Abstract
Vaccinia DNA topoisomerase (vTopo) catalyzes highly specific nucleophilic substitution at a single phosphodiester linkage in the pentapyrimidine recognition sequence 5′- (C/T)+5C+4C+3T+2T+1p↓N−1 using an active site tyrosine nucleophile, thereby expelling a 5′ hydroxyl leaving group of the DNA. Here we report the energetic effects of subtle modifications to the major groove hydrogen bond donor and acceptor groups of the 3′- GGGAA-5′ consensus sequence of the nonscissile strand in the context of duplexes in which the scissile strand length was progressively shortened. We find that the major groove substitutions become energetically more damaging as the scissile strand is shortened from 32 to 24 and 18 nucleotides, indicating that enzyme interactions with the duplex region present in the 32mer, but not the 24 or 18mer, weaken specific interactions with the DNA major groove. Regardless of strand length, the destabilizing effects of the major groove substitutions increase as the reaction proceeds from the Michaelis complex to the transition-state for DNA cleavage, and finally, to the phosphotyrosine-DNA covalent complex. These length-dependent anticooperative interactions involving the DNA major groove and duplex regions 3′ to the cleavage site indicate that major groove binding energy is fully realized late during the reaction for full-length substrates, but that smaller more flexible duplex substrates feel these interactions earlier along the reaction coordinate. Such anticooperative binding interactions may play a role in strand exchange and supercoil unwinding activities of the enzyme.
Cellular DNA is found in a high-energy superhelical state that facilitates many processes including DNA replication, transcription, site-specific recombination, and other genome rearrangements (1). DNA topoisomerases are the essential and remarkable enzyme machines that regulate the topology of supercoiled DNA, thereby influencing all of these DNA transformations. The key mechanistic feature of all topoisomerases and related recombinase enzymes is their reversible formation of a phosphotyrosine linkage between the DNA backbone and an active site tyrosine (Fig. 1A). For eukaryotic Type IB topoisomerases, this covalent linkage is with a single strand of the DNA, and produces a 5′-deoxyribose hydroxyl as the leaving group (Fig. 1A). This linkage provides the molecular hinge that allows these enzymes to perform diverse DNA transformations ranging from supercoil relaxation to strand exchange and Holliday junction resolution reactions (2-7).
Figure 1.
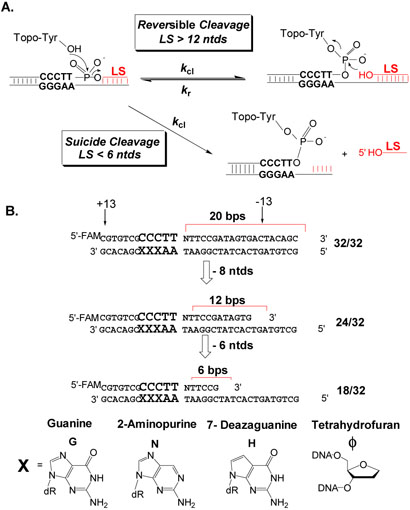
Cleavage reactions and modified substrates for vTopo. (A) Reversible and suicide cleavage reactions. Strands with less than 6 nucleotides 3′ to the cleaved position spontaneously dissociate making the reaction irreversible. (B) Substrate series with truncated scissile strands and base substitutions at the +5, +4 and +3 guanine positions. Nucleotides that extend 3′ to the cleavage site on the scissile strand are given negative numbers and those that extend 5′ are given positive numbers.
The vaccinia virus topoisomerase (vTopo) is a prototypic eukaryotic type IB topoisomerase, but is unique in that it catalyzes reversible site-specific cleavage and religation of the phosphodiester backbone of duplex DNA at 5′-(C/T)CCTT↓N sites (8). vTopo is active with positively or negatively supercoiled DNA (9), or alternatively, with linear DNA duplexes such as those depicted in Figure 1A (10, 11). The reactions with linear DNA fall into two general categories. The first category is cation such that an internal cleavage equilibrium (Kcl) is maintained (Fig. 1A). The second is characterized by irreversible DNA cleavage resulting in the formation of a stable enzyme-DNA phosphotyrosine complex (Fig. 1A). This so-called “suicide” cleavage reaction is facilitated by short scissile DNA strand lengths 3′ to the cleavage site, such that the unstable DNA leaving strand dissociates from the complex preventing strand ligation (12). Despite the structural differences between suicide and equilibrium cleavage substrates, and the observation that the vTopo DNA footprint extends at least 13 nucleotides 3′ to the cleavage site (i.e. beyond the length of typical suicide leaving strand)(13), the single-turnover cleavage rates (kcl) for suicide and equilibrium substrates are similar (11). Thus, additional binding interactions present in the larger equilibrium substrate do not decrease the activation barrier for cleavage.
We recently synthesized a series of 32/32mer equilibrium substrates that displayed different hydrogen bond donor and acceptor groups (X, Figure 1B) in the major groove of the nonscissile strand GGGAA consensus sequence (14). The general effect of these substitutions was to decrease the equilibrium constant for cleavage (Kcl = kcl/kr) by increasing the religation rate (kr) without altering the cleavage rate (kcl). Since reversible cleavage and religation occur through the same transition state, the substitutions must have selectively destabilized the covalent complex, resulting in the selective increases in the religation rate. However, we also noted that the same base substitutions resulted in decreases in the cleavage rate in the context of 18/32mer suicide substrates (14). The distinct effects of major groove alterations using the suicide and equilibrium substrates suggested that the GGGAA major groove interactions were energetically coupled to interactions of the enzyme with the DNA duplex region that is present in the equilibrium substrate but absent in the suicide substrate. That is, the presence of more extensive duplex interactions weakens and delays the formation of the major groove interactions until the covalent complex is reached. These intriguing initial observations prompted us to thoroughly investigate the combined effects of scissile strand length and major groove substitutions on DNA binding, cleavage and religation by vTopo.
