Abstract
Over one-quarter of all plant genes encode proteins of unknown function that can be further classified as proteins with obscure features (POFs), which lack currently defined motifs or domains, or proteins with defined features, which contain at least one previously defined domain or motif. Although empirical data in the form of transcriptome and proteome profiling suggest that many of these proteins play important roles in plants, their functional characterization remains one of the main challenges in modern biology. To begin the functional annotation of proteins with unknown function, which are involved in the oxidative stress response of Arabidopsis (Arabidopsis thaliana), we generated transgenic Arabidopsis plants that constitutively expressed 23 different POFs (four of which were specific to Arabidopsis) and 18 different proteins with defined features. All were previously found to be expressed in response to oxidative stress in Arabidopsis. Transgenic plants were tested for their tolerance to oxidative stress imposed by paraquat or t-butyl hydroperoxide, or were subjected to osmotic, salinity, cold, and heat stresses. More than 70% of all expressed proteins conferred tolerance to oxidative stress. In contrast, >90% of the expressed proteins did not confer enhanced tolerance to the other abiotic stresses tested, and approximately 50% rendered plants more susceptible to osmotic or salinity stress. Two Arabidopsis-specific POFs, and an Arabidopsis and Brassica-specific protein of unknown function, conferred enhanced tolerance to oxidative stress. Our findings suggest that tolerance to oxidative stress involves mechanisms and pathways that are unknown at present, including some that are specific to Arabidopsis or the Brassicaceae.
On average, 20% to 40% of all eukaryotic genomes sequenced to date contain genes that encode for proteins of unknown function (Gollery et al., 2006). We recently used several different bioinformatics approaches to annotate genes of unknown function in Arabidopsis (Arabidopsis thaliana; Gollery et al., 2006, 2007; Horan et al., 2008). One such approach used a hidden Markov model protein family (HMMPFAM) search to identify all proteins that contain no previously defined domains or motifs. These unknown proteins were termed proteins with obscure features (POFs) and were distinguished from proteins with defined features (PDFs), which contained at least one previously defined domain or motif (Gollery et al., 2006, 2007). In a comparison among 10 different eukaryotic proteomes, including yeast (Saccharomyces cerevisiae), Schizosaccharomyces pombe, Arabidopsis, rice (Oryza sativa), Drosophila melanogaster, Anopheles gambiae, Caenorhabditis elegans, Mus musculus, Rattus norvegicus, and Homo sapiens, POFs were found to be similar to PDFs in their relative contribution to biological function, as indicated by their transcript expression, participation in protein-protein interactions, and association with mutant phenotype (Gollery et al., 2006). Surprisingly, 60% of the POFs identified within the different proteomes were, on average, species specific, compared to only 7.5% of the PDFs (Gollery et al., 2006). POFs were also found to contain more disordered structure and were shorter and more hydrophilic than PDFs (Gollery et al., 2006). A comparison among the Arabidopsis, rice, and poplar (Populus trichocarpa) proteomes identified over 2,000 POFs that were specific to Arabidopsis and revealed that POFs were mainly represented as singletons within the different plant proteomes (Gollery et al., 2007).
The identification of over 5,000 proteins of unknown function in Arabidopsis suggests that many of the known pathways and networks currently being studied in Arabidopsis include additional genes and proteins that have an unknown function (Gollery et al., 2007; Horan et al., 2008). Some of these can be identified using correlation studies, which are based on transcriptome profiling analyses (Horan et al., 2008). In addition to the proteins with unknown function that might participate in known pathways and networks, many POFs and PDFs in plants can serve unknown or possibly novel functions involved in basic or specialized processes and might comprise new and undiscovered pathways (Gollery et al., 2006, 2007). The identification of genes with unknown function, such as POFs, which are unique to Arabidopsis, might suggest that some of these proteins are associated with or involved in processes that are unique to Arabidopsis or the Brassicaceae (Gollery et al., 2007). An additional possibility, however, is that some POFs fold and function much like some of the known proteins but do not share any sequence similarity to them (Siew and Fischer, 2004; Gollery et al., 2006). Additionally, some POFs and PDFs might represent misannotated proteins or genes (Gollery et al., 2006). The functional characterization of genes with unknown function might provide an insight into the role of unknown proteins in different organisms and has become a major goal in modern biological research (Fischer and Eisenberg, 1999; Alonso et al., 2003; Chothia et al., 2003; Siew and Fischer, 2003; Roberts, 2004; Gollery et al., 2006, 2007).
To begin the functional characterization of proteins with unknown function in Arabidopsis, we identified 41 different proteins of unknown function that respond to endogenous oxidative stress in Arabidopsis (Davletova et al., 2005a) and constitutively expressed them in transgenic plants. We specifically chose oxidative stress as our source for genes of unknown function, because this stress is considered to be common among many different aerobic organisms, and many of the known pathways and genes involved in the response of different organisms to oxidative stress have overlapping functions and/or structural similarities (Asada and Takahashi, 1987; Halliwell and Gutteridge, 1999; Mittler et al., 2004; Halliwell, 2006; Vandenbroucke et al., 2008). We found that more than 70% of the expressed unknown proteins conferred tolerance to oxidative stress. In contrast, the majority of expressed unknowns (>90%) did not confer tolerance to the other stresses tested, and approximately 50% of the expressed unknown proteins rendered plants more susceptible to osmotic or salinity stress. Two Arabidopsis-specific POFs and an Arabidopsis- and Brassica-specific protein of unknown function, which contained a zinc finger domain, conferred enhanced tolerance to oxidative stress when expressed in transgenic plants. Our findings suggest that tolerance to oxidative stress in Arabidopsis involves different proteins, pathways, and mechanisms that are unknown at present, including some that are specific to Arabidopsis or the Brassicaceae.
RESULTS
Selection of POFs and PDFs and Generation of Transgenic Plants
In a previous study, we conducted a detailed GeneChip (ATH1) time-course analysis comparing wild-type plants to knockout plants lacking the key hydrogen peroxide (H2O2) scavenging enzyme cytosolic Apx1 (ascorbate peroxidase 1) subjected to a moderate treatment of light stress (Davletova et al., 2005a). This treatment resulted in the endogenous accumulation of H2O2 and oxidized proteins in the knockout plants compared to the wild types during light stress (Davletova et al., 2005a). Of the 3,915 transcripts that were found to be significantly altered in their expression in knockout-Apx1 plants, compared to wild types, during this treatment, 119 were designated as unknowns by the GeneChip (ATH1) annotation (Davletova et al., 2005a). Fifty of these were selected based on their mRNA abundance in the knockout plants (exceeding a 2-fold increase threshold), the availability of pUni clones (Yamada et al., 2003), and the lack of internal restriction sites used for cloning into the binary vectors. Of the 50 clones selected, 41 clones were successfully used in the generation of transgenic plants. As shown in Table I, 23 of these proteins were designated as POFs and 18 as PDFs (unknowns that contained a previously defined domain or motif).
Table I.
