extramedullary hematopoiesis was found in the Arid4a−/− spleen and Arid4a−/− liver, which were infiltrated with nucleated elements of blood cells. Ten separate analyses were performed. Scale bars = 20 μm. J) Flow cytometric analysis of cells from spleen in a wild-type mouse and an Arid4a−/− mouse stained with Ter119 surface antigen. The percentages of cells positive for the antigen are indicated. Twenty separate analyses were performed.
Bone marrow from sick Arid4a−/− mice developing monocytosis and severe anemia in the peripheral blood showed reticulin fibrosis and an increased number (average increase = 8%, 95% CI = 6% to 10%, P < .001; the 10.9% increase in Figure 1, E is from one of five separate experiments) of apoptotic cells compared with wild-type mice (Figure 1, D and E). The sick Arid4a−/− mice (n > 50) also developed splenomegaly (Figure 1, F) and hepatomegaly (Figure 1, G) after 5 months of age, with extramedullary hematopoiesis in the spleen (Figure 1, H) and liver (Figure 1, I). A marked increase of erythropoiesis within enlarged spleens of the Arid4a−/− mice was demonstrated by flow cytometry analysis of spleen cells: 65% of spleen cells were erythroid cells (Ter119+) in mutants versus 7% in wild-type mice (difference = 58%, 95% CI = 53% to 63%, P < .001) (Figure 1, J).
In addition to their hematologic abnormalities, female Arid4a−/− mice showed decreased fertility. Litter sizes for the Arid4a−/− females mating with wild-type males were markedly lower than those for wild-type breeding pairs (0.7 vs 7.8, difference = 5.6, 95% CI = 5.2 to 6.0. P < .001). Anemia, hepatosplenomegaly, and systemic illness may have contributed to the decreased fertility, but in addition there was hemorrhage into the ovarian follicles (Supplementary Figure 1, available online), as a result of profound thrombocytopenia (the mean platelet count in these mice was less than 100 × 106/mL, Figure 1, A).
Thus, Arid4a−/− mice initially developed mild cytopenias with substantial thrombocytopenia and later progressed to monocytosis associated with more severe anemia and thrombocytopenia with spontaneous hemorrhage into organs (ie, ovary). Bone marrow failure with myelofibrosis was associated with hepatosplenomegaly due to compensatory extramedullary hematopoiesis. These abnormalities in the Arid4a−/− mice suggest a myelodysplastic/myeloproliferative disorder and are similar to the course of events in humans with CMML derived from myelodysplastic/myeloproliferative diseases.
Development of AML in Arid4a—/— Mice and Arid4a—/—Arid4b +/— Mice
In patients with myelodysplastic/myeloproliferative diseases, progression to AML occurs with a frequency of 5%–30%. Similarly, we found that 5 of 42 (12%) of the Arid4a−/− mice developed AML showing rapid and large increases of WBC counts with an onset between 12 and 22 months of age (data not shown). Hematologic malignancies were more frequent in the Arid4a−/−Arid4b+/− mice. Of the 12 Arid4a−/−Arid4b+/− mice monitored beyond 5 months of age, 10 (83%) developed AML with an earlier age of onset (7–15 months).
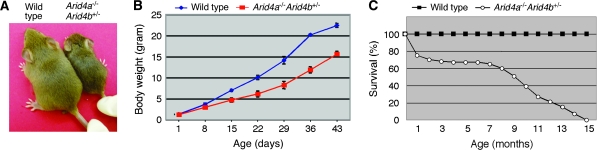
Neither the mice heterozygous for both Arid4a and Arid4b (n = 36) nor wild-type controls (n = 52) developed AML over a 2-year period. littermates (data not shown). Postnatal growth was also delayed (Figure 2, A); body weight of Arid4a−/−Arid4b+/− mice was 30% less than that of wild-type mice at 7 weeks of age (Figure 2, B). The Arid4a−/−Arid4b+/− mice also had increased postnatal mortality: 25% of mutant mice died before attaining 1 month of age (Figure 2, C), and those that survived exhibited increased mortality at 7 months of age (Figure 2, C) that was attributable to the increasing severity of their hematologic malignancies.
Figure 2.
Growth and survival of the Arid4a−/−Arid4b+/− mice. A) Growth retardation in an Arid4a−/−Arid4b+/− mouse compared with a wild-type littermate at 12 days of age. B) Growth curve of the Arid4a−/−Arid4b+/− (n = 7) and wild-type (n = 10) littermates by mean body weight plotted against age with 95% confidence intervals. C) Survival of Arid4a−/−Arid4b+/− (n = 31) and wild-type (n = 25) mice.
