Abstract
By interacting with the cytoplasmic tail of a Golgi-processed form of transforming growth factor-α (TGFα), Naked2 coats TGFα-containing exocytic vesicles and directs them to the basolateral corner of polarized epithelial cells where the vesicles dock and fuse in a Naked2 myristoylation-dependent manner. These TGFα-containing Naked2-associated vesicles are not directed to the subapical Sec6/8 exocyst complex as has been reported for other basolateral cargo, and thus they appear to represent a distinct set of basolaterally targeted vesicles. To identify constituents of these vesicles, we exploited our finding that myristoylation-deficient Naked2 G2A vesicles are unable to fuse at the plasma membrane. Isolation of a population of myristoylation-deficient, green fluorescent protein-tagged G2A Naked2-associated vesicles was achieved by biochemical enrichment followed by flow cytometric fluorescence-activated vesicle sorting. The protein content of these plasma membrane de-enriched, flow-sorted fluorescent G2A Naked2 vesicles was determined by LC/LC-MS/MS analysis. Three independent isolations were performed, and 389 proteins were found in all three sets of G2A Naked2 vesicles. Rab10 and myosin IIA were identified as core machinery, and Na+/K+-ATPase α1 was identified as an additional cargo within these vesicles. As an initial validation step, we confirmed their presence and that of three additional proteins tested (annexin A1, annexin A2, and IQGAP1) in wild-type Naked2 vesicles. To our knowledge, this is the first large scale protein characterization of a population of basolaterally targeted exocytic vesicles and supports the use of fluorescence-activated vesicle sorting as a useful tool for isolation of cellular organelles for comprehensive proteomics analysis.
Transforming growth factor-α (TGFα),1 one of seven mammalian epidermal growth factor receptor ligands, is directed preferentially to the basolateral surface of polarized epithelial cells (1–4). It contains a dileucine basolateral sorting motif in its 39-amino acid cytoplasmic tail, the region of TGFα that is most highly conserved across species (4). We have found that Naked2, but not Naked1, recognizes basolateral sorting determinants in the cytoplasmic tail of Golgi-processed TGFα, coating these TGFα-containing vesicles and directing these vesicles to the basolateral surface of polarized epithelial cells (1). Naked1 and -2 are mammalian members of the Naked family (Naked Cuticle in Drosophila) that are reported to act as inducible antagonists of canonical Wnt signaling in Drosophila, zebrafish, and mammals (5–7).
Based on studies of basolateral trafficking of LDLR and VSVG protein (8, 9), it is thought that basolaterally targeted vesicles and their attendant cargo are directed to a subapical Sec6/8 exocyst targeting patch where the vesicles dock and fuse. In addition, LDLR and VSVG utilize the AP-1 adaptor machinery, either at the TGN and/or recycling endosome, for their successful delivery to the basolateral surface (10, 11). TGFα-containing Naked2-associated vesicles appear to represent a distinct set of basolaterally targeted vesicles based on the following four observations. First, Naked2 contains basolateral sorting information; residues 1–173 of Naked2 fused to Na+/H+ exchanger regulatory factor-1 are able to completely redirect Na+/H+ exchanger regulatory factor-1 from the apical cytoplasm to the basolateral plasma membrane (2). Second, Naked2 vesicles are delivered directly to the lower lateral membrane of polarized MDCK cells and not to a subapical Sec6/8 targeting patch. Third, this delivery does not require AP-1B because Naked2-associated vesicles are found at the basolateral surface of polarized LLC-PK1 cells that lack μ1B, an essential component of the heterotetrameric AP-1B complex (12). Finally, docking and fusion of these vesicles is dependent upon Naked2 myristoylation because Naked2-associated vesicles accumulate asymmetrically at the basolateral corner of MDCK cells stably expressing myristoylation-deficient G2A Naked2. These multiple functions of Naked2 have led us to designate it a cargo recognition and targeting protein (CaRT) for TGFα-containing, basolaterally targeted exocytic vesicles (2).
The distinctive properties of these Naked2 vesicles spurred us to determine the protein composition of these vesicles. Previous attempts to analyze basolaterally targeted vesicles have been unsuccessful likely because of their low abundance and transient nature with rapid fusion to acceptor membranes. Herein we exploited the fact that myristoylation-deficient G2A Naked2-enhanced green fluorescent protein (EGFP)-associated vesicles are “trapped” in time and space to biochemically isolate a population of these vesicles that are de-enriched for plasma membrane, ER, and Golgi constituents. We then used fluorescence-activated vesicle sorting (FAVS) as a novel means to purify a discrete pool of G2A Naked2-containing exocytic vesicles for large scale LC/LC-MS/MS analysis. By application of stringent acceptance criteria, we identified 389 proteins in these G2A Naked2-containing vesicles. We selected six of these proteins to study in wild-type (WT) Naked2-expressing MDCK cells, and we confirmed the presence of all six in WT Naked2-containing vesicles. These included proteins previously linked to basolateral exocytosis (Rab10, myosin IIA, Na+/K+-ATPase α1, IQGAP1, and annexin A2) as well as annexin A1 that has been associated with apical trafficking. We propose that FAVS may be a useful tool for isolation of cellular organelles for comprehensive LC/LC-MS/MS analysis.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies—
All cell culture reagents were from HyClone (Logan, UT). All chemicals, including TCA, anti-mouse myosin IIA antibody, and protease inhibitor mixture, were from Sigma unless otherwise stated. Chemicals for electrophoresis were purchased from Bio-Rad. A rabbit anti-human Naked2 antibody, R44, was made in collaboration with Covance (Princeton, NJ) as described by Li et al. (1). Monoclonal antibodies to annexins A1 and A2 were purchased from Abcam (Cambridge, MA). Anti-Na+/K+-ATPase α1, -E-cadherin, and -IQGAP1 were obtained from Upstate (Temecula, CA), and anti-caveolin-1 was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Golgin97, anti-mouse CD147, anti-rabbit IgG, and DiD (in oil) were purchased from Invitrogen-Molecular Probes. Protein A beads and M-PER (Mammalian Protein Extraction Reagent) were purchased from Pierce. Cy3-conjugated donkey anti-sheep IgG and horseradish peroxidase-conjugated donkey anti-mouse and -rabbit IgG were obtained from Jackson ImmunoResearch Laboratories (West Grove, PA). The Rab11a antibody has been characterized previously (13). The rabbit polyclonal antibody against Rab10 (VU132/134) was raised against a specific peptide sequence at the carboxyl-terminal variable domain, CKTPVKEPNSENVDIS. Keyhole limpet hemocyanin was covalently attached to the amino-terminal cysteine for immunization of the rabbits (Covance). The antiserum displayed a single 23-kDa band upon Western blotting of MDCK and HeLa cell lysates (data not shown). The HaloTag vector, pHT2, was purchased from Promega (Promega, Madison, WI).
Cell Culture and Transfection—
MDCK Tet-Off cells T23 1628 (Clontech) were stably transfected with WT human Naked2 (codons 1–451) or G2A (in which the second residue glycine was mutated to alanine) cDNAs; EGFP was fused to the carboxyl terminus of both Naked2 cDNAs. Cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, glutamine, non-essential amino acids, 100 units/ml penicillin, 100 μg/ml streptomycin, and 500 μg/ml Geneticin or 200 μg/ml hygromycin B (Roche Applied Science). Cells were maintained in doxycycline (1 μg/ml). Experiments were performed 48 h after removal of doxycycline at which time there was maximum expression of transfected Naked2 as described previously (1). HEK 293 cells and HCA-7 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. For polarization experiments with MDCK cells, 1 × 105 cells were seeded on 12-mm Transwell filters (0.4 μm; Corning Costar, Corning, NY) and cultured for 4–5 days with replenishment of medium every other day until trans-epithelial electrical resistance exceeded 200 ohms/cm2 using the Millicell Electrical Resistance System (Millipore Corp., Bedford, MA). MDCK cells were transiently co-transfected with WT Naked2-HaloTag™ and G2A Naked2-EGFP cDNAs, and fluorescence was observed 24 h later. The Goldenring laboratory kindly provided the monomeric dsRed-Rab10 (C2 Clontech vector) plasmid.
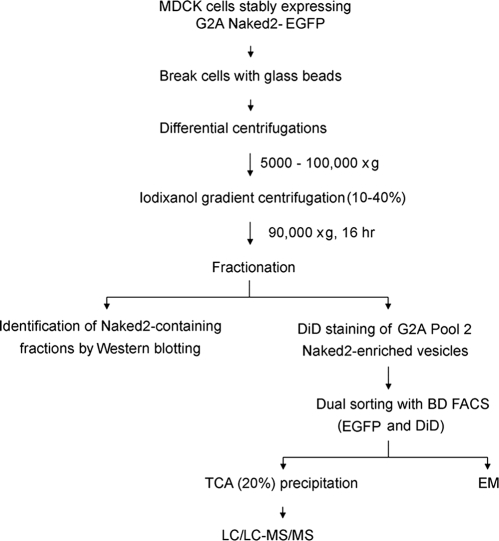
Vesicle Purification and Iodixanol Equilibrium Density Gradient Analysis—
The schema for purifying Naked2-EGFP vesicles from MDCK cells is summarized in Fig. 1. We modified the OptiPrep™ protocol (S37; Axis Shield, Oslo, Norway) as described below. For each preparation, MDCK cells were plated on 24 × 150-mm plastic dishes and grown to 70% confluence at which time doxycycline was removed for 48 h. 5 × 108 cells were rinsed three times in 1× PBS and then washed once in Solution D (78 mm KCl, 4 mm MgCl2, 8 mm CaCl2, 10 mm EGTA, and 50 mm HEPES-KOH, pH 7.0). The cells were scraped off with a rubber policeman in Solution E (0.25 m sucrose, 78 mm KCl, 4 mm MgCl2, 8 mm CaCl2, 10 mm EGTA, and 50 mm HEPES-KOH, pH 7.0). The cell suspension was mixed with an equal volume of acid-washed glass beads (Sigma) and agitated at top speed eight times for 2 min on a BioSpec Mini-Beadbeater-8TM vortex mixer (BioSpec Products Inc., Bartlesville, OK). In preliminary experiments (data not shown), we determined that the efficiency of vesicle isolation was greater using glass beads (80%) than a Dounce homogenizer (30%; Wheaton Science Products, Millville, NJ). Lysates were then diluted 10-fold with Solution E and centrifuged at 4 °C sequentially at 1,000 × g (10 min; SS-34 rotor, Sorvall), 5,000 × g (40 min; SS-34 rotor, Sorvall), and 100,000 × g (2 h; SW 28 rotor, Beckman). This vesicle-enriched pellet was resuspended in 9 ml of digestion buffer (75 mm KCl, 50 mm Tris-HCl, 3 mm MgCl2, and 10 mm DTT, pH 8.3) and digested with RNase A (20 μg/ml) at 37 °C for 30 min at which time the digestion was stopped by the addition of 10 mm EDTA. Approximately 1.5 ml of the resuspended pellet then was placed on top of a discontinuous 10, 20, 25, 30, and 40% iodixanol gradient solution (2 ml each in a 12-ml-volume tube) and centrifuged at 90,000 × g at 4 °C for 16 h in a TH641 rotor (Beckman). Successive 500-μl aliquots were taken from the top of the ultracentrifuge tubes, and 23 fractions were collected. All operations were performed either on ice or at 4 °C. The optical densities were read at 600 and 280 nm. An aliquot from each fraction was used for Western blotting of Naked2 and various cellular markers.
Fig. 1.
Flow chart for identification of Naked2-EGFP-associated vesicles from MDCK cells.