MATERIALS AND METHODS
Enzymes
The cloning and purification of wild-type vaccinia topoisomerase has been previously described (11). The enzyme concentration was determined by UV absorption using an extinction coefficient of 41,797 M−1cm−1 in a buffer containing 20 mM sodium phosphate at pH 6.0.
DNA Substrates
The sequences of the substituted 32/32, 24/32 and 18/32mer duplex DNA substrates containing the consensus cleavage sequence are shown in Figure 1B, where FAM is 6- carboxyfluorescein. The synthesis of the non-scissile strand with modified bases incorporated at various sites was accomplished by substituting the modified base during solid-phase DNA synthesis. All oligonucleotides were synthesized using an ABI 394 synthesizer using nucleoside phosphoramidites obtained from Glen Research. The oligonucleotides were purified using anion exchange HPLC and then desalted using C-18 reverse phase chromatography. The purity of oligonucleotides was confirmed using electrophoresis through a 20 % polyacrylamide gel containing 7 M urea and MALDI-TOF analysis. The DNA duplexes were prepared in buffer A (20 mM Tris-HCl, 200 mM NaCl, 1mM DTT, pH 8.0) by mixing the two strands in a molar ratio of 1:2 (non-scissile strand was in excess).
Equilibrium Binding and Cleavage Measurements
The equilibrium cleavage measurements with the 5′-FAM-labeled 32/32 and 24/32mers were performed using Buffer A by titrating each DNA (60 nM) with increasing concentrations of topo (40-600 nM). The covalent complexes were trapped by the addition of 1 volume of 10% SDS after 1h. The fraction covalent complex at each enzyme concentration [counts in covalent complex/(counts in covalent complex + counts in free DNA)] was quantified using ImageQuant software. The fraction covalent complex was plotted against enzyme concentration to obtain the values of the binding constant (KD) and cleavagereligation equilibrium constant (kcl) using the following equation (eq 1) (11).
| (1) |
where a = 1 + 1/Kcl, b = a[E] + a[S] KD/Kcl, Kcl = (kcl/kr), [E] and [S] are the total enzyme and substrate concentrations respectively. In this analysis, the counts that migrate as free DNA represent the sum of the counts that were bound non-covalently to the enzyme, and those of the unbound DNA. All measurements were repeated two or three times to estimate errors.
Approach to Equilibrium Kinetics
The rate constant for approach-to-equilibrium for the 5′FAM-labeled 32/32 and 24/32mers was measured using a KinTek rapid quench instrument. The final enzyme and DNA concentrations were maintained at 1 μM and 100 nM respectively. The enzyme and DNA were mixed and the reactions were quenched using 10% SDS at time intervals ranging between 2.5 and 750 milliseconds. The samples were subjected to electrophoresis on a 10% SDS-PAGE gel. The FAM fluorophore in the free DNA and enzyme-DNA covalent adduct was quantified using Typhoon scanner. Since the dye front ran along with the free DNA band, giving rise to inner filter effects, the samples were loaded on to the gel without the dye component. The fraction of covalent complex formed at each time was fitted to a first order rate equation to obtain kobs which equals the sum of the cleavage and religation rate constants (i.e kobs = kcl + kr) (11). Thus, using the Kcl obtained from equilibrium cleavage measurements the cleavage and religation rate constants (kr) may be calculated using the equations kcl = kobs/(1/Kcl + 1) and kr = kobs/(kcl + 1). All measurements were repeated two or three times to estimate errors.
Single-turnover Cleavage Measurements
Depending on the 18/32mer under study, either stopped-flow fluorescence or manual chemical quench reactions kinetic methods were used (11, 14). The stopped-flow reactions were performed using an Applied Photophysics stopped-flow fluorescence instrument in the two-syringe mode. Equal volumes of 1 μM vTopo and 50 nM substrate were rapidly mixed, and the increase in 2-aminopurine fluorescence of the substrate was followed using excitation at 315 nm with monitoring at emission wavelengths greater than 360 nm using a cut off filter (11). On average, five or more kinetic transients were averaged to obtain the reported rate constants and errors. Observed rate constant (kobs) were plotted against [vTopo], and KD and kcl were determined form nonlinear least squares fitting to eq 2.
| (2) |
For 18/32mers with fairly slow cleavage rates (<0.5 s−1), cleavage experiments were performed using manual chemical quench single-turnover conditions. The enzyme was present in at least a 4-fold excess over the DNA so that the observed kinetics were pseudo-first order. Experiments were performed by mixing 10 μL of vTopo (0.200-2 μM) with 10 μL DNA (0.05 μM) in an eppendorf tube. The reactions were allowed to proceed for various times and were quenched with 20 μL of 10% SDS. Ten microliters of 5X load buffer (omitting the dye components) was added to each quenched reaction sample and twelve microliters was removed for analysis. These samples were subjected to electrophoresis through a polyacrylamide gel containing 10 % SDS. The fraction covalent complex formed at each time point was determined from fluorescence imaging of the gel, plotted against time, and the cleavage rates (kcl) were determined from nonlinear regression fitting to a first-order rate equation. One exception was the slowly reactive 18/32mer with the ϕ4 substitution, where kobs (eq 2) was calculated from the linear slope of the initial cleavage rate (15).