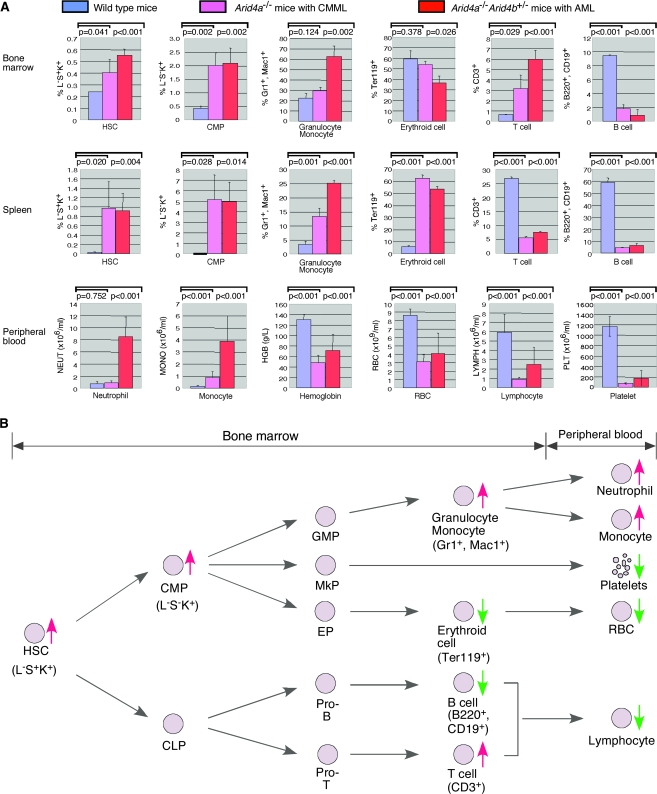
Bone marrow smears from mice with AML (two Arid4a−/− mice and three Arid4a−/−Arid4b+/− mice) demonstrated the presence of a mixture of immature and dysplastic WBC precursors with more than 20% nonlymphoid immature forms and blasts (Figure 3, A). Flow cytometric analysis of cell populations within the bone marrow from the Arid4a−/− mice with CMML-like phenotype and the Arid4a−/−Arid4b+/− mice with AML revealed that the majority of excess leukocytes were granulocytes and monocytes (22% of wild-type mice, 29% of mice with CMML-like phenotype, and 63% of mice with AML were positive for Gr1 and Mac1, difference between wild-type and CMML-like mice = 7%, P = .124; difference between wild-type and AML mice = 41%, P = .002) (Figure 4, A). This increase in granulocytes and monocytes was accompanied by an increase of T lymphoid cells (CD3+, 0.65% T lymphoid cells in wild-type mice, 3.16% in mice with CMML-like phenotype, and 5.99% in AML mice, difference between wild-type and CMML-like mice = 2.15%, P = .029; difference between wild-type and AML mice = 5.34%, P < .001) and by decreases in B lymphoid cells (B220+CD19+, 9.4% in wild-type mice, 1.8% in mice with CMML-like phenotype, and 0.9% in AML mice, difference between wild-type and CMML-like mice = 7.6%, P < .001; difference between wild-type and AML mice = 8.5%, P < .001) and erythroid populations (Ter119+, 60% erythroid cells in wild-type mice, 54% in mice with CMML-like phenotype, and 36% in AML mice, difference between wild-type and CMML-like mice = 6%, P = .378; difference between wild-type and AML mice = 24%, P = .026) (Figure 4, A).
Figure 3.
Acute myeloid leukemia (AML)–like phenotype in Arid4a−/−Arid4b+/− mice. A) Bone marrow smears from a wild-type mouse and an Arid4a−/−Arid4b+/− mouse with AML were stained with Wright–Giemsa. Black arrowheads indicate blasts. White arrowheads indicate red blood cell precursors in both wild-type and AML bone marrow (for both wild-type and the Arid4a−/−Arid4b+/− bone marrow smears, original magnifications are the same). Five separate analyses were performed. Scale bars = 20 μm. B) Wright–Giemsa staining of peripheral blood smears from the AML Arid4a−/−Arid4b+/− mice showing blasts (a, black arrowhead), lymphocytes (a, white arrowhead), increased numbers of immature cells (b–d, arrowheads), and phagocytosis of cells by a macrophage (e). Eight separate analyses were performed. Scale bars = 5 μm.
Figure 4.
Hematopoietic lineage analysis of the Arid4a−/− mice and Arid4a−/−Arid4b+/− leukemic mice. A) Comparison of cell populations in bone marrow and spleen (HSC, CMP, granulocyte, monocyte, erythroid cell, T cell, and B cell) and cell counts in peripheral blood (neutrophil, monocyte, hemoglobin, RBC, lymphocyte, and platelet) between wild-type mice (n = 5), the Arid4a−/− mice with CMML-like phenotype (n = 5), and the Arid4a−/−Arid4b+/− mice with acute myeloid leukemia (AML) (n = 5). Means (and 95% confidence intervals) for all cell populations and cell counts are shown, and P values were calculated using Student t test. B) Hematopoietic lineage tree displaying the combined impact of the Arid4a mutation with or without the Arid4b mutations in bone marrow and peripheral blood of mice with CMML-like or AML phenotype. Increased and decreased cell populations are indicated by red and green, respectively. HSC, hematopoietc stem cell; CMP, common myeloid progenitor; CLP, common lymphoid progenitor; GMP, granulocyte and monocyte progenitor; MkP, megakaryocyte progenitor; EP, erythroid progenitor.
Examination of peripheral blood smears from the Arid4a−/− and Arid4a−/−Arid4b+/− mice with AML showed the presence of more than 20% of atypical cells with morphology consistent with blasts or immature myeloid precursors (Figure 3, B, a–d). There was accompanying monocytosis, and hemophagocytosis was often observed (Figure 3, B, e). Serial monitoring of peripheral blood counts in mice developing AML revealed a rapid increase of WBC counts over a 2- to 4-week interval. The elevated WBC count was attributable to increased neutrophilic and monocytic forms (neutrophil: mean wild-type count = 0.75 × 106/mL, mean AML count = 8.50 × 106/mL, difference = 7.75 × 106/mL, 95% CI = 5.85 × 106/mL to 9.65 × 106/mL, P < .001; monocyte: mean wild-type count = 0.16 × 106/mL, mean AML count = 3.90 × 106/mL, difference = 3.74 × 106/mL, 95% CI = 2.54 × 106/mL to 4.94 × 106/mL, P < .001) (Figure 4, A). Soon after the development of leukocytosis, the mice became moribund and were sacrificed. In most of the Arid4a−/−Arid4b+/− mice, AML occurred before development of severe anemia (peripheral blood in Figure 4, A; hemoglobin: mean wild-type count = 130 g/L, mean Arid4a−/−Arid4b+/− count = 71 g/L, difference = 59 g/L, 95% CI = 40 g/L to 78 g/L, P < .001). Of the few Arid4a−/− mice that progressed to AML, most developed a CMML-like phenotype and became moribund from severe anemia (hemoglobin: mean wild-type count = 130 g/L, mean Arid4a−/− count = 48 g/L, difference = 82 g/L, 95% CI = 76 g/L to 88 g/L, P < .001) (Figures 1, A and 4, A).