FAVS—
DiD is a lipophilic fluorescent dye that nonspecifically partitions into membrane bilayers. Pool 2 (fractions 18–22) from G2A Naked2-EGFP-expressing MDCK cells was subjected to DiD staining (final concentration, 12.5 μg/ml) at 37 °C for 10 min in Solution D. The EGFP and DiD dual-labeled Pool 2 vesicles were pelleted at 100,000 × g for 1 h. The pellet was resuspended in 7 ml of Solution E and successively sheared through 22-, 27- and 30-gauge syringes six times each before FAVS. Sorting was performed on a BD Biosciences FACSAria customized with a forward scatter PMT and standardized for linearity and sensitivity using eight peak beads (Spherotech, Lake Forest, IL). Resolution of particle size was refined using green fluorescent beads ranging in size from 40 to 700 nm (Duke Scientific, Fremont, CA). A custom high salt sheath (Solution D), which was compatible with the vesicles, was filtered through a 100-nm filter prior to installation into the Aria sheath reservoir. Two additional in-line filters (200 nm) were used to assure the sheath had the lowest background possible. Unstained and single stained (DiD or GFP only) vesicles were used to compensate for spectral overlap. Doubly positive DiD-counterstained G2A Naked2-EGFP-tagged vesicles were gated and subjected to pulse processing analysis for doublet discrimination. Extensive preliminary studies were performed to determine and validate that the sorting gates contained the double-labeled vesicles of interest. Briefly vesicles were sorted using a variety of sort gating strategies, and the isolated vesicles were concentrated separately and imaged using confocal microscopy. These preliminary studies allowed us to define a target mask with a linear range of fluorescence intensities that enabled significant enrichment of the double-labeled vesicles. Vesicles falling outside the target mask included significant numbers of doublet vesicles that, even upon repeated sorting, could not yield a single peak and thus were excluded from subsequent analysis. Finally double positive vesicles were gated to remove those vesicles greater than 0.55 SD from their mean fluorescence intensities.
LC/LC-MS/MS Analysis—
Pool 2 from G2A Naked2-sorted vesicles was concentrated using 20% TCA at 4 °C overnight. A transparent pellet was obtained after centrifugation at 15,000 rpm for 20 min at 4 °C. The pellet was washed three times with 10 ml of ice-chilled acetone, dried in a fume hood for 10 min, and then resuspended in 100 μl of 1× lauryl dodecyl sulfate NuPAGE sample buffer (Invitrogen, 4×) with reducing agent DTT (final concentration, 50 μm). The sample was heated for 10 min at 85 °C and then run ∼1.5 cm into a 10% SDS-polyacrylamide gel. After staining with Colloidal Coomassie Blue (Invitrogen), the entire stained protein region was excised, chopped into 1-mm cubes, and placed in 0.5-ml Eppendorf tubes containing 150 μl of 100 mm ammonium bicarbonate. Samples were reduced with 10 μl of 45 mm DTT for 20 min at 55 °C and alkylated with 10 μl of 100 mm iodoacetamide for 20 min at room temperature in the dark. Samples were destained with 100 μl of 50% acetonitrile and 50 mm ammonium bicarbonate. The gel pieces were then dehydrated with 100% acetonitrile and digested with trypsin (10 μl of 0.01 ng/μl Trypsin Gold (Promega) in 25 mm ammonium bicarbonate) overnight at 37 °C. The peptides were extracted with two rounds of 60% acetonitrile and 0.1% trifluoroacetic acid. They were dried down and reconstituted in 0.1% formic acid. The resulting peptides were analyzed by multidimensional chromatography-tandem mass spectrometry using a combination of off-line cation exchange separation and on-line reverse phase separation. The peptides were first fractionated using strong cation exchange chromatography (Luna SCX, Phenomenex, Torrance, CA) with a 100-μm × 10-cm column and using a 0–500 mm ammonium formate, pH 3.0–8.0, gradient in 25% acetonitrile as originally described by Adkins et al. (14) with minor modifications as described previously (13). Ten equal fractions were collected from the SCX fractionation, and fractions 7–9 were combined. The flow-through and seven fractions were subjected to LC-MS analysis using a ThermoFinnigan LTQ ion trap mass spectrometer equipped with a Thermo MicroAS autosampler and Thermo Surveyor HPLC pump, Nanospray source, and Xcalibur 1.4 instrument control. The peptides were separated on a packed capillary tip, 100 μm × 11 cm, with C18 resin (Jupiter C18, 5 μm, 300 Å, Phenomenex, Torrance, CA) using an in-line solid phase extraction column (100 μm × 3 cm) packed with the same C18 resin using a 0.1% formic acid, acetonitrile gradient as described previously (14).
Centroided MS/MS scans were acquired using an isolation width of 2 m/z, an activation time of 30 ms, an activation Q of 0.250, and 30% normalized collision energy using one microscan and maximum injection time of 100 ms for each MS/MS scan. The mass spectrometer was tuned prior to analysis using the synthetic peptide TpepK (AVAGKAGAR) so that some parameters may have varied slightly from experiment to experiment, but typically the tune parameters were as follows: spray voltage of 1.8 KV, a capillary temperature of 160 °C, a capillary voltage of 50 V, and tube lens voltage of 120 V. The MS/MS spectra of the peptides were collected using data-dependent scanning in which one full MS spectrum was followed by three MS/MS spectra. Raw data files for these analyses are available from the ProteomeCommons Tranche network (http://www.proteomecommons.org/data-downloader.jsp?fileName=t7lfD%2BVtLYdFQT8Bhlq7Zok3c52%2BJWOeJKmWUcCBTA/d65bAVtmTiIZkg8QMoCpWGXy/77QFnszqvSxjkDiuCCwNPBgAAAAAAAAT3w==) using hash codes.
Database Searching and Data Analysis—
The “ScanSifter” algorithm read tandem mass spectra stored as centroided peak lists from Thermo RAW files and transcoded them to mzData v1.05 files. Only MS/MS scans were written to the mzData files; MS scans were excluded. If 90% of the intensity of a tandem mass spectrum appeared at a lower m/z than that of the precursor ion, a single precursor charge was assumed; otherwise the spectrum was processed under both double and triple precursor charge assumptions. Proteins were identified using the MyriMatch v1.0.385 algorithm (15) on a cluster of 12 dual core × 86 processors. The database used (available as part of the raw data download above) was based on the Ensembl Canis familiaris sequence database, release 44 (25,568 sequences), with protein descriptions added through links between Ensembl and other protein databases. The database was augmented with 74 common contaminant proteins including five proteases, 42 Ig constant regions, 15 human keratins, and 12 proteins from wool, cotton, and saliva. Green fluorescent protein (GFP_AEQVI) and a sequence for human Naked2 (Q969F2_HUMAN) with the G2A mutation were added; accession numbers are from UniProtKB. Each protein was included in both normal and reversed orientation for a total of 51,288 database entries. The database search encompassed tryptic peptides with any number of missed cleavage sites; in practice, this setting yielded identifications in which one-third of the lysine and arginine residues were found in positions other than the peptide carboxyl terminus. All cysteines were expected to undergo carboxamidomethylation and were assigned a mass of 160 kDa. All methionines were allowed to be oxidized. Precursor ions were required to fall within 1.25 m/z of the position expected from their average masses, and fragment ions were required to fall within 0.5 m/z of their monoisotopic positions. The database searches produced raw identifications in SQT format (16).
Peptide identification filtering and protein assembly were conducted by the IDPicker algorithm (17). Initial filtering took place in multiple stages. First IDPicker filtered raw peptide identifications to a target false discovery rate (FDR) of 5%. The peptide filtering used reversed matching information to determine thresholds that yielded an estimated 5% FDR for the identifications of each charge state by the formula FDR = (2R)/(R + F) where R is the number of passing reversed peptide identifications and F is the number of passing forward (normal orientation) peptide identifications. The second round of filtering removed proteins supported by less than three distinct peptide identifications in the aggregate of the three experiments. As a final criterion, protein identifications were required to be supported by at least one spectrum in each of the three SCX/LC-MS/MS experiments (methionine oxidations were considered sequence differences). Indistinguishable proteins were recognized and grouped. Parsimony rules were applied to generate a minimal list of proteins that explained all of the peptides that passed our entry criteria. No reversed proteins passed the second level criteria so that zero proteins were estimated to be falsely identified in this list, i.e. a 0% FDR.
Immunoprecipitation, Immunofluorescence, and Western Blotting—
Cells were lysed in Pierce lysis buffer (M-PER). Protein A beads were blocked for 10 min in 2% BSA (TBS buffer, pH 8.0). For immunoprecipitations, lysates corresponding to 1 mg of total cellular protein were incubated overnight with primary antibody and then bound to protein A-agarose beads for 2 h at 4 °C. The beads were then washed five times with 1× lysis buffer-protease inhibitor mixture (0.2 mm 4-(2-aminoethyl)benzenesulfonyl fluoride, 0.1 mm EDTA, 13 μm bestatin, 1.4 μm E-64, 0.1 μm leupeptin, and 0.03 μm aprotinin). Immunoprecipitates were resolved by 7–12.5% SDS-PAGE and transferred onto nitrocellulose membranes before Western blotting. For immunofluorescence, MDCK cells cultured on plastic were fixed with 4% paraformaldehyde (PF) for 15 min, permeabilized with 0.5% Triton X-100 for 10 min, and then blocked for 1 h in 2% BSA. For annexin A2 analysis, cells were treated with 0.25% Triton X-100 for 30 s before 4% PF fixation. Immunofluorescence or live cell microscopy was visualized using a Zeiss LSM 510 confocal microscope. Microscopy was performed within the Vanderbilt Cell Imaging Shared Resource. All micrographs were taken using a 63× oil immersion objective lens.
Verification of Myosin IIA Identification and Blebbistatin Treatment—
HCA-7 cells were lysed with M-PER (Pierce) plus protease inhibitors. After 10-s sonication, cell lysates were centrifuged at 13,000 rpm for 10 min at 4 °C. Five μl of R44 (0.5 μg/μl) was used to immunoisolate Naked2. Immunoprecipitates were resolved by SDS-PAGE and stained with Coomassie Blue. Bands were isolated and treated with trypsin (as described above) and identified by peptide mass fingerprinting augmented with MS/MS analysis. MALDI-TOF MS and data-dependent TOF/TOF tandem MS were performed on a Voyager 4700 mass spectrometer (Applied Biosystems, Framingham, MA). Peptide mass maps were acquired at mass accuracy within 10 ppm using trypsin autolytic fragments for internal calibration. MS data were used to search for statistically significant candidate protein matches in the Swiss-Prot database (Sprot_20060322, 212,425 sequences) using GPS Explorer software v3.6 (Applied Biosystems) running the MASCOT search algorithm v1.9 (Matrix Science). Searches were performed without constraining protein molecular weight or isoelectric point and with complete carbamidomethylation of cysteine, partial oxidation of methionine residues, and one missed trypsin cleavage allowed. MDCK cells stably transfected with Naked2-EGFP were grown on coverslips for 2 h and then exposed to 5 μm blebbistatin (Calbiochem) for 16 h.
