Abstract
Gibberellins (GAs) regulate many aspects of plant development, such as germination, growth, and flowering. The barley (Hordeum vulgare) Amy32b α-amylase promoter contains at least five cis-acting elements that govern its GA-induced expression. Our previous studies indicate that a barley WRKY gene, HvWRKY38, and its rice (Oryza sativa) ortholog, OsWRKY71, block GA-induced expression of Amy32b-GUS. In this work, we investigated the functional and physical interactions of HvWRKY38 with another repressor and two activators in barley. HvWRKY38 blocks the inductive activities of SAD (a DOF protein) and HvGAMYB (a R2R3 MYB protein) when either of these proteins is present individually. However, SAD and HvGAMYB together overcome the inhibitory effect of HvWRKY38. Yet, the combination of HvWRKY38 and BPBF (another DOF protein) almost diminishes the synergistic effect of SAD and HvGAMYB transcriptional activators. Electrophoretic mobility shift assays indicate that HvWRKY38 blocks the GA-induced expression of Amy32b by interfering with the binding of HvGAMYB to the cis-acting elements in the α-amylase promoter. The physical interaction of HvWRKY38 and BPBF repressors is demonstrated via bimolecular fluorescence complementation assays. These data suggest that the expression of Amy32b is modulated by protein complexes that contain either activators (e.g. HvGAMYB and SAD) or repressors (e.g. HvWRKY38 and BPBF). The relative amounts of the repressor or activator complexes binding to the Amy32b promoter regulate its expression level in barley aleurone cells.
GAs control many plant developmental processes, such as germination, growth, and flowering (Olszewski et al., 2002; Sun and Gubler, 2004). During germination of cereal grains, GA is secreted from embryos into aleurone cells to promote the expression of hydrolytic enzymes, such as α-amylases, which degrade stored starches within the endosperm for seed germination and postgermination growth (Ritchie and Gilroy, 1998; Lovegrove and Hooley, 2000).
The GA signal is perceived by receptors, such as GID1 (for GIBBERELLIN INSENSITIVE DWARF1) in rice (Oryza sativa) and AtGID1a, AtGID1b, and AtGID1c in Arabidopsis (Arabidopsis thaliana; Ueguchi-Tanaka et al., 2005; Nakajima et al., 2006). The activated GID1 binds to the negative regulator, such as the DELLA protein RGA, triggering the degradation of this DELLA protein by the 26S proteasomes (Itoh et al., 2003; Sun and Gubler, 2004; Griffiths et al., 2006; Nakajima et al., 2006). Recent studies revealed that DELLA proteins modulate GA signaling by establishing GA homeostasis via feedback regulation of GA biosynthetic gene and GA receptors and by promoting the expression of downstream negative regulatory proteins in GA signaling (Zentella et al., 2007). Studies of constitutively activated GA signaling mutants reveal that Arabidopsis SPY and its barley (Hordeum vulgare) ortholog HvSPY encode a Ser/Thr O-linked GlcNAc transferase, which is a repressor of GA signaling (Jacobsen et al., 1996; Robertson et al., 1998). SPY increases the activity of DELLA proteins such as Arabidopsis RGA and rice SLR1, probably by attaching a GlcNAc moiety to the Ser/Thr residues of a targeted protein (Robertson, 2004; Shimada et al., 2006; Silverstone et al., 2007). SPY also physically interacts with two transcriptional repressors of α-amylase expression in aleurone cells (Robertson, 2004). Studies of unresponsive GA signaling mutants have identified positive regulators in GA signaling such as Arabidopsis SLY1 and PICKLE (Ogas et al., 1997; Steber et al., 1998). SLY1 encodes an F-box protein, a component of the SCF (SLY1) E3 UBIQUITIN (UBI) ligase that targets the DELLA protein for degradation (McGinnis et al., 2003; Griffiths et al., 2006). The PICKLE gene encodes a CHD3 chromatin-remodeling factor, which negatively regulates embryo-specific gene transcription (Henderson et al., 2004). It is also known that the activation of G-proteins (Hooley, 1998) and the enhancement of cytoplasmic cGMP (Penson et al., 1996) and Ca2+ concentrations (Gilroy and Jones, 1992) follow GA treatment, although it is not entirely clear how they are linked to upstream and downstream events.
Several types of cis-acting elements for the GA responses of high-pI and low-pI α-amylase genes have been defined (Skriver et al., 1991; Gubler and Jacobsen, 1992; Lanahan et al., 1992; Rogers and Rogers, 1992; Rogers et al., 1994; Tanida et al., 1994). These motifs interact with various transcription factors controlling seed germination. In the low-pI α-amylase promoter, Amy32b, five elements, namely, O2S/W-box, pyrimidine box, GA response element (GARE), amylase box (Amy), and downstream amylase element, are essential for the high level of GA-induced expression (Lanahan et al., 1992; Rogers and Rogers, 1992; Rogers et al., 1994; Gómez-Cadenas et al., 2001). Each of these elements may be bound by one or more transcription factor(s) of R2R3 MYB, R1 MYB, DOF, and zinc finger protein families (Gubler et al., 1995; Raventós et al., 1998; Diaz et al., 2002; Isabel-LaMoneda et al., 2003; Washio, 2003; Mare et al., 2004; Peng et al., 2004; Zhang et al., 2004; Rubio-Somoza et al., 2006; Xie et al., 2006; Moreno-Risueno et al., 2007). However, it still remains unclear how these repressors and activators interact with each other in regulating gene expression.
WRKY proteins mediate plant responses to biotic and abiotic stresses as well as anthocyanin biosynthesis, senescence, and trichome development (Eulgem et al., 2000; Ulker and Somssich, 2004). Our previous studies indicate that a rice WRKY gene, OsWRKY71, repressed GA-induced expression of the barley Amy32b promoter in barley aleurone cells (Zhang et al., 2004). We further showed that OsWRKY71, by forming a complex with OsWRKY51, functionally interferes with OsGAMYB, a transcriptional activator of GA signaling, hence blocking the GA-induced expression of Amy32b-GUS in barley aleurone cells (Xie et al., 2006). We studied those rice WRKY genes in barley because of the technical difficulties of using rice aleurone cells, such as the high background of the GUS reporter driven by the rice GA-inducible promoters and, hence, the low level of induction (Zhang et al., 2004). Later, we showed, unsurprisingly, that the putative barley ortholog of OsWRKY71, HvWRKY38 (Mare et al., 2004), also represses the GA-induced expression of Amy32b-GUS (Xie et al., 2007). This work paved the road to addressing GA signaling in barley aleurone cells using several barley effector genes and the barley GA-inducible Amy32b promoter. Here, we present data showing the biochemical basis of the HvWRKY38 and HvGAMYB interaction. Electrophoretic mobility-shift assay (EMSA) data explain the functional competition of the WRKY and the R2R3 MYB proteins we observed before (Xie et al., 2006). Furthermore, we demonstrate the functional interactions of HvWRKY38 with another transcriptional repressor, HvBPBF, and another transcriptional activator, HvSAD. Together, these data indicate that the ratios of the repressors (e.g. HvWRKY38 and BPBF) to the activators (e.g. HvGAMYB and SAD), regulate the expression level of Amy32b in barley aleurone cells. Also, at the same molar concentration, a repressor is dominant over an activator.
