Abstract
Eukaryotic translation initiation factor 5A (eIF5A) is a highly conserved protein found in all eukaryotic kingdoms. This study demonstrates that plant eIF5A is involved in the development of disease symptoms induced by a common necrotrophic bacterial phytopathogen. Specifically, AteIF5A-2, one of the three eIF5A genes in Arabidopsis (Arabidopsis thaliana), is shown to regulate programmed cell death caused by infection with virulent Pseudomonas syringae pv tomato DC3000 (Pst DC3000). Transgenic Arabidopsis plants with constitutively suppressed AteIF5A-2 exhibited marked resistance to programmed cell death induced by virulent Pst DC3000, and there was a corresponding reduction in pathogen growth and development of disease symptoms in the plant tissue. Constitutive overexpression of AteIF5A-2 circumvented the apparent posttranscriptional regulation of AteIF5A-2 protein expression characteristic of wild-type plants but did not increase susceptibility to virulent Pst DC3000 ingression. The transgenic plants with constitutive AteIF5A-2 overexpression did, however, display phenotypes consistent with precocious cell death. The results indicate that AteIF5A-2 is a key element of the signal transduction pathway resulting in plant programmed cell death.
Eukaryotic translation initiation factor 5A (eIF5A) is a highly conserved protein in eukaryotes and is the only known protein containing the posttranslationally synthesized amino acid hypusine (Park et al., 1981; Cooper et al., 1982; Hanauske-Abel et al., 1994). Studies with mammalian cells and yeast have indicated that the biosynthesis of hypusine occurs on eIF5A precursor protein through a two-step posttranslational modification involving spermidine and a conserved Lys residue of eIF5A. The first step in hypusine formation is catalyzed by the enzyme deoxyhypusine synthase (DHS) and involves the transfer of a 4-amino-butyl moiety from spermidine to the ɛ-amino group of the conserved Lys residue of eIF5A to form deoxyhypusine (Wolff et al., 1997). The second step, hydroxylation of deoxyhypusine to hypusine, is catalyzed by deoxyhypusine hydroxylase (Abbruzzese et al., 1986; Park et al., 2006). It is generally accepted that eIF5A is essential for normal function of mammalian cells (Tome and Gerner, 1997; Caraglia et al., 2001, 2003). There is also evidence that eIF5A may play a role in apoptosis as it accumulates at the onset of cell death (Tome and Gerner, 1997; Tome et al., 1997; Beninati et al., 1998; Caraglia et al., 2003; Li et al., 2004; Taylor et al., 2007). The precise way in which eIF5A functions in cells is not known, but there are some reports based on studies with yeast and mammalian cells that it may selectively recruit specific subsets of mRNA from the nucleus, facilitating their translocation to the cytoplasm for subsequent translation (Shi et al., 1996; Rosorius et al., 1999; Lipowsky et al., 2000; Jao and Chen, 2005).
There is growing evidence that plant eIF5A is encoded by a multigene family (Wang et al., 2001; Chou et al., 2004; Feng et al., 2007). In Arabidopsis (Arabidopsis thaliana), for example, there are three isoforms of eIF5A protein, each encoded by a separate gene and exhibiting distinct expression profiles (Thompson et al., 2004; Feng et al., 2007). Plant eIF5A, like its yeast and mammalian counterparts, also appears to be posttranslationally hypusinated. This is supported by the finding that recombinant plant DHS is capable of catalyzing the formation of deoxyhypusine on plant eIF5A (Ober and Hartmann, 1999; Wang et al., 2001). The sequence of plant eIF5A is highly conserved, especially in the regions bordering the site of hypusination (Thompson et al., 2004), suggesting that all of the isoforms of plant eIF5A are capable of being hypusinated. However, there appears to be only one isoform of DHS in plants (Thompson et al., 2004; Duguay et al., 2006). That this single isoform of DHS is involved in posttranslational modification of more than one isoform of eIF5A is supported by the finding that suppression of DHS has pleiotropic effects. For example, constitutive suppression of DHS in Arabidopsis results in several phenotypes, including delayed natural leaf senescence, delayed bolting, increased rosette leaf and root biomass, and enhanced seed yield (Wang et al., 2003). It is possible that these pleiotropic effects are attributable to inhibition of hypusine formation on different isoforms of eIF5A throughout plant development.
Studies with transgenic plants have provided evidence for the involvement of eIF5A in programmed cell death associated not only with leaf senescence but also fruit senescence and petal senescence (Wang et al., 2001, 2003; Duguay et al., 2006; Hopkins et al., 2007). In addition, a fumonisin B1-resistant12 (fbr12) Arabidopsis mutant was recently determined to be an AteIF5A-2 knockout (Feng et al., 2007). The fbr12 mutant exhibits aberrant growth and development as well as strong resistance to fumonisin- or dark-induced apoptosis. In this study, we describe evidence indicating that Arabidopsis eIF5A-2 potentiates programmed cell death in response to pathogen attack by virulent Pseudomonas syringae pv tomato DC3000 (Pst DC3000). That programmed cell death is induced in the event of both compatible and incompatible plant-pathogen interactions is well established (Greenberg et al., 1994; Beers and McDowell, 2001; Greenberg and Yao, 2004). The incompatible interaction engenders the hypersensitive response (HR), which entails rapid, localized cell death together with the induction of pathogenesis-related genes and ensuing systemic acquired resistance (Heath, 2000; Zhang et al., 2004). Although they are temporally correlated and both elements of HR, there is some evidence that cell death and systemic acquired resistance are independent processes (Hatsugai et al., 2004). In disease-susceptible or compatible interactions, programmed cell death occurs over the course of many days, and there is growing evidence that it is host mediated (Yao et al., 2002; Greenberg and Yao, 2004).
Pst DC3000 is a facultative necrotroph that induces host programmed cell death and utilizes nutrients released from dying host cells to maximize its growth. Initial colonization of Pst DC3000 is dependent upon water and nutrient leakage from the plant cells into the intercellular space, which can be induced by various virulence factors secreted by the pathogen. To gain further access to nutrients, it is thought that Pst DC3000 induces programmed death of host cells during the later stages of infection (Abramovitch and Martin, 2004). The findings presented herein support a role for Arabidopsis eIF5A-2 in Pst DC3000-induced death of host cells.