Annotation, homology, and predicted length of the proteins expressed in transgenic plants
A summary of the different proteins with unknown function expressed and characterized in transgenic plants. The locus, pUni, and predicted length are given on the left for each clone. Homology to other organisms determined by a BLAST cutoff value of 10−6 and annotation based on an HMMPFAM search are given on the right. *, Has homolog in Xanthomonas.
| Locus | pUni Clone | Length (Amino Acids) | Homology | Annotation |
|---|---|---|---|---|
| AT1G11210 | U13249 | 308 | Plant | Contain DUF761 domain |
| AT1G13340 | U51136 | 409 | Plant, microbial | Contain DUF292 domain |
| AT1G14870 | U24063 | 152 | Plant | Contain PLAC8 domain |
| AT1G19020 | U11998 | 86 | Plant | Unknown (POF) |
| AT1G21520 | U50590 | 66 | Arabidopsis | Unknown (POF) |
| AT1G27330 | U22724 | 68 | Plant, animal, microbial | Contain ribosome associated membrane RAMP4 domain |
| AT1G32220 | U17408 | 296 | Plant, algae, fungi | Contain epimerase/dehydratase domain |
| AT1G50170 | U20498 | 225 | Plant, microbial | Contain Cbix domain |
| AT1G50290 | U14460 | 134 | Arabidopsis | Unknown (POF) |
| AT1G52200 | U22606 | 190 | Plant | Contain PLAC8 domain |
| AT1G64360 | U61211 | 85 | Arabidopsis | Unknown (POF) |
| AT1G72060 | U62101 | 81 | Plant | Contain Ser-type endopeptidase inhibitor domain |
| AT1G73120 | U60254 | 109 | Plant | Unknown (POF) |
| AT1G78410 | U21397 | 108 | Plant | Contain VQ motif |
| AT1G80130 | U09684 | 305 | Plant | Unknown (POF) |
| AT2G04795 | U12390 | 95 | Plant | Unknown (POF) |
| AT2G15560 | U61937 | 489 | Plant, animal, microbial | Contain DUF537 domain |
| AT2G19310 | U18059 | 162 | Plant | Contain HSP20/α-crystallin domain |
| AT2G21195 | U23029 | 93 | Plant | Unknown (POF) |
| AT2G22080 | U61331 | 177 | Arabidopsis, Brassica | Contain zinc finger domain |
| AT2G24150 | U10208 | 344 | Plant, animal, microbial | Contain HlyIII domain |
| AT2G40000 | U10524 | 435 | Plant | Contain Hs1pro-1 domain |
| AT2G41650 | S66967 | 66 | Arabidopsis | Unknown (POF) |
| AT2G44240 | U14883 | 402 | Plant* | Contain DUF239 domain |
| AT3G08670 | U61987 | 567 | Plant | Unknown (POF) |
| AT3G10020 | U13057 | 149 | Plant | Unknown (POF) |
| AT3G14430 | U23423 | 76 | Plant | Unknown (POF) |
| AT3G16670 | U22022 | 154 | Plant | Unknown (POF) |
| AT3G20340 | U63078 | 115 | Plant | Unknown (POF) |
| AT3G23910 | U19105 | 421 | Plant | Unknown (POF) |
| AT3G51610 | U24527 | 230 | Plant, algae | Unknown (POF) |
| AT3G60980 | U24736 | 412 | Plant | Contain pentatricopeptide repeat domain |
| AT4G08940 | U61884 | 395 | Plant | Contain ubiquitin hydrolase domain |
| AT4G12000 | U20042 | 306 | Plant, microbial | Contain SNARE domain |
| AT4G17070 | U14323 | 240 | Plant, algae | Unknown (POF) |
| AT5G18040 | U11383 | 252 | Arabidopsis, Brassica | Unknown (POF) |
| AT5G19875 | U61271 | 123 | Plant | Unknown (POF) |
| AT5G27830 | U09971 | 300 | Plant, algae | Unknown (POF) |
| AT5G43750 | U50098 | 212 | Plant | Unknown (POF) |
| AT5G50350 | U24198 | 584 | Plant | Unknown (POF) |
| AT5G59080 | U11769 | 135 | Plant | Unknown (POF) |
All pUni clones obtained from the Arabidopsis Biological Resource Center (Yamada et al., 2003) were PCR cloned into pGEM-T vectors and sequenced. They were then cloned into a modified pGreen binary vector (Hellens et al., 2000; Suzuki et al., 2008) as an in-frame fusion protein upstream to GFP and expressed in transgenic plants under the control of the cauliflower mosaic virus (CaMV) 35S promoter (35S∷POF-GFP; C-terminal fusion). At least 15 independent transgenic lines were initially obtained for each unknown protein. These were scored based on the presence of the selectable marker (hygromycin), GFP imaging, and transcript and protein expression. Two homozygous independent lines with similar expression levels of the POF-GFP transcript/protein were then selected for each gene for further analysis.
To examine the relationship of the different unknown proteins to other organisms, their predicted amino acid sequences were compared with the National Center for Biotechnology Information nonredundant, as well as EST, databases translated in all reading frames, and a BLAST E-value cutoff of 10−6 was used to determine sequence homology with other organisms (http://www.ncbi.nlm.nih.gov/; Gollery et al., 2006, 2007; Table I). The proteins were then classified as Arabidopsis-specific (i.e. homology was found only to Arabidopsis), plant-specific (i.e. homology was found to at least one other plant; with the exception of AT5G18040 and AT2G22080 that had homology only to Arabidopsis and Brassica napus and were designated as such), or found to have homologs in other plants or organisms, including algae, microbes, and/or animals (Table I). Altogether, we identified four Arabidopsis-specific, two Arabidopsis- and Brassica-specific, and 25 plant-specific proteins. Interestingly, despite the relative similarity in transcript expression in response to oxidative stress between plants and animals (Vandenbroucke et al., 2008), only three of the unknown proteins identified and selected for analysis in Arabidopsis had homologs in animals (AT1G27330, AT2G15560, and AT2G24150; Table I). It should be noted, however, that the domain and homology identification for the proteins included in this study is likely to change with time as more plant sequences and HMMPFAM models will be deposited in public databases (Gollery et al., 2006, 2007).
Oxidative and Abiotic Stress Assays
To examine the stress tolerance of the different transgenic lines expressing proteins of unknown function, seedlings of transgenic plants were subjected to different oxidative and abiotic stresses, as described previously (Davletova et al., 2005b; Mittler et al., 2006; Ciftci-Yilmaz et al., 2007; Miller et al., 2007). As controls, we used seedlings of transgenic plants that expressed GFP under the control of the CaMV 35S promoter (35S∷GFP) and seedlings of wild-type plants.