Development of Myeloid Sarcoma in Arid4a—/— and Arid4a—/—Arid4b +/— Mice
In the human World Health Organization classification (29), myeloid sarcoma is considered an alternative presentation of AML. The Arid4a−/− mice and the Arid4a−/−Arid4b+/− mice with leukemia also developed myeloid sarcoma. Soft tissue tumor nodules were found within the enlarged spleens (Figure 5, A) and livers (Figure 5, B) that were infiltrated with aggressive leukemic (malignant) cells (Figure 5, C). Blood vessels in the lungs of the AML mice showed a marked increase of nucleated elements, indicative of leukemic involvement (Figure 5, D). Similar lesions composed of leukemic cells were also found in lymph nodes and kidneys (data not shown). Flow cytometric analysis of cell populations within the splenic tissue showed an increase of leukocytes of granulocytic and monocytic origin (Gr1+ and Mac1+, 3% in wild-type mice, 13% in mice with CMML-like phenotype, and 26% in AML mice, difference between wild-type and CMML-like mice = 10%, P < .001; difference between wild-type and AML mice = 23%, P < .001) (Figure 4, A). This increase in granulocytes and monocytes was accompanied by an increase of erythroid cells (Ter119+, 6% erythroid cells in wild-type mice, 63% in mice with CMML-like phenotype, and 54% in AML mice, difference between wild-type and CMML-like mice = 57%, P < .001; difference between wild-type and AML mice = 48%, P < .001) and relative decreases of the T lymphoid (CD3+, 26.7% in wild-type mice, 5.5% in mice with CMML-like phenotype, and 7.6% in AML mice, difference between wild-type and CMML-like mice = 21.2%, P < .001; difference between wild-type and AML mice = 19.1%, P < .001) and B lymphoid (B220+CD19+, 58.9% in wild-type mice, 4.8% in mice with CMML-like phenotype, and 6.7% in AML mice, difference between wild-type and CMML-like mice = 54.1%, P < .001; difference between wild-type and AML mice = 52.2%, P < .001) populations (Figure 4, A). These features fulfill the criteria for AML (granulocytic and monocytic) and myeloid (granulocytic) sarcoma in mice (Bethesda proposals) (30).
Figure 5.
Development of myeloid sarcoma in Arid4a−/−Arid4b+/− mice. A) Splenomegaly and B) hepatomegaly in the acute myeloid leukemia (AML) Arid4a−/−Arid4b+/− mice relative to spleen and liver from wild-type littermates. C) Histologic analysis of spleen and liver from a wild-type mouse and an AML Arid4a−/−Arid4b+/− mouse. Paraffin sections were stained with hematoxylin and eosin. Black arrowheads indicate mitotic leukemic cells in spleen and liver from the Arid4a−/−Arid4b+/− mouse. White arrowheads indicate hepatocytes. D) Histologic analysis of lungs from a wild-type and an Arid4a−/− mouse. Paraffin sections were stained with hematoxylin and eosin. Blood vessels in the Arid4a−/− lungs showed a marked increase of leukemic cells.
Expansion of HSCs and Downstream Progenitors in Arid4a—/— and Arid4a—/—Arid4b +/— Mice
Because the abnormalities were found in all hematologic lineages, we tested whether deficiency of Arid4a and Arid4b has effects on HSCs and downstream progenitors. Compared with wild-type mice, Arid4a−/− mice had an increased proportion of HSCs (Lin−Sca1+c-Kit+, referred to as L−S+K+, 0.24% in wild-type mice, 0.40% in Arid4a−/− mice, difference = 0.16%, P = .041) in bone marrow (Figure 4, A). In mice that were also heterozygous for the Arid4b mutation, there was greater expansion of the HSCs population in the Arid4a−/−Arid4b+/− bone marrow (L−S+K+, 0.24% in wild-type mice, 0.55% in Arid4a−/−Arid4b+/− mice, difference = 0.31%, P < .001) (Figure 4, A). Furthermore, the proportions of CMPs (Lin−Sca1−c-Kit+, referred as L−S−K+) in the Arid4a−/− and Arid4a−/−Arid4b+/− bone marrow were statistically significantly higher than those from wild-type mice (L−S−K+, 0.4% in wild-type mice, 2.0% in mice with CMML-like phenotype, and 2.1% in AML mice, difference between wild-type and CMML-like mice = 1.6%, P < .001; difference between wild-type and AML mice = 1.7%, P < .001) (Figure 4, A). In the spleen, dramatic increases of the HSC (L−S+K+, 0.03% in wild-type mice, 0.4% in mice with CMML-like phenotype, and 0.55% in AML mice, difference between wild-type and CMML-like mice = 0.37%, P = .02; difference between wild-type and AML mice = 0.52%, P = .004) and CMP (L−S−K+, 0.01% in wild-type mice, 5.13% in mice with CMML-like phenotype, and 5.01% in AML mice, difference between wild-type and CMML-like mice = 5.12%, P = .028; difference between wild-type and AML mice = 5%, P = .014) populations were found in both Arid4a−/− mice and Arid4a−/−Arid4b+/− mice (Figure 4, A), suggesting that extramedullary hematopoiesis in spleen might be due to mobilization of HSCs from bone marrow.