RESULTS
AteIF5A-2 Is Selectively Up-Regulated following Infection with Virulent Pst DC3000 or Mechanical Wounding
There are three genes encoding eIF5A in Arabidopsis, all located on chromosome 1 and numbered according to their relative positions on the chromosome. AteIF5A-1, first cloned from senescing tissue by Wang et al. (2001), is closest to the top of chromosome 1 at gene locus At1g13950. The other two isoforms, AteIF5A-2 and AteIF5A-3, are at loci At1g26630 and At1g69410 of chromosome 1, respectively. The inferred amino acid sequences of these isoforms have a high degree of sequence identity, especially in the vicinity of the Lys residue (K51) that undergoes modification to hypusine.
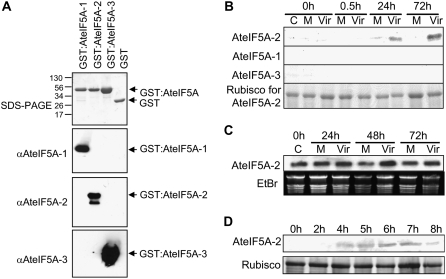
To determine whether any of the AteIF5A isoforms are up-regulated in response to Pst DC3000 ingress, western blots of total protein extracts from infected leaves were probed with isoform-specific antibodies. The antibodies were generated against synthetic peptides corresponding to small domains in the C termini of the AteIF5A proteins that exhibit sequence diversity (Liu et al., 2008), and they proved to be isoform specific (Fig. 1A). Leaves of 4-week-old wild-type Arabidopsis plants were syringe infiltrated with virulent Pst DC3000, which results in a compatible host-pathogen interaction and the development of disease symptoms (Katagiri et al., 2002). There was no protein corresponding to any of the AteIF5A isoforms detectable in uninfected control leaves or in mock-treated leaves (Fig. 1B). AteIF5A-2 showed strong up-regulation within 24 h of infection and even stronger expression 72 h after infection when the infected leaves were chlorotic, whereas AteIF5A-1 and AteIF5A-3 remained undetectable (Fig. 1B). Northern-blot analysis was performed using the 3′-untranslated region (UTR) of AteIF5A-2 as a probe (Fig. 1C). Unlike the case for protein, the transcript of AteIF5A-2 proved to be constitutively expressed and exhibited only slight up-regulation postinfection with virulent Pst DC3000 (Fig. 1C). This apparent posttranscriptional regulation of AteIF5A-2 was also evident when leaves were wounded with a hemostat. Strong expression of AteIF5A-2 protein was evident within 4 h of mechanically wounding the leaves with a hemostat, and this level of expression was sustained for up to 7 h (Fig. 1D). The transcript of AteIF5A-2 showed constitutive expression in a corresponding northern blot (data not shown).
Figure 1.
AteIF5A-2 is up-regulated by mechanical wounding and during compatible host-pathogen interaction. A, Western blots illustrating the isoform specificity of the AteIF5A antibodies. AteIF5A-1, -2, and -3 were expressed as GST fusion proteins in E. coli. The purified GST:AteIF5A proteins were subjected to SDS-PAGE, and the blots were probed with isoform-specific antibodies as indicated. Approximately 1 μg of purified protein was loaded for each sample. The lower band in the blot probed with anti-AteIF5A-2 is presumed to be a proteolytic degradation product. B, Western blots showing expression of the three isoforms of eIF5A, AteIF5A-1, AteIF5A-2, and AteIF5A-3, in Arabidopsis rosette leaves following infection with virulent Pst DC3000. A total of 10 μg of protein was loaded for each sample, and the blot was probed with antibodies raised against synthetic peptides unique to each isoform. Hours after infection are indicated. C, Uninfected control; M, mock infiltrated; Vir, infiltrated with virulent Pst DC3000. The corresponding SDS-PAGE for the AteIF5A-2 blot (large subunit of Rubisco only) is illustrated. C, Northern blot of total leaf RNA showing that AteIF5A-2 transcript is constitutively expressed and only marginally up-regulated following infection with virulent Pst DC3000. Hours after infection are indicated. C, Uninfected control; M, mock infiltrated; Vir, infiltrated with virulent Pst DC3000. The corresponding ethidium bromide-stained agarose gel is shown. D, Western blot of total leaf protein illustrating that AteIF5A-2 is up-regulated following mechanical wounding. A total of 10 μg of protein was loaded for each sample, and the blot was probed with AteIF5A-2-specific antibody. Hours after wounding are indicated. The corresponding SDS-PAGE (large subunit of Rubisco only) is illustrated.
AteIF5A-2 Protein and TUNEL Labeling Are Colocalized in Cells of Virulent Pst DC3000-Infected Leaves
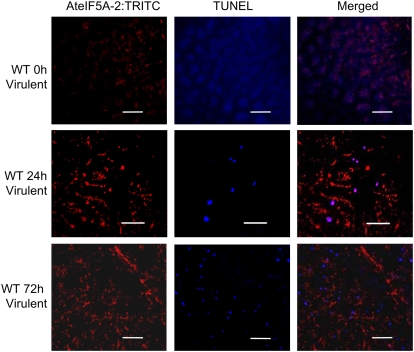
Infection of Arabidopsis leaves with virulent Pst DC3000 engenders symptoms of disease, including extensive chlorosis and necrosis reflecting programmed cell death (Katagiri et al., 2002; Nomura et al., 2005). To determine whether AteIF5A-2 expression is associated with programmed cell death induced by pathogen ingress, its expression following infection with virulent Pst DC3000 was spatially and temporally compared with the incidence of cell death by confocal microscopy. DNA fragmentation, a hallmark of programmed cell death, was visualized by terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) labeling, and AteIF5A-2 protein was detected by immunolocalization using isoform-specific antibodies and a tetramethyl rhodamine isothiocyanate (TRITC)-conjugated secondary antibody. TUNEL labeling, which depicts DNA fragmentation, is used routinely to detect mammalian cells undergoing apoptosis (Gorczyca et al., 1994) and has been adapted for detection of programmed cell death in plants (Wang et al., 1996). For example, TUNEL was recently used to demonstrate the anti-apoptotic phenotype of fumonisin-treated protoplasts derived from the Arabidopsis eIF5A-2 knockout mutant fbr12 (Feng et al., 2007). Leaves of wild-type Arabidopsis were infected with virulent Pst DC3000 by syringe inoculation, and at 24 and 72 h after inoculation, leaf sections were fixed, TUNEL labeled, and stained for AteIF5A-2 protein. Examination of these sections by confocal microscopy revealed strong up-regulation of AteIF5A-2 protein as well as induction of cell death visualized by TUNEL staining (Fig. 2). Moreover, when the images were merged, there was evidence for overlap of AteIF5A-2 protein staining and TUNEL labeling (Fig. 2). Because TUNEL labeling depicts fragmented DNA, this indicates that AteIF5A-2 is at least partially localized in nuclei and is consistent with mammalian cell studies signifying that eIF5A acts as a nucleocytoplasmic shuttling protein (Shi et al., 1996; Rosorius et al., 1999; Lipowsky et al., 2000; Jao and Chen, 2005). The incidence of TUNEL staining increased between 24 and 72 h after infection (Fig. 2), in keeping with the fact that within this time frame, virulent Pst DC3000 becomes necrotrophic (Katagiri et al., 2002). However, the coincidence of AteIF5A-2 and TUNEL signals decreased between 24 and 72 h after infection (Fig. 2), presumably reflecting degradation of eIF5A-2 protein along with other cellular proteins at later stages of cell death. It is apparent, therefore, that the up-regulation of AteIF5A-2 following infection correlates with the onset of programmed cell death attributable to compatible pathogenesis.