Difference Free Energy Calculations
Changes in the free energy of DNA binding (ΔΔGbind), the cleavage activation barriers (ΔΔGclact), and in the overall equilibrium for DNA binding and cleavage (ΔΔGcleq), arising from the base substitutions or deletions were calculated using the measured kinetic and thermodynamic parameters (KD, kcl/KD and Keq = Kcl/KD) using eqs 3-5, where the superscript ″sub′ denotes the substrate with the base substitution.
| (3) |
| (4) |
| (5) |
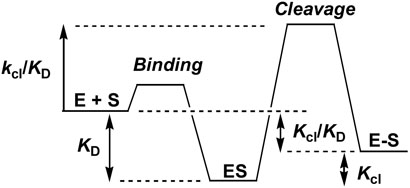
The free energy changes corresponding to the KD, kcl/KD and Keq = Kcl/KD measurements are indicated in [Scheme 1]. The relative free energy levels are drawn arbitrarily and are only meant to visually depict the kinetic or thermodynamic parameter that is measured. As indicated by equations 3-5, we elect to use E + S as the primary reference state for comparison of the base substitution effects. E + S provides a stable state from which to evaluate the energy level changes in ES, ES‡ and E-S arising from each substitution. This analysis assumes that the free energy effect of the base substitution on the free substrate (if any) is approximately equal for each of the three substrates. This assumption is reasonable because (i) the substitutions are distant from the nucleotide deletions, and (ii) the substitutions are very subtle. Accordingly, the substitution effects in moving from 18 to 24 and 32 mer strand lengths only reflect changes in the enzyme bound species ES, ES‡ and E-S.
Scheme 1.
RESULTS
Approach
Unmasking energetic coupling between enzyme interactions with two different regions of a DNA substrate requires both selective and combined perturbations in each binding region. Coupling is indicated by an increase or decrease in the energetic effect of a perturbation to one region in the context of a perturbation to the second region. In the present case, region 1 is the major groove of the nonscissile strand G+5G+4G+3A+2A+1consensus sequence, and region 2 is the duplex region 3′ to the cleavage site on the scissile strand. The subtle perturbations to the major groove involve removal of the N7 and O6 hydrogen bond acceptor groups of each guanine within the consensus sequence, which are replaced with the unnatural 7-deazaguanine and 2-aminopurine analogues (H and N, (Fig. 1B). The importance of these major groove hydrogen bonding groups is highlighted in the recent x-ray structure of the closely related variola virus Topo I bound to DNA, where major groove interactions, and in particular the N7 and O6 positions of the +5 and +4 guanines, play a key role in site recognition (16). We also explored the energetic effects of the more harsh perturbation of removal of the entire base at each guanine position (ϕ, Fig. 1B). The perturbations to the 3′ duplex region are much less subtle and involve 3′ truncations of the scissile strand from an initial length of 32 to 24 and 18 nucleotides (Fig. 1B). The 32/32 and 24/32mers are both equilibrium substrates that exhibit reversible DNA cleavage and religation, while the 18/32mer is a suicide substrate that can only bind and cleave DNA in a single suicide turnover event. The thermodynamic and kinetic parameters for each duplex are reported in Table 1. The parameters for the 32/32mer, with the exception of the substrates with the ϕ3, ϕ4 and ϕ5 substitutions, were reported in our previous study and are reproduced here to facilitate comparison and discussion of the entire data (14). In addition for vTopo, changes in DNA interactions at the cleavage site or consensus sequence do not result in a change in the site of nucleophilic attack (8, 10, 17, 18). Thus, comparisons here reflect cleavage at the same phosphodiester linkage.
Table 1.