RESULTS
HvWRKY38 Represses GA Induction of the Amy32b α-Amylase Promoter in Aleurone Cells
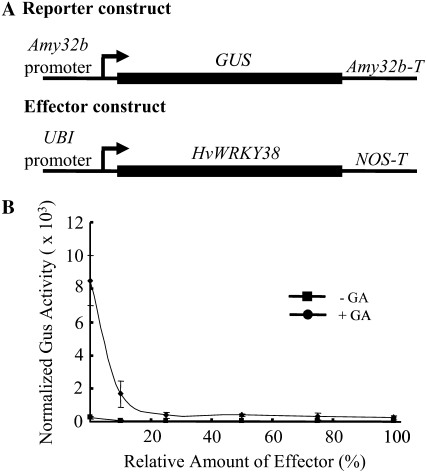
HvWRKY38 is a member of the group II WRKY transcription factors, which contain only one WRKY domain (Mare et al., 2004). It is capable of binding to W-boxes/Box2 in the GA-responsive Amy32b promoter (Mare et al., 2004). We showed that HvWRKY38 suppressed the GA-induced expression of Amy32b-GUS (Xie et al., 2007). This role was confirmed in a dosage experiment in which the reporter construct (Amy32b-GUS) concentration was kept constant, whereas the effector construct (UBI-HvWRKY38) varied from 0% to 100% (Fig. 1). When the Amy32b-GUS construct was transformed alone, treatment with 1 μm GA induced the expression of this gene construct by 37-fold. The expression of Amy32b-GUS in response to GA was reduced to 7-fold in the presence of 10% of the relative amount of effector. When the relative amounts of effector were higher than 25%, GUS expression was reduced to 2-fold or less. These data indicate that the effect of HvWRKY38 on the repression of GA induction of the Amy32b α-amylase gene expression is dosage dependent, similar to what was reported for OsWRKY71 (Xie et al., 2006).
Figure 1.
HvWRKY38 specifically represses GA induction of the Amy32b α-amylase promoter in aleurone cells. A, Schematic diagrams of the reporter and effector constructs used in the cobombardment experiment. B, The effector construct, UBI-HvWRKY38, was cobombarded into barley aleurone cells along with the reporter construct, Amy32b-GUS, and the internal control construct, UBI-Luciferase. The amount of reporter and internal control plasmid DNA was always constant, whereas that of the effector varied with respect to the reporter, as shown on the x axis. One hundred percent means that the same amount of effector and reporter DNA was used. GUS activity was normalized in every independent transformation relative to the same number of luciferase activity units (Lanahan et al., 1992). The lines indicate GUS activities ± se after 24 h of incubation of the bombarded aleurone cells with (+) or without (−) 1 μm GA. Data are means ± se of four replicates.
Loss of HvWRKY38 Activity Leads to Increased Expression of Amy32b in the Absence of GA
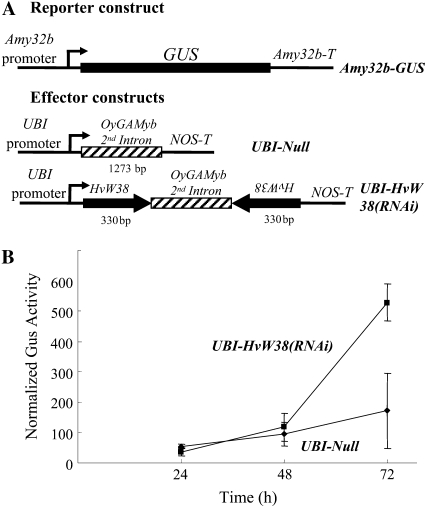
If HvWRKY38 indeed encodes a negative regulator of the GA pathway controlling the expression of Amy32b in aleurone cells, knocking down its expression by double-stranded RNA interference (RNAi) should enhance the expression of the Amy32b promoter in the absence of GA. To test this hypothesis, UBI-Null and UBI-HvW38(RNAi) constructs were prepared (Fig. 2A). The UBI-Null control contains the second intron (1,273 bp) of OsGAMyb, which is flanked by the UBI promoter and the NOS terminator (Zhang et al., 2004). The OsGAMyb intron functions as a loop in the RNAi construct. The UBI-HvW38(RNAi) construct contains two HvWRKY38 fragments in opposite orientations. The Amy32b-GUS reporter construct was cobombarded into aleurone cells along with the UBI-Null or the UBI-HvW38(RNAi) construct. As shown in Figure 2B, coexpression of UBI-Null had little effect on Amy32b-GUS expression at 24, 48, and 72 h after bombardment. In contrast, coexpression of UBI-HvW38(RNAi) led to a 3-fold increase in Amy32b-GUS expression without GA at the 72-h time point. The relative low induction might be due to the presence of WRKY genes with redundant functions, a phenomenon commonly observed for supergene families. Alternatively, the HvWRKY38 protein might be stable in the absence of GA, similar to what we observed for the OsWRKY71-GFP fusion (Zou et al., 2004).
Figure 2.
Loss of HvWRKY38 activity leads to increased expression of Amy32b in the absence of GA. A, Schematic diagrams of gene constructs. Arrowheads indicate the orientations of the gene fragments. Numbers below the effector constructs represent the size (in base pairs) of every segment (not drawn to scale). B, The reporter construct, Amy32b-GUS, and the internal construct, UBI-Luciferase, were cobombarded into barley aleurone cells with the effector construct, UBI-Null or UBI-HvW38(RNAi), using the same molar ratio of effector and reporter constructs. GUS activity was normalized in every independent transformation relative to the luciferase activity. Lines indicate GUS activities ± se after 24, 48, or 72 h of incubation of the bombarded aleurone cells without GA. Data are means ± se of four replicates.
Physical Interactions of HvWRKY38 and HvBPBF Proteins in the Nuclei of Aleurone Cells
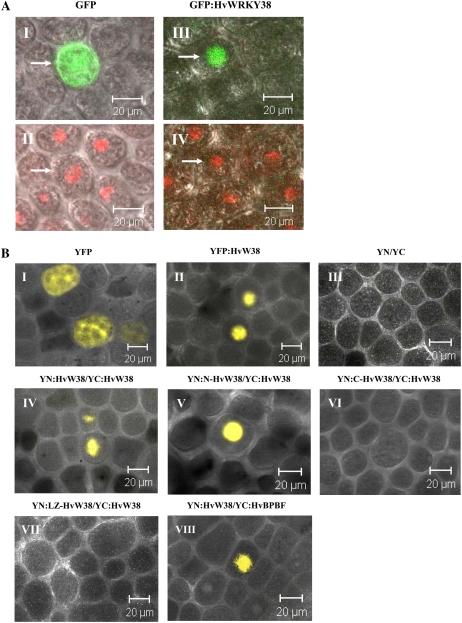
To examine the subcellular localization of the HvWRKY38 protein, the HvWRKY38 gene was fused in frame to the 3′ end of a GFP gene that is driven by the constitutive maize (Zea mays) UBI promoter. The UBI-GFP control or the UBI-GFP:HvWRKY38 plasmid was introduced into barley aleurone cells by particle bombardment, and GFP fluorescence was visualized with confocal microscopy. In the control, GFP fluorescence was observed throughout the cells (Fig. 3A, I). In contrast, GFP:HvWRKY38 fusion proteins were localized exclusively in the nuclei (Fig. 3A, III), as confirmed by staining with the red fluorescent nucleic acid stain SYTO 17 (Fig. 3A, II and IV). These results suggest that HvWRKY38 is targeted to the nuclei of aleurone cells.
