Abstract
The Arabidopsis (Arabidopsis thaliana) MKK1 and MKK2 mitogen-activated protein kinase kinases have been implicated in biotic and abiotic stress responses as part of a signaling cascade including MEKK1 and MPK4. Here, the double loss-of-function mutant (mkk1/2) of MKK1 and MKK2 is shown to have marked phenotypes in development and disease resistance similar to those of the single mekk1 and mpk4 mutants. Because mkk1 or mkk2 single mutants appear wild type, basal levels of MPK4 activity are not impaired in them, and MKK1 and MKK2 are in part functionally redundant in unchallenged plants. These findings are confirmed and extended by biochemical and molecular analyses implicating the kinases in jasmonate- and salicylate-dependent defense responses, mediated in part via the MPK4 substrate MKS1. In addition, transcriptome analyses delineate overlapping and specific effects of the kinases on global gene expression patterns demonstrating both redundant and unique functions for MKK1 and MKK2.
Eukaryotic mitogen-activated protein (MAP) kinase (MPK) cascades act downstream of receptors or sensors to transduce extracellular stimuli, including abiotic and biotic stresses, into adaptive, intracellular responses. The basic assembly of a MPK cascade is a three-kinase module. MPK, the last kinase in the cascade, is activated by dual phosphorylation of Thr and Tyr residues in its kinase catalytic activation loop. This phosphorylation is mediated by a MPK kinase (MPKK or MEK), which is activated by a MPKK kinase (MPKKK or MEKK). Multiple members for each of these three tiers of kinases may be expressed in a cell. This may contribute to the specificity of transmitted signals and permit the integration of signals due to interactions between a kinase of one tier with more than one kinase of another tier (Madhani and Fink, 1998). DNA-binding transcription factors are targets of MPK cascades whose activities are directly or indirectly regulated by MPK phosphorylation. MPK cascades may thereby regulate gene expression in response to extracellular signals. In plants, MPK signaling has been associated inter alia with abiotic stress (Ichimura et al., 2000; Teige et al., 2004; Alzwiy and Morris, 2007) and with basal and systemic acquired resistance (SAR) to pathogens (Asai et al., 2002; Mészáros et al., 2006; Brader et al., 2007; Loake and Grant, 2007; Zhang et al., 2007).
Genetic analyses indicate that Arabidopsis (Arabidopsis thaliana) MPK4 acts as a negative regulator of salicylic acid (SA)-mediated defense against biotrophic pathogens, as illustrated by enhanced resistance to biotrophs and expression of the PATHOGENESIS-RELATED1 (PR1) gene in mpk4 loss-of-function mutants. Conversely, MPK4 is required for ethylene (ET)- and jasmonic acid (JA)-mediated defense against necrotrophic pathogens, as typified by enhanced susceptibility to necrotrophs and reduced expression of the antifungal PLANT DEFENSIN1.2 (PDF1.2) gene in mpk4 mutants (Petersen et al., 2000; Brodersen et al., 2006). MPK4 kinase activity is enhanced by pathogen-derived elicitors (Desikan et al., 2001) and also by abiotic stresses such as cold and wounding (Ichimura et al., 2000).
MPK4 is predominantly localized in nuclei where it interacts with a substrate protein, MKS1. As may be expected for a kinase substrate, MKS1 overexpression partially phenocopies the mpk4 mutant (dwarfism, enhanced SA levels, and constitutive PR1 gene expression), while MKS1 underexpression partially suppresses these mpk4 mutant phenotypes (Andreasson et al., 2005). MKS1 can be multiply phosphorylated by MPK4 (Caspersen et al., 2007) and may function as an adaptor linking MPK4 activity to the two transcription factors WRKY25 and WRKY33. Both of these factors may be involved in regulating gene expression in response to pathogens; both appear to negatively regulate certain SA-mediated responses, while WRKY33 may activate certain ET- and JA-mediated responses (Zheng et al., 2006, 2007).
Arabidopsis MKK1 (Morris et al., 1997) and MKK2 (Ichimura et al., 1998) both interact with MPK4 in yeast two-hybrid assays (Ichimura et al., 1998; Mizoguchi et al., 1998), and MPK4 can be phosphorylated and activated both in vitro and in vivo by MKK1 (Huang et al., 2000; Matsuoka et al., 2002). MKK1 and MKK2 have also both been associated with biotic and abiotic stresses in several studies. For example, MKK1 gene expression is wound induced (Morris et al., 1997), and transiently expressed MKK1 is activated by pathogen-derived elicitors such as flagellin and laminarin, as well as by hydrogen peroxide (Teige et al., 2004). In addition, an MKK1 T-DNA insertion mutant exhibits a small reduction in MPK4 activation in response to flagellin treatment and is slightly more sensitive to infection by Pseudomonas syringae pv tomato (Pst) DC3000 (Mészáros et al., 2006).
Following transient overexpression in protoplasts, MKK2 kinase activity and MPK4 activation have been shown to be enhanced by salt and cold treatment, but not significantly so by elicitor treatment. An MKK2 T-DNA insertion mutant was found to be hypersensitive to cold and salt treatment, with much reduced MPK4 activity in response to cold stress, whereas plants expressing a constitutively active version of MKK2 showed enhanced cold and salt tolerance (Teige et al., 2004). In addition, these latter plants were found to exhibit enhanced resistance to biotrophic pathogens but compromised resistance to necrotrophs (Brader et al., 2007). Interestingly, the single mkk1 or mkk2 mutants do not exhibit the phenotypic characteristics of mpk4. This genetic evidence indicates that MKK1 and MKK2 have partly redundant functions in the activation of MPK4, because complete loss of MPK4 kinase activity results in the severe developmental phenotype of the mpk4 single mutant (Petersen et al., 2000).
Yeast two-hybrid analyses indicated that MPK4, MKK1, and MKK2 also interact with the stress-induced MPKKK, MEKK1 (Ichimura et al., 1998; Mizoguchi et al., 1998), and MEKK1 can activate both MKK1 (Hadiarto et al., 2006) and MKK2 (Teige et al., 2004). Mutations in MEKK1 (Ichimura et al., 2006; Nakagami et al., 2006; Suarez-Rodriguez et al., 2007) result in plants morphologically similar to mpk4 mutants, including dwarfed growth, enhanced PR1 gene expression, and reduced MPK4 activity induction by hydrogen peroxide or flagellin. mekk1 mutants show some differences from mpk4 in root phenotype and premature senescence of primary leaves, and the mekk1-1/mpk4 double mutant has a more severe phenotype than either single mutant, including an absence of root and shoot growth. This suggests that MEKK1 and MPK4 function in more than one pathway, a shared one regulating pathogen responses and another (or others) important for seedling development (Su et al., 2007). However, genetic data have not previously been presented to identify which MPKK (or MPKKs) functions between MEKK1 and MPK4.
To further investigate the roles of MKK1 and MKK2 in plant stress and disease responses, T-DNA insertion mutants of both genes were isolated and analyzed as single and double mutants. Our data show that the morphological, molecular, and defense-related phenotypes of the mkk1/2 double mutant resemble those of the mpk4 and mekk1 single mutants. This indicates that MPK4 is controlled by MEKK1 through the partially redundant activities of MKK1 and MKK2.