Collectively, our data suggested the following disease model for the observed hematologic disorders. Deficiency of Arid4a and Arid4b results in increase of HSCs, CMPs, and Gr1+Mac1+ myeloid cells in bone marrow and spleen, which leads to increases of neutrophils and monocytes in the peripheral blood (Figure 4, B). Despite the compensatory erythropoiesis within the enlarged spleen due to the decrease of erythroid activity in the bone marrow (Figures 1, J and 4, A and B), the enlarged spleen sequestrates RBCs. Although it is not known if the reduction in platelets is due to decreased production or increased destruction, the reduction leads to spontaneous hemorrhage (Supplementary Figure 1, available online). All of these processes contribute to a decrease of RBCs in peripheral blood (Figure 4, B). Although the proportion of T lymphoid cells was increased in the bone marrow, the population of B lymphoid cells was substantially decreased, leading to the lower number of total lymphocytes in the peripheral blood (Figure 4, B). Thus, Arid4a and Arid4b have essential roles in hematopoietic homeostasis and in lineage fate determination.
Disturbed Patterns of Histone Modifications in Arid4a—/— Bone Marrow Cells
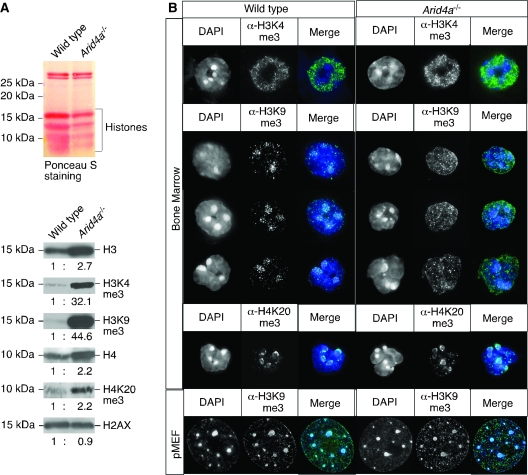
The ARID4A and ARID4B proteins contain a chromodomain and a Tudor domain. Both domains have been reported to mediate binding to methylated lysines of histones H3 and H4 (11–14). Given the bone marrow failure phenotype found in the Arid4a−/− mice, we investigated the expression and methylation status of histones in bone marrow of these mice. By western blot analysis, levels of histones H3 and H4 were elevated by 2.7- and 2.2-fold, respectively (95% CI = 2.3 to 3.1 and 1.9 to 2.5, respectively) in the Arid4a−/− bone marrow constituents compared with the wild-type samples (Figure 6, A). Levels of H2AX were not different. Methylation of lysine in H3 can lead to either repression or activation of gene expression; trimethylation of H3K4 (H3K4me3) is associated with transcriptional activation at euchromatic regions, whereas trimethylation of H3K9 (H3K9me3) is usually associated with repressive states at pericentric heterochromatin. Analysis of H3 lysine methylation revealed very large increases of both H3K4me3 (32-fold, 95% CI = 27 to 37) and H3K9me3 (45-fold, 95% CI = 41 to 49) in the bone marrow cells from the Arid4a−/− mice compared with wild-type mice (Figure 6, A). As detected by immunofluorescence, H3K4me3 was broadly distributed over euchromatic regions but showed speckled patterns in both wild-type and Arid4a−/− bone marrow cells (Figure 6, B). In wild-type bone marrow cells, H3K9me3 was localized in discrete spots in the DAPI-dense regions that correspond to the pericentric heterochromatin structure (Figure 6, B). Arid4a−/− bone marrow cells showed a diffuse pattern of H3K9me3-derived immunofluorescence with small foci spread throughout the nuclei (Figure 6, B). These different patterns of H3K9me3 between wild-type and the Arid4a−/− samples were obvious across the great majority of cells in the marrow even though the cell populations in wild-type and the Arid4a−/− bone marrow samples were different (Figure 4, A). These differences of immunofluorescence staining patterns of H3K9me3 were not seen in primary mouse embryo fibroblasts (pMEFs) (Figure 6, B). Trimethylation of H4K20 (H4K20me3) is another modification usually found on repressed chromatin accumulated at pericentric heterochromatin regions. When analyzed by western blotting, the Arid4a−/− bone marrow cells revealed a slight increase of H4K20me3 (2.2-fold, 95% CI = 1.7 to 2.7), which might reflect increased expression of histone H4 (2.2-fold, 95% CI = 1.9 to 2.5) (Figure 6, A). The fluorescence signals representing H4K20me3 were focally enriched at pericentric heterochromatin in both wild-type and the Arid4a−/− bone marrow cells (Figure 6, B). Collectively, these results suggest that Arid4a participates in regulating lysine trimethylation of histones H3 and H4 in bone marrow.
Figure 6.
Histone modifications in bone marrow cells of mice lacking Arid4a. A) Western blot analysis of acid-extracted proteins from bone marrow of a wild-type and an Arid4a−/− mouse. Total proteins were transferred to a nitrocellulose membrane and stained with Ponceau S (top), followed by staining with antibodies against histones H3, H4, H2AX, H3K4me3, H3K9me3, and H4K20me3 (bottom). Ratios of histones were quantified by densitometry. Five separate experiments were performed. B) Immunofluorescence analysis of bone marrow cells or primary mouse embryo fibroblasts from wild-type and the Arid4a−/− mice using antibodies against H3K4me3, H3K9me3, and H4K20me3. DNA was counterstained with DAPI. Images were analyzed by deconvolution microscopy. Three separate experiments were performed, all with similar results.