Figure 3.
Visualization of interactions between transcription factors of GA signaling in aleurone cells by BiFC. A, Barley half-seeds were bombarded with either UBI-GFP (I and II) or UBI-GFP:HvWRKY38 (III and IV). After incubation for 24 h, the aleurone cells were stained with SYTO17 to localize nuclei (II and IV), followed by examination of GFP fluorescence (I and III). Arrows point to the same cell. Bars = 20 μm. B, Barley aleurone cells were bombarded with UBI-YFP, UBI-YFP:HvWRKY38, or a combination of constructs encoding the indicated fusion proteins. YN is the fragment containing amino acid residues 1 to 154 of YFP, and YC is the fragment containing amino acid residues 155 to 238 of YFP. After incubation at 24°C for 24 h, yellow fluorescence was observed through a confocal microscope.
To understand how HvWRKY38 modulates the expression level of Amy32b, we studied whether HvWRKY38 proteins physically interact with each other in aleurone cells. The HvWRKY38 coding sequences were fused in-frame with sequences encoding the N-terminal fragment of yellow fluorescent protein (YFP; YN) or the C-terminal fragment (YC), respectively. The complementation between different combinations of fusion proteins was tested by introducing the corresponding constructs into barley aleurone cells. Confocal microscopic studies showed that YFP fluorescence was detected in the entire cells bombarded with the UBI-YFP construct (Fig. 3B, I). Consistent with the nuclear localization data in Figure 3A, YFP fluorescence was only observed in the nuclei of the aleurone cells bombarded with UBI-YFP:HvWRKY38 (Fig. 3B, II). No fluorescence was detected when both YN and YC fusion constructs were introduced into aleurone cells (Fig. 3B, III). However, yellow fluorescence was detected in the nuclei of aleurone cells bombarded with UBI-YN:HvWRKY38 and UBI-YC:HvWRKY38 constructs (Fig. 3B, IV), suggesting that HvWRKY38 proteins interact in the nuclei of aleurone cells.
We further studied whether the WRKY domain of HvWRKY38 is essential for the interaction. The N-terminal and C-terminal regions of HvWRKY38 were fused in-frame with UBI-YN to produce UBI-YN:N-HvWRKY38 and UBI-YN:C-HvWRKY38, respectively. The N-terminal region includes the first 199 amino acid residues. The WRKY domain is present in the C-terminal domain, which is 154 amino acids in length. Yellow fluorescence was detected in the nuclei of aleurone cells bombarded with UBI-YN:N-HvWRKY38 and UBI-YC:HvWRKY38 (Fig. 3B, V). However, no fluorescence was detected when UBI-YN:C-HvWRKY38 and UBI-YC:HvWRKY38 (Fig. 3B, VI) were introduced into aleurone cells. These data suggest that the interaction domain is located in the N terminus.
A putative LZ motif from residues Leu-63 to Leu-91 was identified in this N-terminal fragment. To determine whether this putative LZ motif is involved in protein-protein interaction, a double mutant construct was created in which Leu-63 and Leu-77 were changed to Arg and His residues, respectively. The LZ motif mutation of HvWRKY38 was then fused in-frame with UBI-YN to produce UBI-YN:LZ-HvWRKY38. Interestingly, no fluorescence was detected when UBI-YN:LZ-HvWRKY38 and UBI-YC:HvWRKY38 were introduced into aleurone cells in the bimolecular fluorescence complementation (BiFC) experiment (Fig. 3B, VII). These data further suggest that the putative LZ domain is necessary for the interaction of HvWRKY38.
HvBPBF is also a negative regulator of the GA response in aleurone cells (Mena et al., 2002). We hypothesized that HvBPBF and HvWRKY38 physically interact in a repression complex, because they bind to cis-acting elements that are only 14 bp apart. To test this hypothesis, the HvWRKY38 and HvBPBF coding sequences were fused in-frame with sequences coding the YN or the YC, respectively. Yellow fluorescence was detected in the nuclei of aleurone cells bombarded with UBI-YN:HvWRKY38 and UBI-YC:HvBPBF (Fig. 3B, VIII), indicating that HvWRKY38 interacts with HvBPBF in the nuclei of aleurone cells.
HvWRKY38 Interferes with the Binding of HvGAMYB to the Amy32b Promoter
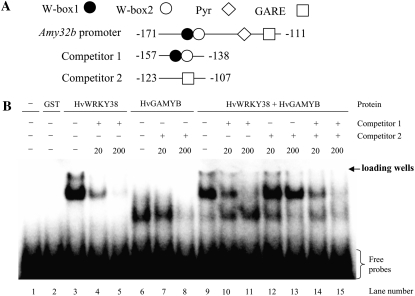
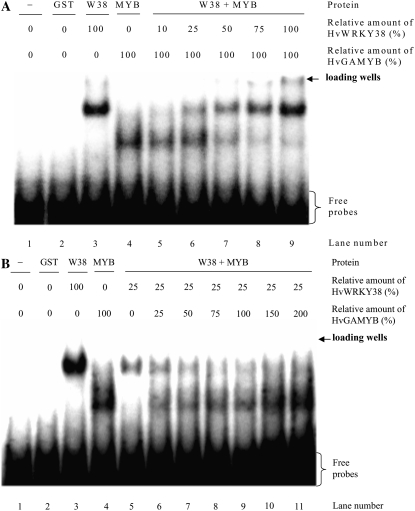
HvGAMYB is a positive regulator mediating the pathway between GA and α-amylase in aleurone cells (Gubler et al., 1995). We showed previously that OsWRKY71 (the putative rice ortholog of HvWRKY38) functionally interferes with the activity of OsGAMYB (Xie et al., 2006). HvWRKY38 behaved similarly to OsWRKY71 in its functional interaction with GAMYB (Supplemental Fig. S1). To understand the biochemical basis of these two regulators in modulating the expression level of Amy32b, we studied whether HvWRKY38 and HvGAMYB bind to the promoter of Amy32b simultaneously or compete in binding to the promoter. EMSAs were performed with glutathione S-transferase (GST)-tagged HvWRKY38 and HvGAMYB. A DNA fragment containing the Amy32b promoter region from −171 to −111 was used as a probe (Fig. 4A). No binding signal was detected in reactions without protein (Fig. 4B, lane 1) or with GST only (lane 2). Both HvWRKY38 and HvGAMYB bound specifically to the Amy32b promoter (lanes 3 and 6), and excess amounts of unlabeled promoter fragment competed off the in vitro binding signal (lanes 4, 5, 7, and 8). It has been demonstrated that the dimerization of HvWRKY38 is essential for high-affinity binding to the W-boxes in the Amy32b promoter (Mare et al., 2004); we also found that HvWRKY38 proteins physically interact in nuclei (Fig. 3B, IV). Here, we show that the GST-HvWRKY38 complex was shifted higher than that of GST-HvGAMYB, although the size of the GST-HvGAMYB protein (approximately 80 kD) is greater than that of the GST-HvWRKY38 protein (approximately 60 kD; Fig. 4B, compare lane 3 with lane 6). These data suggest that GST-HvWRKY38 binds to DNA as a dimer while GST-HvGAMYB binds as a monomer. However, the mobility of the protein-DNA complex in the gel retardation assay depends on both the size and the charge of a protein. The pI values of HvWRKY38 and HvGAMYB were predicted to be 8.2 and 4.7, respectively, using the Sequence Manipulation Suite (http://www.bioinformatics.org/sms2/protein_iep.html). Therefore, it is possible that under the electrophoresis condition (pH 8), the HvGAMYB is more negatively charged and, hence, mobilized faster than HvWRKY38.