Figure 2.
AteIF5A-2 expression and TUNEL labeling in Arabidopsis leaf tissue infected with virulent Pst DC3000. Confocal images of leaf tissue discs harvested 0, 24, and 72 h postinoculation with virulent Pst DC3000 and stained for AteIF5A-2:TRITC and TUNEL, together with the corresponding merged images, are illustrated. Size bars = 50 μm.
Generation of Plants with Altered AteIF5A-2 Expression
To investigate further the role of AteIF5A-2 in programmed cell death induced by infection with virulent Pst DC3000, transgenic plants with suppressed and enhanced expression of AteIF5A-2 were generated. Antisense-suppressed (AS) lines were obtained by constitutive expression of the 3′-UTR of AteIF5A-2 in the antisense orientation. Overexpression (OE) lines were obtained by constitutive expression of the full-length AteIF5A-2 gene. There was strong inhibition of AteIF5A-2 up-regulation normally triggered by infection with virulent Pst DC3000 in the AS lines. Specifically, by 72 h postinfection, there was an abundance of AteIF5A-2 protein in infected leaves of wild-type plants, but the protein was not detectable in infected leaves of AS1 and AS2 and only barely detectable in infected leaves of AS3 and AS4 (Fig. 3A). Western blots of AteIF5A-2 OE9A and OE3F revealed strong expression of AteIF5A-2 protein in uninfected leaves compared to wild-type uninfected leaves (Fig. 3B). In addition, the levels of AteIF5A-2 protein in leaves of OE lines, both before and following infection with virulent Pst DC3000, were comparable to those in corresponding infected leaves of wild-type plants (Fig. 3C). These findings indicate that the apparent posttranscriptional regulation of AteIF5A-2 protein synthesis in wild-type plants is overcome by constitutive overexpression of AteIF5A-2. Of particular note as well is the fact that eIF5A-2 protein from infected wild-type and OE leaves migrates slightly faster than its counterpart in uninfected OE leaves (Fig. 3C), suggesting that it may be posttranslationally modified following infection.
Figure 3.
Modulation of AteIF5A-2 expression in transgenic plants. A, Western blot of total leaf protein extracted from wild-type (WT) plants and plants with suppressed AteIF5A-2 (lines AS1, AS2, AS3, and AS4) 72 h after infection with virulent Pst DC3000. A total of 10 μg of protein was loaded for each sample, and the blot was probed with AteIF5A-2-specific antiserum. The corresponding SDS-PAGE (large subunit of Rubisco only) is illustrated. B, Western blot of total leaf protein for wild-type (WT) plants and AteIF5A-2-overexpressing lines OE9A and OE3F showing constitutive expression of AteIF5A-2 protein in the OE lines. A total of 10 μg of protein was loaded for each sample, and the blot was probed with AteIF5A-2-specific antiserum. The corresponding SDS-PAGE (large subunit of Rubisco only) is illustrated. C, Western blot of total leaf protein illustrating AteIF5A-2 protein levels in wild-type leaves and leaves of the OE lines OE9A and OE3F at various times after infection with virulent Pst DC3000. A total of 10 μg of protein was loaded for each sample, and the blot was probed with AteIF5A-2-specific antiserum. The corresponding SDS-PAGE (large subunit of Rubisco only) is illustrated.
Down-Regulation of AteIF5A-2 Suppresses Bacterial Growth and Abrogates Disease Symptoms
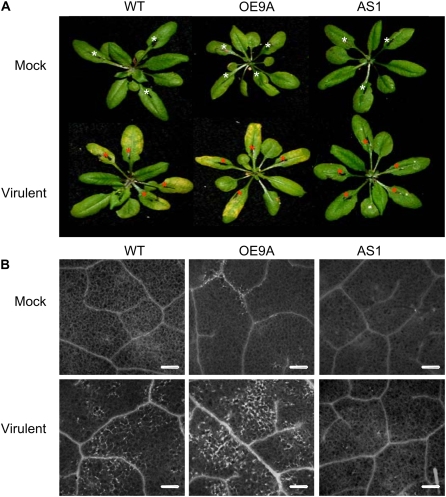
Suppression of AteIF5A-2 protein in the AS lines correlated with marked resistance to colonization by virulent Pst DC3000 (Fig. 4). Colonization was quantified by determining colony-forming units (CFU) per leaf disc immediately following infiltration of virulent Pst DC3000 and at 24 and 72 h after infection. At 24 h postinoculation, there was no significant difference in colonization of wild-type and transgenic plants (Fig. 4). The level of colonization 24 h after infection reflects initial growth of the pathogen within the apoplast (Katagiri et al., 2002). Subsequent rapid growth of the pathogen over the next 48 h is accompanied by host programmed cell death (Tao et al., 2003; Pozo et al., 2004; Nomura et al., 2005). This pattern of colonization was evident in both wild-type plants and AteIF5A-2-overexpressing plants in this study (Fig. 4). Indeed, the degree of colonization for the OE lines OE9A and OE3F was not materially different from that for wild-type plants at either 24 h or 72 h after infection, which is in keeping with the fact that the levels of eIF5A-2 expression were comparable in infected OE leaves and infected wild-type leaves (Fig. 3C). In both cases, CFU increased between 24 and 72 h after infection (Fig. 4), and this correlated with the development of chlorotic regions in the infected leaves reflecting programmed cell death (Fig. 5A). Of particular note, however, is the finding that necrotrophic growth of virulent Pst DC3000 was strongly inhibited in leaves of AteIF5A-2-suppressed lines. At 72 h after inoculation, CFU/leaf disc values were >94% lower for infected AS1 leaves (P < 0.01), infected AS2 leaves (P < 0.05), and infected AS3 leaves (P < 0.05) than for infected wild-type leaves (Fig. 4). Moreover, this inhibition of virulent Pst DC3000 colonization in the AS leaves correlated with curtailed development of chlorosis reflecting cell death. There was clear evidence of chlorosis in wild-type leaves within 72 h of infection with virulent Pst DC3000, whereas over the same time interval, there was no visual evidence of chlorosis in infected AS leaves (Fig. 5A). As well, necrotic regions visualized by fluorescence microscopy were clearly evident in infected wild-type leaves and infected OE leaves within 72 h of ingression of the pathogen, but were not detectable in infected AS leaves (Fig. 5B). The necrotic regions in infected OE leaves were similar to those in infected wild-type leaves but more extensive (Fig. 5B), possibly reflecting greater sensitization to the induction of programmed cell death due to constitutive expression of AteIF5A-2. Colonization of the leaves of empty vector control plants by virulent Pst DC3000 was comparable to that for leaves of wild-type plants, and infected leaves of empty vector (BIN) plants also became chlorotic (data not shown).