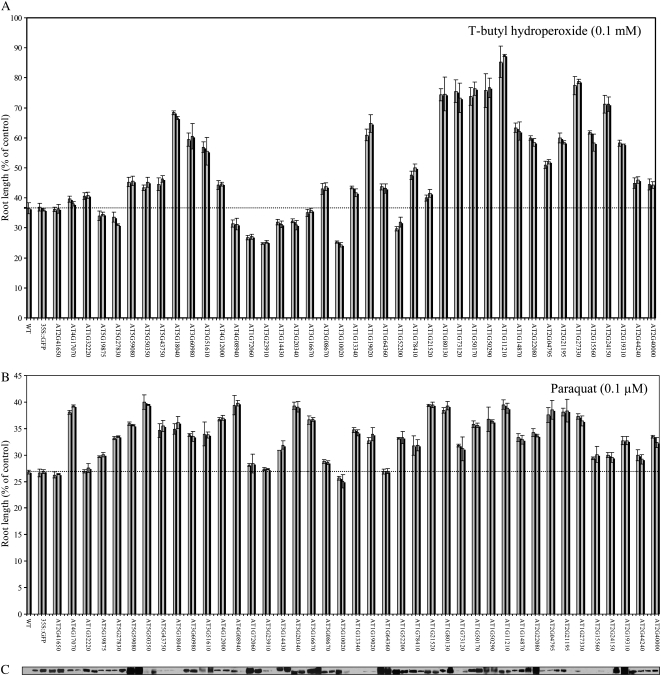
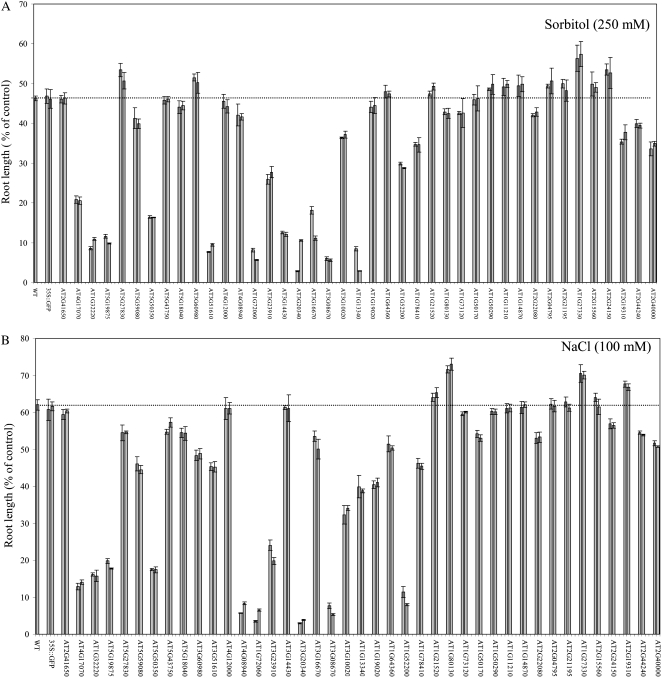
As shown in Figure 1 and Supplemental Table S1, more than 70% of the lines expressing proteins of unknown function were found to be more tolerant to oxidative stress imposed by paraquat or t-butyl hydroperoxide (35 or 29 out of 41, respectively). In contrast, as shown in Figure 2 and Supplemental Table S1, the majority of expressed unknowns did not confer tolerance to osmotic or salinity stresses (>92%; 38 and 38 out of 41, respectively), and approximately 50% of the lines expressing protein of unknown function were found to be more sensitive to osmotic or salinity stresses (21 or 27 out of 41, respectively). An inverse correlation between tolerance to oxidative stress imposed by paraquat and salinity was identified in 46% of the lines (a total of 19 different lines; Figs. 1B and 2B; Supplemental Table S1). This finding was in agreement with our previous observation made with knockout-Apx1 plants that showed an inverse correlation between tolerance to salinity and oxidative stress (Ciftci-Yilmaz et al., 2007).
Figure 1.
Tolerance of transgenic Arabidopsis seedlings expressing proteins of unknown function to oxidative stress. A, Root growth assays showing tolerance to t-butyl hydroperoxide in seedlings of transgenic plants. B, Root growth assays showing tolerance to paraquat in seedlings of transgenic plants. C, A composite protein gel blot showing the expression level of the different fusion proteins using GFP antibody. Molecular size of the different proteins is indicated in Supplemental Table S1. Two independent lines were tested for each construct. WT, Control wild-type seedlings; AT2G41650 to AT2G40000, transgenic seedlings expressing different proteins of unknown function under the control of the CaMV 35S promoter and fused in frame to the N terminal of GFP.
Figure 2.
Tolerance of transgenic Arabidopsis seedlings expressing proteins of unknown function to osmotic and salinity stresses. A, Root growth assays showing tolerance to osmotic stress in seedlings of transgenic plants. B, Root growth assays showing tolerance to salinity in seedlings of transgenic plants. Two independent lines were tested for each construct. WT, Control wild-type seedlings; AT2G41650 to AT2G40000, transgenic seedlings expressing different proteins of unknown function under the control of the CaMV 35S promoter and fused in frame to the N terminal of GFP.
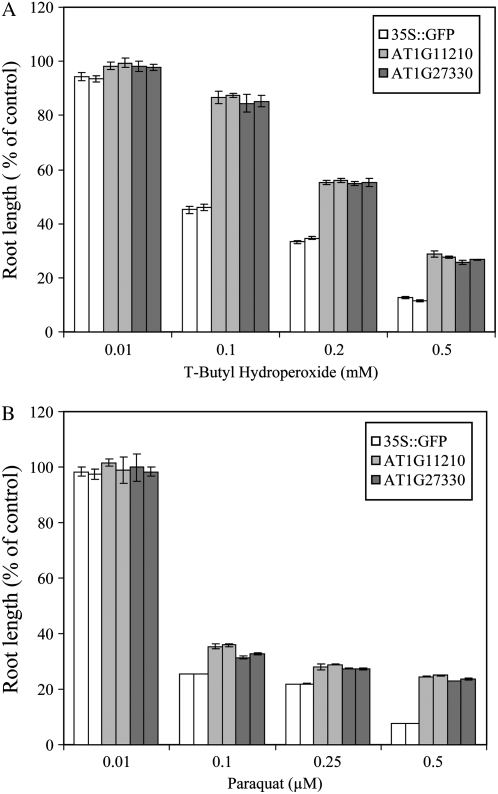
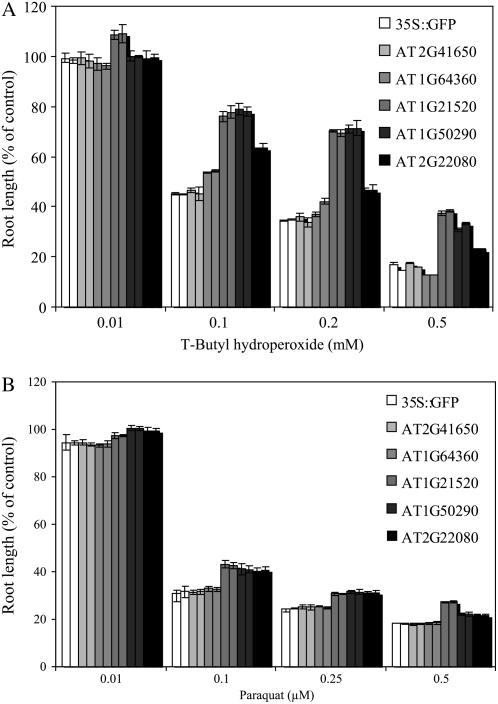
As shown in Figure 3 and below for Arabidopsis and/or Arabidopsis- and Brassica-specific proteins, the tolerance of selected lines was tested against a range of paraquat and t-butyl hydroperoxide concentrations to confirm the results shown in Figure 1 (obtained with one concentration of paraquat and t-butyl hydroperoxide). As shown in Supplemental Figure S1 and compared to the results obtained with oxidative, salinity, and osmotic stresses (Figs. 1 and 2; Supplemental Table S1), tolerance to cold or heat stress was not dramatically enhanced in the transgenic lines. With the exception of AT5G19875 and AT1G11210, no significant effects were also observed in the germination of the different lines in the presence or absence of 0.5 μm abscisic acid (Supplemental Fig. S2; Supplemental Table S1).