RESULTS
The mkk1/2 Double Mutant Is Severely Dwarfed
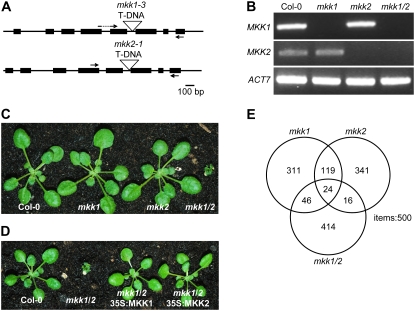
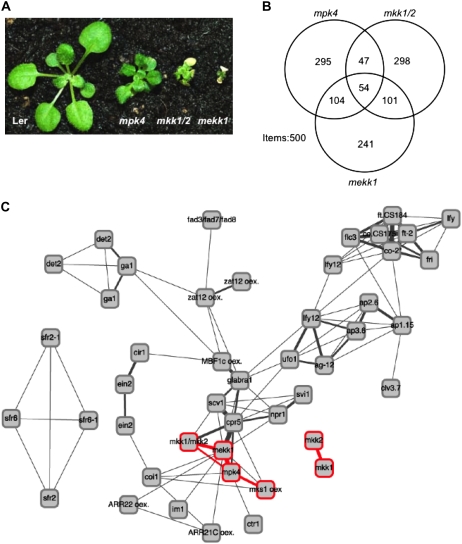
T-DNA insertion mutants of MKK1 (Salk line 027645; Alonso et al., 2003) and MKK2 (SAIL line 511_H 01; Sessions et al., 2002) were backcrossed to wild-type Columbia (Col-0). Plants homozygous for the insertions were isolated by PCR screening with gene-specific and T-DNA left border primers. Sequence analysis of the left border PCR products showed an insertion 1,332 bp after the initiation ATG of the major open reading frame in intron 5 of mkk1 and at 1,387 bp, also in intron 5, for mkk2 (Fig. 1A). Because two previous insertion mutations have already been described for MKK1 (Mészáros et al., 2006; Xing et al., 2007), this new allele may be designated mkk1-3 but is referred to simply as mkk1 below. The mkk2-1 allele is identical to that analyzed by Teige et al. (2004). Reverse transcription (RT)-PCR of RNA from the homozygous mkk1/mkk2 double mutant (hereafter referred to as mkk1/2) did not detect full-length transcripts for either MKK1 or MKK2 (Fig. 1B). Neither single mutant showed any obvious morphological or developmental abnormality compared to wild-type Col-0 plants (Fig. 1C), including flowering time under long or short days, root growth, root hair length and morphology, as well as guard cell spacing and morphology (data not shown). Germination and seedling growth of the single mutants were not significantly different from wild type after treatments with auxin, cytokinin, 1-aminocyclopropane-1-carboxylic acid (ACC), GA, or JA, nor was etiolation or de-etiolation affected (data not shown). The mkk2 mutant, but not mkk1, exhibited slightly enhanced abscisic acid (ABA) and salt sensitivity in germination tests, confirming previous findings (Supplemental Fig. S1; Teige et al., 2004). In contrast to the single mutants, mkk1/2 was severely dwarfed with curled, dark green leaves, and exhibited early senescence of the cotyledons and the first few true leaves (Fig. 1C).
Figure 1.
Mutant phenotypes. A, Gene structure of MKK1 and MKK2. Exons are represented by black boxes, and introns and untranslated regions by black lines. Triangles indicate T-DNA insertions; the T-DNA was located 1,332 bp downstream of the translational initiation codon in mkk1 and 1,387 bp downstream of the initiation codon in mkk2. Primers used for RT-PCR in B are indicated by arrows. B, MKK1 and MKK2 mRNA levels in wild type, mkk1, and mkk2 detected by RT-PCR. C, Morphology of Col-0 wild type, mkk1, mkk2, and the mkk1/2 double mutant. D, Complementation of mkk1/2 by CaMV 35S promoter-driven MKK1 or MKK2 cDNA. E, Venn diagram of global gene expression signatures of mkk1, mkk2, and mkk1/2 mutants compared to wild type. Here, intersections are between the top 500 most significantly differentially expressed genes as measured by the Affymetrix GeneChip, ATH1 (22,810 probe sets). If at random, 11 genes are expected to overlap between any two gene sets and approximately 0.2 genes between three gene sets.
Wild-type MKK1 or MKK2 cDNA sequences driven by the cauliflower mosaic virus (CaMV) 35S promoter were used to transform wild-type-like plants heterozygous for mkk1 and homozygous for mkk2. Of 12 primary transformants shown to contain the MKK1 cDNA by PCR, five were found to be homozygous for mkk1/2 and these were all fully wild type in morphology. Similarly, of 12 MKK2 transformed lines, four were found to be homozygous for mkk1/2 and also wild type in morphology (Fig. 1D). These results confirm that the dwarfism and premature senescence phenotypes of mkk1/2 are due to loss-of-function of both MKK1 and MKK2.
To extend this comparative phenotypic analysis at the molecular level, gene expression profiling was also performed on mkk1, mkk2, and mkk1/2. As summarized in a Venn diagram, of the most significantly differentially regulated genes compared to wild type (Fig. 1E; Supplemental Table S1), most of the changes in transcript levels in the mkk1/2 double mutant were not shared by mkk1 or mkk2 single mutants. The large overlap between genes differentially regulated in mkk1 and mkk2 of almost 30% (Fig. 1E) indicates that a common function of basal levels of MKK1 or MKK2 is required for wild-type expression of these shared genes. In keeping with the morphological phenotypes of the single and double mutants, this indicates that MKK1 and MKK2 have partially redundant functions.
mkk1/2 and the Defense-Related Hormones SA, ET, and JA
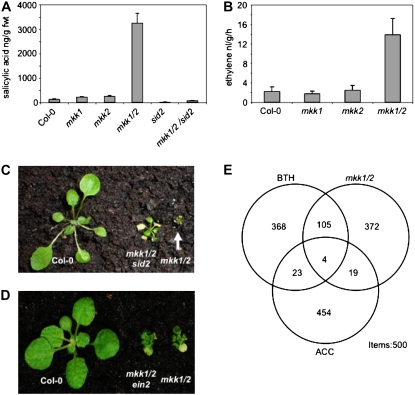
Both single mkk1 and mkk2 mutants were found to have close to wild-type levels of free SA, whereas mkk1/2 exhibited more than a 20-fold increase in SA levels compared to wild type (Fig. 2A). The salicylic acid induction-deficient2 (sid2) mutant prevents SA accumulation during the development of SAR (Wildermuth et al., 2001), and the mkk1/2/sid2 triple mutant showed much reduced levels of SA (Fig. 2A). In keeping with these findings, the dwarf habit, but not the early senescence phenotype, of mkk1/2 was partially alleviated by sid2 mutation (Fig. 2C). This indicates that the severe growth phenotype of mkk1/2 is in part dependent upon SA, as is the case for both the mpk4 (Petersen et al., 2000) and the mekk1 mutants (Suarez-Rodriguez et al., 2007).
Figure 2.
Hormone levels and growth phenotypes of mkk1/2/sid2 and mkk1/2/ein2 triple mutants. A and B, Free SA (A) and ET (B) levels. Error bars represent se of the mean. C and D, Phenotypes of mkk1/2/sid2 (C) and mkk1/2/ein2 triple mutants (D). E, Venn diagram showing overlapping gene expression signatures between BTH- or ACC-treated wild-type plants and the mkk1/2 double mutant. Overlap is between the top 500 most significantly differentially expressed genes as measured by the ATH1 Affymetrix GeneChip.