Figure 4.
Necrotrophic growth of virulent Pst DC3000 is inhibited in AteIF5A-2-suppressed lines. Pathogen growth was scored as CFU/leaf disc for wild-type plants, three AteIF5A-2 antisense lines (AS1, AS2, and AS3), and two AteIF5A-2-overexpressing lines (OE9A and OE3F) following infection with virulent Pst DC3000. Time after inoculation with the pathogen is indicated. CFU/leaf disc at 24 h reflects apoplastic growth. The increase in CFU/leaf disc between 24 and 72 h reflects necrotrophic growth. The insert illustrates the data for 72 h postinfection plotted on a linear scale. **, Significant difference at P < 0.01 compared to the mean for wild-type plants. *, Significant difference at P < 0.05 compared to the mean for wild-type plants.
Figure 5.
Photographs and fluorescence micrographs of leaves from wild-type and transgenic plants infected with virulent Pst DC3000. A, Photographs of representative rosettes illustrating chlorosis associated with Pst DC3000-induced disease in wild-type plants (WT), AteIF5A-2-overexpressing plants (line OE9A), and AteIF5A-2-suppressed plants (line AS1). Several leaves of each rosette were syringe infiltrated with a mock treatment (white asterisks) or virulent Pst DC3000 (red asterisks). The photographs were taken 72 h postinfection. B, Representative micrographs illustrating autofluorescence in necrotic leaf tissue 48 h after mock treatment (M) or Pst DC3000 infection of wild-type plants (WT), AteIF5A-2-overexpressing plants (line OE9A), and AteIF5A-2-suppressed plants (line AS1). Bars = 200 μm.
Transgenic Plants Overexpressing AteIF5A-2 Are Developmentally Compromised
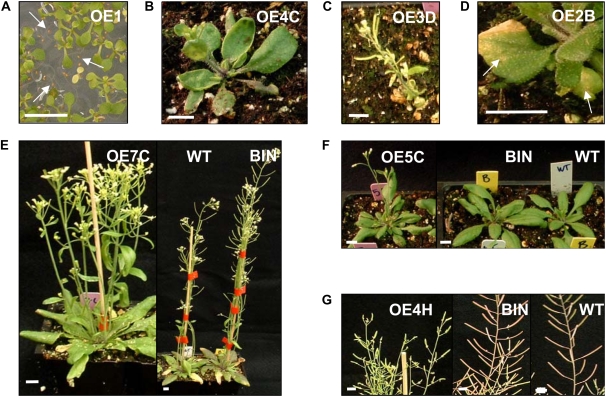
Transgenic plants with suppressed AteIF5A-2 proved to be morphologically indistinguishable from wild-type plants throughout growth and development. Inasmuch as fbr12, a knockout mutant of AteIF5A-2, exhibits defective growth (Feng et al., 2007), this presumably reflects the fact that AteIF5A-2 is only partially suppressed in the antisense lines developed in this study. For example, although eIF5A-2 protein is not visible in the western blots for infected AS1 and AS2 plants, it is clearly visible in corresponding blots for infected AS4 plants and slightly visible for infected AS3 plants (Fig. 3A). AteIF5A-2-overexpressing lines, however, exhibited a strong developmental phenotype. The growth and developmental patterns of 14 OE lines were examined. For most of these lines, the T1 plants were severely stunted, and for one-third of the lines, the T1 plants did not survive to produce seed. For those that did survive, the stunted, developmentally compromised phenotypes were of varying intensity (Fig. 6). Lines OE1, OE4, OE6, OE8, OE10, OE11, OE12, OE13, and OE14 were severely stunted, being <50% the size of wild-type or BIN controls (Fig. 6A), and lines OE6, OE8, OE10, OE13, and OE14 did not grow past the rosette stage and did not produce any seed (Fig. 6B). Line OE12, although severely stunted, was able to send up a bolt and produce a few seeds (Fig. 6B). Lines OE2 and OE3 were moderately stunted (between 50% and 75% the size of wild-type or BIN controls), and lines OE5, OE7, and OE9 grew similarly to wild-type or BIN control plants (Fig. 6A). Plants exhibiting stunted growth also had yellow and curled leaves, purple cotyledons, and abnormal flower morphology. The developmentally compromised phenotype continued in T2 plants for those lines that produced seed in the T1 generation (Fig. 7). However, 30% to 40% of the T2 seeds for lines OE1, OE2, OE3, OE4, and OE12 did not germinate. This is illustrated for line OE1 in Figure 7A and likely reflects ovule abortion or inhibited embryo growth. Several phenotypes were also evident in the vegetative phase of growth for T2 generation plants, including curled leaves (Fig. 7B), severe stunting (Fig. 7C), and chlorosis and spontaneous lesion formation (Fig. 7D). Other phenotypes included an increase in basal branches (Fig. 7E), early bolting (Fig. 7F), and small stunted siliques (Fig. 7G).
Figure 6.
Photographs of T1 plants overexpressing AteIF5A-2. A, Four-week-old wild-type plants (WT), empty binary vector control plants (BIN), and T1 plants of AteIF5A-2-overexpressing lines OE1 through OE14. B, Four-week-old plants of AteIF5A-2-overexpressing lines that showed severe stunting at 4 weeks of age. Bar = 1 cm.
Figure 7.