Figure 3.
Tolerance of transgenic Arabidopsis seedlings expressing proteins of unknown function to oxidative stress. A, Root growth assays showing enhanced tolerance to a range of t-butyl hydroperoxide concentrations in seedlings of transgenic plants. B, Root growth assays showing enhanced tolerance to a range of paraquat concentrations in seedlings of transgenic plants. Two independent lines were tested for each construct.
As shown in Figure 1C, the expression level of the different unknown proteins between the different constructs did not correlate with the resistance phenotype (i.e. some constructs with a low expression level in both lines tested showed enhanced tolerance to oxidative stress compared to other lines with a high level of expression in both lines tested). This result suggested that the mechanism of action of the different unknown genes is different and that for some genes, even a low expression level is sufficient to result in high tolerance to oxidative stress.
Analysis of Arabidopsis-Specific POFs
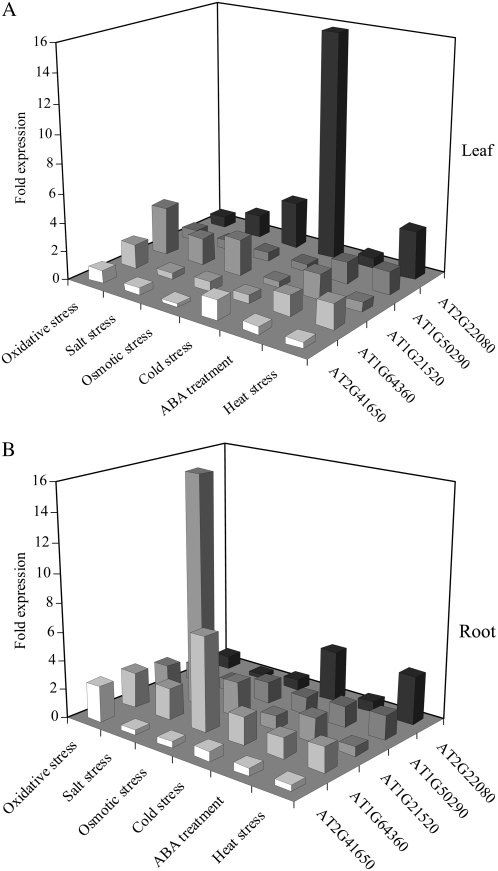
To further characterize the four Arabidopsis-specific POFs identified in this study (Table I), we examined their relative mRNA abundance in response to different abiotic stresses, as monitored by microarray (Affymetrix GeneChip) experiments and deposited in public databases (Zimmermann et al., 2004; Miller and Mittler, 2006). As shown in Figure 4, AT2G41650 was mainly expressed in roots in response to oxidative stress, AT1G64360 was mainly expressed in roots in response to osmotic stress, AT1G21520 was mainly expressed in roots in response to salt stress, AT2G50290 was mainly expressed in roots and leaves in response to heat stress, and AT2G22080 (an Arabidopsis- and Brassica-specific protein used as a control) was mainly expressed in leaves in response to cold stress. At least based on their transcript expression patterns (Fig. 4; Davletova et al., 2005a), the different Arabidopsis-specific POFs appear to have a putative function in Arabidopsis during stress.
Figure 4.
A and B, Expression of four Arabidopsis-specific POFs and an Arabidopsis- and Brassica-specific protein of unknown function in response to different abiotic treatments in leaves (A) and roots (B) of wild-type plants. Microarray expression data was obtained from Genevestigator (Zimmermann et al., 2004) and is presented as fold expression compared to control untreated (Miller and Mittler, 2006).
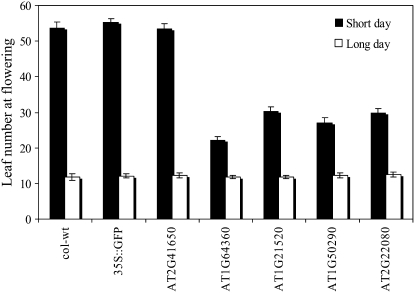
As shown in Figure 5, constitutive expression of four of the five proteins selected for further analysis (AT1G64360, AT1G21520, AT2G50290, and AT2G22080) resulted in an accelerated flowering time phenotype when grown under short-day conditions. An altered flowering time phenotype was previously observed for certain oxidative stress mutants grown under controlled growth conditions (Pnueli et al., 2003; Rizhsky et al., 2003; Miller et al., 2007).
Figure 5.
Early flowering phenotype in transgenic plants expressing Arabidopsis-specific POFs and an Arabidopsis- and Brassica-specific protein of unknown function. Measurements of leaf number at time of flowering under short- and long-day conditions are shown in transgenic plants expressing Arabidopsis-specific POFs.
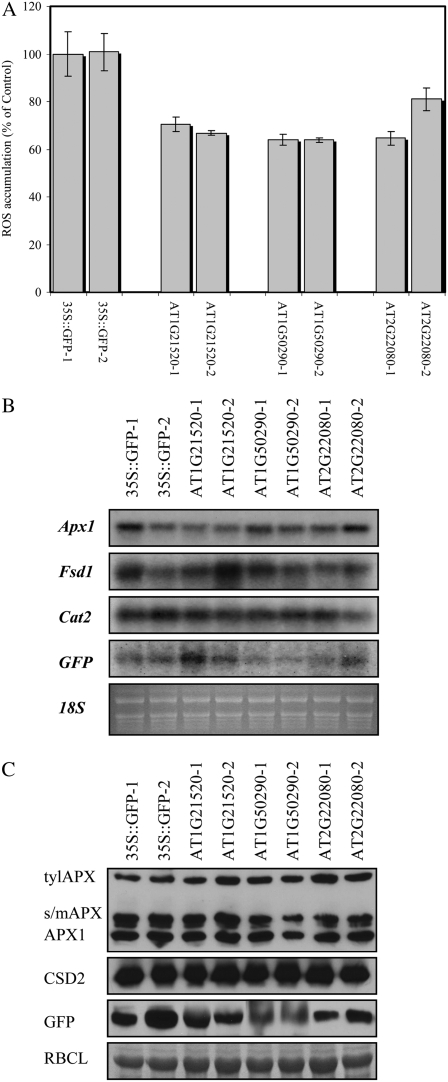
As shown in Figure 6, constitutive expression of two of the Arabidopsis-specific POFs (AT1G21520, AT1G50290) and the Arabidopsis- and Brassica-specific protein with unknown function, which contained a zinc finger domain (AT2G22080), conferred enhanced tolerance to oxidative stress imposed by a range of different concentrations of paraquat or t-butyl hydroperoxide. To test whether the enhanced tolerance of transgenic plants expressing these proteins was associated with a general decrease in the level of reactive oxygen species (ROS), we used Amplex Red to measure H2O2 in 5-d-old seedlings of transgenic plants expressing GFP or AT1G21520, AT1G50290, and AT2G22080 fused to GFP grown on agar plates in the presence or absence of 0.1 μm paraquat. As shown in Figure 7A, transgenic seedlings expressing AT1G21520, AT1G50290, or AT2G22080 accumulated less H2O2 compared to control plants when grown on agar plates that contained paraquat. In contrast, no differences were observed between the levels of H2O2 in control seedlings or seedlings of transgenic plants expressing AT1G21520, AT1G50290, or AT2G22080 grown on agar plates in the absence of paraquat (Supplemental Fig. S3).