Electron Microscopy—
5 μl of the post-sorted Pool 2 G2A Naked2-associated vesicles were dried on a grid overnight at room temperature, applied to Formvar carbon-coated grids, and negative stained by exposure for 20 s to 2% aqueous sodium phosphotungstic acid, pH 6.1. The negative stain solution was removed by wicking onto filter paper, and the dried sample was viewed with a Philips/FEI CM-12 transmission electron microscope operating at 80 keV (Fig. 4C, left panel; magnification, 53,000; bar, 100 nm). For thin section transmission electron microscopy, post-sorted vesicles were fixed and stained by incubation for 1 h with 1% aqueous osmium tetroxide. The osmium-treated post-sorted vesicles were then extensively washed and spun down at 100,000 × g for 1 h three times. The washed postsorted vesicles were then mixed with 10% BSA using a 27-gauge needle, 0.5-ml syringe to facilitate embedding. The vesicle-BSA mixtures were fixed in 2.5% glutaraldehyde in 0.1 m cacodylate buffer overnight at 4 °C, dehydrated through a graded series of ethanol washes, and embedded in Spurr resin. Thin sections of the embedded pellet were viewed at 80 keV using a Philips/FEI CM-12 transmission electron microscope (Fig. 4C, right panel; magnification, 88,000; bar, 100 nm).
Fig. 4.
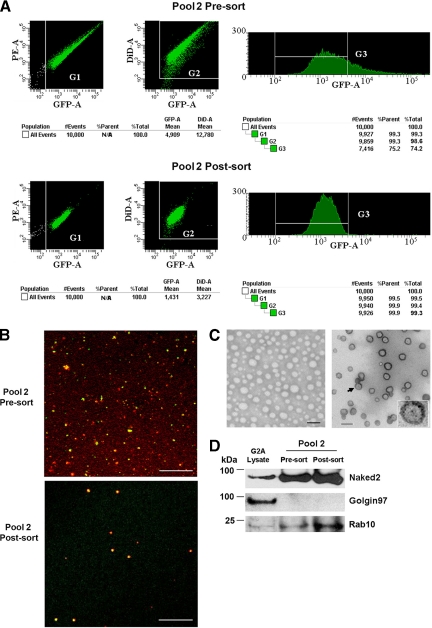
FAVS of Pool 2 G2A Naked2-EGFP vesicles. A, purification of GFP and DiD dual-labeled G2A Naked2-associated Pool 2 vesicles is described in detail under “Experimental Procedures.” Sorting was performed on a BD Biosciences FACSAria customized with a forward scatter PMT and standardized for linearity. Unstained and single stained (GFP or DiD) vesicles were used for compensation of spectral overlap. Doubly positive DiD-counterstained EGFP-tagged Pool 2 G2A Naked2 fluorescent vesicles were gated and subjected to pulse processing analysis for doublet discrimination. Finally single doubly positive fluorescent vesicles were gated to remove GFP and DiD vesicles that were greater than 0.55 SD from their mean fluorescent intensities, respectively. Partially purified vesicles (top panel) exhibited, at best, 74% double positivity (dual fluorescence for DiD and GFP), whereas post-sorted vesicles (bottom panel) displayed greater than 99% double positivity (see “Experimental Procedures” for details). B, merged images of DiD (red) and Naked2-EGFP (green) of pre-sorted (top) and post-sorted (bottom) Pool 2 G2A Naked2-associated vesicles (bar, 10 μm). C, 5 μl of post-sorted vesicles was dried on a grid overnight at room temperature for negative staining (left panel; bar, 100 nm). For positive staining (right panel; bar, 100 nm), post-sorted vesicles were stained with 1% aqueous osmium tetroxide for 1 h. The negative staining grids or positive staining thin sections then were observed at 80 kV on a Philips FEI CM-12 electron microscope. The arrow points to a vesicle shown at higher magnification in the inset. This vesicle exhibits a single membrane bilayer. D, immunoblots showing relative abundance of Naked2, Golgin97, and Rab10 in whole cell lysates and pre-sort and post-sort Pool 2 G2A Naked2-associated vesicles. Equal amounts of protein (15 μg/lane) were loaded and separated in NuPAGE 4–12% Bis-Tris gels (Invitrogen).
RESULTS AND DISCUSSION
Isolation of Naked2-associated Vesicles—
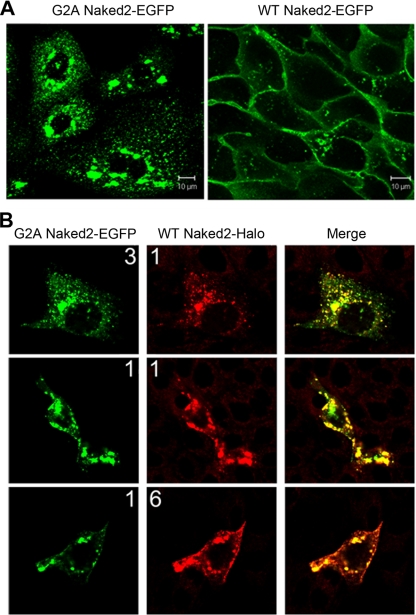
We demonstrated previously that WT Naked2-EGFP decorates the basolateral membrane of polarized MDCK cells (1). In contrast, myristoylation-deficient G2A Naked2-EGFP is not found at the plasma membrane but rather accumulates in the cytoplasm; by electron microscopy, this fluorescence was shown to represent cytoplasmic accumulation of G2A Naked2-associated vesicles (1). We considered that these trapped G2A Naked2-associated vesicles might provide a source of vesicles for proteomics analysis. Fig. 2A shows punctate vesicle-like fluorescence in the cytoplasm of MDCK cells stably expressing G2A Naked2 cultured on plastic, whereas WT Naked2 fluorescence was observed at the plasma membrane as well as in the cytoplasm. There was co-localization of fluorescence in MDCK cells transiently expressing both Halo-tagged WT Naked2 and EGFP-tagged G2A Naked2 (Fig. 2B). Of interest, the distribution of co-localization depends upon the input ratio of the two constructs. When the G2A Naked2 input exceeded that of WT Naked2, there was co-localization in vesicles. However, there was plasma membrane and vesicle co-localization when WT Naked2 input exceeded that of G2A Naked2. The preponderance of cytosolic vesicle-like co-staining at equal input ratios may reflect the longer half-life of myristoylation-deficient G2A Naked2 compared with WT Naked2 (1). The co-localization of WT and G2A Naked2 in co-expressing MDCK cells suggests that these trapped G2A Naked2-associated vesicles are bona fide transport intermediates and supports using them as the starting material to isolate basolaterally targeted vesicles in sufficient quantity to perform proteomics analysis.
Fig. 2.
G2A Naked2 does not localize to the plasma membrane. A, MDCK cells stably expressing G2A Naked2-EGFP (left panel) and WT Naked2-EGFP (right panel) were cultured on plastic. Punctate vesicle-like fluorescence was observed in the cytoplasm of G2A Naked2-EGFP-expressing cells, whereas WT Naked2 fluorescence was observed at the plasma membrane as well as in the cytoplasm. B, there was co-localization of vesicle-like fluorescence in MDCK cells transiently co-expressing G2A Naked2-EGFP and Halo-tagged WT Naked2. The input ratio of the respective cDNAs is shown for each panel with 1:1 representing 0.5 μg of DNA.
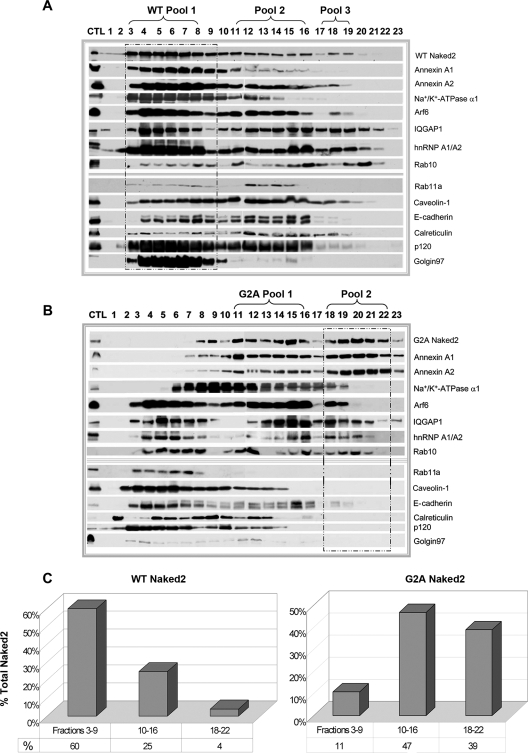
Initial attempts to immunoisolate G2A Naked2-associated vesicles were unsuccessful (data not shown), leading us to devise an alternate strategy. Our schema to isolate these trapped vesicles is shown in Fig. 1 and is fully described under “Experimental Procedures.” We initially compared our ability to isolate Naked2-containing fractions from lysates of WT and G2A Naked2-expressing MDCK cells resolved on 10–40% discontinuous iodixanol gradients. In WT Naked2-expressing MDCK cells, 60% of Naked2 was found in fractions 3–9 (WT Pool 1, Fig. 3A, framed in dashed box, and quantified in Fig. 3C). Pool 1 also contained markers for plasma membrane (E-cadherin, p120, and Na+/K+-ATPase α1) as well as ER- (calreticulin) and Golgi-resident (Golgin97) proteins (Fig. 3A). Although WT Naked2 was found predominantly in the lighter fractions of the iodixanol gradient, there was a small proportion of WT Naked2 found in the densest part of the gradient (fractions 18–22). In contrast, G2A Naked2 shifted to denser fractions of the gradient (fractions 11–16 and 18–22), and in particular, G2A Naked2 Pool 2 (Fig. 3B, framed in dashed box) was largely depleted of plasma membrane, ER, and Golgi constituents. For example, there was no Golgin97 or calreticulin in G2A Naked2 Pool 2, and only a small amount of E-cadherin was detected by Western blotting. Na+/K+-ATPase α1 was observed in G2A Naked2 Pool 2; however, this proved to be an additional cargo in Naked2-associated vesicles (see Fig. 6 and supplemental table, worksheet 5). Although WT Naked2 was found in fractions 18–22, there was a nearly 10-fold increase of Naked2 in the corresponding fractions of G2A Naked2-expressing MDCK cells (39 versus 4%; Fig. 3C).
Fig. 3.
Western blot analysis of iodixanol gradient fractions from WT and G2A Naked2-expressing MDCK cells. Cell lysates were prepared from stably expressing WT and G2A Naked2-expressing MDCK cells as described under “Experimental Procedures.” After a series of sequential centrifugations (see “Experimental Procedures”), a crude vesicle preparation was passed over a 10–40% discontinuous iodixanol gradient. Twenty-three successive 500-μl fractions were collected, and 30-μl individual aliquots were separated by 7–12.5% SDS-PAGE and probed with antibodies to the markers indicated. The dashed boxes indicate fractions designated Pool 1 in WT Naked2-expressing cells (A) and those containing Pool 2 in G2A Naked2-expressing cells (B) (see text for details). C, quantification of corresponding fractions between G2A and WT Naked2-expressing vesicles in iodixanol gradient fractions. CTL, control; hnRNP, heterogeneous nuclear ribonucleoprotein.
Fig. 6.
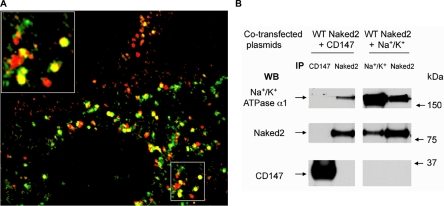
A, WT Naked2-expressing MDCK cells were cultured on glass coverslips, fixed, and stained as described under “Experimental Procedures.” Naked2 (green fluorescence) co-localized with endogenous Na+/K+-ATPase α1 (red fluorescence) with one example shown in higher power in the upper left corner. B, HEK 293T cells transiently co-transfected with human WT Naked2 and Na+/K+-ATPase α1 or CD147 were lysed, and reciprocal co-immunoprecipitation (IP) and Western blotting (WB) were performed between Naked2 and Na+/K+-ATPase α1 or CD147.