The mkk1/2 double mutant also superficially resembles the dark green, dwarf phenotype of the constitutive ET response mutant constitutive triple response1 (Kieber et al., 1993) and of plants overproducing ET (Guzman and Ecker, 1990). ET measurements showed that levels produced by mkk1/2 were some 4-fold higher than in wild type (Fig. 2B). However, several lines of evidence indicate that mkk1/2 dwarfism and early senescence are not ET related. First, no plants were found to exhibit a constitutive triple response among a large population (>200) segregating for mkk1/2 doubly homozygous seedlings. Second, although the triple mutants mkk1/2/ein2 and mkk1/2/etr1-1 were ET insensitive, as judged by the absence of a triple response when dark germinated in the presence of the ET precursor ACC, they were morphologically indistinguishable from mkk1/2 mutants when grown in the light (Fig. 2D). Similar experiments have shown that the morphological phenotype of the mpk4 mutant is independent of ET signaling (Petersen et al., 2000).
To further compare mkk1/2 responses with those to SA or ET treatments, gene expression profiling of mkk1/2 was compared with that of wild type treated with the SA analog benzo(1,2,3)thiadiazole-7-carbothioic acid S-methyl ester (BTH) or the ET precursor ACC. As shown in a Venn diagram (Fig. 2E; Supplemental Table S2), only 23 genes (4.6%) from the 500 most differentially regulated genes were shared between ACC-treated wild type and mkk1/2 mutants, which is comparable to that shared between ACC-treated and BTH-treated wild-type plants (27 genes, 5.4%). In contrast, 109 genes (21.8%; P value approximately 10−78) were shared by BTH-treated wild type and mkk1/2 mutants. Significantly, these genes include the “classic” pathogenesis-related proteins PR1, PR2, two PR3-type chitinases, and PR5 (Supplemental Table S2). A gene ontology analysis of the differentially regulated genes further supports this, as defense-related terms in this overlap were overrepresented (Supplemental Table S4). These results indicate that the molecular phenotype of the mkk1/2 mutant is independent of ET but dependent upon SA and affects, like mpk4, the expression of genes required for SAR.
Endogenous JA levels were also determined in leaves of mutant plants. Basal JA levels were slightly higher in mkk1, mkk2, and mkk1/2 plants in comparison to the wild type. JA levels were also observed to increase after wounding in the single mutants and mkk1/2 plants, in both wounded leaves and systemically (Supplemental Fig. S2). Thus, wound-induced JA biosynthesis is not impaired, either in the single mutants or in the mkk1/2 double mutant.
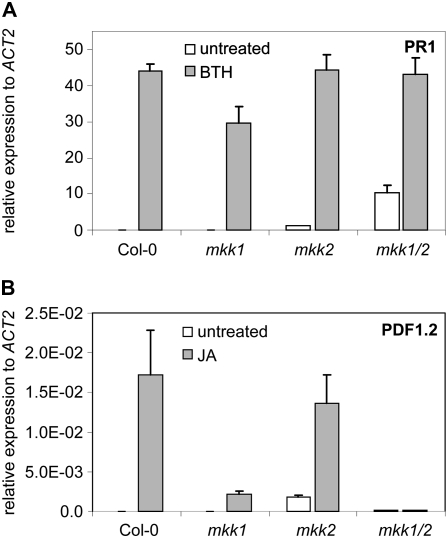
Real-time PCR was used to examine the expression of the SA- and ET/JA-responsive genes PR1 and PDF1.2 in response to treatment with BTH or JA, respectively. mRNA of the SA-responsive PR1 gene (Uknes et al., 1992) was barely detectable in wild-type and mkk1 plants, while PR1 mRNA levels were slightly raised in mkk2 and markedly so in untreated mkk1/2 double mutants (Fig. 3A). BTH treatment enhanced PR1 expression to higher levels in all genotypes. As the mkk1/2 mutant exhibited constitutive PR1 expression, this suggests that these two genes function redundantly as negative regulators of SAR. However, PR1 mRNA accumulation was still increased by BTH in mkk1/2. This is in keeping with earlier findings that signaling through MPK4 primarily affects basal PR1 expression levels (Andreasson et al., 2005).
Figure 3.
Defense gene expression in the mutants. PR1 (A) and PDF1.2 (B) mRNA levels were monitored by real-time RT-PCR after treatment of the mutants with BTH or JA, respectively. Means ± sd are shown.
PDF1.2 is an antifungal protein whose expression is induced by JA and ET (Manners et al., 1998). PDF1.2 expression was strongly enhanced by JA in wild type and in mkk2 plants, but much less so in the mkk1 and mkk1/2 backgrounds (Fig. 3B). Given that JA levels are normal in all mutant lines, this suggests that MKK1 is an intermediary in JA signaling. Consistent with this, wounding induced systemic VEGETATIVE STORAGE PROTEIN1 (VSP1) mRNA accumulation in wild type and in mkk2, but much less so in mkk1 and mkk1/2 plants (Supplemental Fig. S3). These results indicate that the insensitivity of mkk1/2 to JA and wounding is mainly due to loss-of-function of MKK1.
Disease Responses of mkk1/2
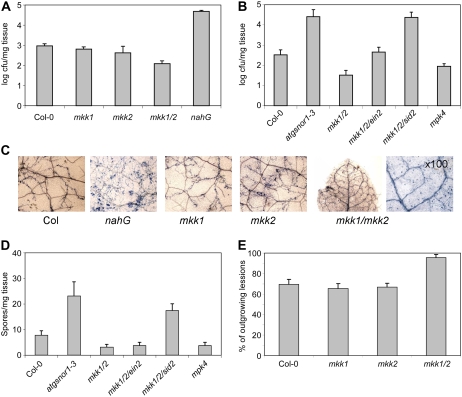
The response of both single and double mutants to biotic stress was investigated. mkk1 and mkk2 showed no difference to wild type with respect to bacterial growth 5 d after challenge with a virulent strain of Pst DC3000 (Fig. 4A). However, mkk1/2 and mpk4 supported significantly less bacterial growth, whereas the positive controls NahG (Ryals et al., 1995) and atgsnor1-3 (Feechan et al., 2005) showed higher levels of bacterial growth. The resistance of mkk1/2 to Pst DC3000 can be partly attributed to the enhanced ET production of the double mutant but more obviously by its SA content, because both the mkk1/2/ein2 (ET insensitive) and the mkk1/2/sid2 (reduced SA production) triple mutants carried a greater bacterial load than mkk1/2 alone (Fig. 4B).
Figure 4.
Disease resistance in mutants. A, Response to infection by P. syringae of wild-type Col-0, mkk1, mkk2, and mkk1/2 compared to nahG transgenic. B, Response to P. syringae of mkk1/2 compared to mkk1/2/ein2 and mkk1/2/sid2 triple mutants, atgsnor1-3, and mpk4. C and D, Growth of H. parasitica detected by trypan blue staining (C) and corresponding conidial counts/milligram tissue (D). E, Growth of B. cinerea in mutants assayed as the number of outgrowing lesions developed 3 d after inoculation. Experiments were repeated three times with similar results and error bars represent se.
The mutants were also challenged with the virulent oomycete pathogen Hyaloperonospora parasitica (Noco2). At 8 d postinoculation, extensive hyphal growth and oospore production was seen on the wild type and the single mutants. Increased fungal growth for NahG plants and atgsnor1-3 was observed, but very little fungal development on leaves of mkk1/2 could be detected (Fig. 4, C and D). Spore counts on the triple mutants mkk1/2/ein2 and mkk1/2/sid2 showed that the enhanced SA level of mkk1/2 is a major determinant of resistance.