Figure 6.
Tolerance of transgenic Arabidopsis seedlings expressing Arabidopsis-specific POFs or an Arabidopsis- and Brassica-specific protein of unknown function to oxidative stress. A, Root growth assays showing enhanced tolerance to a range of t-butyl hydroperoxide concentrations in seedlings of transgenic plants. B, Root growth assays showing enhanced tolerance to a range of paraquat concentrations in seedlings of transgenic plants. Two independent lines were tested for each construct.
Figure 7.
Accumulation of H2O2 and protein and transcript expression in transgenic Arabidopsis seedlings expressing Arabidopsis-specific POFs or an Arabidopsis- and Brassica-specific protein of unknown. A, Accumulation of H2O2 in roots of 5-d-old seedlings grown in the presence of 0.1 μm paraquat. B, RNA gel-blot analysis showing the expression of transcripts encoding the reactive oxygen scavenging enzymes APX1, CAT2, and FSD1 in transgenic plants grown under controlled growth conditions. Ribosomal RNA was used to control for RNA loading. C, Protein gel-blot analysis showing the expression of the reactive oxygen scavenging enzymes tylAPX, s/mAPX, CSD2, and GFP in transgenic plants grown under controlled growth conditions. Rubisco large subunit (RBCL) was used to control for protein loading. Size and loading factor for the GFP fusion proteins in C is: 35S∷GFP, 46 kD, 1× loading; AT1G21520, 53 kD 1× loading; AT1G50290, 61.7 kD, 20× (concentrated); AT2G22080, 65 kD, 1× loading.
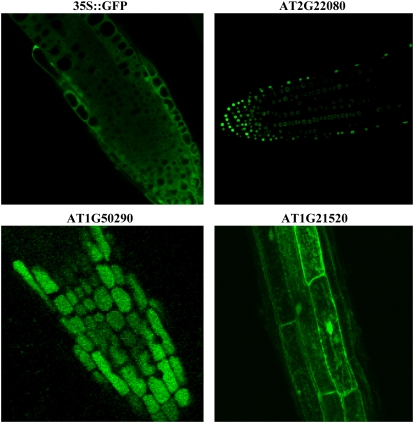
To test whether the enhanced tolerance of transgenic plants expressing AT1G21520, AT1G50290, or AT2G22080 to oxidative stress was associated with enhanced expression of known reactive oxygen scavenging enzymes, we conducted RNA and protein gel-blot analysis on these transgenic plants grown under controlled growth conditions. As shown in Figure 7, B and C, this analysis revealed that expression of the key reactive oxygen scavenging enzymes Apx1, thylakoid APX (tylAPX), stromal mitochondrial APX (s/mAPX), catalase 2 (Cat2), copper-zinc superoxide dismutase 2 (CSD2), or iron superoxide dismutase 1 (FSD1) in transgenic plants grown under controlled growth conditions was not elevated or did not correlate with enhanced tolerance to oxidative stress (Fig. 6) or accumulation of H2O2 (Fig. 7A; Supplemental Fig. S3). Because elevated expression of Apx1, tylAPX, s/mAPX, Cat2, CSD2, or FSD1 could be used as a measure for endogenous oxidative stress in plants (Mittler et al., 2004), these results might also indicate that, at least with respect to the classical markers for internal oxidative stress, the transgenic plants tested did not suffer from an internal oxidative stress. As shown in Figure 8, AT2G22080 appeared to be localized to nuclei, AT1G50290 appeared to be localized to the cytosol but excluded from nuclei, and AT1G21520 appeared to be localized to the endoplasmic reticulum and perhaps other cellular membrane systems.
Figure 8.
Imaging of GFP in root tips of transgenic seedlings expressing Arabidopsis-specific POFs or an Arabidopsis- and Brassica-specific protein of unknown function, which conferred tolerance to oxidative stress, grown under controlled growth conditions. The different unknown proteins or POFs were expressed in plants under the control of the CaMV 35S promoter and fused in frame to the N terminal of GFP.
Expression of AT1G50290 in Yeast
To determine whether constitutive expression of some Arabidopsis proteins with unknown function will confer enhanced tolerance against oxidative stress to other organisms, we transformed and tested the oxidative stress tolerance of yeast expressing the following proteins: AT2G41650, AT1G72060, and AT3G10020 (proteins that did not confer enhanced tolerance to oxidative stress in transgenic plants; Fig. 1), and AT5G19875, AT5G59080, AT5G43750, AT5G18040, AT3G51610, AT4G12000, AT1G52200, AT1G78410, AT1G21520, AT1G80130, AT1G73120, AT1G50170, AT2G04795, AT1G27330, At1g50290, AT2G19310, AT2G44240, and AT2G40000 (proteins that did confer enhanced tolerance to oxidative stress in transgenic plants; Fig. 1). Two different yeast strains were used for these assays: wild-type yeast (BY4743) and a yeast mutant lacking the transcription factor Yap1 that is essential for tolerance to oxidative stress (in the BY4743 background; termed Yap1Δ; Kuge and Jones, 1994). Transformed and untransformed yeast were subjected to oxidative stress imposed by paraquat or t-butyl hydroperoxide as described by Tiên Nguyên-nhu and Knoops (2003) and Davies et al. (1995). In contrast to the results obtained with plants (Fig. 1), none of the genes expressed in yeast was able to enhance the tolerance of transgenic yeast (Yap1Δ) to oxidative stress (Supplemental Fig. S4). These results might suggest that, at least with respect to their functionality in yeast, the majority of proteins identified in this study did not have a general antioxidative function.
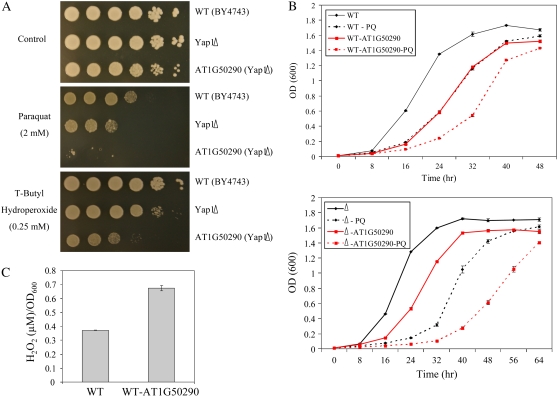
One of the proteins tested, AT1G50290, an Arabidopsis-specific POF, was found to have a negative impact on the oxidative stress tolerance of yeast. As shown in Figure 9, A and B, expression of AT1G50290 in yeast rendered yeast (wild type or Yap1Δ) more susceptible to oxidative stress imposed by paraquat or t-butyl hydroperoxide. Expression of AT1G50290 in yeast (wild type or Yap1Δ) was also found to have a deleterious effect on growth in the absence of oxidative stress (Fig. 9B). As shown in Figure 9C, expression of AT1G50290 in wild-type yeast resulted in enhanced accumulation of H2O2 in the growth media of cells grown under controlled growth conditions. In contrast to these findings, expression of AT1G50290 did not result in accumulation of H2O2 in transgenic plants grown under controlled growth conditions (Supplemental Fig. S3).