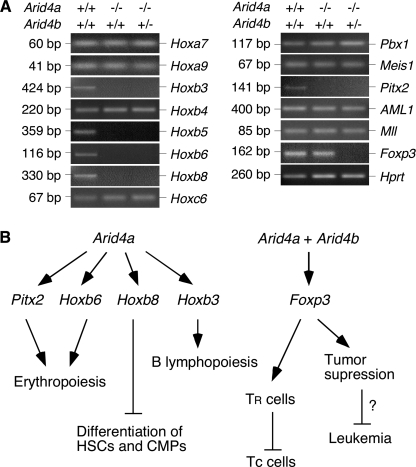
Decreased Expression of the Hox and Fox Genes in Bone Marrow Cells with Arid4a and Arid4b +/— Mutations
To investigate the downstream genes regulated by Arid4a and Arid4b, we first compared gene expression of wild-type pMEFs and Arid4a−/−Arid4b+/− pMEFs using gene expression microarrays. We identified genotype-specific differences in expression of Hox and Fox genes (data not shown). To further elucidate the regulatory mechanisms by which Arid4a and Arid4b are involved in acquisition of the leukemic phenotype, we compared expression of several genes important for normal hematopoiesis and leukemogenesis. These included the homeobox genes in the Hox clusters (Hoxa, Hoxb, and Hoxc) (31,32), homeodomain transcription factors Pbx1 (33), Meis1 (34), and Pitx2 (35), AML1 (acute myeloid leukemia 1) (36), Mll (mixed-lineage leukemia) (37), and the forkhead box gene Foxp3 (38,39). We compared the expression of these genes in bone marrow from wild-type, Arid4a−/−, and Arid4a−/−Arid4b+/− mice. To minimize variation arising from differences in cell populations, we collected bone marrow cells from mice before development of illness. We monitored relative gene expression levels using reverse transcription–PCR. The expression of several Hoxb genes, including Hoxb3, Hoxb5, Hoxb6, and Hoxb8, but not Hoxb4, was decreased in both Arid4a−/− and Arid4a−/−Arid4b+/− bone marrow compared to wild-type mice. This decrease was specific for Hoxb genes because the expression of Hoxa7, Hoxa9, Hoxc6, Pbx1, and Meis1 was not altered (Figure 7, A). The expression of Pitx2 that is required for normal hematopoiesis was also reduced in Arid4a−/− and Arid4a−/−Arid4b+/− bone marrow (Figure 7, A). In contrast, expression of AML1 and Mll, genes involved in leukemogenesis, was similar in all the bone marrow samples (Figure 7, A). These results suggested that Arid4a regulates hematopoiesis by controlling the expression of specific homeodomain genes, such as Pitx2 and a subset of Hoxb genes.
Figure 7.
Gene expression analysis of bone marrow cells from wild-type, Arid4a−/−, and Arid4a−/−Arid4b+/− mice. A) Reverse transcriptase–polymerase chain reaction was performed to analyze the genes indicated, with Hprt serving as the control gene. Three separate experiments were performed. B) Pathways by which Arid4a and Arid4b might regulate hematopoiesis through control of the Hox and Fox genes. In the scenario shown, Arid4a controls erythropoiesis, possibly by positively regulating Pitx2 and Hoxb6 genes. Arid4a also controls the expression of Hoxb8, whose product blocks differentiation of hematopoietic stem cells and common myeloid progenitors. Control of B lymphopoiesis by Arid4a may be achieved by increasing expression of Hoxb3. Arid4a, together with Arid4b, increases expression of Foxp3, which acts on regulatory T (TR) cells to suppress conventional T (Tc) cells. Foxp3 also functions as a tumor suppressor gene. However, it is unclear whether Foxp3 suppresses leukemia malignancies.
The FOXP3 protein plays an important role in control of the regulatory T-cell lineage (40), and Foxp3 was recently identified as a tumor suppressor gene (41). The expression of Foxp3 was reduced specifically in the Arid4a−/−Arid4b+/− bone marrow but not in the Arid4a−/− bone marrow (Figure 7, A), suggesting that downregulation of Foxp3 may be involved in the mechanisms underlying the increased numbers of T cells in the Arid4a−/−Arid4b+/− bone marrow (Figure 4, A) and the high frequency of leukemia malignancies in the Arid4a−/−Arid4b+/− mice.
Discussion
Arid4a and Arid4b knockout mice provide a suitable animal model for better understanding the progression of a premalignant hematologic disorder and the eventual transformation to AML. Using this model, we found disruption of myeloid homeostasis in the Arid4a−/− mice due to the increase of HSCs and downstream progenitors, which might be the principal components of the expansion of a transformed leukemic stem cell compartment with aggressive self-renewal properties. We have obtained some insight into the molecular mechanisms by defining the downstream regulatory impact on the Hox and Fox genes. Moreover, our results suggest that Arid4a controls chromatin modification necessary to support normal hematopoiesis, and the data are consistent with an important role of epigenetic regulation in cancer development.
H3K4me3 is a mark of transcriptionally active chromatin states, whereas the H3K9me3 and H4K20me3 modifications are usually found at repressive chromatin domains. However, we found that H3K4me3, H3K9me3, and H4K20me3 were all increased in bone marrow cells from mice deleted for Arid4a. We envision that these histone methylations will reach a combinatorial steady state that may mediate activation or repression of specific genomic loci or chromosome domains. We propose that ARID4A is a critical determinant for distinct histone methylation states and that it plays an important role in coordination between the modification marks at different chromatin regions. Further analysis of chromatin-specific components associated with ARID4A and/or ARID4B could more clearly address the contribution of these two proteins to the combinatorial pattern of histone modifications. A goal of future research should be to determine whether ARID4A and ARID4B contain any intrinsic enzyme activity and whether they mediate their effects either through direct interaction with chromatin complexes or through intermediate steps involving various pathways that are modulated through chromatin remodeling.