Kinetic and thermodynamic parameters for variable length vTopo substrates with guanine base substitutions and deletionsa
| DNA | Sequence |
KD (nM) |
kcl |
kcl (S−1) |
kr (S−1) |
kcl/KDb (107 M−1s−1) |
kcl/KDb (107 M−1) |
|---|---|---|---|---|---|---|---|
| 32/32 | GGGAA | 118 ± 20 | 1.6 ± 0.3 | 2.2 ± 0.2 | 1.4 ± 0.2 | 1.9 | 1.4 |
| N5 | NGGAA | 148 ± 30 | 0.5 ± 0.05 | 2.3 ± 0.1 | 4.8 ± 0.1 | 1.6 | 0.34 |
| N4 | GNGAA | 171 ± 20 | 0.5 ± 0.03 | 2.4 ± 0.1 | 5.1 ± 0.1 | 1.4 | 0.3 |
| N3 | GGNAA | 55 ± 10 | 1 ± 0.2 | 3.2 ± 0.2 | 3.1 ± 0.2 | 5.8 | 1.8 |
| H5 | HGGAA | 75 ± 20 | 0.4 ± 0.03 | 1.5 ± 0.1 | 4.2 ± 0.1 | 2.0 | 0.53 |
| H4 | GHGAA | 59 ± 10 | 0.5 ± 0.03 | 1.6 ± 0.1 | 3.4 ± 0.1 | 2.7 | 0.85 |
| H3 | GGHAA | 77 ± 20 | 0.7 ± 0.03 | 3.2 ± 0.2 | 4.4 ± 0.2 | 4.2 | 0.91 |
| N5N4 | NNGAA | 87 ± 10 | 0.1 ± 0.01 | 0.2 ± 0.1 | 2.5 ± 0.3 | 0.2 | 0.11 |
| N5N3 | NGNAA | 48 ± 10 | 0.2 ± 0.02 | 1.1 ± 0.2 | 4.8 ± 0.2 | 2.2 | 0.42 |
| H5H4 | HHGAA | 67 ± 20 | 0.5 ± 0.06 | 2.7 ± 0.1 | 5.8 ± 0.1 | 4.0 | 0.75 |
| H5H3 | HGHAA | 110 ± 20 | 0.3 ± 0.02 | 0.9 ± 0.1 | 3.1 ± 0.2 | 0.8 | 0.27 |
| ϕ3 | ϕGGAA | 64 ± 20 | 0.015 ± 0.002 | 0.02 | |||
| ϕ4 | GϕGAA | 55 ± 20 | 0.01 ± 0.002 | 0.07 ± 0.04 | 73 ± 45 | 0.13 | 0.018 |
| ϕ5 | GGϕAA | 98 ± 20 | 0.019 ± 0.003 | 0.019 | |||
| 24/32 | GGGAA | 20 ± 5 | 1.5 ± 0.1 | 5.6 ± 0.1 | 3.7 ± 0.1 | 28 | 7.5 |
| N5 | NGGAA | 105 ± 20 | 0.6 ± 0.1 | 3.5 ± 0.2 | 5.9 ± 0.2 | 3.3 | 0.57 |
| N4 | GNGAA | 150 ± 30 | 0.5 ± 0.05 | 3.1 ± 0.1 | 6.4 ± 0.1 | 2.1 | 0.33 |
| N3 | GGNAA | 44 ± 7 | 2.1 ± 0.2 | 1.9 ± 0.1 | 0.9 ± 0.1 | 4.3 | 4.8 |
| H5 | HGGAA | 62 ± 10 | 0.5 ± 0.03 | 1.7 ± 0.1 | 3.8 ± 0.1 | 2.7 | 0.8 |
| H4 | GHGAA | 34 ± 8 | 0.7 ± 0.05 | 2.1 ± 0.1 | 3.3 ± 0.1 | 6.1 | 2.0 |
| H3 | GGHAA | 87 ± 20 | 1 ± 0.1 | 2.7 ± 0.1 | 2.7 ± 0.1 | 3.1 | 1.1 |
| N5N4 | NNGAA | 110 ± 20 | 0.08 ± 0.01 | 0.8 ± 0.4 | 9.4 ± 0.5 | 0.73 | 0.073 |
| N5N3 | NGNAA | 192 ± 20 | 0.21 ± 0.03 | 1.6 ± 0.1 | 7.9 ± 0.1 | 0.83 | 0.11 |
| H5H4 | HHGAA | 54 ± 10 | 0.25 ± 0.02 | 1.8 ± 0.1 | 7.2 ± 0.1 | 3.3 | 0.46 |
| H5H3 | HGHAA | 113 ± 20 | 0.23 ± 0.03 | 1.2 ± 0.2 | 5.1 ± 0.2 | 1.1 | 0.2 |
| ϕ3 | ϕGGAA | 84 ± 20 | 0.011 ± 0.001 | 0.012 | |||
| ϕ4 | GϕGAA | 7 ± 2 | 0.007 ± 0.001 | 0.1 | |||
| ϕ5 | GGϕAA | 64 ± 15 | 0.014 ± 0.003 | 0.02 | |||
| 18/32 | GGGAA | 95 ± 20 | 2.1 ± 0.03 | 2.2 | |||
| N5 | NGGAA | 380 ± 50 | 0.59 ± 0.05 | 0.15 | |||
| N4 | GNGAA | 95 ± 20 | 0.65 ± 0.05 | 0.68 | |||
| N5N4 | NNGAA | 167 ± 20 | 0.09 ± 0.02 | 0.054 | |||
| N5N3 | NGNAA | 134 ± 20 | 0.12 ± 0.04 | 0.084 | |||
| H5H4 | HHGAA | 347 ± 50 | 0.29 ± 0.11 | 0.083 | |||
| H5H3 | HGHAA | 590 ± 70 | 0.24 ± 0.10 | 0.04 | |||
| ϕ4 | GϕGAA | 129 ± 20 | 0.0003 ± 0.0001 |
0.0002 | |||
See Fig. 2 for structures of H, N and ϕ. Except for the ϕ substitutions, the data for the 32/32mer have been previously reported and are recapitulated here for comparative purposes.
Propagated errors in kcl/KD and kcl/KD are less than ± 30% .