Phenotypes of T2 plants overexpressing AteIF5A-2. A, Compromised germination of 10-d-old plants of the OE line OE1. The arrows indicate seeds that did not germinate. B, Curled leaf phenotype of 3-week-old plants of the OE line OE4C. C, Severely stunted phenotype of 5-week-old plants of the OE line OE3D. D, Necrotic lesions (indicated by white arrows) and premature chlorosis of 5-week-old plants of the OE line OE2B. E, Enhanced basal inflorescence branching in 5-week-old plants of the OE line OE7C. F, Early bolting (by a week) in 3-week-old plants of the OE line OE5C. G, Short stunted siliques of 7-week-old plants of the OE line OE4H. Bars = 1 cm.
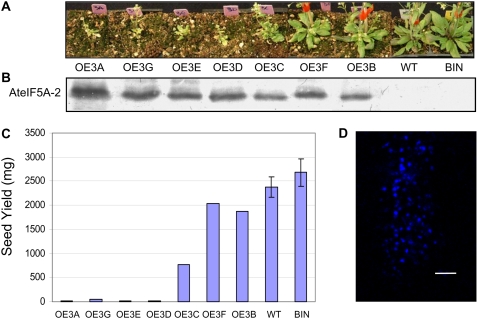
Western-blot analysis confirmed that the intensity of the stunted phenotype is correlated with the degree of AteIF5A-2 overexpression. Specifically, higher levels of overexpression resulted in more severely stunted growth (Fig. 8, A and B). There was also a correlation between degree of AteIF5A-2 overexpression and reduction in seed yield. Plants with high levels of AteIF5A-2 had severely compromised fecundity and produced very little seed (Fig. 8, B and C). TUNEL labeling depicting cell death in uninfected AteIF5A-2-overexpressing leaf tissue (OE3F) is illustrated in Figure 8D, an observation that is consistent with the anti-apoptotic phenotype of the AteIF5A-2 knockout mutant characterized by Feng et al. (2007).
Figure 8.
Photographs of plants overexpressing AteIF5A-2 and corresponding levels of AteIF5A-2 expression. A, Photographs of plants at 5 weeks of age overexpressing AteIF5A-2. Plants are ordered from the most stunted to the least stunted, with the wild-type (WT) and binary vector (BIN) controls on the far right. B, Western blot of total leaf protein for each of the lines illustrated in A. Five micrograms of protein was loaded for each sample, and the blot was probed with AteIF5A-2-specific antibody. C, Seed yield for each of the lines illustrated in A. ses of the means are shown for wild-type and BIN plants; n = 10. D, Uninfected OE1 leaves stained with TUNEL. Bar = 50 μm.
DISCUSSION
It is generally recognized that signaling associated with a compatible host-pathogen interaction induces programmed cell death (Beers and McDowell, 2001; Greenberg and Yao, 2004; Pozo et al., 2004). Two lines of evidence indicate that AteIF5A-2 plays a role in the induction of programmed cell death following ingression of virulent Pst DC3000 into rosette leaves of Arabidopsis. First, TUNEL fluorescence reflecting the onset of cell death and TRITC fluorescence reflecting the presence of AteIF5A-2 both increased with time after infection and proved to be spatially coincident in the cells of infected leaves. TUNEL labeling depicts DNA fragmentation and is considered to be a better index of programmed cell death than markers such as fluorescein diacetate or Evan's Blue (Ning et al., 2002). Up-regulation of AteIF5A-2 and its colocalization with TUNEL fluorescence were evident within 24 h of infection, before chlorosis was apparent, suggesting that AteIF5A-2 plays a role in the induction of cell death. Levels of the protein continued to increase between 24 and 72 h after infection, coincident with the progression of cell death in the infected leaves manifesting as diffuse chlorosis and necrotic lesions, which are the typical symptoms of virulent Pst DC3000-induced disease. That immunolabeled AteIF5A-2 protein colocalizes with TUNEL-positive nuclei in infected cells is consistent with the proposal emanating from studies with mammalian cells that eIF5A may function as a nucleocytoplasmic shuttle, recruiting and translocating specific mRNAs out of the nucleus into the cytoplasm for subsequent translation (Shi et al., 1996; Rosorius et al., 1999; Lipowsky et al., 2000; Jao and Chen, 2005). It is also consistent with reports that there is an accumulation of eIF5A within the nuclei of mammalian cells undergoing apoptosis (Tome and Gerner, 1997; Tome et al., 1997; Beninati et al., 1998; Caraglia et al., 2003; Jin et al., 2003; Taylor et al., 2007).
The second line of evidence supporting a role for AteIF5A-2 in the induction of programmed cell death following a compatible host-pathogen interaction is the finding that antisense suppression of AteIF5A-2 inhibited colonization of virulent Pst DC3000 in infected leaves and curtailed the development of disease symptoms. Suppression of AteIF5A-2 had no effect on the initial limited growth of the pathogen during the first 24 h after syringe infiltration into leaves. Virulent Pst DC3000 colonization reached comparable levels for both control plants and AteIF5A-2-suppressed transgenic plants during this period. However, subsequent more rapid growth of the pathogen, which leads to the development of disease symptoms and is thought to be attributable to the induction of host cell death (Abramovitch and Martin, 2004), was curtailed in the AteIF5A-2-suppressed plants. For infected wild-type and empty vector control plants, colonization of virulent Pst DC3000 in infected leaves increased strongly between 24 and 72 h after infection, whereas for the transgenic lines, this increase in colonization attributable to necrotrophic growth was inhibited by >90%. In keeping with this, infected leaves of the transgenic plants did not become chlorotic, indicating that the development of disease symptoms accompanying necrotrophic growth of the pathogen had been abrogated. Virulent Pst DC3000 colonization of AteIF5A-2-overexpressing lines was comparable to that for wild-type plants, indicating that necrotrophic growth of the pathogen in wild-type plants is not limited by the availability of endogenous AteIF5A-2. Consistent with this, the development of chlorosis was comparable in infected wild-type leaves and infected AteIF5A-2-overexpressing leaves.