FAVS of Dual-labeled G2A Naked2-associated Vesicles—
This plasma membrane-de-enriched G2A Naked2 Pool 2 was then subjected to further purification by flow cytometry. Pool 2 vesicles were stained with the nonspecific lipophilic tracer DiD and then flow-sorted based on DiD and GFP fluorescence using a BD Biosciences FACSAria customized with a forward scatter PMT and standardized for linearity and sensitivity using eight peak beads (see “Experimental Procedures”). As described under “Experimental Procedures,” we performed extensive preliminary experiments to determine the sorting gates (G) that contained single peaks of double-labeled vesicles. Iodixanol gradient-fractionated Pool 2 vesicles exhibited 64–74% purity dual fluorescence for DiD and GFP (Fig. 4A, upper panel, G3), whereas post-sorted vesicles exhibited 99% double fluorescence (lower panel, G3). Note the dispersion of vesicles within G1 and G2 for the pre-sorted vesicles compared with the tight grouping of G1 and G2 post-sorted vesicles. The pre-sorted vesicles ranged in size from 30 to 300 nm (Fig. 4B, upper panel). In addition to dual fluorescence, we set stringent vesicle size inclusion criteria for post-sorted vesicles. Vesicles had to conform to a specified electrical signal pulse geometry to assure removal of doublets (see “Experimental Procedures”). A combination of light scatter pulse width versus fluorescence pulse height was used to recover doubly positive vesicles of the appropriate pulse geometry. In a representative experiment, 24 150-mm dishes of confluent G2A Naked2-EGFP-expressing MDCK cells were used. From Pool 2 alone, 30–50 × 109 vesicles were sorted over 24–40 h. By the criteria described above, 100–170 × 106 doubly positive vesicles that ranged in size from 40 to 75 nm were collected (Fig. 4B, lower panel). Thus, greater than 95% of the input vesicles were excluded to achieve 99% doubly positive, flow-sorted vesicles.
By negative stain EM (Fig. 4C, left panel), post-sorted Pool 2 contained membrane-delimited vesicles of a uniform size similar to that detected by confocal microscopy (Fig. 4B). The smaller size of these vesicles compared with what we reported previously for G2A Naked2-associated vesicles (1) likely represents a process of selection during the isolation procedure. The negative stain preparations also confirmed that we had isolated a population of smooth membrane-delimited vesicles with no evidence of contamination from mitochondria, nucleus, or rough microsomes. The strength of the negative stain procedure is that it required little additional sample manipulation and so represented a sample very similar to that analyzed by flow cytometry. However, to confirm and extend the negative stain analysis, we osmicated and embedded a sorted sample in Spurr resin (as described under “Experimental Procedures”) for thin section transmission EM analysis (Fig. 4C, right panel). This confirmed that the sample consisted primarily of vesicles with smooth membranes with no evidence of contamination with other intact organelles. Moreover by transmission EM, the vesicles did not have the appearance of lysosomes, multivesicular bodies, or late endosomes consistent with our conclusion that these structures represent exocytic vesicles. We cannot exclude the presence of Golgi membranes as the Golgi stacks may have been disrupted during the isolation procedure, and they will resemble smooth membranes; however, a trans Golgi marker, Golgin97, was not detected in pre-sort or post-sort Pool 2 vesicles (see below; Fig. 4D).
To further assess our enrichment strategy, we compared Naked2 levels by Western blotting of whole cell lysates and the pre-sort and post-sort Pool 2 vesicles (Fig. 4D). There was a marked increase in Naked2 levels in the pre-sort and post-sort compared with whole cell lysates. Despite the marked reduction in the total number of vesicles in the post-sort compared with pre-sort (Fig. 4B, compare upper and lower panels), Naked2 levels were increased in the post-sort compared with the pre-sort. Golgin97 was present in the whole cell lysates but was not detected in Pool 2 pre-sort (consistent with results in Fig. 3B) or the post-sort. In contrast, Rab10 was enriched in the pre-sort and further enriched in the post-sort. As shown below (see Fig. 7 and supplemental movie), Rab10, but not Golgin97, is a constituent of G2A Naked2 vesicles and co-localized with Naked2 in WT Naked2-expressing MDCK cells. It is instructive to note that total Rab10, unlike Naked2, did not shift in the gradient when comparing fractions from wild-type and G2A Naked2-expressing MDCK cells (Fig. 3). Rab10 has been implicated in regulating multiple post-Golgi and recycling pathways. Thus, it is likely that only a subset of total cellular Rab10 associates with Naked2 vesicles. Moreover live cell imaging indicated that the association of Rab10 with Naked2 vesicles is dynamic (see supplemental movie). To our knowledge, this combined biochemical and flow cytometric strategy is the first to successfully isolate basolaterally targeted exocytic vesicles in sufficient quantities to perform large scale proteomics analysis.
Fig. 7.
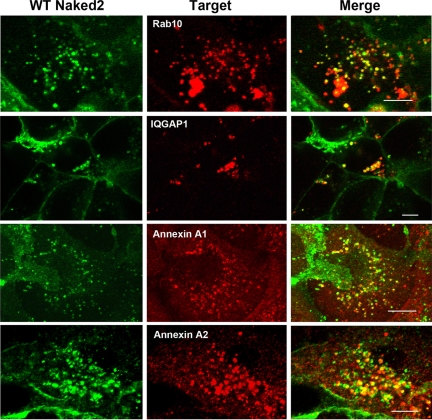
Proteins identified in G2A Naked2 vesicles are also found in WT Naked2 vesicles. MDCK cells stably expressing WT Naked2-EGFP were cultured on glass coverslips, fixed with 4% PF, and then permeabilized with 0.5% Triton X-100. Ds-Red Rab10 and endogenous IQGAP1, annexin A1, and annexin A2 were visualized using a Zeiss LSM 510 confocal microscope (see “Experimental Procedures” for details). Micrographs were taken with a 63× oil objective lens. Bars, 10 μm.
Protein Identification in G2A Naked2-associated Vesicles by LC/LC-MS/MS—
After optimizing our vesicle isolation and purification strategy, we performed three independent experiments to identify constituents of this population of exocytic vesicles. Details of the methods for peptide identification and protein assignment are provided under “Experimental Procedures.” Key features of the analysis included the use of a well annotated sequence database (Ensembl for C. familiaris), a high discrimination peptide identifier (15), and a parsimonious protein assembly algorithm (17) to improve the reliability of protein identification from these samples. Once the data were filtered by our second criterion to include at least three unique peptide identifications in any of the three experiments, we identified 1067 proteins (see supplemental table, worksheet 1) at a 0% FDR. As a final stringent criterion, it was required that three distinct peptide sequences in aggregate were identified with at least one spectrum in each of the three experiments (labeled Exp1/G8, Exp2/4t, and Exp3/G10).
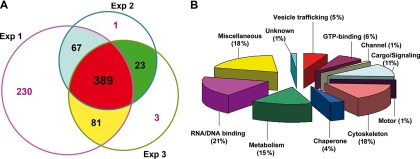
By these criteria, 389 proteins were identified (Fig. 5A) that were classified into 11 functional groups (Fig. 5B) (see supplemental Table, worksheet 2, for a complete list of all 389 proteins). We were able to achieve a greater number of protein identifications in Experiment 1 (Exp1/G8) utilizing twice as many vesicles as in Exp2/4t and Exp3/G10 (see supplemental table). Although many of the proteins identified in Experiment 1 may indeed include valid protein identifications not included in this final list, we chose to focus only on those proteins we identified in each of three separate biological isolations to increase our confidence that they were specific to the basolaterally targeted exocytotic vesicles. Proteins reported in the supplemental table are hyperlinked via their cluster IDs to peptide, group, and cluster information supplied by IDPicker via their individual descriptive html pages. These html pages for all protein clusters are provided in the supplemental IDPicker source folder (prow_files) that also contains a PDF tutorial for navigation of the IDPicker reports.
Fig. 5.
Summary of the proteins identified in G2A Naked2-associated vesicles. A, 389 proteins were identified in Pool 2 G2A Naked2-associated vesicles by LC/LC-MS/MS in three independent experiments. A total of 767, 480, and 496 proteins were identified in the three experiments from 1.75 × 108, 6.5 × 107, and 9.4 × 107 flow-sorted vesicles, respectively. The numbers 67, 23, and 81 represent overlapping proteins between two different experiments, respectively. B, functional classification of the 389 proteins found in all three experiments.
The supplemental table lists all 1067 proteins found in any of the three experiments (worksheet 1). The 389 proteins found in all three experiments are listed in the supplemental table (worksheet 2) by their functional classification (worksheets 3–13). An overall summary of the proteomics data is provided in worksheet 15. Assignment of a protein to a specific category can be somewhat arbitrary and restrictive because many proteins may perform multiple functions. For example, GRP75, a chaperone, may also participate in trafficking of FGF1 (18) and voltage-dependent anion-selective channel 1 (VDAC1) (19), the latter protein also found to associate with G2A Naked2 vesicles. It is important to emphasize that this is not intended to be an exhaustive or definitive list of proteins associated with basolateral trafficking but rather an initial characterization of a subpopulation of myristoylation-deficient Naked2-associated vesicles trapped in the cytoplasm. In the following section, we make some general comments about the proteins that were identified before discussing the six proteins whose presence was confirmed in WT Naked2 vesicles.
As expected, a total of 20 proteins were identified in the vesicle trafficking category including Naked2 (Table I and supplemental table, worksheet 3). We consistently identified human G2A Naked2 (eight peptides, 18 spectra) and GFP (UniProtKB accession number P42212|GFP_AEQVI) (seven peptides, nine spectra). Other vesicle-related proteins identified that have been linked to basolateral trafficking include caveolin-1 (20), vesicular integral membrane protein 36 precursor (21), vesicle-associated membrane protein A (VAP-33) (22), annexin A2 (23, 24), and annexin A13a (25).
Table I.
Identification of proteins linked to vesicle trafficking
Proteins are listed by canine Ensembl accession numbers with the exception of Naked2 and GFP. SID (sequence ID) represents arbitrary values differentiating indiscernible protein sets; GID (global ID) represents indiscernible protein group identifier; CID (cluster ID) represents protein groups that have common peptides (in the supplemental table, these identifiers are underlined and hyperlinked to their IDPicker Web pages); Unique peptides are the total number of unique peptides that were identified. Spectra list the total number of spectra in the three independent experiments: Exp1/G8 (Experiment 1), Exp2/4t (Experiment 2), and Exp3/G10 (Experiment 3). Acc/ID indicates other accession numbers and IDs. Numbers in the last column are the references reporting a link to basolateral trafficking.