DNA Binding (KD) and Cleavage Equilibrium (Kcl)
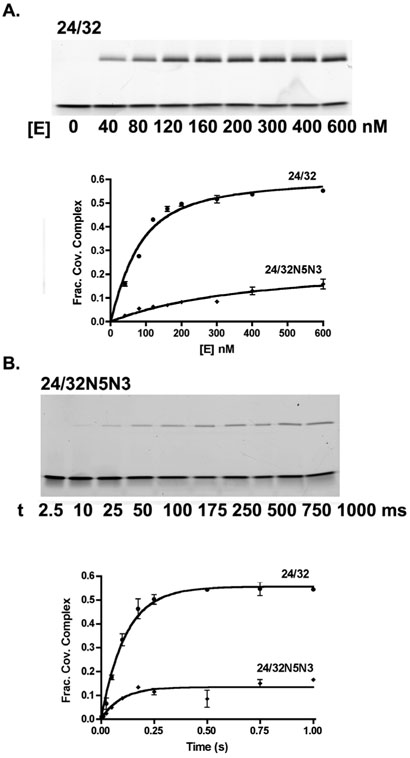
For the 32/32 and 24/32mer equilibrium substrates the KD and Kcl values are determined from a single experiment by titrating a limiting amount of DNA with excess vTopo and fitting to eq 1 (Fig. 2A). The unmodified 24/32mer substrate binds about 6-fold more tightly to vTopo than the longer 32/32mer but shows an identical Kcl ~ 1.5 (Table 1). The KD values for the substituted 24/32mers are increased by about two to five-fold relative to the unmodified sequence, except for the ϕ4 sequence that shows a three-fold lower value. The weakened binding induced by the 24/32mer substitutions differs from the 32/32mer context in which small increases in binding affinity were generally observed for the same substitutions. The substitutions produce similar decreases in Kcl for both the 24/32 and 32/32mers, and the base deletion variants cause the largest damaging effects, typically decreasing the magnitude of Kcl by 100-fold or more (Table 1). The overall effect of the substitutions on the free energy change corresponding to moving from the free enzyme and substrate to the covalent complex (-RT ln Kcl/KD, Scheme 1) parallels the trends observed for Kcl alone, although the magnitudes are sometimes larger due to the additional damaging effects on KD.
Figure 2.
Reversible DNA cleavage of unmodified 24/32mer and its N5N3 analogue. (A) Determination of the equilibrium constants for DNA binding (KD) and cleavage (Kcl) by titrating DNA with increasing concentrations of vTopo. The covalently bound DNA (upper band) was separated from noncovalent and free DNA using SDS-PAGE, and the gel was imaged for determination of the fraction covalent complex. A representative gel image for the 24/32mer is shown. (B) Determination of the observed rate constant for approach-to-equilibrium cleavage using saturation conditions. A representative gel image for the 24/32N5N3mer is shown. The kobs was used to calculate the cleavage and religation rates according to kobs = kcl + kr and Kcl = kcl/kr.
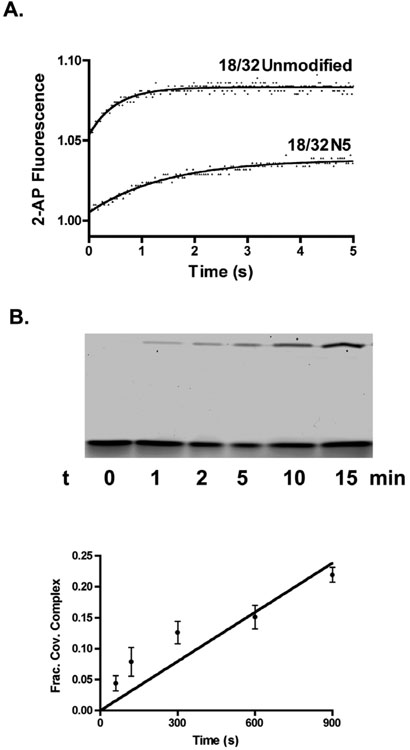
Based on the largest effects observed for the 32/32 and 24/32mers, we selected a subset of major groove substitutions to be investigated in the context of an 18/32mer. In this case, the KD values are measured from the concentration dependence of the single-turnover cleavage (kcl) rates determined in stopped-flow or chemical quench experiments (Figure 3A, 3B)). As generally found with the 24/32mer, but not the 32/32mer, the 18/32mer major groove substitutions resulted in 1.4 to 6-fold weakening of DNA binding (Table 1).
Figure 3.
Suicide cleavage of 18/32mer and its N5 and ϕ4 analogues. (A) The increase in 2-aminopurine fluorescence of the 18/32mer and 18/32N5mer upon suicide DNA strand cleavage by vTopo is followed using stopped-flow fluorescence. (B) Initial rate of 18/32ϕ4 strand cleavage was measured using manual chemical quench methods with excess enzyme. The time dependence for formation of the covalent complex (upper band) provides the rate of suicide cleavage.
Cleavage Kinetics (kcl, kcl/DD)
The major groove substitutions produce the most disparate effects on the first-order (kcl) and second-order (kcl/KD) rate constants for cleavage of each substrate (Table 1). The cleavage rate constant for the 32/32 and 24/32mer equilibrium cleavage substrates were measured using a rapid mix-chemical quench instrument. The enzyme and substrate were rapidly mixed and the reactions were quenched with 10% SDS at various time intervals before separation of the covalently bound DNA product by denaturing polyacrylamide gel electrophoresis (Fig. 2B). In this analysis, the fraction of covalent complex increases over time until the equilibrium level is reached. The reversible nature of the reaction allows the calculation of kcl and kr from a single observed rate constant because kobs = kcl + kr and Kcl = kcl/kr (14). Except for the N5N3 substituted substrate, and the more severe base deletions, the 32/32mer shows small or modest increases or decreases in kcl or kcl/KD. In contrast, substitutions in the 24/32mer context give rise to consistent decreases in kcl of as much as sevenfold, and in the second-order rate constant kcl/KD of as much as 38-fold for N5N4 (Table 1). Of the three substrates, the 18/32mer shows on average the largest decreases in kcl and kcl/KD arising from major groove substitutions (Figure 3A, 3B). Examples include a 55-fold decrease in kcl/KD for the H5H3 substitution and an 11,000-fold decrease for the ϕ4 substrate (Figure 3B, Table 1).