There is evidence that the timing of host programmed cell death induced by facultative necrotrophic pathogens such as Pst DC3000 is a critical determinant of the ensuing disease state (Abramovitch and Martin, 2004; Pozo et al., 2004). For example, early HR-based host cell death and the associated development of systemic acquired resistance following infection with avirulent Pst DC3000 prevent the development of disease symptoms (Katagiri et al., 2002; Pozo et al., 2004). This study indicates that AteIF5A-2 may regulate the induction of this later onset of host cell death that underlies the development of disease symptoms. Moreover, recent evidence indicates that host cell death leading to development of the disease state has features in common with mammalian cell apoptosis (Abramovitch and Martin, 2004; Pozo et al., 2004). This contention is supported by the finding in this study that the nuclei of dying cells in leaves infected with virulent Pst DC3000 are TUNEL-positive. TUNEL staining depicts DNA fragmentation, which is a signature feature of mammalian cell apoptosis (Gorczyca et al., 1994). In light of this, the growing evidence that human eIF5A induces apoptosis (Tome and Gerner, 1997; Tome et al., 1997; Beninati et al., 1998; Caraglia et al., 2003; Li et al., 2004; Taylor et al., 2007) lends further support to the contention in this study that AteIF5A-2 regulates the induction of programmed cell death associated with the ingression of virulent Pst DC3000 in Arabidopsis leaves.
AteIF5A-2-suppressed transgenic plants were indistinguishable from wild-type plants during growth and development. However, AteIF5A-2-overexpressing plants were severely stunted and exhibited delayed development suggestive of hormonal imbalance. Moreover, the intensity of this phenotype correlated with the degree of AteIF5A-2 overexpression. Specifically, the lines that were most stunted had the highest level of AteIF5A-2 expression and produced very few seeds, whereas lines that were less stunted had lower levels of AteIF5A-2 protein and were capable of producing more seed. First-generation OE lines exhibited a 33% lethal phenotype. Plants that survived displayed phenotypes seemingly attributable to constitutive initiation of cell death that resemble those of spontaneous cell death mutants (Dietrich et al., 1994; Pilloff et al., 2002). These phenotypes included decreased fitness, early bolting, severe stunting, spontaneous necrotic lesions, and increased basal inflorescence branching, developmental phenotypes documented previously for plants that are chronically infected with Pseudomonas (Korves and Bergelson, 2003) or Rhodococcus fascians (Manes et al., 2004). Inasmuch as suppression of DHS has been shown to delay leaf senescence (Wang et al.,2003), it is conceivable that the phenotype of the AteIF5A-2-overexpressing plants is attributable to the hypusinated form of the protein. However, unlike certain cell death mutants (Dietrich et al., 1994; Pilloff et al., 2002; Senda and Ogawa, 2004), the AteIF5A-2-overexpressing lines did not show resistance to pathogen-induced disease. Indeed, the AteIF5A-2-overexpressing lines were equally as susceptible to virulent Pst DC3000-induced disease as wild-type plants. There is some overlap of resistance and cell death signaling pathways (Heath, 2000), but it would appear that AteIF5A-2 has no direct role in resistance. Rather, it appears to be involved in potentiating programmed cell death accompanying compatible host-pathogen interaction. Other positive regulators of cell death have also been implicated in the development of plant disease. For example, MAPKKKα-suppressed tobacco (Nicotiana tabacum) leaves show resistance to virulent P. syringae pv tabaci-induced disease that correlates with reduced pathogen colonization and reduced host cell death (Pozo et al., 2004).
The expression of AteIF5A-2 appears to be posttranscriptionally regulated in that, although its mRNA is constitutively present in Arabidopsis leaves, the protein is only expressed in the event of pathogen ingression or wounding. However, in AteIF5A-2-overexpressing plants, high levels of leaf AteIF5A-2 protein were constitutively present. It is not clear why the apparent posttranscriptional control of AteIF5A-2 expression in wild-type plants is overridden in AteIF5A-2-overexpressing lines, but it may reflect the fact that the regulatory mechanisms inhibiting its translation are overwhelmed in the OE plants. That AteIF5A-2 was shown to be up-regulated following mechanical injury of Arabidopsis leaves in this study suggests that it may also be involved in wounding-induced cell death. Wounding is a common stress engendered by weather or insect feeding (Cheong et al., 2002). Consistent with the fact that damaged plant tissue provides entrance points for pathogen invasion, wounding and pathogen responses share common signaling elements (Maleck and Dietrich, 1999; Mayrose et al., 2006). The finding that AteIF5A-2 is up-regulated within 4 h of mechanical wounding and also following ingression with virulent Pst DC3000 suggests that it may be part of the common signaling network invoked by wounding and pathogenesis.
MATERIALS AND METHODS
Plant Material
Seeds of wild-type Arabidopsis (Arabidopsis thaliana) ecotype Columbia were sown in soil (Premier Pro-Mix BX; Premier Horticulture) and maintained in a growth chamber at 20°C under a long-day photoperiod (16 h light and 8 h darkness) at 75% relative humidity. Four-week-old plants were infected with the bacterial pathogen Pseudomonas syringae pv tomato DC3000 pVSP61 (virulent Pst DC3000), a kind gift from Dr. Robin Cameron, McMaster University, Hamilton, ON, Canada. For inoculation, bacterial cultures were grown to mid-log growth phase (OD600 = 0.15–0.3) at 28°C in King's B media containing kanamycin (50 μg/mL), pelleted, and resuspended at a concentration of 106 CFU/mL in 10 mm MgCl2. Rosette leaves of 4-week-old plants were inoculated with 10 mm MgCl2 (mock inoculation) or virulent Pst DC3000 (106 CFU/mL in 10 mm MgCl2) by pressure infiltration into the abaxial surface using a 1-mL syringe without a needle until the apoplast was filled. Inoculated leaves were labeled with a permanent marker. The treated plants were maintained in the growth chamber until used for analysis.
In Planta Bacterial Growth Assays and Mechanical Wounding
In planta bacterial growth in infected leaves was quantified according to Katagiri et al. (2002). The leaves were surface-sterilized in 50% ethanol and rinsed twice with sterile water. Eight leaf discs, 0.4 cm in diameter, per replicate were excised using a cork borer and placed in an Eppendorf tube. The leaf discs were homogenized in 500 μL of 10 mm MgCl2 using a drill fitted with a plastic bit, and the resulting homogenates were diluted 10−2 and 10−4 with 10 mm MgCl2. A 0.1-mL sample of each dilution was distributed evenly on a King's B rifampicin/kanamycin plate. Each assay was replicated three times. The plates were maintained in darkness at 25°C, and the CFU were determined after 2 d.
Three-week-old rosette leaves were mechanically wounded by crushing with a hemostat alongside the midvein (Orozco-Cardenas and Ryan, 1999). The wounded leaves were collected at 0, 2, 4, 5, 6, 7, and 8 h after wounding. All tissue was flash-frozen in liquid nitrogen and stored at −80°C until used for protein extraction for western blotting.