| Protein | SID | GID | CID | Unique peptides | Spectra | Exp1/G8 | Exp2/4t | Exp3/G10 | Description | Acc/ID | Linked to basolateral trafficking |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Q969F2|0969F2_HUMAN | 1 | 794 | 196 | 8 | 18 | 6 | 5 | 7 | Naked cuticle homolog 2 (Naked2) (Homo sapiens) | UniRef100_Q9BSN0 | 1, 2 |
| P42212|GFP_AEQVI | 1 | 793 | 253 | 7 | 9 | 2 | 2 | 5 | GFP (Aequorea victoria) (jellyfish) | ||
| ENSCAFP00000026045 | 1 | 685 | 9 | 51 | 86 | 75 | 5 | 6 | Similar to clathrin heavy chain 1 (CLH-17, splice isoform 2) | ID: LOC480578 (Entrez Gene) | |
| ENSCAFP00000024537 | 1 | 638 | 21 | 36 | 103 | 56 | 18 | 29 | Annexin A2 | Acc: NP_001002961 | 23, 24, 52 |
| ENSCAFP00000002591 | 1 | 91 | 24 | 32 | 123 | 117 | 2 | 4 | Annexin Al (fragment) | Acc:G6JDN3 | 13, 52 |
| ENSCAFP00000018474 | 1 | 468 | 19 | 20 | 30 | 17 | 8 | 5 | Similar to coatomer α subunit (α-coat protein) (α-COP) (HEPCOP) | ID: LOC478973 (Entrez Gene) | |
| ENSCAFP00000015809 | 1 | 406 | 247 | 7 | 10 | 4 | 1 | 5 | Similar to SEC22 vesicle trafficking protein-like 1 (Sec22b) | ID: LOC475816 (Entrez Gene) | |
| ENSCAFP00000025081 | 1 | 653 | 230 | 7 | 13 | 6 | 3 | 4 | Similar to transmembrane trafficking protein 21 (transmembrane emp24-like trafficking protein 10) | ID: LOC610559 (Entrez Gene) | |
| ENSCAFP00000005595 | 1 | 168 | 391 | 5 | 5 | 1 | 3 | 1 | Similar to translocation protein SEC63 homolog | ID: LOC475016 (Entrez Gene) | |
| ENSCAFP00000009784 | 1 | 264 | 343 | 5 | 8 | 5 | 1 | 2 | Similar to ADP-ribosylation factor-like 6-interacting protein 5 | ID: LOC476559 (Entrez Gene) | |
| ENSCAFP00000011273 | 1 | 306 | 365 | 5 | 6 | 3 | 2 | 1 | Similar to coatomer protein complex, subunit β 2 | ID: LOC477088 (Entrez Gene) | |
| ENSCAFP00000027637 | 1 | 718 | 334 | 5 | 14 | 4 | 3 | 7 | Similar to vesicle-associated membrane protein-associated protein A (VAMP-associated protein A) (VAMP-A) (VAP-A) (33-kDa vamp-associated protein) (VAP-33) | ID: LOC480208 (Entrez Gene) | 22 |
| ENSCAFP00000001437 | 1 | 58 | 423 | 4 | 6 | 1 | 2 | 3 | Annexin A13 (annexin XIII) (annexin, intestine-specific) (ISA) | Acc: Q29471 | 25 |
| ENSCAFP00000005060 | 1 | 152 | 396 | 4 | 11 | 5 | 3 | 3 | Caveolin-1 (vesicular integral membrane protein VIP21) | Acc: P33724 | 20 |
| ENSCAFP00000006404 | 1 | 191 | 357 | 4 | 6 | 3 | 2 | 1 | Similar to coatomer protein complex, subunit γ 1 | ID: LOC476517 (Entrez Gene) | |
| ENSCAFP00000012445 | 1 | 330 | 498 | 4 | 4 | 2 | 1 | 1 | Similar to general vesicular transport factor p115 (transcytosis-associated protein) (TAP) (vesicle docking protein) | ID: LOC478430 (Entrez Gene) | |
| ENSCAFP00000024181 | 1 | 629 | 409 | 4 | 7 | 5 | 1 | 1 | Vesicular integral membrane protein VIP36 precursor (lectin, mannose-binding 2) | Acc: P49256 | 21 |
| ENSCAFP00000000331 | 1 | 29 | 521 | 3 | 5 | 2 | 2 | 1 | Transmembrane emp24 domain-containing protein 9 precursor (glycoprotein 25L2) | Acc: Q9BVK6 | |
| ENSCAFP00000002070 | 1 | 80 | 357 | 3 | 4 | 1 | 2 | 1 | Similar to coatomer γ-2 subunit (γ-2 coat protein) (γ-2 COP) | ID: LOC475189 (Entrez Gene) | |
| ENSCAFP00000012933 | 1 | 345 | 547 | 3 | 4 | 1 | 1 | 2 | Similar to transmembrane 9 superfamily protein member 3 precursor | ID: LOC612786 (Entrez Gene) |
We identified a number of Rabs and Arfs (Table II and supplemental table, worksheet 4). These included Rab10, Rab8a (26, 27), IQGAP1, and Arf 6 (28) that have been reported to be involved in basolateral trafficking. Forty-seven unique IQGAP1 peptides from 65 spectra were identified in Pool 2 G2A Naked2 flow-sorted vesicles. IQGAP1 is a widely expressed protein that has been linked to basolateral trafficking (29, 30). Of potential interest, we also identified RACK1, which has been shown to interact with IQGAP1 (31). We also found potential cargo molecules (supplemental table, worksheet 6) that have been linked to basolateral trafficking, including cathepsin D (32), integrin β1 (33), and solute carrier family 3 member 2 isoform a (CD98HC) (34).
Table II.
Identification of small GTP-binding proteins
SID (sequence ID) represents arbitrary values differentiating indiscernible protein sets; GID (global ID) represents indiscernible protein group identifier; CID (cluster ID) represents protein groups that have common peptides (in the supplemental table, these identifiers are hyperlinked to their IDPicker Web pages); Unique peptides is the total number of unique peptides that were identified. Spectra lists the total number of spectra in the three independent experiments: Exp1/G8 (Experiment 1), Exp2/4t (Experiment 2), and Exp3/G10 (Experiment 3). Acc/ID indicates other accession numbers and IDs. Numbers in the last column are the references reporting a link to basolateral trafficking.
| Protein | SID | GID | CID | Unique peptides | Spectra | Exp1/G8 | Exp2/4t | Exp3/G10 | Description | Acc/ID | Linked to basolateral trafficking |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ENSCAFP00000006309 | 1 | 187 | 50 | 6 | 11 | 3 | 2 | 6 | Ras-related protein Rab10 | Acc: P24409 | 26 |
| ENSCAFP00000023207 | 1 | 587 | 50 | 3 | 6 | 1 | 1 | 4 | Ras-related protein Rab8A (oncogene c-mel) | Acc: P61007 | 26 |
| ENSCAFP00000018867 | 1 | 482 | 11 | 47 | 65 | 50 | 8 | 7 | Similar to Ras GTPase-activating-like protein IQGAP1 (p195) | ID: LOC479050 (Entrez Gene) | 29, 30 |
| ENSCAFP00000032857 | 1 | 784 | 490 | 4 | 4 | 1 | 2 | 1 | Similar to ADP-ribosylation factor 6 | ID: LOC490679 (Entrez Gene) | 28 |
| ENSCAFP00000018770 | 1 | 479 | 85 | 14 | 37 | 14 | 16 | 7 | Similar to guanine nucleotide-binding protein β subunit 2-like 1 (receptor of activated protein kinase C 1) (RACK1) (receptor for activated C kinase) | ID: LOC480818 (Entrez Gene) | |
| ENSCAFP00000021319 | 1 | 543 | 53 | 19 | 50 | 21 | 12 | 17 | Prohibitin | ID: NP_009204 | |
| ENSCAFP00000008328 | 1 | 237 | 127 | 12 | 16 | 13 | 2 | 1 | Similar to RAN-binding protein 5 | ID: LOC485528 (Entrez Gene) | |
| ENSCAFP00000024890 | 1 | 650 | 148 | 10 | 27 | 10 | 4 | 13 | Similar to prohibitin | ID: LOC480547 (Entrez Gene) | |
| ENSCAFP00000000123 | 1 | 21 | 77 | 9 | 15 | 11 | 2 | 2 | Similar to RAB5B, member of RAS oncogene family | IDLOC474394 (Entrez Gene) | |
| ENSCAFP00000014941 | 1 | 387 | 156 | 9 | 14 | 9 | 3 | 2 | Interferon-induced GTP-binding protein Mx1 | Acc: Q9N0Y3 | |
| ENSCAFP00000012918 | 1 | 343 | 117 | 8 | 14 | 10 | 2 | 2 | Similar to ADP-ribosylation factor 3 | ID: LOC486562 (Entrez Gene) | |
| ENSCAFP00000006177 | 1 | 182 | 226 | 7 | 14 | 4 | 4 | 6 | Ras-related protein Rab7 | Acc: P18067 | |
| ENSCAFP00000011521 | 1 | 312 | 117 | 7 | 12 | 7 | 3 | 2 | Similar to ADP-ribosylation factor 4 | ID: LOC607073 (Entrez Gene) | |
| ENSCAFP00000018936 | 1 | 484 | 50 | 7 | 11 | 5 | 2 | 4 | Similar to RAB1B, member RAS oncogene family | ID: LOC610762 (Entrez Gene) | |
| ENSCAFP00000022977 | 1 | 582 | 77 | 7 | 13 | 7 | 1 | 5 | Ras-related protein Rab5C | Acc:P51147 | |
| ENSCAFP00000002515 | 1 | 90 | 117 | 6 | 10 | 6 | 2 | 2 | Similar to ADP-ribosylation factor 5 | ID: LOC475206 (Entrez Gene) | |
| ENSCAFP00000004753 | 1 | 143 | 50 | 6 | 9 | 6 | 1 | 2 | Ras-related protein Rab1A | Acc: P62822 | |
| ENSCAFP00000008690 | 1 | 245 | 77 | 6 | 9 | 5 | 1 | 3 | Ras-related protein Rab5A | Acc: P18066 | |
| ENSCAFP00000025408 | 1 | 669 | 49 | 6 | 13 | 3 | 4 | 6 | Ras-related protein Rab11A (Rab11) | Acc: P62490 | |
| ENSCAFP00000000596 | 1 | 35 | 564 | 3 | 4 | 2 | 1 | 1 | Similar to RAS-related protein 1b | ID: LOC608981 (Entrez Gene) | |
| ENSCAFP00000013676 | 1 | 362 | 11 | 3 | 5 | 2 | 2 | 1 | Similar to Ras GTPase-activating-like protein IQGAP2 | ID: LOC479177 (Entrez Gene) | |
| ENSCAFP00000014935 | 1 | 386 | 156 | 3 | 6 | 3 | 1 | 2 | Interferon-induced GTP-binding protein Mx2 | Acc: Q9N0Y2 | |
| ENSCAFP00000015807 | 1 | 405 | 348 | 3 | 6 | 2 | 1 | 3 | Guanine nucleotide-binding protein Gi, α-2 subunit (adenylate cyclase-inhibiting Gα protein) | Acc: P38400 | |
| ENSCAFP00000022215 | 1 | 569 | 539 | 3 | 4 | 1 | 2 | 1 | Similar to atlastin-like | ID: LOC476044 (Entrez Gene) |
Five channel proteins were identified (supplemental table, worksheet 5): Na+/K+-ATPase α1 and α2 and VDAC1, -2, and -3. Na+/K+-ATPase is a ubiquitous transmembrane heterodimer composed of a catalytic α subunit and a glycosylated β subunit. It is known to be trafficked to the basolateral surface of polarized epithelial cells (35). Na+/K+-ATPase α1 also has been found as a cargo in Aquaporin-2-containing vesicles and identified from a pool of synaptic vesicles (36, 37). Although not directly linked to basolateral trafficking, VDACs can be part of the exocytic machinery and have been associated with the natural killer cell secretory lysosome (38), H+/K+-ATPase-containing tubulovesicles (13), and zymogen granules isolated from pancreatic acinar cells (39).
Myosin IIA (identified as myosin, heavy polypeptide 9, non-muscle in supplemental table, worksheet 7) (40, 41) and kinesin receptor (kinectin 1) (42) are two motor-related proteins that have been associated with basolateral trafficking. Although kinesin receptor is thought to reside in the ER, recent work has shown that it can be transported to focal adhesions (43, 44). Basolateral linked cytoskeletal proteins identified include β-catenin (45), plakoglobin (desmoglein 3) (46), and β-spectrin (47).