Pst DC3000 is semi-biotrophic and H. parasitica is a biotrophic pathogen. Thus, the resistance of mkk1/2 to the nectrophic fungal pathogen Botrytis cinerea was also tested by quantifying the number of outgrowing lesions following infection. As shown in Figure 4E, the resistance to B. cinerea of mkk1 and mkk2 was not significantly different than wild type, while mkk1/2 was statistically significantly more susceptible than wild type.
MKK1 and MKK2 Function Upstream of MPK4
Because the mkk1/2 double mutant resembles mpk4 both morphologically and at the molecular level, MPK4 kinase activity was monitored in the single and double MKK mutants. After flagellin elicitor peptide flg22 treatment, MPK4 was strongly activated in the wild type, mkk1, and mkk2. However, the signal for MPK4 activity in the mkk1/2 double mutant was similar to that of the untreated control and to mpk4; western blotting showed that MPK4 protein levels remained constant after flg22 treatment and in the MKK mutants (Fig. 5A). After treatment with the SA analog BTH, no difference was seen in MPK4 kinase activity between the wild type and the single mkk1 mutant. In contrast, neither mkk2 nor the mkk1/2 double mutant showed induction in MPK4 activity, although MPK4 levels stayed constant (Fig. 5B). These results indicate that the requirement of MKK1 and MKK2 for the enhancement of MPK4 activity is dependent on the upstream stimulus. In at least one instance, both MKKs are required, but in another MKK2 will suffice.
Figure 5.
MKK1 and MKK2 function upstream of MPK4. A and B, MPK4 kinase activity in single and double mutants. MPK4 protein was immunoprecipitated out of total protein extracts from plants treated for 30 min with 10 μm flg22 (A) or 100 μm BTH (B) and used in an in vitro kinase assay with myelin basic protein as substrate. Duplicate gels were subjected to western blotting to monitor MPK4 protein levels. C, Phenotype of mkk1/2/mks1 triple mutant in comparison to mkk1/2 and wild type.
It has been reported that MPK3 and MPK6 activation by flg22 is also modified in an mkk1 mutant (Mészáros et al., 2006) and that MPK6 activity is reduced in the mkk2 mutant when cold stressed (Teige et al., 2004). Therefore, MPK3 and MPK6 activity was also monitored in the same samples used for flg22-induced MPK4 activity assays (Supplemental Fig S5). Both MPK3 and MPK6 kinase activities were reduced in mkk1, mkk2, and in mkk1/2 relative to that seen for flg22-treated wild-type plants. However, as no significant difference in MPK3 or MPK6 activity was seen between the single and double mutants, it is unlikely that either of these two kinases is involved in the differences in phenotype observed between the single mutants and mkk1/2.
Because the mpk4 mutant is partially rescued by knockdown of the MPK4 substrate MKS1 (Andreasson et al., 2005), an mks1 knockout mutant (transposon insertion line GT5.108403; J-L. Qiu, unpublished data) was crossed into the mkk1/2 double mutant. mkk1/2/mks1 triple mutants selected by genotyping exhibited partial alleviation of the dwarf phenotype but retained the early senescence phenotype (Fig. 5C). This partial suppression provides genetic evidence confirming that MPK4 and MKS1 act downstream of MKK1 and MKK2.
MEKK1, MKK1/2, and MPK4 Exhibit a Gradient of Phenotypic Severities
The mkk1/2 double mutant morphologically resembles not only mpk4 but also the mekk1 mutant (Fig. 6A) in that both mekk1 and mkk1/2 are severely dwarfed, show premature senescence of cotyledons and leaves, have stubby root hairs, and are practicably infertile when grown at 24°C, although mkk1/2 pollen was viable when used in crosses. Both mkk1/2 dwarfism and infertility were partially rescued by growth at temperatures >30°C (Supplemental Fig. S4A), as has been reported for mekk1 and mpk4 (Petersen et al., 2000; Andreasson et al., 2005; Ichimura et al., 2006; Nakagami et al., 2006; Su et al., 2007; Suarez-Rodriguez et al., 2007). However, the dwarf phenotype of mekk1 is more pronounced than that of mkk1/2 and, in contrast to mekk1, the root length and branching pattern of mkk1/2 appeared normal, as did that of mpk4 (Supplemental Fig. S4, B and C). Thus, our data appears to show that mutations in genes encoding proteins that are biochemically linked in signaling cascades show decreasing complexities of phenotype “down” through the pathway.
Figure 6.
MEKK1, MKK1/2, and MPK4 function in the same pathway. A, Morphological phenotypes of Landsberg erecta wild type, mekk1, mkk1/2, and mpk4. B, Venn diagram showing overlapping gene expression changes in mekk1, mkk1/2, and mpk4. Overlap is between the top 500 most significantly differentially expressed genes as measured by the ATH1 Affymetrix GeneChip. C, FARO genotype graph illustrating the intersections between gene expression signatures of a series of mutants and overexpressors. The thickness of the edges illustrates the size of the intersections between transcriptional signatures of the mutants (nodes). Highlighted in red are mkk1, mkk2, mkk1/2, mekk1, mpk4, and the MKS1 overexpressor. Only response signature intersections exceeding 250 genes are drawn. Mutants assayed multiple times by independent laboratories (i.e. det2, ein2, ga1, zat12 oex., and lfy12) are represented by a node for each assay.
This relationship was further assessed by comparative analysis of differentially regulated genes among mekk1, mkk1/2, and mpk4 (Fig. 6B; Supplemental Table S3). This revealed that >10% of genes overlap for all three mutants, while 20% to 30% of genes are shared by any two of the mutants. Significantly, the same PR1, PR3 chitinase, and PR5 genes with shared differential expression in mkk1/2 and BTH treatment (Fig. 2E) were among the 54 genes differentially expressed in the three mutants. In addition, three enzymes implicated in synthesis of the phytoalexin camalexin (PAD3, CYP71A13, and CYP79B2; Nafisi et al., 2007), and three genes involved in the synthesis or maintenance of SA (ICS1, EDS1, and PAD4) are shared by mekk1 and mpk4 and are also up-regulated in mkk1/2, although they are not among the 500 most significantly differentially regulated genes in mkk1/2. Gene ontology analysis of the overlapping genes in mkk1/2, mekk1, and mpk4 revealed SAR to be the most significantly overrepresented term among many other terms implicated in defense response (Supplemental Table S4). Thus, these data confirm the strong phenotypic harmony of the mutants and indicate that they are all involved in signaling defense responses despite the distinct transcriptome signatures of each mutant.
To further assess the relationships between mkk1, mkk2, and mkk1/2 relative to other mutants, their global transcriptional profiles were compared to those of other mutants, as described previously (Nielsen et al., 2007). To this end, the gene expression signature of each mutant, consisting of their 1,200 most significantly differentially expressed genes versus wild type, was compared to a compendium of 44 public, mutant transcriptome profiles (Nielsen et al., 2007). Figure 6C illustrates these comparisons as a graphic, wherein the thickness of the lines (edges) denotes the relative strengths of the intersections between transcriptional signatures of the mutants. The mkk1 and mkk2 single mutants both have moderate transcriptional phenotypes and do not associate strongly to other mutants. In contrast, the transcriptional phenotype of the mkk1/2 double mutant deviates strongly from wild type and Figure 6C shows that their signatures intersect strongly with those of the mekk1, cpr5, and mpk4 mutants containing 468, 419, and 365 genes, respectively. With respect to the MPK4 substrate MKS1, the intersection is weaker for both mkk1/2 (225 genes) and mekk1 (274 genes) than for mpk4 (454 genes). This is consistent with the function of MKS1 as a direct MPK4 substrate.