Altered Expression of AteIF5A-2 in Transgenic Plants
Suppression of endogenous AteIF5A-2 (At1g26630) was achieved by constitutively expressing the 3′-UTR of the AteIF5A-2 gene in the antisense orientation in transgenic plants. A cDNA fragment corresponding to the 3′-UTR of AteIF5A-2 was obtained by PCR using genomic DNA as a template and the upstream primer 5′-GAGCTCGGTGGCAAGTAAACAAGTATCATTCG-3′ containing a SacI restriction site (underlined) and the downstream primer 5′-CTCGAGAAGAATAACATCTCATAAGAAAC-3′ containing a XhoI restriction site (underlined). Genomic DNA was isolated using a Promega Wizard Genomic DNA Purification kit according to the manufacturer's instructions. The PCR fragment was digested with SacI and XhoI and subcloned into the binary vector pKYLX71-35S2 (Schardl et al., 1987) in the antisense orientation under the regulation of two copies of the constitutive cauliflower mosaic virus promoter 35S. The pKYLX71-35S2 vector contains a tetracycline-resistance gene, a bacterial replication region, as well as a kanamycin-resistance gene (NPT II) within the T-DNA region. Binary vector expressing antisense 3′-UTR of AteIF5A-2 as well as binary vector alone were introduced into Agrobacterium tumefaciens GV3010 by electroporation, and 4-week-old Arabidopsis plants were infected with transformed A. tumefaciens by vacuum infiltration (Bechtold et al., 1993). The vacuum-infiltrated plants were grown to maturity and the seed harvested. Homozygous transgenic lines were obtained by screening seed from successive generations on kanamycin-containing media. AteIF5A-2-overexpressing lines were obtained in the same manner using full-length AteIF5A-2 genomic DNA in the sense orientation as the transgene. Full-length AteIF5A-2 DNA was obtained by PCR using genomic DNA as the template and the upstream primer 5′-CTCGAGTGCTCACTTCTCTCTCTTAGG-3′ containing a XhoI restriction site (underlined) and the downstream primer 5′-GAGCTCAAGAATAACATCTCATAAGAAAC-3′ containing a SacI restriction site (underlined).
Molecular and Biochemical Analyses
For northern blotting, total leaf RNA was isolated using a Qiagen kit according to manufacturer's instructions, fractionated on 1.0% denaturing formaldehyde agarose gels, and immobilized on Hybond-N+ nylon membrane (Amersham-Pharmacia Biotech). The blots were probed with cDNA corresponding to the 3′-UTR of AteIF5A-2 (GenBank accession no. At1g26630) using hybridization conditions described previously (Wang et al., 2001).
For western blotting, total leaf protein was isolated according to Kaup et al. (2002), fractionated on 12% SDS polyacrylamide gels, and the separated proteins transferred to polyvinylidene difluoride membrane (Bio-Rad). Immunoblotting was carried out using AteIF5A isoform-specific antisera from rabbit as the primary antibody and alkaline phosphatase-conjugated secondary antibody (Roche). There are three eIF5A gene family members in the Arabidopsis genome: AteIF5A-1 (At1g13950), AteIF5A-2 (At1g26630), and AteIF5A-3 (At1g69410). Antisera were raised against a unique synthetic peptide for each of the three isoforms (Liu et al., 2008). The peptides were synthesized in the laboratory of Dr. G. Lajoie (University of Western Ontario). A Cys residue was added to the N terminus of each peptide to enable conjugation to a carrier protein, keyhole limpet hemocyanin, prior to being used as an antigen for polyclonal antibody production (Drenckhahn et al., 1993). Antibodies were raised in rabbits using Freund's complete adjuvant for the first immunization and Freund's incomplete adjuvant for two booster injections, and purified using an affinity SulfoLink coupling gel column (Pierce) according to the manufacturer's instructions.
Specificity of the AteIF5A antibodies was confirmed by conducting western blots of purified glutathione S-transferase (GST):AteIF5A-1, GST:AteIF5A-2, and GST:AteIF5A-3 recombinant proteins. cDNAs corresponding to AteIF5A-1, AteIF5A-2, and AteIF5A-3 were cloned into pGEX-5X-3 as described previously (Wang et al., 2001). The GST fusion proteins were overexpressed in Escherichia coli DH5α and purified by glutathione-Sepharose 4B chromatography.
Fluorescence Microscopy
Leaves harvested at specified time points were fixed in 4% (w/v) paraformaldehyde in phosphate buffered saline (PBS), pH 7.0, under vacuum for 1 h. The fixed leaf tissue was then rinsed in PBS twice and gradually dehydrated to 100% ethanol and rehydrated back to 100% water. The leaf tissue was then mounted on slides in 70% glycerol. Autofluorescence of necrotic lesions was visualized by fluorescence microscopy as done by Dietrich et al. (1994).
Confocal Microscopy
Leaf discs were cut with a cork borer (0.4 cm diameter), vacuum infiltrated with fixing buffer (4% paraformaldehyde in PBS) for 10 min, stained for DNA fragmentation by TUNEL, and immunostained for AteIF5A-2 expression. For TUNEL labeling, the Promega DeadEnd fluorometric TUNEL system was used according to the manufacturer's specifications. The samples were then washed with PBS twice for 10 min at room temperature, treated with primary antibody against AteIF5A-2 (1:50) in 1% bovine serum albumin (v/w) followed by goat anti-rabbit secondary antibody conjugated to TRITC (Sigma; 1:100 in 1% bovine serum albumin [w/v] in PBS), and mounted on slides in 70% glycerol. For optimal visualization by confocal microscopy, number 1.5 cover slips were used. The samples were observed using a Zeiss LSM 510 confocal laser scanning microscope attached to an axiovert-inverted microscope.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers Q93VP3 and BE039424.
Acknowledgments
We are grateful to Dr. Robin Cameron, Department of Biology, McMaster University, for providing cultures of Pseudomonas syringae and for reviewing this manuscript.
This work was supported by the Natural Sciences and Engineering Research Council of Canada.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: John E. Thompson (jet@sciborg.uwaterloo.ca).
Open Access articles can be viewed online without a subscription.