Figure 4.
Electrophoretic mobility shift assays of GST:HvGAMYB and GST:Hv-WRKY38 fusion proteins. A, The 61-bp probe (−171 to −111) and a 20- or 28-bp synthetic oligonucleotide competitor containing W-boxes or GARE were used in EMSA. Circles denote W-boxes, the diamond represents the pyrimidine box, and the rectangle represents GARE in the Amy32b promoter. The DNA probe was end labeled with [α-32P]dATP. B, EMSA with recombinant GST:HvWRKY38 or GST:HvGAMYB proteins without (−) or with excess amounts of competitors (20- or 200-fold).
Interestingly, HvWRKY38 and HvGAMYB bind to two different cis-acting elements that are 24 bp apart. If both of the transcription factors could bind to the DNA simultaneously, one would see a third band corresponding to the protein-DNA complex that mobilized at a rate slower than those of the HvWRKY38-DNA and HvGAMYB-DNA complexes. However, this was not what we observed; when both HvWRKY38 and HvGAMYB were present in the reactions, HvGAMYB binding to the Amy32b promoter was inhibited (Fig. 4B, lane 9). As HvWRKY38 binding was competed off by an excess amount of the competitor 1, which contains two W-boxes (the WRKY binding site), HvGAMYB was able to bind to the promoter (lanes 10 and 11). As expected, competitor 2, which contains GARE only (the HvGAMYB binding site), did not compete with the binding of HvWRKY38 (lanes 12 and 13). When both competitors 1 and 2 were added, the binding signals of both proteins were decreased (lanes 14 and 15).
HvWRKY38 and HvGAMYB Functionally Compete with Each Other to Regulate the Expression of Amy32b
EMSA data suggested that binding of HvWRKY38 to the W-boxes prevented HvGAMYB from binding to the Amy32b promoter. Dosage experiments were performed to study the functional interactions of these two regulators. Varied amounts of UBI-HvWRKY38 (from 0% to 100% relative to the reporter construct) were introduced into aleurone cells along with a constant amount of UBI-HvGAMYB (100%) and Amy32b-GUS (100%). The HvGAMYB induction of Amy32b was gradually suppressed by increasing the amounts of UBI-HvWRKY38 (Supplemental Fig. S1B). It also showed that 10% of UBI-HvWRKY38 is sufficient to repress the HvGAMYB transactivating activity on the Amy32 promoter. We further performed a similar dosage experiment with a fixed amount of UBI-HvWRKY38 (10%) and Amy32b-GUS (100%) and varied amounts of UBI-HvGAMYB (from 0% to 200%). The GUS activity was gradually increased as the amount of UBI-HvGAMYB was augmented in this experiment (Supplemental Fig. S1C).
We then performed EMSA to investigate the biochemical basis of the dosage effect observed in the functional assays. Varied amounts of GST-HvWRKY38 (from 0% to 100% relative to GST-HvGAMYB) with a fixed amount of GST-HvGAMYB (100%) were used in the competition study. Increasing amounts of HvWRKY38 protein gradually decreased the binding of HvGAMYB to the Amy32b promoter (Fig. 5A, lanes 4–9). On the other hand, increasing amounts of HvGAMYB enhanced the binding signal of HvGAMYB to the Amy32b promoter (Fig. 5B, lanes 6–11). Together, these data suggest that the relative amount of a repressor or activator binding to the Amy32b promoter determines its expression level.
Figure 5.
HvWRKY38 competes with HvGAMYB in binding to the Amy32b promoter. A, EMSA with a fixed amount of GST:HvGAMYB proteins and different amounts of GST:HvWRKY38 proteins. The relative amount of GST:HvWRKY38 is indicated as a percentage compared with the amount of GST:HvGAMYB (0.5 μg) used in the competition reactions. B, EMSA with a fixed amount of GST:HvWRKY38 and different amounts of GST:HvGAMYB proteins. The relative amount of GST:HvGAMYB is indicated as a percentage compared with the amount of GST:HvWRKY38 (0.125 μg or 25%) used in the competition reactions.
Interactions of HvWRKY38, BPBF, SAD, and HvGAMYB in the Expression of the Amy32b Promoter
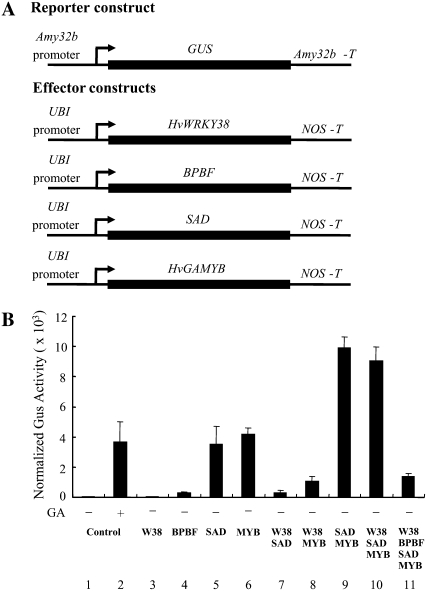
The five cis-acting elements involved in GA responses of Amy32b could be bound by either a transcriptional activator or a repressor of the same or a related gene family. For example, we have shown that the W-box (O2S) is essential for the activity of the Amy32b promoter (Zhang et al., 2004). However, this element is bound by the OsWRKY71/HvWRKY38 repressor (Mare et al., 2004; Zhang et al., 2004) and possibly by another WRKY or other zinc finger proteins that could function as activators (Zhang et al., 2004). Similarly, SAD (an activator) and BPBF (a repressor) can bind to the same cis-acting element (i.e. the pyrimidine box; Mena et al., 2002; Isabel-LaMoneda et al., 2003). The HvGAMYB transcriptional activator interacts with the third element, GARE (Gubler et al., 1995). Here, we tested the functional interactions among HvWRKY38, BPBF, SAD, and HvGAMYB on regulating the expression of the Amy32b promoter. As expected, little expression of Amy32b-GUS was detected when HvWRKY38 and BPBF were overexpressed in the absence of GA (Fig. 6B, columns 1, 3, and 4). In contrast, cotransformation of UBI-SAD or UBI-HvGAMYB alone resulted in 71-fold or 85-fold induction of the Amy32b promoter, respectively (columns 5 and 6). The inductive activity of SAD and HvGAMYB is repressed by HvWRKY38 (columns 7 and 8). Coexpression of SAD and HvGAMYB dramatically increased the expression of the Amy32b promoter to 201-fold induction (column 9). Interestingly, this combined effect of SAD and HvGAMYB was not suppressed by HvWRKY38 (column 10). However, HvWRKY38 and BPBF together dramatically reduced the combined effect of SAD and HvGAMYB on the induction of the Amy32b promoter (column 11). Taken together, these data suggest that the expression level of Amy32b is governed by ratios of a set of transcriptional repressors and a set of transcriptional activators. Because each of the cis elements in the Amy32b promoter could be bound by a repressor or an activator of the same or a related family, it is easy to envision their competition in regulating the Amy32b promoter. Our data also indicated the competition of transcriptional regulators whose binding sites are different (e.g. HvWRKY38 and HvGAMYB).