Our results suggest plausible molecular mechanisms for the hematologic disorder in Arid4a-deficient mice based on downregulation of a number of homeobox genes (Pitx2 and a cluster of Hoxb genes including Hoxb3, Hoxb5, Hoxb6, and Hoxb8) in Arid4a−/− bone marrow cells with or without Arid4b haploinsufficiency. A homeobox sequence encodes a protein domain that binds DNA. Homeobox genes encode transcription factors and are classified into two subgroups: Hox and non–Hox genes. Abnormal expression of both groups of homeobox genes plays a role in the pathogenesis of myeloid malignancies (31,32). Little is known about the molecular mechanisms of homeobox gene regulation, but the MLL gene is reported to positively regulate multiple Hox genes (28). MLL encodes a histone methyltransferase that methylates H3K4 (42). Leukemogenic MLL translocations define a specific group of leukemias (mixed lineage leukemia) (43). We suggest that Arid4a and Arid4b play a prominent role in leukemic transformation by controlling the trimethylation of H3K4 and H3K9. Although we did not find that mutations of Arid4a and Arid4b affected the expression of Mll, our results suggest plausible molecular mechanisms whereby Arid4a and Arid4b might regulate hematopoiesis by controlling the expression of specific homeodomain genes, such as Pitx2, required for normal erythropoiesis (44), and a subset of Hoxb genes, including Hoxb6, whose disruption results in an increase of early erythrocyte progenitors (45); Hoxb8, which affects lineage-specific development of hematopoietic progenitor cells (46,47); and Hoxb3, whose deficiency impairs B lymphopoiesis (48) (Figure 7, B).
Our results also imply that Arid4a and Arid4b are involved in the regulation of a forkhead box gene Foxp3 (Figure 7, B), an X chromosome–encoded forkhead transcription factor family member that plays an important role in the development and function of natural regulatory T cells (40). FOXP3 protein interacts with AML1 to control regulatory T-cell function (39). Although Arid4a and Arid4b mutations did not affect the expression of AML1, we found decreased expression of Foxp3 in the Arid4a−/−Arid4b+/− mice. FOXP3 functions as a transcriptional regulator by assembling chromatin remodeling complexes involved in histone modification (49). In humans, mutations of FOXP3 leads to an X-linked fatal autoimmune disease known as IPEX (immune dysregulation, polyendocrinopathy, enteropathy, X-linked) syndrome and an analogous lymphoproliferative disease (50). Deficiency of Foxp3 in mice leads to an early onset, highly aggressive and fatal autoimmune disease characterized by excessive proliferation of CD4+ T cells and extensive infiltration by leukocytes in multiple organs (38,51,52). Recently, it has been reported that FOXP3 protein functions as a tumor suppressor involved in the development of breast cancer (41). Decreased expression of Foxp3 may be involved in the disease mechanisms through epigenetic effects in the Arid4a−/−Arid4b+/− mice, which develop an increased population of T cells in bone marrow and progress to AML and myeloid sarcoma with various tissues infiltrated by leukemic cells. It will be of interest to examine whether decreased expression of Foxp3 contributes to the high frequency of leukemia malignancies in the Arid4a−/−Arid4b+/− mice.
We present evidence that ARID4A and ARID4B function as tumor suppressors and that a myelodysplastic/myeloproliferative disorder in mice carrying the Arid4a or/and Arid4b mutations progresses to hematologic malignancies, resembling human CMML and AML. However, we have not investigated whether mutations in ARID4A and ARID4B participate in genetic and/or epigenetic mechanisms of human CMML and AML, or other cancers. Although the ARID4A and ARID4B proteins have been identified as breast cancer–associated antigens (3,20,21) and Arid4a and Arid4b are involved in the regulation of Foxp3, which functions as a breast cancer suppressor gene (41), we have not observed primary solid tumors in mice with ARID4 family deficiency, (although we have not performed a detailed search for breast cancer). Further study of the Arid4 gene family may advance our understanding of the connection between gene regulation, epigenetic control, disease development, and cancer formation. We also suggest that gene regulation by ARID4A and ARID4B should be examined for potential disease-related roles, not only in human malignancies, but also in other complex disease traits.
Funding
National Institutes of Health (HD-37283 to A.L.B.).
Supplementary Material
Footnotes
We are particularly grateful to Silvia Briones, Catherine Tran, and Minnie Freeman for technical assistance. We thank Ray-Chang Wu, Orla M. Conneely, Shuo Zhang, Alice J. Chen, Stuart M. Chambers, and Neal Copeland for critical reading of the manuscript.
The sponsor had no role in the study design, data collection and analysis, interpretation of the results, the preparation of the manuscript, or the decision to submit the manuscript for publication.
The authors declare no competing financial interests.