Unexpectedly we found a considerable number of RNA/DNA-binding proteins. In fact, RNA/DNA-binding proteins comprised 21% of the proteins identified, making it the largest category of proteins with known function. These included splicing factor/proline- and glutamine-rich protein, 40 or 60 S ribosomal proteins, heterogeneous nuclear ribonucleoproteins, and DEAD box protein 3. It is intriguing that mRNAs have been found in different subcellular compartments where they enable the rapid translation of needed proteins; for example, mRNA for actin has been found in lamellipodia (48, 49). Mann and co-workers (50) recently identified a number of RNA-binding proteins in spreading initiating centers by quantitative proteomics-based analysis of adherent versus non-adherent lung fibroblasts. These newly described transient structures precede the development of focal adhesions. These authors postulated that these RNA-binding proteins may control the rate of cell spreading by binding to RNA in a protective or stabilizing role. It is tempting to speculate that Naked2 transports components of nascent focal adhesions to their destination because we found a number of components of focal adhesions in our proteomics analysis (e.g. integrins α2 and β1 as well as vimentin). Studies are underway to determine whether these proteins are present in WT Naked2 vesicles or whether they represent artifactual curiosities.
Confirmation of the Presence of All Six Proteins Tested from G2A Naked2 Vesicles in WT Naked2 Vesicles—
The α1 subunit of Na+/K+-ATPase was identified by 32 spectra comprising 17 unique peptides. We observed that endogenous Na+/K+-ATPase α1 partially co-localized with Naked2 in a subpopulation of vesicles in MDCK cells stably expressing WT Naked2-EGFP (Fig. 6A). To determine whether these two proteins were in a complex, we transiently overexpressed their respective cDNAs into HEK 293 cells and performed co-immunoprecipitation experiments. As a negative control, we co-transfected WT Naked2-EGFP with CD147, a basolaterally trafficked cell surface glycoprotein that was not found in G2A Naked2 vesicles by LC/LC-MS/MS. A significant amount of total cellular Na+/K+-ATPase α1 was immunoprecipitated with anti-Naked2 antibody, and Naked2 was immunoprecipitated using a Na+/K+-ATPase α1 antibody (Fig. 6B). Anti-CD147 antibody, however, did not pull down either Naked2 or Na+/K+-ATPase α1 (Fig. 6B).
Confirmation of Rab10 in TGFα-containing Naked2-associated vesicles would be noteworthy in that TGFα would represent the first cargo identified for Rab10. Rab10 is a small molecular weight GTPase that facilitates trafficking from the TGN during early stages of epithelial polarization (26, 51) and is involved in basolateral transport and endocytic recycling in polarized MDCK cells (26). We transiently transfected dsRed-Rab10 cDNA into MDCK cells that stably expressed WT Naked2-EGFP and observed partial co-localization of dsRed-Rab10 and Naked2-EGFP in a subset of vesicles (Fig. 7). We evaluated the behavior of these two epitope-tagged proteins by live cell microscopy (see supplemental movie). Both proteins associated with vesicles. Some vesicles exhibited only dsRed-Rab10 fluorescence, and others exhibited only Naked2-EGFP fluorescence; however, numerous vesicles displayed dual fluorescence. The patterns of fluorescence in these dual-labeled vesicles were dynamic and variable. For example, one portion of a vesicle displayed Naked2 fluorescence, and another portion displayed Rab10 fluorescence, whereas in other cases, there was clear co-localization of Naked2 and Rab10. Some vesicles moved quickly and others slowly; some appeared to collide with rapid changes in the intensity of their fluorescence and morphology, prompting speculation that there may be an active exchange of cargo and machinery along motor-based trafficking routes. The same phenomenon was observed in the live polarized MDCK cells (data not shown). As mentioned above, IQGAP1 has been linked to basolateral trafficking (29, 30). Endogenous IQGAP1 partially co-localized on Naked2-associated vesicles in MDCK cells stably expressing wild-type Naked2-EGFP (Fig. 7).
Annexin A2, like the other validated proteins, has been linked to basolateral trafficking. In contrast, annexin A1 might be associated with apical trafficking (13, 52). The distribution of annexin A2 and annexin A1 in iodixanol gradient fractions showed a migration pattern similar to that of WT and G2A Naked2, shifting in the gradient to correspond to the migration of these proteins (Fig. 3, A and B). We observed endogenous annexin A2 immunoreactivity in most of the fluorescent vesicles in MDCK cells stably expressing WT Naked2-EGFP (Fig. 7). Furthermore we were able to co-immunoprecipitate annexin A2 and Naked2 in transiently co-transfected HEK 293 cells (data not shown). In addition, endogenous annexin A1 co-localized with Naked2 fluorescence in WT Naked2-expressing MDCK cells (Fig. 7).
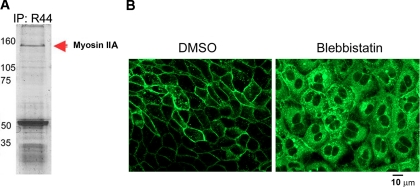
Conventional non-muscle myosin II (myosin, heavy polypeptide 9, non-muscle) is comprised of two genetically distinct isoforms referred to as myosins IIA and IIB (53). Myosin IIA is concentrated at the TGN consistent with its role in vesicle trafficking during cell membrane repair (54, 55). We identified 75 unique myosin IIA peptides from 104 spectra. To determine whether Naked2 was found in a complex with myosin IIA, we immunoprecipitated endogenous Naked2 from a human colorectal cancer cell line, HCA-7; separated the products by SDS-PAGE; and stained the gel with Coomassie Blue. A 160-kDa band was isolated and identified as myosin IIA by MALDI-TOF/TOF analysis (Fig. 8A and supplemental MALDI-TOF/TOF data); this band likely represents a partially degraded form of full-length 220-kDa myosin IIA (56). Moreover treatment of WT Naked2-EGFP-expressing MDCK cells with the specific myosin II inhibitor blebbistatin resulted in an accumulation of Naked2-associated vesicles in the cytoplasm (Fig. 8B).
Fig. 8.
Evidence that myosin IIA is a functional motor for Naked2-associated vesicles. A, Naked2 antibody R44 immunoprecipitation (IP) products were resolved using SDS-PAGE and stained with Coomassie Blue. Bands were cut out, treated with modified trypsin, and identified by peptide mass fingerprinting using the MASCOT software (Matrix Science). Mass spectra were obtained as described under “Experimental Procedures.” Protein scores greater than 74 were significant (p < 0.05). The score for this band was 349, and the predicted molecular weight of the identified protein was consistent with the corresponding region of the gel as determined by the molecular weight markers. B, MDCK cells stably transfected with Naked2-EGFP were grown on coverslips for 2 h, and then either DMSO or blebbistatin was applied at a concentration of 5 μm for 16 h. Bar, 10 μm.
Use of FAVS to Isolate Cellular Organelles—
Rapid advances in the field of organelle proteomics have been facilitated by the increased sensitivity of mass spectrometers, expanding genome data sets, and improved search algorithms (57–61). Previous studies have compiled inventories for a number of secretory organelles including TGN-derived carrier vesicles (40), synaptic vesicles (62–64), clathrin-coated vesicles (62, 65), lysosomes (38), Aquaporin-2-containing vesicles (36), zymogen granules (39), H+/K+-ATPase-containing vesicles (13), constituents of the ER and Golgi (66–69), and epidermal growth factor receptor endosomes (70). Most of these studies have relied on biochemical enrichment and immunoisolation (37). However, large scale proteomics analysis of basolaterally targeted exocytic vesicles has not been performed previously.
These studies introduce a novel method of isolating basolaterally targeted exocytic vesicles using tandem fluorescent probes and FAVS. It should be noted that Wolkoff and co-workers (71) used flow cytometry to prepare highly purified Alexa 488-labeled endocytic vesicles, but they did not perform large scale proteomics analysis. We used a nonspecific lipid bilayer-partitioning probe (DiD) to ensure that the vesicle membranes were intact and a specific probe consisting of a fusion protein (G2A-Naked2-EGFP) lacking an essential myristoylation site required for fusion of Naked2-associated vesicles at the basolateral surface of polarized MDCK cells. The specific probe design was critical to allow efficient concentration of the vesicles in a “frozen state” (i.e. unable to fuse with the cell membrane) for partial purification over a discontinuous 10–40% iodixanol gradient. Typically we used 24 150-mm dishes containing ∼500 million cells to generate cell lysates for the iodixanol gradients to isolate G2A Naked2-containing Pool 2 vesicles that ranged in purity from 64–75% as assessed by FAVS. Generally 3–5 billion vesicles were sorted over 24–40 h of sort time to recover 100–170 million doubly positive vesicles in a final volume of ∼300 ml. This sorting strategy was designed to eliminate greater than 95% of the vesicles that lacked double positivity and appropriate pulse geometry. The electronic pulse geometry of the vesicles was another critical consideration in this sort strategy because of their extremely small size (30–300 nm). The sorting strategy was validated using confocal and electron microscopy of the sorted vesicles and ultimately yielded double positivity in excess of 99%. A major limitation of this method is the significant amount of time required to physically purify the vesicles. Although every precaution was maintained during the sorting (i.e. samples were chilled at all times and collection buffers contained protease inhibitors), we cannot exclude the possible loss and/or degradation of some proteins.
The customized fluorescence-activated cell sorter used to develop this method has three critical design features necessary for resolution of particles significantly smaller than the wavelength of light used to interrogate the particles. First is the slow transit through the laser intercepts that allows passive integration of the fluorescence signal. Second is the high efficiency of the light collection optics (numerical aperture, ∼1.2 (near the theoretical limit of 1.4)), and third is the digital signal processing electronics that enable high sensitivity and low background. Future refinements to this method will include the addition of other markers for added dimensionality of the data (up to 12 colors could be used simultaneously). The added dimensionality of the data will allow better phenotyping of the specific vesicles of interest. Improvements to the sorter will include more efficient light collection optics, a higher speed digital signal processor, and higher laser excitation powers. The addition of fluorescence polarization and anisotropy measurements could allow added resolution without requiring additional reagents. The inclusion of different lipid dyes might allow further refinement of vesicle subpopulations that not only differ in protein content but also have different lipid composition. This method could also be improved by depleting dominant protein species that obscure rare proteins of interest in the LC/LC-MS/MS analysis.
FAVS allows purification and subsequent proteomics analysis of basolaterally targeted vesicles that could not be purified using other more conventional methodology and opens the door to refined phenotypic subsetting and functional analysis of other cellular organelles. It should be noted that FAVS relies on an epitope-tagged protein for vesicle isolation. One must keep in mind that this modification could result in altered protein-protein interactions leading to false positive or negative protein identifications. However, in our case, we have shown previously that WT EGFP-tagged Naked2 does not interfere with the basolateral trafficking of TGFα, and that this epitope-tagged Naked2 has a cellular distribution identical to that of endogenous Naked2 (1).
Conclusions—
The present study provides the first large scale proteomics characterization of a population of basolaterally targeted exocytic vesicles. To do this, we exploited our previous observations that myristoylation of Naked2 is required for Naked2-associated vesicles to fuse at the plasma membrane and that myristoylation-deficient G2A Naked2-associated vesicles are trapped in the cytoplasm. We combined biochemical enrichment and FAVS to purify a discrete pool of G2A Naked2-containing exocytic vesicles for large scale LC/LC-MS/MS analysis. By the application of stringent acceptance criteria, we identified 389 proteins in these G2A Naked2-containing vesicles by LC/LC-MS/MS and confirmed the presence of six of these in WT Naked2-containing vesicles. This report is not intended to be the definitive, or even an exhaustive, analysis of basolaterally targeted vesicles but rather the beginnings of an effort that may be helpful to the field of basolateral trafficking to define the identity of basolateral exocytic vesicles. Finally we propose that FAVS may be a useful tool for isolation of cellular organelles for comprehensive LC/LC-MS/MS analysis.