Figure 6.
Interactions of HvWRKY38, BPBF, SAD, and HvGAMYB in regulating the expression of the Amy32b promoter. A, Schematic diagram of the reporter and effector constructs used in the cobombardment experiment. B, The reporter construct, Amy32b-GUS, and the internal construct, UBI-Luciferase, were cobombarded into barley aleurone cells either with (+) or without (−) the effector construct using the same molar amount of effector and reporter constructs. GUS activity was normalized in every independent transformation relative to the luciferase activity. Bars indicate GUS activities ± se after 24 h of incubation of the bombarded aleurone cells with (+) or without (−) 1 μm GA. Data are means ± se of four replicates.
DISCUSSION
In this work, we confirmed that HvWRKY38 repressed the GA-induced expression of Amy32b-GUS in barley aleurone cells (Fig. 1). The role of HvWRKY38 as a transcriptional repressor in the GA response was further demonstrated by the RNAi experiment (Fig. 2). We also found that HvWRKY38 repressed the transactivating activity of HvGAMYB (Supplemental Fig. S1) by competing with the binding of HvGAMYB to the Amy32b α-amylase promoter (Figs. 4 and 5). This inhibitory effect of HvWRKY38 was overcome by the SAD and HvGAMYB activators in combination (Fig. 6). However, combination of WRKY38 and BPBF, which appeared to physically interact (Fig. 3), suppressed the combined effect of SAD and HvGAMYB on inducing the Amy32b promoter in the absence of GA (Fig. 6).
HvWRKY38 is inducible by cold and drought treatments in barley leaves and roots (Mare et al., 2004). It is also induced by abscisic acid and salicylic acid but suppressed by GA in aleurone cells (Mare et al., 2004; Xie et al., 2007). HvWRKY38 represses GA induction of Amy32b (Fig. 1; Xie et al., 2007). Consistent with its function as a repressor, coexpression of a UBI-HvW38(RNAi) construct derepressed the expression of the Amy32b promoter, leading to increased expression of Amy32b in the absence of GA (Fig. 2). The effect of UBI-HvW38(RNAi) was significant, although it was lower than that of UBI-HvSLN1 RNAi (Zentella et al., 2002). There are two possibilities accounting for the lower effect of UBI-HvW38(RNAi). The preexisting HvWRKY38 protein might be more stable than the SLN1 protein; HvWRKY38 RNAi might only knock out HvWRKY38 but not other WRKY genes. The WRKY family has 74 members (Eulgem et al., 2000) in Arabidopsis and about 100 members in rice (Zhang and Wang, 2005; Ross et al., 2007). As many as 45 WRKY genes have been identified in barley; among these genes, HvWRKY38 (also called HvWRKY1) is closely related to HvWRKY2, -3, and -23 (Mangelsen et al., 2008). Like OsWRKY71 and AtWRKY40, these four barley genes belong to group 2a of the WRKY gene superfamily (Eulgem et al., 2000). Several members could be involved in the same processes. For example, Arabidopsis WRKY18, WRKY40, and WRKY60 have partially redundant roles in plant responses to pathogens (Xu et al., 2006). Rice WRKY24, WRKY51, and WRKY71 blocked GA induction of the Amy32b promoter (Zhang et al., 2004; Xie et al., 2006). It will be interesting to study the function of other barley WRKY family members in GA signaling.
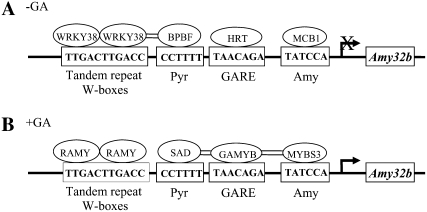
Mounting evidence suggests that each of the five cis-acting elements essential for GA induction of Amy32b can be bound by both transcriptional repressors and activators in barley aleurone cells. The pyrimidine box can be bound by the SAD activator and the BPBF and HvDOF19 repressors, which are DOF proteins (Diaz et al., 2002, 2005; Isabel-LaMoneda et al., 2003; Moreno-Risueno et al., 2007). In addition to the transcriptional activator GAMYB, the repressor HRT, a zinc finger protein, can also bind to GARE (Raventós et al., 1998). The Amy box can interact with the HvMCB1 repressor and the HvMYBS3 activator (Rubio-Somoza et al., 2006). We show here that HvWRKY38 interacts with the W-boxes (Fig. 6). An activator for this element has not been reported, although RAMY, another zinc finger protein, also binds to this element (Peng et al., 2004). Furthermore, physical interactions between two repressors, HvWRKY38 and HvBPBF, and two activators, HvGAMYB and HvSAD or HvGAMYB and HvMYBS3, have been demonstrated by BiFC (Fig. 3; Diaz et al., 2005; Rubio-Somoza et al., 2006). In light of these results, we propose a hypothetical model for the control of Amy32b gene expression in aleurone cells (Fig. 7). In the absence of GA, negative regulators such as HvWRKY38, BPBF, HRT, and HvMCB1 bind to corresponding cis-acting elements and form a “repressosome,” which diminishes the binding or transactivating activities of positive regulators to the promoter, thereby preventing Amy32b transcription. In the presence of GA, positive regulators such as RAMY, SAD, HvGAMYB, and HvMYBS3 bind to their respective DNA sequences and form an “enhanceosome,” leading to a high level of Amy32b gene expression.
Figure 7.
Hypothetical model for the control of Amy32b α-amylase gene expression in aleurone cells. A, Negative regulators (HvWRKY38, BPBF, HRT, and HvMCB1) bind to their corresponding cis-acting elements in the absence of GA. B, Positive regulators (RAMY, SAD, HvGAMYB, and HvMYBS3) bind to corresponding cis-acting elements in the presence of GA. Double lines between proteins indicate that their physical interactions have been detected by BiFC. The arrow denotes the transcription start site; X over the arrow means the transcription of Amy32b is off or at a very low level in the absence of GA.