DISCUSSION
The Arabidopsis MPK4 has been reported to function as a negative regulator of SA-dependent systemic required resistance (Petersen et al., 2000), as a positive regulator of ET- and JA-mediated defenses (Brodersen et al., 2006), and has also been implicated in responses to abiotic stress (Teige et al., 2004). Biochemical and yeast two-hybrid evidence suggests that MPK4 is part of signaling cascades, including MKK1 and MKK2 and MEKK1 (Ichimura et al., 1998; Ichimura et al., 2006). Here, we present genetic, molecular, and biochemical evidence confirming this model. In addition, the results presented show that each kinase has specific functions in regulating other defense-related, morphological, and developmental processes.
A key genetic observation is that while the mkk1 and mkk2 single mutants appear morphologically normal, the mkk1/2 double mutant is similar to the mpk4 mutant, as both: (1) are dark green dwarves with curled leaves; (2) constitutively overproduce SA and exhibit enhanced resistance to biotrophic pathogens and insensitivity to JA; and (3) have enhanced basal expression of PR1 and other SA-dependent defense genes. Importantly, the mkk1/2 phenotype is partially complemented by loss-of-function of the MPK4 substrate MKS1, indicating that MKS lies downstream of MPK4, MKK1, and MKK2. In addition, the mkk1/2 double mutant also resembles the mekk1 mutant (Ichimura et al., 2006; Nakagami et al., 2006; Suarez-Rodriguez et al., 2007), as both are more severely dwarfed than mpk4, both are infertile and have curled, prematurely senescing leaves and stubby root hairs, and both can be, in part, rescued by high temperature. However, mekk1 is even more severely dwarfed than is mkk1/2, has a greater incidence of short root hairs, and exhibits much reduced root branching, not seen in mkk1/2 or mpk4 (Nakagami et al., 2006; Su et al., 2007). Thus, the order of severity of morphological phenotypes is mekk1 > mkk1/2 > mpk4, which supports their proposed functional order in a signaling cascade, consistent with a hypothesis that other pathways branch off at each level of signaling, although other mechanisms might also explain these phenotypic differences.
The morphological similarities between mekk1, mkk1/2, and mpk4 mutants are mirrored in their transcriptome profiles. While the single mkk1 and mkk2 mutants have modest transcriptional phenotypes relative to wild-type plants, mkk1/2 exhibits a gene expression signature more similar to both mekk1 and mpk4. This close relationship is confirmed by comparison of the transcript profiles with those of more than 40 other mutants and transgenics. In this analysis, mekk1, mkk1/2, mpk4, and transgenics overexpressing MKS1 associate closely. Interestingly, although both are wild type in appearance, the transcriptome profiles of the single mkk1 and mkk2 mutants differ significantly, as they share <30% of genes differentially expressed versus wild type. This provides molecular evidence that MKK1 and MKK2 have both overlapping and specific functions.
These genetic and transcriptional observations concur with biochemical and physiological data presented here and previously. Although MPK4 activity in untreated wild-type Col-0 is barely detectable by in vitro kinase assays, the mpk4 mutant shows a pronounced phenotype. This phenotype cannot be complemented by expressing an inactive (kinase-dead) MPK4 transgene in mpk4, indicating that a basal level of MPK4 activity is necessary to maintain the wild-type phenotype (Petersen et al., 2000). Because the mkk1 and mkk2 mutants appear morphologically wild type, this indicates that in these single mutants, basal level MPK4 activity is not impaired and that in the unchallenged condition MKK1 and MKK2 are at least in part functionally redundant. This is supported by the finding that for mkk1/2, SA levels are high, and there is enhanced resistance to the biotrophs Pst DC3000 and H. parasitica. Consistent with MPK4's role as a positive regulator of ET- and JA-mediated defenses, mkk1/2 is less resistant to the necrotrophic pathogen B. cinerea. Because both the mkk1 single mutant and mkk1/2 were found to be less sensitive to JA, but mkk1 was not significantly more susceptible to B. cinerea than the wild type, this suggests that resistance to B. cinerea is mediated by factors other than just by JA.
As well as maintaining ground-state MPK4 activity, the data presented here show that MKK1 and MKK2 are both involved in the enhancement of MPK4 activity by elicitor treatment. Treatment with the flg22 elicitor results in strong MPK4 activation in wild type, mkk1, and mkk2, but not in mkk1/2. In this case, both MKK1 and MKK2 appear to be required to activate MPK4. In contrast, BTH treatment resulted in MPK4 activation in wild type and in mkk1, but not in mkk2. This suggests that different stimuli can induce MPK4 activation by different degrees of upstream activation of MKK1 and/or MKK2. This hypothesis might explain why a small reduction in MPK4 activity at certain time points in flagellin-treated mkk1 mutants was noted by Mészáros et al. (2006), because only 1 μm flg22 was used in their experiments, whereas 10 μm flg22 was used here. As treatment with the flg22 elicitor does not activate MPK4 in mekk1 (Suarez-Rodriguez et al., 2007), this is consistent with MEKK1 functioning upstream of both MKK1 and MKK2.
As noted above, phenotypic complexity appears reduced in mutants at each level of the MPK4 signaling pathway, which suggests that other signaling pathways branch off at these points. This is also indicated by analysis of triple mutants, as a knockout of the gene encoding the MPK4 substrate MKS1 only rescues the dwarf phenotype, not the early senescence phenotype. Thus, MPK4 affects only one subset of a pathway downstream of MEKK1, MKK1, and MKK2. Similarly, the sid2 mutant only partially complements mkk1/2, as the triple mkk1/2/sid2 mutant is still relatively small and exhibits early senescence. This indicates that MKK1 and MKK2 affect signaling pathways independently of MPK4 and SA and is consistent with our findings that BTH can still induce PR1 expression in the mkk1/2 and mpk4 mutants, indicating that MKK1, MKK2, and MPK4 are not the sole players regulating SA-mediated plant defense (Fig. 3A). Evidence for distinct functions for MKK1 and MKK2, some of which may be associated with MPK4 activation but others of which might be linked to activation of alternative MPKs, is also presented here and elsewhere (Teige et al., 2004; Mészáros et al., 2006; Brader et al., 2007; Xing et al., 2007). For example, the mkk1 mutant exhibited JA insensitivity, a trait shared by mpk4 but not by mkk2. In contrast to mkk1, the mkk2 mutant showed salt and ABA insensitivity, traits not shared with mpk4, which may be associated with the activation of MPK6 by MKK2.
There are interesting parallels, but also some differences, in the regulation of defense signaling in other species, such as tobacco (Nicotiana tabacum). Biotic and abiotic stress in this tobacco will activate SIPK and WIPK, orthologs of Arabidopsis MPK3 and MPK6, respectively, resulting in JA accumulation and hypersensitive responses (Yang et al., 2001). Plants in which SIPK or WIPK are silenced show reduced wound-induced JA production, and SA levels after wounding are enhanced (Seo et al., 2007). This is in contrast to the situation in Arabidopsis in which JA levels are not affected in the mutants studied here, whereas SA levels are enhanced in mkk1/2 in the unchallenged state. The tobacco ortholog of MPK4 is NtMPK4, which is also activated by wounding and is required for expression of JA-responsive genes (Gomi et al., 2005). As in Arabidopsis, stress-related MPK activation in tobacco involves multiple upstream regulators; SIPKK will activate NtMPK4 and SIPK, but not WIPK (Gomi et al., 2005), whereas MEK2 will activate both SIPK and WIPK (Yang et al., 2001).