Supplementary Material
Acknowledgments
We thank Vivian Siegel for reviewing the manuscript and Zhao Zuo (Department of Biomedical Informatics, Vanderbilt University) for assistance in the organization of the data in the supplemental table.
Footnotes
Published, MCP Papers in Press, May 25, 2008, DOI 10.1074/mcp.M700155-MCP200
The abbreviations used are: TGFα, transforming growth factor-α; DiD, 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine perchlorate; FAVS, fluorescence-activated vesicle sorting; FDR, false discovery rate; MDCK, Madin-Darby canine kidney; TGN, trans Golgi network; GFP, green fluorescent protein; EGFP, enhanced green fluorescent protein; ER, endoplasmic reticulum; WT, wild-type; M-PER, Mammalian Protein Extraction Reagent; HEK, human embryonic kidney; SCX, strong cation exchange; G, sorting gate; EM, electron microscopy; Exp, experiment; ID, identity; VDAC, voltage-dependent anion-selective channel; PMT, photomultiplier tube; Bis-Tris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; PF, paraformaldehyde.
This work was supported, in whole or in part, by National Institutes of Health Grants CA 46413, RO1 CA126218 (to D. L. T.), U24 CA126479 (to D. L. T.), and CA 95103 (Specialized Program of Research Excellence; to R. J. C.) from the NCI; U01 084239 (Mouse Models of Human Cancers Consortium; to R. J. C.); and DK070856 (to J. R. G.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Li, C., Franklin, J. L., Graves-Deal, R., Jerome, W. G., Cao, Z., and Coffey, R. J. ( 2004) Myristoylated Naked2 escorts transforming growth factor α to the basolateral plasma membrane of polarized epithelial cells. Proc. Natl. Acad. Sci. U. S. A. 101, 5571–5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li, C., Hao, M., Cao, Z., Ding, W., Graves-Deal, R., Hu, J., Piston, D. W., and Coffey, R. J. ( 2007) Naked2 acts as a cargo recognition and targeting protein to ensure proper delivery and fusion of TGF-α-containing exocytic vesicles at the lower lateral membrane of polarized MDCK cells. Mol. Biol. Cell 18, 3081–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris, R. C., Chung, E., and Coffey, R. J. ( 2003) EGF receptor ligands. Exp. Cell Res. 284, 2–13 [DOI] [PubMed] [Google Scholar]
- 4.Dempsey, P. J., Meise, K. S., and Coffey, R. J. ( 2003) Basolateral sorting of transforming growth factor-α precursor in polarized epithelial cells: characterization of cytoplasmic domain determinants. Exp. Cell Res. 285, 159–174 [DOI] [PubMed] [Google Scholar]
- 5.Zeng, W., Wharton, K. A., Jr., Mack, J. A., Wang, K., Gadbaw, M., Suyama, K., Klein, P. S., and Scott, M. P. ( 2000) nakedcuticle encodes an inducible antagonist of Wnt signalling. Nature 403, 789–795 [DOI] [PubMed] [Google Scholar]
- 6.Ishikawa, A., Kitajima, S., Takahashi, Y., Kokubo, H., Kanno, J., Inoue, T., and Saga, Y. ( 2004) Mouse Nkd1, a Wnt antagonist, exhibits oscillatory gene expression in the PSM under the control of Notch signaling. Mech. Dev. 121, 1443–1453 [DOI] [PubMed] [Google Scholar]
- 7.Van Raay, T. J., Coffey, R. J., and Solnica-Krezel, L. ( 2007) Zebrafish Naked1 and Naked2 antagonize both canonical and non-canonical Wnt signaling. Dev. Biol. 309, 151–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grindstaff, K. K., Yeaman, C., Anandasabapathy, N., Hsu, S. C., Rodriguez-Boulan, E., Scheller, R. H., and Nelson, W. J. ( 1998) Sec6/8 complex is recruited to cell-cell contacts and specifies transport vesicle delivery to the basal-lateral membrane in epithelial cells. Cell 93, 731–740 [DOI] [PubMed] [Google Scholar]
- 9.Yeaman, C., Grindstaff, K. K., and Nelson, W. J. ( 2004) Mechanism of recruiting Sec6/8 (exocyst) complex to the apical junctional complex during polarization of epithelial cells. J. Cell Sci. 117, 559–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyer, C., Zizioli, D., Lausmann, S., Eskelinen, E. L., Hamann, J., Saftig, P., von Figura, K., and Schu, P. ( 2000) mu1A-adaptin-deficient mice: lethality, loss of AP-1 binding and rerouting of mannose 6-phosphate receptors. EMBO J. 19, 2193–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Traub, L. M., and Kornfeld, S. ( 1997) The trans-Golgi network: a late secretory sorting station. Curr. Opin. Cell Biol. 9, 527–533 [DOI] [PubMed] [Google Scholar]
- 12.Sugimoto, H., Sugahara, M., Folsch, H., Koide, Y., Nakatsu, F., Tanaka, N., Nishimura, T., Furukawa, M., Mullins, C., Nakamura, N., Mellman, I., and Ohno, H. ( 2002) Differential recognition of tyrosine-based basolateral signals by AP-1B subunit μ1B in polarized epithelial cells. Mol. Biol. Cell 13, 2374–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lapierre, L. A., Avant, K. M., Caldwell, C. M., Ham, A. J., Hill, S., Williams, J. A., Smolka, A. J., and Goldenring, J. R. ( 2007) Characterization of immunoisolated human gastric parietal cells tubulovesicles: identification of regulators of apical recycling. Am. J. Physiol. 292, G1249–G1262 [DOI] [PubMed] [Google Scholar]
- 14.Adkins, J. N., Varnum, S. M., Auberry, K. J., Moore, R. J., Angell, N. H., Smith, R. D., Springer, D. L., and Pounds, J. G. ( 2002) Toward a human blood serum proteome: analysis by multidimensional separation coupled with mass spectrometry. Mol. Cell. Proteomics 1, 947–955 [DOI] [PubMed] [Google Scholar]
- 15.Tabb, D. L., Fernando, C. G., and Chambers, M. C. ( 2007) MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J. Proteome Res. 6, 654–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDonald, W. H., Tabb, D. L., Sadygov, R. G., MacCoss, M. J., Venable, J., Graumann, J., Johnson, J. R., Cociorva, D., and Yates, J. R., III ( 2004) MS1, MS2, and SQT—three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun. Mass Spectrom. 18, 2162–2168 [DOI] [PubMed] [Google Scholar]
- 17.Zhang, B., Chambers, M. C., and Tabb, D. L. ( 2007) Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J. Proteome Res. 6, 3549–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizukoshi, E., Suzuki, M., Loupatov, A., Uruno, T., Hayashi, H., Misono, T., Kaul, S. C., Wadhwa, R., and Imamura, T. ( 1999) Fibroblast growth factor-1 interacts with the glucose-regulated protein GRP75/mortalin. Biochem. J. 343, 461–466 [PMC free article] [PubMed] [Google Scholar]
- 19.Schwarzer, C., Barnikol-Watanabe, S., Thinnes, F. P., and Hilschmann, N. ( 2002) Voltage-dependent anion-selective channel (VDAC) interacts with the dynein light chain Tctex1 and the heat-shock protein PBP74. Int. J. Biochem. Cell Biol. 34, 1059–1070 [DOI] [PubMed] [Google Scholar]
- 20.Breton, S., Lisanti, M. P., Tyszkowski, R., McLaughlin, M., and Brown, D. ( 1998) Basolateral distribution of caveolin-1 in the kidney. Absence from H+-ATPase-coated endocytic vesicles in intercalated cells. J. Histochem. Cytochem. 46, 205–214 [DOI] [PubMed] [Google Scholar]
- 21.Hara-Kuge, S., Ohkura, T., Ideo, H., Shimada, O., Atsumi, S., and Yamashita, K. ( 2002) Involvement of VIP36 in intracellular transport and secretion of glycoproteins in polarized Madin-Darby canine kidney (MDCK) cells. J. Biol. Chem. 277, 16332–16339 [DOI] [PubMed] [Google Scholar]
- 22.Lapierre, L. A., Tuma, P. L., Navarre, J., Goldenring, J. R., and Anderson, J. M. ( 1999) VAP-33 localizes to both an intracellular vesicle population and with occludin at the tight junction. J. Cell Sci. 112, 3723–3732 [DOI] [PubMed] [Google Scholar]
- 23.Oliferenko, S., Paiha, K., Harder, T., Gerke, V., Schwarzler, C., Schwarz, H., Beug, H., Gunthert, U., and Huber, L. A. ( 1999) Analysis of CD44-containing lipid rafts: recruitment of annexin II and stabilization by the actin cytoskeleton. J. Cell Biol. 146, 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konig, J., Prenen, J., Nilius, B., and Gerke, V. ( 1998) The annexin II-p11 complex is involved in regulated exocytosis in bovine pulmonary artery endothelial cells. J. Biol. Chem. 273, 19679–19684 [DOI] [PubMed] [Google Scholar]
- 25.Lecat, S., Verkade, P., Thiele, C., Fiedler, K., Simons, K., and Lafont, F. ( 2000) Different properties of two isoforms of annexin XIII in MDCK cells. J. Cell Sci. 113, 2607–2618 [DOI] [PubMed] [Google Scholar]
- 26.Schuck, S., Gerl, M. J., Ang, A., Manninen, A., Keller, P., Mellman, I., and Simons, K. ( 2007) Rab10 is involved in basolateral transport in polarized Madin-Darby canine kidney cells. Traffic 8, 47–60 [DOI] [PubMed] [Google Scholar]
- 27.Au, J. S., Puri, C., Ihrke, G., Kendrick-Jones, J., and Buss, F. ( 2007) Myosin VI is required for sorting of AP-1B-dependent cargo to the basolateral domain in polarized MDCK cells. J. Cell Biol. 177, 103–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El-Annan, J., Brown, D., Breton, S., Bourgoin, S., Ausiello, D. A., and Marshansky, V. ( 2004) Differential expression and targeting of endogenous Arf1 and Arf6 small GTPases in kidney epithelial cells in situ. Am. J. Physiol. 286, C768–C778 [DOI] [PubMed] [Google Scholar]
- 29.Zhou, R., Guo, Z., Watson, C., Chen, E., Kong, R., Wang, W., and Yao, X. ( 2003) Polarized distribution of IQGAP proteins in gastric parietal cells and their roles in regulated epithelial cell secretion. Mol. Biol. Cell 14, 1097–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chew, C. S., Okamoto, C. T., Chen, X., and Qin, H. Y. ( 2005) IQGAPs are differentially expressed and regulated in polarized gastric epithelial cells. Am. J. Physiol. 288, G376–G387 [DOI] [PubMed] [Google Scholar]
- 31.Meng, X., Krokhin, O., Cheng, K., Ens, W., and Wilkins, J. A. ( 2007) Characterization of IQGAP1-containing complexes in NK-like cells: evidence for Rac 2 and RACK1 association during homotypic adhesion. J. Proteome Res. 6, 744–750 [DOI] [PubMed] [Google Scholar]
- 32.Lkhider, M., Castino, R., Bouguyon, E., Isidoro, C., and Ollivier-Bousquet, M. ( 2004) Cathepsin D released by lactating rat mammary epithelial cells is involved in prolactin cleavage under physiological conditions. J. Cell Sci. 117, 5155–5164 [DOI] [PubMed] [Google Scholar]