The intriguing question is how the repressosome is replaced by the enhanceosome during the transition of Amy32b from the repressed state to the activated state or vice versa. Two possible mechanisms can be envisioned. (1) GA promotes the degradation of repressors, as reported for OsWRKY71 (Zhang et al., 2004), allowing transcriptional activators to occupy the cis-acting elements. (2) GA induces the expression of activators such as GAMYB (Gubler et al., 1995). These activators physically interact with repressors, eventually leading to the dissociation of repressors from and association of activators to the corresponding cis-acting elements cooperatively. Interactions of repressors and activators have been observed in mammalian systems. For example, B-Myb, a cell cycle-regulated transcriptional repressor, physically interacts with the transcriptional activators specificity protein 1 and CCAAT-binding factor and interferes with their binding to the promoter (Cicchillitti et al., 2004). As a result, COL1A1 (for type I collagen) transcription in human scleroderma fibroblasts is decreased. The barley repressors BPBF, HvDOF17, and HvDOF19 physically interact with HvGAMYB, as demonstrated in the nucleus of onion (Allium cepa) cells using the BiFC approach (Diaz et al., 2005; Moreno-Risueno et al., 2007). HvDOF17 represses the expression of Al21, a thiol protease gene, probably by decreasing the binding affinity of HvGAMYB to GARE in the Al21 promoter (Moreno-Risueno et al., 2007). Our data showed that the binding of HvGAMYB to the Amy32b promoter was decreased in the presence of increasing amounts of HvWRKY38 (Fig. 5), suggesting that HvWRKY38 interferes with the binding of HvGAMYB to the Amy32b promoter. Accordingly, the induction of Amy32b by HvGAMYB was reduced gradually with increasing amounts of HvWRKY38 (Supplemental Fig. S1). The repression effect of HvWRKY38 was overcome by the coexpression of two transcriptional activators, SAD and HvGAMYB. WRKY38 and BPBF together block the combined effect of SAD and HvGAMYB on inducing the Amy32b promoter in the absence of GA (Fig. 6). Therefore, the ratios of repressors to activators, and more importantly, the cooperative binding of repressors or activators to the Amy32b promoter, determine the levels of Amy32b expression (Fig. 7).
In summary, the data presented in this study support the hypothesis that the GA induction of Amy32b is modulated by two protein complexes, one for activation and the other for repression. Further work is needed to isolate these endogenous complexes using immunoprecipitation with antibodies against different regulators or the technique that combines tandem affinity purification with mass spectrometry (Dziembowski and Seraphin, 2004; Van Leene et al., 2007). With these methods, we will be able to further dissect the interactions among transcription factors in regulating GA-controlled gene expression.
MATERIALS AND METHODS
RNA Isolation and RT-PCR
Total RNA was isolated from barley (Hordeum vulgare) aleurone cells with the LiCl precipitation method as described (Zhang et al., 2004). The first-strand cDNAs were synthesized using ImProm-II reverse transcriptase in a 50-μL reaction containing 2.5 μm oligo(dT) primers, 2.5 μm random hexamer, and 2.5 μg of total RNA, according to the manufacturer's instructions. Five microliters of each reaction mixture was used as a template for PCR amplification in a 25-μL mixture containing 1.5 μm MgCl2, 200 μm dNTPs, 5% dimethyl sulfoxide, 2.5 units of Taq DNA polymerase, and 0.4 μm primers.
Effector Construct Preparations
Three types of DNA constructs were used in the transient expression experiments: reporter, effector, and internal control. Plasmid Amy32b-GUS (Lanahan et al., 1992), HVA1-GUS, and HVA22-GUS (Shen et al., 1993) were used as the reporter constructs. Plasmid pAHC18 (UBI-Luciferase), which contains the luciferase reporter gene driven by the constitutive maize (Zea mays) ubiquitin promoter (Bruce et al., 1989), was used as an internal control construct to normalize GUS activities of the reporter construct. The full-length cDNA of HvWRKY38 and HvGAMYB was amplified from total RNA of barley aleurone cells by reverse transcription-PCR and cloned into the AscI site of the intermediate construct containing the UBI promoter and the NOS terminator (Zhang et al., 2004) using primers P1 and P2 for the preparation of UBI-HvWRKY38 and primers P3 and P4 for the preparation of UBI-HvGAMYB (Supplemental Table S1). The full-length cDNAs of BPBF and SAD were amplified and cloned into the AscI site of the expression vector using the following primers: P5 and P6 for the preparation of UBI-BPBF and P7 and P8 for the preparation of UBI-SAD. To construct UBI-HvW38(RNAi), a 335-bp gene-specific fragment was obtained using primers P9 and P10 and cloned into BlpI and Bsu36I sites, respectively, using the symmetrical directional cloning method (Zhang et al., 2004). The substitution mutants were prepared by oligonucleotide-directed mutagenesis (Kunkel et al., 1987). Single-stranded DNA from plasmid UBI-HvWRKY38 was used as a template. Primers P11 and P12 were used to mutate the putative LZ motif.
Particle Bombardment and Transient Expression Assays
Detailed descriptions of the transient expression procedure with the barley aleurone system and the particle bombardment technique have been published before (Shen et al., 1993). Briefly, deembryonated half-seeds of ‘Himalaya’ barley were soaked for 2.5 to 3 d, and then the pericarp and testa were removed. The DNA mixture (in a 1:1 molar ratio) of HVA1-GUS and UBI-Luciferase or Amy32b-GUS and UBI-Luciferase, along with or without an effector construct, was bombarded into barley embryoless half-seeds (four replicates per test construct). After incubation for 24 h with various treatments, GUS assays and luciferase assays were performed as described previously (Shen et al., 1996).
Subcellular Localization and BiFC
The HvWRKY38 gene was inserted into the AscI site in UBI-GFP (Zhang et al., 2004), UBI-YFP, UBI-YN, and UBI-YC (Xie et al., 2006) to generate UBI-GFP:HvWRKY38, UBI-YFP:HvWRKY38, UBI-YN:HvWRKY38, and UBI-YC:HvWRKY38. The N-terminal region of HvWRKY38 was produced by introducing a stop codon at amino acid 200, which is upstream from the WRKY domain. The rest (i.e. the C-terminal region) was amplified using primers P13 and P14 (Supplemental Table S1). The N-terminal and C-terminal regions of HvWRKY38 were inserted into the AscI site in UBI-YN to produce UBI-YN:N-HvWRKY38 and UBI-YN:C-HvWRKY38. The LZ motif-mutated HvWRKY38 was inserted into the AscI site of UBI-YN to produce UBI-YN:LZ-HvWRKY38. The HvBPBF gene was amplified and inserted into the AscI site in UBI-YC to generate UBI-YC:HvBPBF. After incubation at 24°C for 24 h, the aleurone layers were peeled from barley half-seeds and soaked in a 5 μm SYTO17 solution (Molecular Probes). The stained samples were observed, and images of GFP fluorescence and SYTO17 staining were obtained simultaneously through a LSM 510 laser scanning microscope (Carl Zeiss), with 488-nm excitation and 505- to 530-nm emission wavelengths for the green fluorescence and 633-nm excitation and 650-nm emission wavelengths for the red fluorescence in separate channels. The yellow fluorescence was observed with 514-nm excitation and 530-nm emission wavelengths. At least 8,000 cells were checked in each sample, and fluorescence was observed in more than 5% of total cells in positive results. The acquired images were processed using Paint Shop Pro 7 (Jasc Software).