In summary, several lines of evidence provided here strongly support a pathway including MEKK1, MKK1, MKK2, and MPK4 in defense against pathogens. Some 60 MPKKKs, but only 10 MKKs and 20 MPKs, are predicted to be encoded in the Arabidopsis genome (Ichimura et al., 2002). While it therefore seems likely that multiple MPKs can be activated by one MKK, it is more surprising to find that a single MPK is regulated by more than one MKK. This might be explained if different combinations of MKKs maintain the specificity of different stimuli to certain responses. While such signaling specificity may be effected in individual cells, it is also likely that different cell type and temporal expression or activation patterns of the three tiers of kinases contribute to the integration of signaling specificities within plants.
MATERIALS AND METHODS
Plant Materials and Treatments
Plants were grown in a controlled-environment room at 22°C constant temperature and a 16-h-light, 8-h-dark photoperiod (200 mE m−2 s−1). For treatment prior to RT-PCR or immunoprecipitation, plants were sprayed with BTH (Bion 50WG, 50% active ingredient) or methyl jasmonate (Sigma) as 100 μm solutions in water. Pst DC3000 inoculations were conducted by spraying 4-week-old plants with virulent bacteria at 6 × 108 cfu/mL and assaying for bacterial growth after 5 d. Hyaloperonospora parasitica (Noco2) inoculations were conducted spraying with conidia at 4 × 104/mL and condidial production was assayed after 10 d. Botrytis cinerea infections were performed as described by Bessire et al. (2007). Treatments with plant growth substances during germination and wounding of leaves were as described previously (Alzwiy and Morris, 2007).
T-DNA Insertional Mutants, Crosses, and Complementation
T-DNA insertion mutants were for mkk1 (At4g26070; Salk line 027645; Alonso et al., 2003) and mkk2 (At4g29810; SAIL line 511_H 01; Sessions et al., 2002). Mutants were back-crossed to Col-0 and homozygous mutants isolated from the F2 progeny by means of PCR analysis using gene-specific and T-DNA-specific primers. These were as follows: MKK1-126, 5′-GGAAGCTTATGCCCTAATCCC-3′; MKK1-1022, 5′-CACCATTGCGAGATGAAGGAG-3′; PROK-LB, 5′-GCGTGGACCGCTTGCTGCAACT-3′; MKK2-2, 5′-GCGAAGAACGAAGTAGACGAA-3′; MKK2-1889, 5′-GAAGTAGGACGCGAGATTGAT-3′; and Garlic-LB, 5′-TAGCATCTGAATTTCATAACCAATCTCGATACAC-3′.
MKK1 and MKK2 are closely linked; to isolate the double mutant, F2 plants were screened on kanamycin to select the MKK1-specific T-DNA and 200 resistant seedlings treated with Basta to select for the MKK2-specific T-DNA. Two doubly resistant plants were found and the mkk1/2 genotype confirmed by PCR analysis. These plants were heterozygous for mkk1 and segregated out homozygous mkk1/2 plants in the F3 in an approximate 1:3 ratio.
Triple mutants were produced by using wild-type-looking plants heterozygous for mkk1 and homozygous for mkk2. The ET-insensitive mkk1/2/ein2 and mkk1/2/etr1-1 mutants were selected by germination of the F2 on ACC-containing media, selection of ET-insensitive seedlings, and PCR analysis for mkk1/2. The mkk1/2/sid2 mutants were selected by PCR analysis using the following SID2-specific primers: SID2-F, 5′-ACATCTATATTCTCAATTGGC-3′; and SID2-R, 5′-CCAATAGGTCCCGCATACAT-3′. The mkk1/2/mks1 triple mutant was also selected for by PCR analysis using the following mks1-specific primers: MKS1gt-For, 5′-GGTACATTGATAGGAGGAACTTGGAAC-3′; MKS1gt-Rev, 5′-TCCTTCCGATCAACAGAACCAGAAG-3′; and DS1, 5′-GTTTTCGTTTCCGTCCCGCAAG-3′.
The mkk1/2 mutant was complemented by transforming plants heterozygous for mkk1 and homozygous for mkk2 with constructs based on pGPTV-HPT (Becker et al., 1992) and bearing CaMV 35S promoter-driven cDNAs for MKK1 or MKK2 and selecting for transformants with hygromycin. Transformed plants were analyzed by PCR.
SA, ET, and JA Measurements
ET was measured by gas chromatography as described by Locke et al. (2000). SA and JA were measured by gas chromatography-mass spectrometry as described by Engelberth et al. (2003) and Lee et al. (2004), using propyl paraben and dihydrojasmonic acid as internal standards.
RNA Isolation, RT-PCR, and Real-Time RT-PCR
Total RNA was extracted by Tri-Reagent-based RiboPure kit (Ambion). RT-PCR was performed as before (Qiu et al., 2002) using the following primers: MKK1-F, 5′-AGCGAGTTCTTCGAGGTCTTTG-3′; MKK1-R, 5′-AGCAAGTGGGGGAATCAAAGATC-3′; MKK2-F, 5′-CATCCCTGACTCCTATCTTTCTG-3′; MKK2-R, 5′-CGATCCTGCATCTGTGAAGTAG-3′; ACT7-F, 5′-GTGAGGATATTCAGCCACTTGTCTG-3′; and ACT7-R, 5′-TGTGAGATCCCGACCCGCAAGATC-3′.
For quantitative PCR analysis, RNA samples were first treated with RQ1 DNase (Promega). Quantitative RT-PCR was performed using the Superscript III Platinum SYBR Green One-Step qRTPCR kit (Invitrogen) with 10 pmol of each primer and approximately 100 ng of total RNA in a 20-μL reaction. Reactions were run on icycler IQ (Bio-Rad). Quantitative PCR reactions were performed in triplicate for each individual line and quantification of the threshold cycle values obtained by quantitative PCR analysis was achieved by calculating means of normalized expressions using the Q-gene software (Muller et al., 2002). Primers for RT-PCR were as follows: ACT2-F, 5′-GGTAACATTGTGCTCAGTGGTGG-3′; ACT2-R, 5′-AACGACCTTAATCTTCATGCTGC-3′; PR1-F, 5′-GTGGGTTAGCGAGAAGGCTA-3′; PR1-R, 5′ACTTTGGCACATCCGAGTCT-3′; PDF1.2-F, 5′-TGGTGGAAGCACAGAAGTTG-3′; and PDF1.2-R, 5′-GATCCATGTTTGGCTCCTTC-3′.
Northern blotting with digoxigenin-labeled VSP probes was carried out as described by Alzwiy and Morris (2007).
Immunoprecipitation and Western Blotting
Two milligrams total protein extract in immunoprecipitation buffer (40 mm Tris/HCl, pH7.5, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 10 mm β-glycerophosphate, 0.5% Triton, 0.5% NP-40, 1 mm dithiothreitol, phosphatase inhibitor cocktail, and protease inhibitor cocktail from Roche) was incubated with 3 μg of antibodies for 3 h at 4°C followed by precipitation with protein A-Sepharose CL-4B or protein G-agarose beads (GE-Healthcare) for 2 h. After washing the beads four times with immunoprecipitation buffer, the proteins were eluted by boiling in 40 μL of Laemmli loading buffer for 10 min. Samples were subsequently processed by western blotting using the One-Step IP-western kit from Genscript following the protocol described by the manufacturer.