Figure 9.
Enhanced sensitivity to oxidative stress in yeast cells expressing AT1G50290. A, Plate growth assays of wild-type yeast, Yap1Δ, and Yap1Δ transformed with AT1G50290, showing enhanced sensitivity to oxidative stress induced by paraquat and t-butyl hydroperoxide in Yap1Δ cells transformed with AT1G50290. B, Liquid media growth assays of wild-type yeast, wild-type yeast transformed with AT1G50290 (top), Yap1Δ, and Yap1Δ transformed with AT1G50290 (bottom), showing suppressed growth and enhanced sensitivity to oxidative stress induced by paraquat and t-butyl hydroperoxide in yeast cells transformed with AT1G50290. C, Accumulation of H2O2 in the growth media of wild-type yeast cells transformed with AT1G50290. H2O2 was determined in the media of log phase cells as described in “Materials and Methods.” [See online article for color version of this figure.]
DISCUSSION
ROS are produced in cells in response to many different abiotic or biotic conditions, and their uncontrolled accumulation can lead to oxidative stress (Asada and Takahashi, 1987; Halliwell and Gutteridge, 1999; Kovtun et al., 2000; Mittler, 2002; Apel and Hirt, 2004; Foyer and Noctor, 2005; Asada, 2006; Halliwell, 2006; Van Breusegem and Dat, 2006). Enhanced tolerance to oxidative stress, brought about by constitutive expression of a single protein (Figs. 1, 3, and 6) might result from a number of different mechanisms: (1) the expressed protein might have a direct scavenging activity that can detoxify certain species of reactive oxygen; (2) the expressed protein might be part of a cellular network, or signal transduction pathway that protects the cell from damage caused by oxidative stress or is involved in the detoxification of reactive oxygen, and its constitutive expression enhances the activity of this network or pathway; or (3) the expressed protein might alter plant metabolism, causing the accumulation of reactive oxygen and indirectly activating the cells' scavenging, protection, and/or repair mechanisms against oxidative stress similar to some transgene-induced lesion mimics (Mittler and Rizhsky, 2000).
Previous studies have shown a direct correlation between enhanced tolerance to oxidative stress and enhanced tolerance to different abiotic stresses, leading to the assumption that enhancing the ability of plants to scavenge reactive oxygen would also enhance the ability of plants to tolerate other stresses (e.g. Roxas et al., 1997; Deak et al., 1999; Kovtun et al., 2000; Rivero et al., 2007). Interestingly, in our hands, the majority of transgenic plants expressing proteins of unknown function and showing enhanced tolerance to oxidative stress (Fig. 1) did not show enhanced tolerance to osmotic, salinity, heat, or cold stresses (Fig. 2; Supplemental Fig. S1; Supplemental Table S1). This finding could suggest that the expressed unknown proteins do not have a general reactive oxygen scavenging activity that would have made them more tolerant to other stresses (Roxas et al., 1997; Deak et al., 1999; Kovtun et al., 2000; Rivero et al., 2007). A direct scavenging activity would have also made at least some of the yeast cells expressing the unknown proteins more tolerant to oxidative stress. However, none of the 18 different proteins with unknown function that enhanced the tolerance of transgenic plants to oxidative stress enhanced the tolerance of yeast cells to oxidative stress, suggesting that these proteins do not function to directly scavenge ROS, such as superoxide radicals or lipid peroxides.
The finding that the expressed proteins with unknown function did not confer enhanced tolerance to other abiotic stresses might also suggest that they did not alter plant metabolism in a way that will indirectly trigger the cells' scavenging, protection, and/or repair mechanisms against general or oxidative stress (Roxas et al., 1997; Deak et al., 1999; Kovtun et al., 2000; Mittler and Rizhsky, 2000). Moreover, with the exception of AT1G50290 (Fig. 9), none of the unknown proteins tested in yeast exhibited deleterious effects on growth or caused yeast to have an altered susceptibility to oxidative stress (Supplemental Fig. S4). This finding supports the possibility that constitutive expression of proteins with unknown function in plants did not have an indirect effect on cellular metabolism causing a generalized stress tolerance response (Mittler and Rizhsky, 2000).
At least two different findings, therefore, point to a high specificity in the function of the expressed proteins with unknown function: (1) enhanced tolerance to oxidative stress caused by their constitutive expression did not cause a general cellular effect that made plants more tolerant to other stresses (Figs. 1 and 2; Supplemental Fig. S1); and (2) an inversed correlation was found for at least 19 of them between tolerance to oxidative stress imposed by paraquat and tolerance to salinity (Figs. 1 and 2; Supplemental Table S1). We previously found that knockout plants lacking Apx1 were more susceptible to oxidative stress but were more tolerant to salinity stress (Ciftci-Yilmaz et al., 2007). The finding that constitutive expression of 19 proteins with unknown function, which were identified in knockout-Apx1 plants as responsive to internal oxidative stress (Davletova et al., 2005a), caused plants to become more tolerant to oxidative stress but more susceptible to salinity or osmotic stress could suggest that these proteins have very specific functions that are tied to the pathway(s) activated in knockout-Apx1 plants. The differences observed between the tolerance of the different lines to paraquat, which functions by enhancing the cellular rate of superoxide radical production, and tolerance to t-butyl hydroperoxide, which functions by inducing lipid peroxidation (Figs. 1, A and B), could also suggest that the different unknown genes have specific functions within the ROS gene network (Mittler et al., 2004).
Because the majority of expressed proteins with unknown function appear to have a specific oxidative stress tolerance phenotype when expressed in transgenic plants (Figs. 1 and 2; Supplemental Figs. S1 and S2), they could be involved in different networks that protect cells from specific aspects of oxidative stress. This hypothesis is based on the finding that they do not enhance tolerance to other abiotic stresses, many of them cause plants to become more susceptible to osmotic or salinity stresses, and many of them do not protect yeast against oxidative stress. These findings are in agreement with our previous hypothesis that genes of unknown function are highly specific to different organisms and function as part of specific cellular networks (Gollery et al., 2006, 2007). Our findings, therefore, demonstrate that the study of proteins with unknown function could unravel new and specialized functions that could be phylogenetically specific.
Our characterization of Arabidopsis- and Arabidopsis- and Brassica-specific proteins of unknown function revealed that three of these proteins enhanced the tolerance of plants to oxidative stress (Fig. 6). The constitutive expression of these proteins did not enhance the production of H2O2 in plants grown under controlled growth conditions (Supplemental Fig. S3), did not result in the enhanced expression of known reactive oxygen scavenging mechanisms (Fig. 7, B and C), and did not cause plants to be more tolerant to salinity or osmotic stresses (Fig. 2), suggesting that at least in plants, they did not cause the enhanced production of reactive oxygen, thereby triggering a general oxidative stress response. Because of their high specificity to reactive oxygen stress, it is possible that these proteins function as part of an Arabidopsis- or Arabidopsis- and Brassica-specific network that is involved in cellular repair and/or protection against oxidative stress. It appears, therefore, that Arabidopsis and/or Brassica plants could contain specific pathways, unknown at present, that are involved in the protection of cells against oxidative stress. These pathways could function in different cellular compartments (Fig. 8) and could interact with different developmental pathways (Fig. 5). The possible existence of repair and/or protection pathways against oxidative stress that are specific for Arabidopsis or the Brassicaceae is very interesting, because oxidative stress is considered to be a general type of stress common to many different organisms (Asada and Takahashi, 1987; Halliwell and Gutteridge, 1999; Mittler et al., 2004; Halliwell, 2006; Vandenbroucke et al., 2008). Our study, therefore, highlights the need to characterize phylogenetic-specific proteins of unknown function in different organisms, because these could shed light on new and possibly novel pathways involved in different aspects of plant metabolism.