- 33.Gut, A., Balda, M. S., and Matter, K. ( 1998) The cytoplasmic domains of a β1 integrin mediate polarization in Madin-Darby canine kidney cells by selective basolateral stabilization. J. Biol. Chem. 273, 29381–29388 [DOI] [PubMed] [Google Scholar]
- 34.Chillaron, J., Roca, R., Valencia, A., Zorzano, A., and Palacin, M. ( 2001) Heteromeric amino acid transporters: biochemistry, genetics, and physiology. Am. J. Physiol. 281, F995–F1018 [DOI] [PubMed] [Google Scholar]
- 35.Caplan, M. J., Anderson, H. C., Palade, G. E., and Jamieson, J. D. ( 1986) Intracellular sorting and polarized cell surface delivery of (Na+,K+)ATPase, an endogenous component of MDCK cell basolateral plasma membranes. Cell 46, 623–631 [DOI] [PubMed] [Google Scholar]
- 36.Barile, M., Pisitkun, T., Yu, M. J., Chou, C. L., Verbalis, M. J., Shen, R. F., and Knepper, M. A. ( 2005) Large scale protein identification in intracellular aquaporin-2 vesicles from renal inner medullary collecting duct. Mol. Cell. Proteomics 4, 1095–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morciano, M., Burre, J., Corvey, C., Karas, M., Zimmermann, H., and Volknandt, W. ( 2005) Immunoisolation of two synaptic vesicle pools from synaptosomes: a proteomics analysis. J. Neurochem. 95, 1732–1745 [DOI] [PubMed] [Google Scholar]
- 38.Casey, T. M., Meade, J. L., and Hewitt, E. W. ( 2007) Organelle proteomics: identification of the exocytic machinery associated with the natural killer cell secretory lysosome. Mol. Cell. Proteomics 6, 767–780 [DOI] [PubMed] [Google Scholar]
- 39.Chen, X., Walker, A. K., Strahler, J. R., Simon, E. S., Tomanicek-Volk, S. L., Nelson, B. B., Hurley, M. C., Ernst, S. A., Williams, J. A., and Andrews, P. C. ( 2006) Organellar proteomics: analysis of pancreatic zymogen granule membranes. Mol. Cell. Proteomics 5, 306–312 [DOI] [PubMed] [Google Scholar]
- 40.Musch, A., Cohen, D., and Rodriguez-Boulan, E. ( 1997) Myosin II is involved in the production of constitutive transport vesicles from the TGN. J. Cell Biol. 138, 291–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou, R., Watson, C., Fu, C., Yao, X., and Forte, J. G. ( 2003) Myosin II is present in gastric parietal cells and required for lamellipodial dynamics associated with cell activation. Am. J. Physiol. 285, C662–C673 [DOI] [PubMed] [Google Scholar]
- 42.Hofer, D., Jons, T., Kraemer, J., and Drenckhahn, D. ( 1998) From cytoskeleton to polarity and chemoreception in the gut epithelium. Ann. N. Y. Acad. Sci. 859, 75–84 [DOI] [PubMed] [Google Scholar]
- 43.Palazzo, A. F., and Gundersen, G. G. ( 2002) Microtubule-actin cross-talk at focal adhesions. Sci. STKE 2002, PE31. [DOI] [PubMed] [Google Scholar]
- 44.Tran, H., Pankov, R., Tran, S. D., Hampton, B., Burgess, W. H., and Yamada, K. M. ( 2002) Integrin clustering induces kinectin accumulation. J. Cell Sci. 115, 2031–2040 [DOI] [PubMed] [Google Scholar]
- 45.Miyoshi, K., Shillingford, J. M., Le Provost, F., Gounari, F., Bronson, R., von Boehmer, H., Taketo, M. M., Cardiff, R. D., Hennighausen, L., and Khazaie, K. ( 2002) Activation of β-catenin signaling in differentiated mammary secretory cells induces transdifferentiation into epidermis and squamous metaplasias. Proc. Natl. Acad. Sci. U. S. A. 99, 219–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DePasquale, J. A., Samsonoff, W. A., and Gierthy, J. F. ( 1994) 17-β-Estradiol induced alterations of cell-matrix and intercellular adhesions in a human mammary carcinoma cell line. J. Cell Sci. 107, 1241–1254 [DOI] [PubMed] [Google Scholar]
- 47.Dubreuil, R. R., Wang, P., Dahl, S., Lee, J., and Goldstein, L. S. ( 2000) Drosophila β spectrin functions independently of α spectrin to polarize the Na,K ATPase in epithelial cells. J. Cell Biol. 149, 647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ross, A. F., Oleynikov, Y., Kislauskis, E. H., Taneja, K. L., and Singer, R. H. ( 1997) Characterization of a β-actin mRNA zipcode-binding protein. Mol. Cell. Biol. 17, 2158–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lawrence, J. B., and Singer, R. H. ( 1986) Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell 45, 407–415 [DOI] [PubMed] [Google Scholar]
- 50.de Hoog, C. L., Foster, L. J., and Mann, M. ( 2004) RNA and RNA binding proteins participate in early stages of cell spreading through spreading initiation centers. Cell 117, 649–662 [DOI] [PubMed] [Google Scholar]
- 51.Babbey, C. M., Ahktar, N., Wang, E., Chen, C. C., Grant, B. D., and Dunn, K. W. ( 2006) Rab10 regulates membrane transport through early endosomes of polarized Madin-Darby canine kidney cells. Mol. Biol. Cell 17, 3156–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bensalem, N., Ventura, A. P., Vallee, B., Lipecka, J., Tondelier, D., Davezac, N., Dos Santos, A., Perretti, M., Fajac, A., Sermet-Gaudelus, I., Renouil, M., Lesure, J. F., Halgand, F., Laprevote, O., and Edelman, A. ( 2005) Down-regulation of the anti-inflammatory protein annexin A1 in cystic fibrosis knock-out mice and patients. Mol. Cell. Proteomics 4, 1591–1601 [DOI] [PubMed] [Google Scholar]
- 53.Simons, M., Wang, M., McBride, O. W., Kawamoto, S., Yamakawa, K., Gdula, D., Adelstein, R. S., and Weir, L. ( 1991) Human nonmuscle myosin heavy chains are encoded by two genes located on different chromosomes. Circ. Res. 69, 530–539 [DOI] [PubMed] [Google Scholar]
- 54.Togo, T., and Steinhardt, R. A. ( 2004) Nonmuscle myosin IIA and IIB have distinct functions in the exocytosis-dependent process of cell membrane repair. Mol. Biol. Cell 15, 688–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bustos, R., Kolen, E. R., Braiterman, L., Baines, A. J., Gorelick, F. S., and Hubbard, A. L. ( 2001) Synapsin I is expressed in epithelial cells: localization to a unique trans-Golgi compartment. J. Cell Sci. 114, 3695–3704 [DOI] [PubMed] [Google Scholar]
- 56.Heimann, K., Percival, J. M., Weinberger, R., Gunning, P., and Stow, J. L. ( 1999) Specific isoforms of actin-binding proteins on distinct populations of Golgi-derived vesicles. J. Biol. Chem. 274, 10743–10750 [DOI] [PubMed] [Google Scholar]
- 57.Fiedler, K., Kellner, R., and Simons, K. ( 1997) Mapping the protein composition of trans-Golgi network (TGN)-derived carrier vesicles from polarized MDCK cells. Electrophoresis 18, 2613–2619 [DOI] [PubMed] [Google Scholar]
- 58.Andersen, J. S., and Mann, M. ( 2006) Organellar proteomics: turning inventories into insights. EMBO Rep. 7, 874–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gilchrist, A., Au, C. E., Hiding, J., Bell, A. W., Fernandez-Rodriguez, J., Lesimple, S., Nagaya, H., Roy, L., Gosline, S. J., Hallett, M., Paiement, J., Kearney, R. E., Nilsson, T., and Bergeron, J. J. ( 2006) Quantitative proteomics analysis of the secretory pathway. Cell 127, 1265–1281 [DOI] [PubMed] [Google Scholar]
- 60.Brunet, S., Thibault, P., Gagnon, E., Kearney, P., Bergeron, J. J., and Desjardins, M. ( 2003) Organelle proteomics: looking at less to see more. Trends Cell Biol. 13, 629–638 [DOI] [PubMed] [Google Scholar]
- 61.Yates, J. R., III, Gilchrist, A., Howell, K. E., and Bergeron, J. J. ( 2005) Proteomics of organelles and large cellular structures. Nat. Rev. Mol. Cell Biol. 6, 702–714 [DOI] [PubMed] [Google Scholar]
- 62.Blondeau, F., Ritter, B., Allaire, P. D., Wasiak, S., Girard, M., Hussain, N. K., Angers, A., Legendre-Guillemin, V., Roy, L., Boismenu, D., Kearney, R. E., Bell, A. W., Bergeron, J. J., and McPherson, P. S. ( 2004) Tandem MS analysis of brain clathrin-coated vesicles reveals their critical involvement in synaptic vesicle recycling. Proc. Natl. Acad. Sci. U. S. A. 101, 3833–3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Coughenour, H. D., Spaulding, R. S., and Thompson, C. M. ( 2004) The synaptic vesicle proteome: a comparative study in membrane protein identification. Proteomics 4, 3141–3155 [DOI] [PubMed] [Google Scholar]
- 64.Takamori, S., Holt, M., Stenius, K., Lemke, E. A., Gronborg, M., Riedel, D., Urlaub, H., Schenck, S., Brugger, B., Ringler, P., Muller, S. A., Rammner, B., Grater, F., Hub, J. S., De Groot, B. L., Mieskes, G., Moriyama, Y., Klingauf, J., Grubmuller, H., Heuser, J., Wieland, F., and Jahn, R. ( 2006) Molecular anatomy of a trafficking organelle. Cell 127, 831–846 [DOI] [PubMed] [Google Scholar]
- 65.Borner, G. H., Harbour, M., Hester, S., Lilley, K. S., and Robinson, M. S. ( 2006) Comparative proteomics of clathrin-coated vesicles. J. Cell Biol. 175, 571–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taylor, S. W., Fahy, E., and Ghosh, S. S. ( 2003) Global organellar proteomics. Trends Biotechnol. 21, 82–88 [DOI] [PubMed] [Google Scholar]
- 67.Foster, L. J., de Hoog, C. L., Zhang, Y., Zhang, Y., Xie, X., Mootha, V. K., and Mann, M. ( 2006) A mammalian organelle map by protein correlation profiling. Cell 125, 187–199 [DOI] [PubMed] [Google Scholar]
- 68.de Souza, N. ( 2007) Cartography of an organelle. Nat. Methods 4, 116. [DOI] [PubMed] [Google Scholar]
- 69.70Drahos, K. L., Tran, H. C., Kiri, A. N., Lan, W., McRorie, D. K., and Horn, M. J. ( 2005) Comparison of Golgi apparatus and endoplasmic reticulum proteins from livers of juvenile and aged rats using a novel technique for separation and enrichment of organelles. J. Biomol. Tech. 16, 347–355 [PMC free article] [PubMed] [Google Scholar]
- 70.Stasyk, T., Schiefermeier, N., Skvortsov, S., Zwierzina, H., Peranen, J., Bonn, G. K., and Huber, L. A. ( 2007) Identification of endosomal epidermal growth factor receptor signaling targets by functional organelle proteomics. Mol. Cell. Proteomics 6, 908–922 [DOI] [PubMed] [Google Scholar]
- 71.Bananis, E., Nath, S., Gordon, K., Satir, P., Stockert, R. J., Murray, J. W., and Wolkoff, A. W. ( 2004) Microtubule-dependent movement of late endocytic vesicles in vitro: requirements for Dynein and Kinesin. Mol. Biol. Cell 15, 3688–3697 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.