In Vitro Kinase Assay
Arabidopsis (Arabidopsis thaliana) plants were treated with 10 μm flg22 and 100 μm BTH for 30 min. Total proteins were extracted and immunoprecipitated with anti-MPK3, anti-MPK4, and anti-MPK6 (Sigma) as described above, and kinase assays were performed as previously described (Caspersen et al., 2007) by combining immunoprecipitations with 5 μg of myelin basic protein, 3 μCi [32P]ATP, and kinase assay buffer (50 mm ATP, 80 mm Tris/HCl, pH 7.5, 4 mm EGTA, 120 mm MgCl2, 4 mm Na3VO4, 4 mm dithiothreitol), incubated with shaking for 30 min at 30°C. The reactions were stopped by adding Laemmli loading buffer and heating at 100°C for 10 min. The resultant supernatants were resolved on 12% SDS-PAGE gels, and dried gels were exposed using phosphoimaging screens.
Microarray and Data Analysis
Total RNA was extracted from three independent batches of plants using Trizol Reagent (Ambion). From each batch of wild type (Col-0) and mutants mkk1, mkk2, and mkk1/2, whole plants were harvested before and 24 h after spraying with 100 μm BTH. Total RNA from the resulting 12 samples were amplified and labeled using the MessageAmp II-Biotin Enhanced kit (Ambion). A total of 15 μg of RNA for each sample was fragmented, labeled, and hybridized to Affymetrix ATH1 GeneChips according to the standard Affymetrix protocol. The microarray data was preprocessed by RMA (Irizarry et al., 2003a, 2003b) and annotated according to the Arabidopsis genome TAIR7 version (released April 11, 2007). Venn diagrams were generated with the top 500 genes sorted by P value (mkk1, mkk2, mkk1/2, mpk4, and BTH treatment) or fold change (mekk1). Gene expression signatures consisting of the top most significant genes as assessed by t-statistics versus wild type (Col-0) plants or extracted from the functional association(s) by response overlap (FARO)-compendium generated by Nielsen et al. (2007) were generated for the Venn diagrams (top 500 genes) in Figures 1D, 2E, and 6B, and the FARO graph (top 1,209) in Figure 6C. The FARO genotype graph (Fig. 6C) includes gene expression signatures from mkk1, mkk2, and mkk1/2 and a subset the FARO-compendium, consisting of all mutants and overexpressors (n = 47, of which five exist as duplicates assayed by two independent laboratories). The graph was drawn using Cytoscape 2.5.1 (Shannon et al., 2003). Only gene expression signature intersections exceeding 250 genes are drawn. The thickness of the edges indicates the size (in no. of common genes) of the intersection. The nodes have been manually placed in respect to the intersection sizes. All the microarray data are deposited at the National Center for Biotechnology Information GEO with the accession number GSE10646 (www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GSE10646). Gene Ontology analysis was done using the BINGO plug-in (Maere et al., 2005) for Cytoscape (Shannon et al., 2003).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The mkk2 mutant shows enhanced salt and ABA sensitivity.
Supplemental Figure S2. JA content of wild type, mkk1, mkk2, and mkk1/2 in control, local, and systemic leaves 2 h after wounding.
Supplemental Figure S3. Northern blot of VSP1 gene expression in wild type, mkk1, mkk2, and mkk1/2 in control, local, and systemic leaves 2 h after wounding.
Supplemental Figure S4. Morphological phenotypes of mutants.
Supplemental Figure S5. MPK3 and MPK6 kinase activity in wild-type, mkk1, mkk2, and mkk1/2 backgrounds.
Supplemental Tables S1 to S4. Microarray data from Figures 1, 2, and 6.
Supplementary Material
Acknowledgments
We thank Dr. Gordon Simpson (SCRI, Dundee) for flowering time analysis, and the Nottingham Arabidopsis Stock Centre, Syngenta, and the Salk Institute for providing mutants.
This work was supported by a Heriot-Watt Ph.D. fellowship to L.Z., by the Danish Research Councils (grant nos. 23–03–0076, 272–06–0049, and 272–05–0367) and by the European Union (grant no. LSHG–CT–2004–511983) to J.M., and by the Biotechnology and Biological Sciences Research Council (grant no. BBD011809/1) to G.J.L.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Peter C. Morris (p.c.morris@hw.ac.uk).
The online version of this article contains Web-only data.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Alzwiy IA, Morris PC (2007) A mutation in the Arabidopsis MAP kinase kinase 9 gene results in enhanced seedling stress tolerance. Plant Sci 173 302–308 [Google Scholar]
- Andreasson E, Jenkins T, Brodersen P, Thorgrimsen S, Petersen NH, Zhu S, Qiu JL, Micheelsen P, Rocher A, Petersen M, et al (2005) The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J 24 2579–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415 977–983 [DOI] [PubMed] [Google Scholar]
- Becker D, Kemper E, Schell J, Masterson R (1992) New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol 20 1195–1197 [DOI] [PubMed] [Google Scholar]
- Bessire M, Chassot C, Jacquat AC, Humphry M, Borel S, Petetot JM, Metraux JP, Nawrath C (2007) A permeable cuticle in Arabidopsis leads to a strong resistance to Botrytis cinerea. EMBO J 26 2158–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brader G, Djamei A, Teige M, Palva ET, Hirt H (2007) The MAP kinase kinase MKK2 affects disease resistance in Arabidopsis. Mol Plant Microbe Interact 20 589–596 [DOI] [PubMed] [Google Scholar]
- Brodersen P, Petersen M, Bjorn Nielsen H, Zhu S, Newman MA, Shokat KM, Rietz S, Parker J, Mundy J (2006) Arabidopsis MAP kinase 4 regulates salicylic acid- and jasmonic acid/ethylene-dependent responses via EDS1 and PAD4. Plant J 47 532–546 [DOI] [PubMed] [Google Scholar]
- Caspersen MB, Qiu JL, Zhang X, Andreasson E, Naested H, Mundy J, Svensson B (2007) Phosphorylation sites of Arabidopsis MAP kinase substrate 1 (MKS1). Biochim Biophys Acta 1774 1156–1163 [DOI] [PubMed] [Google Scholar]
- Desikan R, Hancock JT, Ichimura K, Shinozaki K, Neill SJ (2001) Harpin induces activation of the Arabidopsis mitogen-activated protein kinases AtMPK4 and AtMPK6. Plant Physiol 126 1579–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelberth J, Schmelz EA, Alborn HT, Cardoza YJ, Huang J, Tumlinson JH (2003) Simultaneous quantification of jasmonic acid and salicylic acid in plants by vapor-phase extraction and gas chromatography-chemical ionization-mass spectrometry. Anal Biochem 312 242–250 [DOI] [PubMed] [Google Scholar]
- Feechan A, Kwon E, Yun BW, Wang Y, Pallas JA, Loake GJ (2005) A central role for S-nitrosothiols in plant disease resistance. Proc Natl Acad Sci USA 102 8054–8059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomi K, Ogawa D, Katou S, Kamada H, Nakajima N, Saji H, Soyano T, Sasabe M, Machida Y, Mitsuhara I, et al (2005) A mitogen-activated protein kinase NtMPK4 activated by SIPKK is required for jasmonic acid signaling and involved in ozone tolerance via stomatal movement in tobacco. Plant Cell Physiol 46 1902–1914 [DOI] [PubMed] [Google Scholar]
- Guzman P, Ecker JR (1990) Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadiarto T, Nanmori T, Matsuoka D, Iwasaki T, Sato K, Fukami Y, Azuma T, Yasuda T (2006) Activation of Arabidopsis MAPK kinase kinase (AtMEKK1) and induction of AtMEKK1-AtMEK1 pathway by wounding. Planta 223 708–713 [DOI] [PubMed] [Google Scholar]