DNA Religation (kr)
Religation rates can only be measured for substrates like the 32/32 and 24/32mers that exhibit reversible cleavage reactions. For both substrates, almost without exception, the substitutions result in 1.5 to 4-fold increases in the religation rates (Table 1). Because of the extremely small equilibrium constant for cleavage it is difficult to accurately measure the religation rates with the severely damaged ϕ substrates. Nevertheless, we performed a rapid quench-flow experiment using the 32/32mer ϕ4 substrate where a substantial 52-fold increase in ϕkr was estimated (Table 1).
DISCUSSION
The activated phosphotyrosine covalent intermediate is the key species that allows topo I to perform a large repertoire of reactions that includes supercoil relaxation, DNA strand exchange, site specific recombination, and even resolution of synthetic Holliday junctions. The key property of this intermediate that allows these reactions to proceed with high efficiency is its relatively long kinetic lifetime (7). For supercoil relaxation, the lifetime of the complex and the noncovalent “grip” on the DNA 3′ to the cleavage site, determines the number of supercoils that are removed for each cleavage and religation cycle (7). For the efficient strand transfer and Holliday junction resolution activities, it seems likely that topo I must destabilize the DNA 3′ downstream of the cleavage site, or minimally, that the enzyme complex “breaths” to allow binding and invasion of the incoming DNA strand. Thus the dynamics and/or stability of the DNA region 3′ to the cleavage site play a fundamental role in every aspect of the vTopo action.
Anticooperative DNA Binding
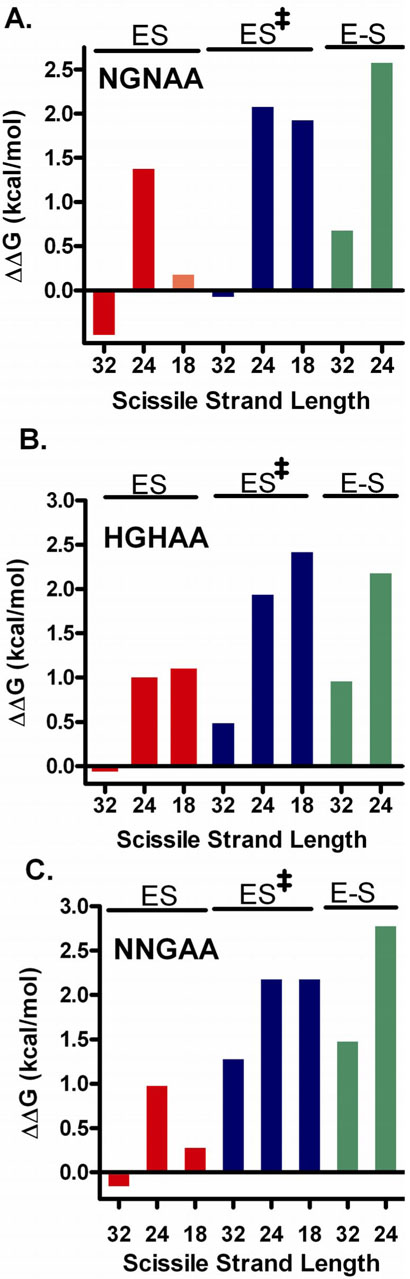
In light of the above described reactions, it is intriguing that we have uncovered anticooperative binding interactions between the DNA major groove and the 3′ DNA region. Although many single and double major groove substitutions were investigated, the overall findings are generally reflected by considering the effects of the three substitutions N5N3, H5H3, and N5N4 (Table 1). (The more damaging base deletion effects are considered separately below). In Figure 4 we have plotted the difference free energies (ΔΔG) as defined in eqs 3-5 for formation of the ES, ES‡ and E-S complexes from a 1 M reference state of free enzyme and substrate.
Figure 4.
Free energy effects of major groove substitutions for each DNA substrate. The ΔΔG = ΔGsub - ΔGunsub values for each DNA are defined in eqs 3-5. Free energy effects of the (A) N5N3, (B) H5H3, and (C) N5N4 substitutions for substrates with 32, 24 and 18 nucleotide scissile strands are shown. We note that changes in the activation barrier (ES → ES‡) arising from each modification may be gleaned by simply adding the bar heights corresponding to ES and ES‡.