References
- 1.Defeo-Jones D, Huang PS, Jones RE, et al. Cloning of cDNAs for cellular proteins that bind to the retinoblastoma gene product. Nature. 1991;352(6332):251–254. doi: 10.1038/352251a0. [DOI] [PubMed] [Google Scholar]
- 2.Fattaey AR, Helin K, Dembski MS, et al. Characterization of the retinoblastoma binding proteins RBP1 and RBP2. Oncogene. 1993;8(11):3149–3156. [PubMed] [Google Scholar]
- 3.Cao J, Gao T, Stanbridge EJ, Irie R. RBP1L1, a retinoblastoma-binding protein-related gene encoding an antigenic epitope abundantly expressed in human carcinomas and normal testis. J Natl Cancer Inst. 2001;93(15):1159–1165. doi: 10.1093/jnci/93.15.1159. [DOI] [PubMed] [Google Scholar]
- 4.Wilsker D, Probst L, Wain HM, Maltais L, Tucker PW, Moran E. Nomenclature of the ARID family of DNA-binding proteins. Genomics. 2005;86(2):242–251. doi: 10.1016/j.ygeno.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 5.Wu MY, Tsai TF, Beaudet AL. Deficiency of Rbbp1/Arid4a and Rbbp1l1/Arid4b alters epigenetic modifications and suppresses an imprinting defect in the PWS/AS domain. Genes Dev. 2006;20(20):2859–2870. doi: 10.1101/gad.1452206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lai A, Lee JM, Yang WM, et al. RBP1 recruits both histone deacetylase-dependent and -independent repression activities to retinoblastoma family proteins. Mol Cell Biol. 1999;19(10):6632–6641. doi: 10.1128/mcb.19.10.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patsialou A, Wilsker D, Moran E. DNA-binding properties of ARID family proteins. Nucleic Acids Res. 2005;33(1):66–80. doi: 10.1093/nar/gki145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bannister AJ, Zegerman P, Partridge JF, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410(6824):120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 9.Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17(15):1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, III, Grant PA. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433(7024):434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- 11.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410(6824):116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 12.Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17(15):1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119(5):603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 14.Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312(5774):748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- 15.Lai A, Marcellus RC, Corbeil HB, Branton PE. RBP1 induces growth arrest by repression of E2F-dependent transcription. Oncogene. 1999;18(12):2091–2100. doi: 10.1038/sj.onc.1202520. [DOI] [PubMed] [Google Scholar]
- 16.Lai A, Kennedy BK, Barbie DA, et al. RBP1 recruits the mSIN3-histone deacetylase complex to the pocket of retinoblastoma tumor suppressor family proteins found in limited discrete regions of the nucleus at growth arrest. Mol Cell Biol. 2001;21(8):2918–2932. doi: 10.1128/MCB.21.8.2918-2932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleischer TC, Yun UJ, Ayer DE. Identification and characterization of three new components of the mSin3A corepressor complex. Mol Cell Biol. 2003;23(10):3456–3467. doi: 10.1128/MCB.23.10.3456-3467.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Binda O, Roy JS, Branton PE. RBP1 family proteins exhibit SUMOylation-dependent transcriptional repression and induce cell growth inhibition reminiscent of senescence. Mol Cell Biol. 2006;26(5):1917–1931. doi: 10.1128/MCB.26.5.1917-1931.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Classon M, Harlow E. The retinoblastoma tumour suppressor in development and cancer. Nat Rev Cancer. 2002;2(12):910–917. doi: 10.1038/nrc950. [DOI] [PubMed] [Google Scholar]
- 20.Cao J, Gao T, Giuliano AE, Irie RF. Recognition of an epitope of a breast cancer antigen by human antibody. Breast Cancer Res Treat. 1999;53(3):279–290. doi: 10.1023/a:1006115922401. [DOI] [PubMed] [Google Scholar]
- 21.Cui D, Jin G, Gao T, et al. Characterization of BRCAA1 and its novel antigen epitope identification. Cancer Epidemiol Biomarkers Prev. 2004;13(7):1136–1145. [PubMed] [Google Scholar]
- 22.Takahashi T, Cao J, Hoon DS, Irie RF. Cytotoxic T lymphocytes that recognize decameric peptide sequences of retinoblastoma binding protein 1 (RBP-1) associated with human breast cancer. Br J Cancer. 1999;81(2):342–349. doi: 10.1038/sj.bjc.6690698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meehan WJ, Samant RS, Hopper JE, et al. Breast cancer metastasis suppressor 1 (BRMS1) forms complexes with retinoblastoma-binding protein 1 (RBP1) and the mSin3 histone deacetylase complex and represses transcription. J Biol Chem. 2004;279(2):1562–1569. doi: 10.1074/jbc.M307969200. [DOI] [PubMed] [Google Scholar]
- 24.Nikolaev AY, Papanikolaou NA, Li M, Qin J, Gu W. Identification of a novel BRMS1-homologue protein p40 as a component of the mSin3A/p33(ING1b)/HDAC1 deacetylase complex. Biochem Biophys Res Commun. 2004;323(4):1216–1222. doi: 10.1016/j.bbrc.2004.08.227. [DOI] [PubMed] [Google Scholar]
- 25.Seraj MJ, Samant RS, Verderame MF, Welch DR. Functional evidence for a novel human breast carcinoma metastasis suppressor, BRMS1, encoded at chromosome 11q13. Cancer Res. 2000;60(11):2764–2769. [PubMed] [Google Scholar]
- 26.Wang GG, Allis CD, Chi P. Chromatin remodeling and cancer, part I: covalent histone modifications. Trends Mol Med. 2007;13(9):363–372. doi: 10.1016/j.molmed.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 27.Miremadi A, Oestergaard MZ, Pharoah PD, Caldas C. Cancer genetics of epigenetic genes. Hum Mol Genet. 2007;16(Spec No. 1):R28–R49. doi: 10.1093/hmg/ddm021. [DOI] [PubMed] [Google Scholar]