- Huang Y, Li H, Gupta R, Morris PC, Luan S, Kieber JJ (2000) ATMPK4, an Arabidopsis homolog of mitogen-activated protein kinase, is activated in vitro by AtMEK1 through threonine phosphorylation. Plant Physiol 122 1301–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura K, Casais C, Peck SC, Shinozaki K, Shirasu K (2006) MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. J Biol Chem 281 36969–36976 [DOI] [PubMed] [Google Scholar]
- Ichimura K, Mizoguchi T, Irie K, Morris P, Giraudat J, Matsumoto K, Shinozaki K (1998) Isolation of ATMEKK1 (a MAP kinase kinase kinase)-interacting proteins and analysis of a MAP kinase cascade in Arabidopsis. Biochem Biophys Res Commun 253 532–543 [DOI] [PubMed] [Google Scholar]
- Ichimura K, Mizoguchi T, Yoshida R, Yuasa T, Shinozaki K (2000) Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J 24 655–665 [DOI] [PubMed] [Google Scholar]
- Ichimura K, Shinozaki K, Tena G, Sheen J, Henry Y, Champion A, Kreis M, Zhang SQ, Hirt H, Wilson C, et al (2002) Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci 7 301–308 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP (2003. a) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003. b) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4 249–264 [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72 427–441 [DOI] [PubMed] [Google Scholar]
- Lee A, Cho K, Jang S, Rakwal R, Iwahashi H, Agrawal GK, Shim J, Han O (2004) Inverse correlation between jasmonic acid and salicylic acid during early wound response in rice. Biochem Biophys Res Commun 318 734–738 [DOI] [PubMed] [Google Scholar]
- Loake G, Grant M (2007) Salicylic acid in plant defence: the players and protagonists. Curr Opin Plant Biol 10 466–472 [DOI] [PubMed] [Google Scholar]
- Locke JM, Bryce JH, Morris PC (2000) Contrasting effects of ethylene perception and biosynthesis inhibitors on germination and seedling growth of barley (Hordeum vulgare L.). J Exp Bot 51 1843–1849 [DOI] [PubMed] [Google Scholar]
- Madhani HD, Fink GR (1998) The riddle of MAP kinase signaling specificity. Trends Genet 14 151–155 [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21 3448–3449 [DOI] [PubMed] [Google Scholar]
- Manners JM, Penninckx IA, Vermaere K, Kazan K, Brown RL, Morgan A, Maclean DJ, Curtis MD, Cammue BP, Broekaert WF (1998) The promoter of the plant defensin gene PDF1.2 from Arabidopsis is systemically activated by fungal pathogens and responds to methyl jasmonate but not to salicylic acid. Plant Mol Biol 38 1071–1080 [DOI] [PubMed] [Google Scholar]
- Matsuoka D, Nanmori T, Sato K, Fukami Y, Kikkawa U, Yasuda T (2002) Activation of AtMEK1, an Arabidopsis mitogen-activated protein kinase kinase, in vitro and in vivo: analysis of active mutants expressed in E. coli and generation of the active form in stress response in seedlings. Plant J 29 637–647 [DOI] [PubMed] [Google Scholar]
- Mészáros T, Helfer A, Hatzimasoura E, Magyar Z, Serazetdinova L, Rios G, Bardóczy V, Teige M, Koncz C, Peck S, et al (2006) The Arabidopsis MAP kinase kinase MKK1 participates in defence responses to the bacterial elicitor flagellin. Plant J 48 485–498 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Ichimura K, Irie K, Morris P, Giraudat J, Matsumoto K, Shinozaki K (1998) Identification of a possible MAP kinase cascade in Arabidopsis thaliana based on pairwise yeast two-hybrid analysis and functional complementation tests of yeast mutants. FEBS Lett 437 56–60 [DOI] [PubMed] [Google Scholar]
- Morris PC, Guerrier D, Leung J, Giraudat J (1997) Cloning and characterisation of MEK1, an Arabidopsis gene encoding a homologue of MAP kinase kinase. Plant Mol Biol 35 1057–1064 [DOI] [PubMed] [Google Scholar]
- Muller PY, Janovjak H, Miserez AR, Dobbie Z (2002) Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques 32 1372–1379 [PubMed] [Google Scholar]
- Nafisi M, Goregaoker S, Botanga CJ, Glawischnig E, Olsen CE, Halkier BA, Glazebrook J (2007) Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 19 2039–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H, Soukupova H, Schikora A, Zarsky V, Hirt H (2006) A Mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. J Biol Chem 281 38697–38704 [DOI] [PubMed] [Google Scholar]
- Nielsen HB, Mundy J, Willenbrock H (2007) Functional Associations by Response Overlap (FARO), a functional genomics approach matching gene expression phenotypes. PLoS ONE 2 e676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE, et al (2000) Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103 1111–1120 [DOI] [PubMed] [Google Scholar]
- Qiu JL, Jilk R, Marks MD, Szymanski DB (2002) The Arabidopsis SPIKE1 gene is required for normal cell shape control and tissue development. Plant Cell 14 101–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals J, Lawton KA, Delaney TP, Friedrich L, Kessmann H, Neuenschwander U, Uknes S, Vernooij B, Weymann K (1995) Signal transduction in systemic acquired resistance. Proc Natl Acad Sci USA 92 4202–4205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Katou S, Seto H, Gomi K, Ohashi Y (2007) The mitogen-activated protein kinases WIPK and SIPK regulate the levels of jasmonic and salicylic acids in wounded tobacco plants. Plant J 49 899–909 [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su SH, Suarez-Rodriguez MC, Krysan P (2007) Genetic interaction and phenotypic analysis of the Arabidopsis MAP kinase pathway mutations mekk1 and mpk4 suggests signaling pathway complexity. FEBS Lett 581 3171–3177 [DOI] [PubMed] [Google Scholar]
- Suarez-Rodriguez MC, Adams-Phillips L, Liu Y, Wang H, Su SH, Jester PJ, Zhang S, Bent AF, Krysan PJ (2007) MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol 143 661–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teige M, Scheikl E, Eulgem T, Doczi R, Ichimura K, Shinozaki K, Dangl JL, Hirt H (2004) The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol Cell 15 141–152 [DOI] [PubMed] [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J (1992) Acquired resistance in Arabidopsis. Plant Cell 4 645–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414 562–565 [DOI] [PubMed] [Google Scholar]
- Xing Y, Jia W, Zhang J (2007) AtMEK1 mediates stress-induced gene expression of CAT1 catalase by triggering H2O2 production in Arabidopsis. J Exp Bot 58 2969–2981 [DOI] [PubMed] [Google Scholar]
- Yang KY, Liu Y, Zhang S (2001) Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc Natl Acad Sci USA 98 741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Dai Y, Xiong Y, Defraia C, Li J, Dong X, Mou Z (2007) Overexpression of Arabidopsis MAP kinase kinase 7 leads to activation of plant basal and systemic acquired resistance. Plant J 52 1066–1079 [DOI] [PubMed] [Google Scholar]
- Zheng Z, Mosher SL, Fan B, Klessig DF, Chen Z (2007) Functional analysis of Arabidopsis WRKY25 transcription factor in plant defense against Pseudomonas syringae. BMC Plant Biol 7 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Qamar SA, Chen Z, Mengiste T (2006) Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J 48 592–605 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.