With respect to noncovalent binding to form ES, the substitutions have little effect in the context of a 32/32mer, but for the 24/32mer, result in a consistent 1-1.4 kcal/mol weakening of the observed binding affinity. The 18/32mer shows a mixed response to these three substitutions, with little binding effects observed for N5N3 and N5N4, but a 1 kcal/mol weakening for H5H3. The lesser damaging effects of these major groove substitutions on noncovalent binding of the 32/32mer as compared to the 24/32mer are in the range 1 to 2 kcal/mol and reveal strain or anticooperativity between the major groove and the additional duplex region present in the longer substrate. These binding effects may be interpreted in light of the DNA footprint of vTopo (13), which extends from +13 to −13 base pairs in each direction from the scissile phosphate (Fig. 1B), and the enzyme-DNA interaction surface observed in the recent crystal structure of variola virus Topo I bound to DNA, which reaches from the +8 to −9 base pairs (16). Both the 32/32 and 24/32mers encompass all of this binding surface, while the 18/32mer lacks seven nucleotides on the 3′ side of the scissile phosphate (i.e. the −7 to −13 nucleotides). A reasonable interpretation is that the anticooperative energetic component to the observed binding affinity with the 32/32mer depends on the −13 to −20 nucleotides which are only present in this substrate. Since these nucleotides extend beyond the direct enzyme interaction surface, this region of base pairing may impart its effects indirectly by increasing duplex rigidity or altering its average conformation.2
The major groove substitutions generally have greater damaging effects in the transitionstate (ES‡) than in the ES complex, but once again the effects are smallest in the context of the 32/32mer (Fig. 4). The greater effect of the major groove substitutions in the transition-state indicates structural rearrangements leading to enhanced binding energy with this region of the consensus sequence. Despite the overall increases in the damaging effects of the major groove substitutions upon moving to the transition-state complex, the anticooperative binding component attributed to the −13 to −20 nucleotides of the 32/32mer is still in the range 0.75 to 2.0 kcal/mol, which may discerned by comparing the differences in the ΔΔGES‡ values for the 32/32mer and the two other truncated substrates (Fig. 4). In other words, strain energy in the 32/32mer persists equally in the ground state ES complex and in the transition-state.
When the final covalent complex is reached there still persists a 1 to 2 kcal/mol strain energy as judged by comparing the damaging effects of the major groove substitutions for the 32/32 and 24/32mers (Fig. 4). However, as compared to the ground state ES complex where virtually no effects of the substitutions are observed, the damaging effects of these changes have increased by as much as 1.5 kcal/mol for the 32/32mer in the E-S complex. Similarly, the 24/32mer shows the largest major groove effects on the stability of the E-S complex. One unifying interpretation for these findings is that when the DNA strand is nicked, some duplex strain is released and the major groove interactions then become stronger. Extending this energetic interpretation, the greater flexibility (or lesser duplex strain) present in the 24/32 and 18/32mers allows the strength of the major groove interactions to be felt earlier along the reaction coordinate, whereas the 32/32mer must first have the strain relieved during formation of the covalent complex to take full advantage of these interactions.
It is of interest to ask if the anticooperative effects extend to the base deletion substrates (ϕ). With the ϕ substitutions it is almost impossible to make cleavage rate measurements with the reversible substrates due to the extremely small equilibrium constant for cleavage. Thus, we generally obtained only KD, Kcl and Kcl/KD values (Table 1). Since ϕ substitutions decreased Kcl to equal extents for both the 32/32 and 24/32mers (100 to 200-fold), to obtain a general estimate of the deletion effects on kcl and kr we performed kinetic measurements using the ϕ4 substrate in the context of the 32/32mer and 18/32mer. Similar to the other more subtle base substitution effects, we find that the ϕ4 deletion gives rise to a 14-fold decrease in kcl/KD for the 32/32mer (an increase in the energy of the ES‡ complex of 1.6 kcal/mol), but a greater 73-fold increase in kr, indicating an even larger 4.2 kcal/mol [= −RT ln (73)(14)] destabilization of the covalent complex (E-S). For the 18/32mer, the ϕ4 deletion produces an 11,000-fold decrease in kcl/KD corresponding to an increase in the energy of the ES‡ complex of 5.6 kcal/mol. Thus, the extended duplex of the 32/32mer weakens the energetic effect of the ϕ4 deletion in the transition state by 4 kcal/mol compared to the 18/32mer, but after formation of the covalent complex, the 32/32mer realizes 75% of this interaction energy. These findings support our previous conclusion that major groove interactions are used to position the phosphotyrosine-DNA linkage an optimal distance from the 5′ hydroxyl leaving group. In other words, vTopo uses these interactions to prevent extremely rapid and efficient religation at consensus sites by decreasing the probability for religation (14).
Implications
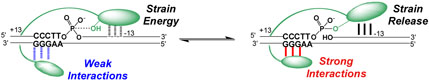
A model that depicts energetic coupling between duplex strain and the DNA major groove of the consensus sequence is shown in Figure 5. The crystal structure of the variola virus Topo bound to DNA has elucidated the nature of the specific interactions between the enzyme with the major groove. These interactions involve hydrogen bonds from the β5 strand from the 10 kDa amino terminal domain to the +2 adenine, as well as helix α5 from the ~30 kDa catalytic domain, which interacts with the +4 and +5 guanines (16). Thus, duplex strain is communicated to disparate regions of the protein. As depicted in Figure 5, strain is greatest in the noncovalent complex and diminishes as the reaction progresses to the covalent complex. Because vTopo does not directly interact with the −13 to −20 region of the 32/32mer, strain may involve a property of the 32/32mer duplex that is indirectly transmitted to the enzyme-DNA binding surface. Thus, vTopo may indirectly sense the stability, dynamics or subtle conformational properties of the 3′ downstream region of the DNA substrate. Further investigations will be required to fully elucidate these effects, but a functional role for duplex strain in promoting strand invasion or responding to DNA superhelicity and relaxation can be imagined.
Figure 5.
Anticooperative DNA binding by vTopo. For substrates with long 3′ base paired regions, a strain component to the observed binding energy is indicated. The duplex strain in the noncovalent complex results in weakening of the interactions in the major groove. Upon formation of the covalent species the strain is at least partially released allowing the major groove interactions to become stronger. Although not shown for clarity, the α5 helix of the larger catalytic domain also interacts with the major groove along with the β5 strand of the amino terminal domain.