The approach of using microarray expression data as a tool to predict function for proteins of unknown function (Horan et al., 2008) appears to be a good approach, because over 70% of the proteins tested in this study showed an oxidative stress response phenotype (Figs. 1 and 2). We are in the process of generating a database that includes the abiotic stress-phenotypic characterization of over 1,000 knockout mutants (Alonso et al., 2003) for proteins of unknown function (Gollery et al., 2007; unpublished data) with the goal of identifying additional pathways and mechanisms involving proteins of unknown function in Arabidopsis.
MATERIALS AND METHODS
Plant Growth, Transformation, and Molecular Analysis
Arabidopsis (Arabidopsis thaliana ‘Columbia’) plants were grown under controlled conditions (21°C, 100 μmol m−2 s−1; Suzuki et al., 2005) and monitored for growth and flowering time as described by Miller et al. (2007). RNA and protein were isolated and analyzed by gel-blot analysis (Rizhsky et al., 2003; Davletova et al., 2005a, 2005b). cDNA probes corresponding to the following Arabidopsis genes were used for the RNA gel blots shown in Figure 7: APX1, AT1G07890; Cat2, AT4G35090; FSD1, AT4G25100. Antibodies for APX1, tylAPX, s/mAPX, and RBCL were obtained as described by Miller et al. (2007). Antibodies to CSD2 were obtained from Agrisera, and antibodies to GFP were obtained from CLONTECH. pUNI clones (Yamada et al., 2003) were obtained from the ABRC, PCR cloned into pGEM-T (Promega) using gene-specific primers (Supplemental Table S2), and sequenced. The resulting plasmids were then digested and cloned in frame to GFP (C-terminal fusion) into a modified pGreen vector (Suzuki et al., 2008). GFP, or GFP fused in-frame to the C terminal of the different unknown proteins, were expressed in plants under the control of the 35S CaMV promoter (Hellens et al., 2000; Suzuki et al., 2008). Transgenic plants were generated using the floral dip method (Zhang et al., 2006), and homozygous lines were selected using hygromycin resistance and RNA and protein blots. In addition, plants were visualized for GFP using a Nikon Eclipse E400 epifluorescence microscope or an Olympus IX 81 FV 1000 confocal microscope, as described by Suzuki et al. (2008). Analysis of microarray data available from https://www.genevestigator.ethz.ch (Zimmermann et al., 2004) was normalized and performed as previously described (Miller and Mittler, 2006).
Stress Assays
For the analysis of stress tolerance, seeds of the wild type and two independent homozygous transgenic lines for 35S∷GFP or 35S∷POFs-GFP were surface-sterilized with bleach and placed in rows on 1% agar plates (0.5× Murashige and Skoog medium) containing different concentrations of paraquat, t-butyl hydroperoxide, NaCl, sorbitol, or abscisic acid (Sigma-Aldrich), as described by Davletova et al. (2005b), Mittler et al. (2006), Ciftci-Yilmaz et al. (2007), and Miller et al. (2007). Each row of seeds (25–30 seedlings) placed on a plate was divided into two parts: control seeds and seeds of transgenic plants expressing the different proteins of unknown function. Thus, the different seeds were placed side-by-side on the same plate. Plates were maintained vertically in a growth chamber (21–22°C, constant light, 100 μmol m−2 s−1) and percent germination and root length were scored 5 d after seed plating. Four- or 5-d-old seedlings grown on 0.5× Murashige and Skoog agar plates were also subjected to heat stress (38°C; 24 h) or cold stress (10°C; 48 h) and scored for percent germination and root length as described by Davletova et al. (2005b), Mittler et al. (2006), Ciftci-Yilmaz et al. (2007), and Miller et al. (2007). All experiments were repeated at least three different times, each with at least three different technical repeats. Results are shown as mean and se bars. Statistical analysis was performed as described in Suzuki et al. (2008). Significant difference between each construct (35S∷POFs-GFP) and the 35S∷GFP control was assigned only when both lines tested for each of the constructs were t test significant at P < 0.05. To image H2O2 accumulation in seedlings subjected to oxidative stress, 5-d-old seedlings grown in the presence of paraquat or t-butyl hydroperoxide were treated with 0.2 μm Amplex Red (Molecular Probes) for 1 h and imaged with a Kodak 2000MM image station (Davletova et al., 2005b).
Characterization of Proteins with Unknown Function in Yeast Cells
pUni clones were digested with EcoRI and NotI, cloned into pESC-His vector (Stratagene) under the GAL10 promoter, and transformed into wild-type BY4743 ([4741/4742] MATa/MATα his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2Δ0/+met15Δ0/+ ura3Δ0/ura3Δ0; American Type Culture Collection no. 201390) and Yap1Δ mutant (MATa/MATα his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2Δ0/+met15Δ0/+ ura3Δ0/ura3Δ0 ΔYAP1; American Type Culture Collection no. 4030569) yeast (Saccharomyces cerevisiae) strains. Empty plasmids were also transformed into the yeast strains as controls. Transformants were selected on solid minimal media that contained yeast nitrogen base (Difco) and −His Dropout supplement (per 20 g/L of Gal; SG-His; Clontech). Transformants grown overnight in liquid SG-His media were subjected to stress on solid SG-His media supplied with either 2 mm methyl viologen (Acros Organics) or 0.25 mm t-butyl hydroperoxide (Sigma-Aldrich) as serial dilutions (Tiên Nguyên-nhu and Knoops, 2003) or subjected to the same oxidative stress treatment in liquid SG-His media (Davies et al., 1995).
Amplex Red Hydrogen Peroxide/Peroxidase Assay kit (Molecular Probes) was used for the detection of H2O2 in the growth media of yeast. Yeast were grown overnight in SG-His liquid media up to mid-log phase, and 50 μL of the growth media was used to detect H2O2 according to the manufacturer's instructions. For each sample, four replicates were used, and 50 μL of working solution (100 μm Oxired probe [MBL International], 0.2 units/mL horseradish peroxidase, Type II [Sigma-Aldrich]) supplied with 0.05 m sodium phosphate, pH 7.4, was added to each sample. Reactions were incubated at room temperature as recommended, and fluorescence was measured with a VICTOR3 V Multilabel Counter (model 1420; PerkinElmer) using excitation at 531 nm and emission at 595 nm.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Effect of heat or cold on root elongation of transgenic plants expressing genes of unknown function.
Supplemental Figure S2. Effect of abscisic acid on root elongation of transgenic plants expressing genes of unknown function.
Supplemental Figure S3. Accumulation of H2O2 in roots of 5-d-old seedlings.