- 28.Ernst P, Mabon M, Davidson AJ, Zon LI, Korsmeyer SJ. An Mll-dependent Hox program drives hematopoietic progenitor expansion. Curr Biol. 2004;14(22):2063–2069. doi: 10.1016/j.cub.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 29.Vardiman JW, Harris NL, Brunning RD. The World Health Organization (WHO) classification of the myeloid neoplasms. Blood. 2002;100(7):2292–2302. doi: 10.1182/blood-2002-04-1199. [DOI] [PubMed] [Google Scholar]
- 30.Kogan SC, Ward JM, Anver MR, et al. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood. 2002;100(1):238–245. doi: 10.1182/blood.v100.1.238. [DOI] [PubMed] [Google Scholar]
- 31.Eklund EA. The role of HOX genes in malignant myeloid disease. Curr Opin Hematol. 2007;14(2):85–89. doi: 10.1097/MOH.0b013e32801684b6. [DOI] [PubMed] [Google Scholar]
- 32.Rice KL, Licht JD. HOX deregulation in acute myeloid leukemia. J Clin Invest. 2007;117(4):865–868. doi: 10.1172/JCI31861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamps MP, Baltimore D. E2A-Pbx1, the t(1;19) translocation protein of human pre-B-cell acute lymphocytic leukemia, causes acute myeloid leukemia in mice. Mol Cell Biol. 1993;13(1):351–357. doi: 10.1128/mcb.13.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moskow JJ, Bullrich F, Huebner K, Daar IO, Buchberg AM. Meis1, a PBX1-related homeobox gene involved in myeloid leukemia in BXH-2 mice. Mol Cell Biol. 1995;15(10):5434–5443. doi: 10.1128/mcb.15.10.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arakawa H, Nakamura T, Zhadanov AB, et al. Identification and characterization of the ARP1 gene, a target for the human acute leukemia ALL1 gene. Proc Natl Acad Sci USA. 1998;95(8):4573–4578. doi: 10.1073/pnas.95.8.4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyoshi H, Shimizu K, Kozu T, Maseki N, Kaneko Y, Ohki M. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc Natl Acad Sci USA. 1991;88(23):10431–10434. doi: 10.1073/pnas.88.23.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ziemin-van der Poel S, McCabe NR, Gill HJ, et al. Identification of a gene, MLL, that spans the breakpoint in 11q23 translocations associated with human leukemias. Proc Natl Acad Sci USA. 1991;88(23):10735–10739. doi: 10.1073/pnas.88.23.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brunkow ME, Jeffery EW, Hjerrild KA, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27(1):68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- 39.Ono M, Yaguchi H, Ohkura N, et al. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature. 2007;446(7136):685–689. doi: 10.1038/nature05673. [DOI] [PubMed] [Google Scholar]
- 40.Zheng Y, Rudensky AY. Foxp3 in control of the regulatory T cell lineage. Nat Immunol. 2007;8(5):457–462. doi: 10.1038/ni1455. [DOI] [PubMed] [Google Scholar]
- 41.Zuo T, Wang L, Morrison C, et al. FOXP3 is an X-linked breast cancer suppressor gene and an important repressor of the HER-2/ErbB2 oncogene. Cell. 2007;129(7):1275–1286. doi: 10.1016/j.cell.2007.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milne TA, Briggs SD, Brock HW, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10(5):1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 43.Huret JL, Dessen P, Bernheim A. An atlas of chromosomes in hematological malignancies. Example: 11q23 and MLL partners. Leukemia. 2001;15(6):987–989. doi: 10.1038/sj.leu.2402135. [DOI] [PubMed] [Google Scholar]
- 44.Zhang HZ, Degar BA, Rogoulina S, et al. Hematopoiesis following disruption of the Pitx2 homeodomain gene. Exp Hematol. 2006;34(2):167–178. doi: 10.1016/j.exphem.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 45.Kappen C. Disruption of the homeobox gene Hoxb-6 in mice results in increased numbers of early erythrocyte progenitors. Am J Hematol. 2000;65(2):111–118. doi: 10.1002/1096-8652(200010)65:2<111::aid-ajh4>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 46.Krishnaraju K, Hoffman B, Liebermann DA. Lineage-specific regulation of hematopoiesis by HOX-B8 (HOX-2.4): inhibition of granulocytic differentiation and potentiation of monocytic differentiation. Blood. 1997;90:1840–1849. [PubMed] [Google Scholar]
- 47.Knoepfler PS, Sykes DB, Pasillas M, Kamps MP. HoxB8 requires its Pbx-interaction motif to block differentiation of primary myeloid progenitors and of most cell line models of myeloid differentiation. Oncogene. 2001;20(39):5440–5448. doi: 10.1038/sj.onc.1204710. [DOI] [PubMed] [Google Scholar]
- 48.Bjornsson JM, Larsson N, Brun AC, et al. Reduced proliferative capacity of hematopoietic stem cells deficient in Hoxb3 and Hoxb4. Mol Cell Biol. 2003;23(11):3872–3883. doi: 10.1128/MCB.23.11.3872-3883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li B, Saouaf SJ, Samanta A, Shen Y, Hancock WW, Greene MI. Biochemistry and therapeutic implications of mechanisms involved in FOXP3 activity in immune suppression. Curr Opin Immunol. 2007;19(5):583–588. doi: 10.1016/j.coi.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 50.Chatila TA, Blaeser F, Ho N, et al. JM2, encoding a fork head-related protein, is mutated in X-linked autoimmunity-allergic disregulation syndrome. J Clin Invest. 2000;106(12):R75–R81. doi: 10.1172/JCI11679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bennett CL, Christie J, Ramsdell F, et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27(1):20–21. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- 52.Wildin RS, Ramsdell F, Peake J, et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat Genet. 2001;27(1):18–20. doi: 10.1038/83707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.