FOOTNOTES
Abbreviations: vTopo, vaccinia DNA topoisomerase IB; FAM, 6-carboxyfluorescein; N, 2-aminopurine; H, deazaguanine; buffer A, 20 mM Tris-HCl, 200 mM NaCl, 1mM DTT, pH 8.0. 2Functional interpretation of the observed coupling energy between the major groove and the downstream DNA duplex does not require detailed knowledge of the coupling mechanism.
Most, but not all, of the measurements in this extensive study can be clustered into the general model of anticooperative binding of the major groove and the downstream region. For instance, a comparison of the changes in activation barrier for cleavage between substituted and unsubstituted substrates show more complexity arising from compensating changes in ground state and transition-state energies. In addition, some of the increased affinities for substrate binding shown in Table 1 are not well-understood.
This work was supported by National Institutes of Health Research Grant GM 68626 (J.T.S.).
REFERENCES
- 1.Champoux JJ. DNA topoisomerases: Structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 2.Shuman S. Vaccinia DNA topoisomerase I promotes illegitimate recombination in Escherichia coli. Proc Natl Acad Sci U S A. 1989;86:3489–93. doi: 10.1073/pnas.86.10.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shuman S. Novel approach to molecular cloning and polynucleotide synthesis using vaccinia DNA topoisomerase. J Biol Chem. 1994;269:32678–84. [PubMed] [Google Scholar]
- 4.Petersen BO, Shuman S. DNA strand transfer reactions catalyzed by vaccinia topoisomerase: Hydrolysis and glycerololysis of the covalent protein-DNA intermediate. Nucleic Acids Res. 1997;25:2091–7. doi: 10.1093/nar/25.11.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shuman S. Polynucleotide ligase activity of eukaryotic topoisomerase I. Mol Cell. 1998;1:741–8. doi: 10.1016/s1097-2765(00)80073-8. [DOI] [PubMed] [Google Scholar]
- 6.Sekiguchi J, Cheng C, Shuman S. Resolution of a holliday junction by vaccinia topoisomerase requires a spacer DNA segment 3′ of the CCCTT/ cleavage sites. Nucleic Acids Res. 2000;28:2658–63. doi: 10.1093/nar/28.14.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stivers JT, Harris TK, Mildvan AS. Vaccinia DNA topoisomerase I: Evidence supporting a free rotation mechanism for DNA supercoil relaxation. Biochemistry. 1997;36:5212–22. doi: 10.1021/bi962880t. [DOI] [PubMed] [Google Scholar]
- 8.Shuman S. Site-specific DNA cleavage by vaccinia virus DNA topoisomerase I. Role of nucleotide sequence and DNA secondary structure. J Biol Chem. 1991;266:1796–803. [PubMed] [Google Scholar]
- 9.Shuman S, Golder M, Moss B. Characterization of vaccinia virus DNA topoisomerase I expressed in Escherichia coli. J Biol Chem. 1988;263:16401–7. [PubMed] [Google Scholar]
- 10.Shuman S. Site-specific interaction of vaccinia virus topoisomerase I with duplex DNA. Minimal DNA substrate for strand cleavage in vitro. J Biol Chem. 1991;266:20576–7. [PubMed] [Google Scholar]
- 11.Kwon K, Stivers JT. Fluorescence spectroscopy studies of vaccinia type IB DNA topoisomerase. Closing of the enzyme clamp is faster than DNA cleavage. J Biol Chem. 2002;277:345–52. doi: 10.1074/jbc.M109449200. [DOI] [PubMed] [Google Scholar]
- 12.Stivers JT, Shuman S, Mildvan AS. Vaccinia DNA topoisomerase I: Single-turnover and steady-state kinetic analysis of the DNA strand cleavage and ligation reactions. Biochemistry. 1994;33:327–39. doi: 10.1021/bi00167a043. [DOI] [PubMed] [Google Scholar]
- 13.Sekiguchi J, Shuman S. Requirements for noncovalent binding of vaccinia topoisomerase I to duplex DNA. Nucleic Acids Res. 1994;22:5360–5. doi: 10.1093/nar/22.24.5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagarajan R, Stivers JT. Major groove interactions of vaccinia topo I provide specificity by optimally positioning the covalent phosphotyrosine linkage. Biochemistry. 2006;45:5775–82. doi: 10.1021/bi060133i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagarajan R, Kwon K, Nawrot B, Stec WJ, Stivers JT. Catalytic phosphoryl interactions of topoisomerase IB. Biochemistry. 2005;44:11476–85. doi: 10.1021/bi050796k. [DOI] [PubMed] [Google Scholar]
- 16.Perry K, Hwang Y, Bushman FD, Van Duyne GD. Structural basis for specificity in the poxvirus topoisomerase. Mol Cell. 2006;23:343–54. doi: 10.1016/j.molcel.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 17.Stivers JT, Jagadeesh GJ, Nawrot B, Stec WJ, Shuman S. Stereochemical outcome and kinetic effects of Rp- and Sp-phosphorothioate substitutions at the cleavage site of vaccinia type I DNA topoisomerase. Biochemistry. 2000;39:5561–72. doi: 10.1021/bi992429c. [DOI] [PubMed] [Google Scholar]
- 18.Shuman S, Prescott J. Specific DNA cleavage and binding by vaccinia virus DNA topoisomerase I. J Biol Chem. 1990;265:17826–36. [PubMed] [Google Scholar]