Supplemental Figure S4. Yeast growth assays performed with wild type, Yap1 mutants, and Yap1 mutants transformed with different Arabidopsis genes.
Supplemental Table S1. Summary of stress tolerance of the different lines.
Supplemental Table S2. Primers used for the cloning of the different proteins with unknown function.
Supplementary Material
This work was supported by the National Science Foundation (grant nos. IBN–0420033, NSF–0431327, and IOS–0743954), by the Nevada Agricultural Experimental Station publication, and by the National Institutes of Health IDeA Network of Biomedical Research Excellence (grant no. RR–03–008).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ron Mittler (ronm@unr.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55 373–399 [DOI] [PubMed] [Google Scholar]
- Asada K (2006) Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol 141 391–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada K, Takahashi M (1987) Production and scavenging of active oxygen in photosynthesis. In DJ Kyle, CB Osmond, CJ Arntzen, eds, Photoinhibition. Elsevier, Amsterdam, pp 227–287
- Chothia C, Gough J, Vogel C, Teichmann SA (2003) Evolution of the protein repertoire. Science 300 1701–1703 [DOI] [PubMed] [Google Scholar]
- Ciftci-Yilmaz S, Morsy MR, Song L, Coutu A, Krizek BA, Lewis MW, Warren D, Cushman J, Connolly EL, Mittler R (2007) The EAR-motif of the Cys2/His2-type zinc finger protein Zat7 plays a key role in the defense response of Arabidopsis to salinity stress. J Biol Chem 282 9260–9268 [DOI] [PubMed] [Google Scholar]
- Davies JM, Lowry CV, Davies KJ (1995) Transient adaptation to oxidative stress in yeast. Arch Biochem Biophys 317 1–6 [DOI] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R (2005. a) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S, Schlauch K, Coutu J, Mittler R (2005. b) The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol 139 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deak M, Horvath GV, Davletova S, Torok K, Sass L, Vass I, Barna B, Kiraly Z, Dudits D (1999) Plants ectopically expressing the iron-binding protein, ferritin, are tolerant to oxidative damage and pathogens. Nat Biotechnol 17 192–196 [DOI] [PubMed] [Google Scholar]
- Fischer D, Eisenberg D (1999) Finding families for genomic ORFans. Bioinformatics 15 759–762 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G (2005) Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17 1866–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollery M, Harper J, Cushman J, Mittler T, Girke T, Zhu JK, Bailey-Serres J, Mittler R (2006) What makes species unique? The contribution of proteins with obscure features. Genome Biol 7 757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollery M, Harper J, Cushman J, Mittler T, Mittler R (2007) POFs: what we don't know can hurt us. Trends Plant Sci 12 492–496 [DOI] [PubMed] [Google Scholar]
- Halliwell B (2006) Reactive species and antioxidants. Redox biology is a fundamental theme of aerobic life. Plant Physiol 141 312–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JMC (1999) Free Radicals in Biology and Medicine, Ed 3. Clarendon, UK
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42 819–832 [DOI] [PubMed] [Google Scholar]
- Horan K, Jang C, Bailey-Serres J, Mittler R, Shelton C, Harper JF, Zhu JK, Cushman JC, Gollery M, Girke T (2008) Annotating genes of known and unknown functions by large-scale co-expression analysis. Plant Physiol 147 41–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge S, Jones N (1994) YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J 13 655–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun Y, Chiu WL, Tena G, Sheen J (2000) Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc Natl Acad Sci USA 97 2940–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Mittler R (2006) Could heat shock transcription factors function as hydrogen peroxide sensors in plants? Ann Bot (Lond) 98 279–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Rizhsky L, Hegie A, Koussevitzky S, Mittler R (2007) Double mutants deficient in cytosolic and thylakoid ascorbate peroxidase reveal a complex mode of interaction between reactive oxygen species, plant development and response to abiotic stresses. Plant Physiol 144 1777–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R (2002) Oxidative stress, antioxidants, and stress tolerance. Trends Plant Sci 7 405–410 [DOI] [PubMed] [Google Scholar]
- Mittler R, Rizhsky L (2000) Transgene-induced lesion mimics. Plant Mol Biol 44 335–344 [DOI] [PubMed] [Google Scholar]
- Mittler R, Song L, Coutu J, Coutu A, Ciftci S, Kim YS, Lee H, Stevenson B, Zhu JK (2006) Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett 580 6537–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) The reactive oxygen gene network of plants. Trends Plant Sci 9 490–498 [DOI] [PubMed] [Google Scholar]
- Pnueli L, Liang H, Mittler R (2003) Growth suppression, abnormal guard cell response, and augmented induction of heat shock proteins in cytosolic ascorbate peroxidase (Apx1)-deficient Arabidopsis plants. Plant J 34 187–203 [DOI] [PubMed] [Google Scholar]
- Rivero RM, Kojima M, Gepstein A, Sakakibara H, Mittler R, Gepstein S, Blumwald E (2007) Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc Natl Acad Sci USA 104 19631–19636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Mittler R (2003) The water-water cycle is essential for chloroplast protection in the absence of stress. J Biol Chem 278 38921–38925 [DOI] [PubMed] [Google Scholar]
- Roberts RJ (2004) Identifying protein function: a call for community action. PLoS Biol 2 293–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roxas VP, Smith RK Jr, Allen ER, Allen RD (1997) Overexpression of glutathione S-transferase/glutathione peroxidase enhances the growth of transgenic tobacco seedlings during stress. Nat Biotechnol 15 988–991 [DOI] [PubMed] [Google Scholar]
- Siew N, Fischer D (2003) Analysis of singleton ORFans in fully sequenced microbial genomes. Proteins 53 241–251 [DOI] [PubMed] [Google Scholar]
- Siew N, Fischer D (2004) Structural biology sheds light on the puzzle of genomic ORFans. J Mol Biol 342 369–373 [DOI] [PubMed] [Google Scholar]
- Suzuki N, Rizhsky L, Liang H, Shuman J, Shulaev V, Mittler R (2005) Enhanced tolerance to environmental stress in transgenic plants expressing the transcriptional coactivator multiprotein bridging factor 1c. Plant Physiol 139 1313–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Bajad S, Shuman J, Shulaev V, Mittler R (2008) The transcriptional co-activator MBF1C is a key regulator of thermotolerance in Arabidopsis thaliana. J Biol Chem 283 9269–9275 [DOI] [PubMed] [Google Scholar]
- Tiên Nguyên-nhu N, Knoops B (2003) Mitochondrial and cytosolic expression of human peroxiredoxin 5 in Saccharomyces cerevisiae protect yeast cells from oxidative stress induced by paraquat. FEBS Lett 544 148–152 [DOI] [PubMed] [Google Scholar]
- Van Breusegem F, Dat JF (2006) Reactive oxygen species in plant cell death. Plant Physiol 141 384–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbroucke K, Robbens S, Vandepoele K, Inzé D, Van de Peer Y, Van Breusegem F (2008) Hydrogen peroxide-induced gene expression across kingdoms: a comparative analysis. Mol Biol Evol 25 507–516 [DOI] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, et al (2003) Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302 842–846 [DOI] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin SS, Niu QW, Chua NH (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protocols 1 641–646 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.