EMSA
The full-length cDNA of HvWRKY38 was cloned into the AscI site of the modified pGEX-KG (Zhang et al., 2004) to generate GST:HvWRKY38. The fusion constructs were then introduced into Escherichia coli strain Origami B DE3 (Novagen). The full-length cDNA of HvGAMYB was cloned into the AscI site of the modified pGEX-KG (Zhang et al., 2004) to generate GST:HvGAMYB. The fusion constructs were then introduced into E. coli BL-21 (DE3) pLysS (Novagen). Overexpression of the fusion proteins was induced by 1 mm isopropylthio-β-galactoside at 25°C in 2× YT medium for 3 h. The cell suspension was passed three times through an SLM-Aminco French pressure cell press at 1,600 psig. The GST fusion proteins were purified using glutathione-agarose beads (Zhang et al., 2004).
A 61-bp fragment of the Amy32b promoter (Lanahan et al., 1992) that spans positions −171 to −111 was used as a probe. The DNA probe was labeled with [α-32P]dATP by a Klenow fill-in reaction. Unless otherwise indicated, binding reactions (20 μL) contained 1.5 ng of probe, 1 μg of poly(dIdC), 10 mm Tris-HCl (pH 7.6), 50 mm KCl, 0.5 mm EDTA, 5 μm ZnCl2, 0.07% Igepal CA-630, and 10% glycerol. Equal amounts of recombinant proteins (0.5 μg for each) were added into reactions and incubated at 4°C for 20 min with labeled DNA probes in the absence or presence of competitors. After incubation, the reactions were resolved by electrophoresis on a 5% polyacrylamide gel in 0.5× TBE buffer (45 mm Tris, 45 mm boric acid, and 1 mm EDTA, pH 8) for 2 h. The signals were scanned with a Typhoon 9400 phosphorimager (Amersham Biosciences).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Functional interactions between HvWRKY38 and HvGAMYB.
Supplemental Table S1. Primers used for construct preparation.
Supplementary Material
Acknowledgments
We thank Dr. Pilar Carbonero for kindly providing the BPBF and DOF cDNA clones. We also thank Dr. Russell R. Johnson and members of our laboratory for advice or contributions to the improvement of this paper.
This work was supported by the U.S. Department of Agriculture (grant no. 2007–35304V18297 to Q.J.S.), the National Institutes of Health Biomedical Research Infrastructure Network (grant no. P20 RR–16464 to the state of Nevada), and the National Science Foundation (EPSCoR IAAS fellowship no. EPS–0132556 to X.Z.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Qingxi J. Shen (jeffery.shen@ccmail.nevada.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Bruce WB, Christensen AH, Klein T, Fromm M, Quail PH (1989) Photoregulation of a phytochrome gene promoter from oat transferred into rice by particle bombardment. Proc Natl Acad Sci USA 86 9692–9696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicchillitti L, Jimenez SA, Sala A, Saitta B (2004) B-Myb acts as a repressor of human COL1A1 collagen gene expression by interacting with Sp1 and CBF factors in scleroderma fibroblasts. Biochem J 378 609–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz I, Martinez M, Isabel-LaMoneda I, Rubio-Somoza I, Carbonero P (2005) The DOF protein, SAD, interacts with GAMYB in plant nuclei and activates transcription of endosperm-specific genes during barley seed development. Plant J 42 652–662 [DOI] [PubMed] [Google Scholar]
- Diaz I, Vicente-Carbajosa J, Abraham Z, Martinez M, Isabel-La Moneda I, Carbonero P (2002) The GAMYB protein from barley interacts with the DOF transcription factor BPBF and activates endosperm-specific genes during seed development. Plant J 29 453–464 [DOI] [PubMed] [Google Scholar]
- Dziembowski A, Seraphin B (2004) Recent developments in the analysis of protein complexes. FEBS Lett 556 1–6 [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5 199–206 [DOI] [PubMed] [Google Scholar]
- Gilroy S, Jones RL (1992) Gibberellic acid and abscisic acid coordinately regulate cytoplasmic calcium and secretory activity in barley aleurone protoplasts. Proc Natl Acad Sci USA 89 3591–3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Cadenas A, Zentella R, Sutliff TDH, Ho THD (2001) Involvement of multiple cis-elements in the regulation of GA responsive promoters: definition of a new cis-element in the Amy32b gene promoter of barley (Hordeum vulgare). Physiol Plant 112 211–216 [DOI] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, Gong F, Phillips AL, Hedden P, Sun TP, et al (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis Plant Cell 18 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Jacobsen JV (1992) Gibberellin-responsive elements in the promoter of a barley high-pI alpha-amylase gene. Plant Cell 4 1435–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Kalla R, Roberts JK, Jacobsen JV (1995) Gibberellin-regulated expression of a myb gene in barley aleurone cells: evidence for Myb transactivation of a high-pI alpha-amylase gene promoter. Plant Cell 7 1879–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson JT, Li HC, Rider SD, Mordhorst AP, Romero-Severson J, Cheng JC, Robey J, Sung ZR, de Vries SC, Ogas J (2004) PICKLE acts throughout the plant to repress expression of embryonic traits and may play a role in gibberellin-dependent responses. Plant Physiol 134 995–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooley R (1998) Plant hormone perception and action: a role for G-protein signal transduction? Philos Trans R Soc Lond B Biol Sci 353 1425–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabel-LaMoneda I, Diaz I, Martinez M, Mena M, Carbonero P (2003) SAD: a new DOF protein from barley that activates transcription of a cathepsin B-like thiol protease gene in the aleurone of germinating seeds. Plant J 33 329–340 [DOI] [PubMed] [Google Scholar]
- Itoh H, Matsuoka M, Steber CM (2003) A role for the ubiquitin-26S-proteasome pathway in gibberellin signaling. Trends Plant Sci 8 492–497 [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Binkowski KA, Olszewski NE (1996) SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc Natl Acad Sci USA 93 9292–9296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Roberts JD, Zakour RA (1987) Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol 154 367–382 [DOI] [PubMed] [Google Scholar]
- Lanahan MB, Ho THD, Rogers SW, Rogers JC (1992) A gibberellin response complex in cereal alpha-amylase gene promoters. Plant Cell 4 203–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovegrove A, Hooley R (2000) Gibberellin and abscisic acid signalling in aleurone. Trends Plant Sci 5 102–110 [DOI] [PubMed] [Google Scholar]
- Mangelsen E, Kilian J, Berendzen KW, Kolukisaoglu UH, Harter K, Jansson C, Wanke D (2008) Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. BMC Genomics 9 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mare C, Mazzucotelli E, Crosatti C, Francia E, Stanca AM, Cattivelli L (2004) Hv-WRKY38: a new transcription factor involved in cold- and drought-response in barley. Plant Mol Biol 55 399–416 [DOI] [PubMed] [Google Scholar]
- McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun TP, Steber CM (2003) The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15 1120–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena M, Cejudo FJ, Isabel-Lamoneda I, Carbonero P (2002) A role for the DOF transcription factor BPBF in the regulation of gibberellin-responsive genes in barley aleurone. Plant Physiol 130 111–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Diaz I, Carrillo L, Fuentes R, Carbonero P (2007) The HvDOF19 transcription factor mediates the abscisic acid-dependent repression of hydrolase genes in germinating barley aleurone. Plant J 51 352–365 [DOI] [PubMed] [Google Scholar]
- Nakajima M, Shimada A, Takashi Y, Kim YC, Park SH, Ueguchi-Tanaka M, Suzuki H, Katoh E, Iuchi S, Kobayashi M, et al (2006) Identification and characterization of Arabidopsis gibberellin receptors. Plant J 46 880–889 [DOI] [PubMed] [Google Scholar]
- Ogas J, Cheng JC, Sung ZR, Somerville C (1997) Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science 277 91–94 [DOI] [PubMed] [Google Scholar]
- Olszewski N, Sun TP, Gubler F (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell (Suppl) 14 S61–S80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng R, Yao Q, Xiong A, Fan H, Li X, Peng Y, Cheng ZM, Li Y (2004) A new rice zinc-finger protein binds to the O2S box of the alpha-amylase gene promoter. Eur J Biochem 271 2949–2955 [DOI] [PubMed] [Google Scholar]
- Penson SP, Schuurink RC, Fath A, Gubler F, Jacobsen JV, Jones RL (1996) cGMP is required for gibberellic acid-induced gene expression in barley aleurone. Plant Cell 8 2325–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raventós D, Skriver K, Schlein M, Karnahl K, Rogers SW, Rogers JC, Mundy J (1998) HRT, a novel zinc finger, transcriptional repressor from barley. J Biol Chem 273 23313–23320 [DOI] [PubMed] [Google Scholar]
- Ritchie S, Gilroy S (1998) Gibberellins: regulating genes and germination. New Phytol 140 363–383 [DOI] [PubMed] [Google Scholar]
- Robertson M (2004) Two transcription factors are negative regulators of gibberellin response in the HvSPY-signaling pathway in barley aleurone. Plant Physiol 136 2747–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson M, Swain SM, Chandler PM, Olszewski NE (1998) Identification of a negative regulator of gibberellin action, HvSPY, in barley. Plant Cell 10 995–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers JC, Lanahan MB, Rogers SW (1994) The cis-acting gibberellin response complex in high pI alpha-amylase gene promoters: requirement of a coupling element for high-level transcription. Plant Physiol 105 151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers JC, Rogers SW (1992) Definition and functional implications of gibberellin and abscisic acid cis-acting hormone response complexes. Plant Cell 4 1443–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, Liu Y, Shen QJ (2007) The WRKY gene family in rice (Oryza sativa). Journal of Integrative Plant Biology 49 827–842 [Google Scholar]
- Rubio-Somoza I, Martinez M, Diaz I, Carbonero P (2006) HvMCB1, a R1MYB transcription factor from barley with antagonistic regulatory functions during seed development and germination. Plant J 45 17–30 [DOI] [PubMed] [Google Scholar]
- Shen Q, Uknes SJ, Ho THD (1993) Hormone response complex of a novel abscisic acid and cycloheximide inducible barley gene. J Biol Chem 268 23652–23660 [PubMed] [Google Scholar]
- Shen Q, Zhang P, Ho THD (1996) Modular nature of abscisic acid (ABA) response complexes: composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley. Plant Cell 8 1107–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A, Ueguchi-Tanaka M, Sakamoto T, Fujioka S, Takatsuto S, Yoshida S, Sazuka T, Ashikari M, Matsuoka M (2006) The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. Plant J 48 390–402 [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Tseng TS, Swain SM, Dill A, Jeong SY, Olszewski NE, Sun TP (2007) Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol 143 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skriver K, Olsen FL, Rogers JC, Mundy J (1991) Cis-acting DNA elements responsive to gibberellin and its antagonist abscisic acid. Proc Natl Acad Sci USA 88 7266–7270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber CM, Cooney SE, McCourt P (1998) Isolation of the GA-response mutant sly1 as a suppressor of ABI1-1 in Arabidopsis. Genetics 149 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP, Gubler F (2004) Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Physiol Plant Mol Biol 55 197–223 [DOI] [PubMed] [Google Scholar]
- Tanida I, Kim JK, Wu R (1994) Functional dissection of a rice high-pI alpha-amylase gene promoter. Mol Gen Genet 244 127–134 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, et al (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437 693–698 [DOI] [PubMed] [Google Scholar]
- Ulker B, Somssich IE (2004) WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol 7 491–498 [DOI] [PubMed] [Google Scholar]
- Van Leene J, Stals H, Eeckhout D, Persiau G, Van De Slijke E, Van Isterdael G, De Clercq A, Bonnet E, Laukens K, Remmerie N, et al (2007) A tandem affinity purification-based technology platform to study the cell cycle interactome in Arabidopsis. Mol Cell Proteomics 6 1226–1238 [DOI] [PubMed] [Google Scholar]
- Washio K (2003) Functional dissections between GAMYB and Dof transcription factors suggest a role for protein-protein associations in the gibberellin-mediated expression of the RAmy1A gene in the rice aleurone. Plant Physiol 133 850–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Zhang ZL, Hanzlik S, Cook E, Shen QJ (2007) Salicylic acid inhibits gibberellin-induced alpha-amylase expression and seed germination via a pathway involving an abscisic-acid-inducible WRKY gene. Plant Mol Biol 64 293–303 [DOI] [PubMed] [Google Scholar]
- Xie Z, Zhang ZL, Zou X, Yang G, Komatsu S, Shen QJ (2006) Interactions of two abscisic-acid induced WRKY genes in repressing gibberellin signaling in aleurone cells. Plant J 46 231–242 [DOI] [PubMed] [Google Scholar]
- Xu X, Chen C, Fan B, Chen Z (2006) Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18 1310–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R, Yamauchi D, Ho THD (2002) Molecular dissection of the gibberellin/abscisic acid signaling pathways by transiently expressed RNA interference in barley aleurone cells. Plant Cell 14 2289–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R, Zhang ZL, Park M, Thomas SG, Endo A, Murase K, Fleet CM, Jikumaru Y, Nambara E, Kamiya Y, et al (2007) Global analysis of della direct targets in early gibberellin signaling in Arabidopsis Plant Cell 19 3037–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang L (2005) The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol Biol 5 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZL, Xie Z, Zou X, Casaretto J, Ho TH, Shen QJ (2004) A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol 134 1500–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X, Seemann JR, Neuman D, Shen QJ (2004) A WRKY gene from creosote bush encodes an activator of the abscisic acid signaling pathway. J Biol Chem 279 55770–55779 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.