References
- Abbruzzese A, Park MH, Folk JE (1986) Deoxyhypusine hydroxylase from rat testis: partial purification and characterization. J Biol Chem 261 3085–3089 [PubMed] [Google Scholar]
- Abramovitch RB, Martin GB (2004) Strategies used by bacterial pathogens to suppress plant defenses. Curr Opin Plant Biol 7 356–364 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. Life Sci 316 1194–1199 [DOI] [PubMed] [Google Scholar]
- Beers EP, McDowell JM (2001) Regulation and execution of programmed cell death in response to pathogen, stress and developmental cues. Curr Opin Plant Biol 4 561–567 [DOI] [PubMed] [Google Scholar]
- Beninati S, Gentile V, Caraglia M, Lentini A, Tagliaferri P, Abbruzzese A (1998) Tissue transglutaminase expression affects hypusine metabolism in BALB/c 3T3 cells. FEBS Lett 437 34–38 [DOI] [PubMed] [Google Scholar]
- Caraglia M, Marra M, Giuberti G, D'Alessandro AM, Baldi A, Tassone P, Venuta S, Tagliaferri P, Abbruzzese A (2003) The eukaryotic initiation factor 5A is involved in the regulation of proliferation and apoptosis induced by interferon-alpha and EGF in human cancer cells. J Biochem (Tokyo) 133 757–765 [DOI] [PubMed] [Google Scholar]
- Caraglia M, Marra M, Giuberti G, D'Alessandro AM, Budillon A, Prete SD, Lentini A, Beninati S, Abbruzzese A (2001) The role of eukaryotic initiation factor 5A in the control of cell proliferation and apoptosis. Amino Acids 20 91–104 [DOI] [PubMed] [Google Scholar]
- Cheong YH, Chang HS, Gupta R, Wang X, Zhu T, Luan S (2002) Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol 129 661–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou WC, Huang YW, Tsay WS, Chiang TY, Huang DD, Huang HJ (2004) Expression of genes encoding the rice translation initiation factor, eIF5A, is involved in developmental and environmental responses. Physiol Plant 121 50–57 [DOI] [PubMed] [Google Scholar]
- Cooper HL, Park MH, Folk JE (1982) Posttranslational formation of hypusine in a single major protein occurs generally in growing cells and is associated with activation of lymphocyte growth. Cell 29 791–797 [DOI] [PubMed] [Google Scholar]
- Dietrich RA, Delaney TP, Uknes SJ, Ward ER, Ryals JA, Dangl JL (1994) Arabidopsis mutants simulating disease resistance response. Cell 77 565–577 [DOI] [PubMed] [Google Scholar]
- Drenckhahn D, Joens T, Schmitz F (1993) Production of polyclonal antibodies against proteins and peptides. In DJ Asai, ed, Methods in Cell Biology, Vol 37. Academic Press, New York, pp 7–56 [DOI] [PubMed]
- Duguay J, Jamal S, Liu Z, Wang TW, Thompson JE (2006) Leaf-specific suppression of deoxyhypusine synthase in Arabidopsis thaliana enhances growth without negative pleiotropic effects. J Plant Physiol 164 408–420 [DOI] [PubMed] [Google Scholar]
- Feng H, Chen Q, Feng J, Zhang J, Yang X, Zuo J (2007) Functional characterization of the Arabidopsis eukaryotic translation initiation factor 5A-2 that plays a crucial role in plant growth and development by regulating cell division, cell growth, and cell death. Plant Physiol 144 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorczyca W, Tuziak T, Kram A, Melamed MR, Darzynkiewicz Z (1994) Detection of apoptosis-associated DNA strand breaks in fine-needle aspiration biopsies by in situ end labeling of fragmented DNA. Cytometry 15 169–175 [DOI] [PubMed] [Google Scholar]
- Greenberg JT, Gao A, Klessig DF, Ausubel FM (1994) Programmed cell death in plants: a pathogen-triggered response activated coordinately with multiple defense functions. Cell 77 551–563 [DOI] [PubMed] [Google Scholar]
- Greenberg JT, Yao N (2004) The role and regulation of programmed cell death in plant-pathogen interactions. Cell Microbiol 6 201–211 [DOI] [PubMed] [Google Scholar]
- Hanauske-Abel HM, Park MH, Hanauske AR, Popowicz AM, Lalande M, Folk JE (1994) Inhibition of the G1-S transition of the cell cycle by inhibitors of deoxyhypusine hydroxylation. Biochim Biophys Acta 1221 115–124 [DOI] [PubMed] [Google Scholar]
- Hatsugai N, Kuroyanagi M, Yamada K, Meshi T, Tsuda S, Kondo M, Nishimura M, Hara-Nishimura I (2004) A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science 305 855–858 [DOI] [PubMed] [Google Scholar]
- Heath MC (2000) Hypersensitive response-related death. Plant Mol Biol 44 321–334 [DOI] [PubMed] [Google Scholar]
- Hopkins MT, Taylor C, Liu Z, Ma F, McNamara L, Wang TW, Thompson JE (2007) Regulation and execution of molecular disassembly and catabolism during senescence. New Phytol 175 201–214 [DOI] [PubMed] [Google Scholar]
- Jao DL, Chen KY (2005) Tandem affinity purification revealed the hypusine-dependent binding of eukaryotic initiation factor 5A to the translating 80S ribosomal complex. J Cell Biochem 97 583–598 [DOI] [PubMed] [Google Scholar]
- Jin BF, He K, Wang HX, Wang J, Zhou T, Lan Y, Hu MR, Wei KH, Yang SC, Shen BF, et al (2003) Proteomic analysis of ubiquitin-proteasome effects: insight into the function of eukaryotic initiation factor 5A. Oncogene 22 4819–4830 [DOI] [PubMed] [Google Scholar]
- Katagiri F, Thilmony R, He SY (2002) The Arabidopsis thaliana-Pseudomonas syringae interaction. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, pp 1–35 [DOI] [PMC free article] [PubMed]
- Kaup MT, Froese CD, Thompson JE (2002) A role for diacylglycerol acyltransferase during leaf senescence. Plant Physiol 129 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korves TM, Bergelson J (2003) A developmental response to pathogen infection in Arabidopsis. Plant Physiol 133 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li AL, Li HY, Jin BF, Ye QN, Zhou T, Yu XD, Pan X, Man JH, He K, Yu M, et al (2004) A novel eIF5A complex functions as a regulator of p53 and p53-dependent apoptosis. J Biol Chem 279 49251–49258 [DOI] [PubMed] [Google Scholar]
- Lipowsky G, Bischoff FR, Schwarzmaier P, Kraft R, Kostka S, Hartmann E, Kutay U, Gorlich D (2000) Exportin 4: a mediator of a novel nuclear export pathway in higher eukaryotes. EMBO J 19 4362–4371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Duguay J, Ma F, Wang TW, Tshin R, Hopkins MT, McNamara L, Thompson J (2008) Modulation of eIF5A1 expression alters xylem abundance in Arabidopsis thaliana. J Exp Bot 59 939–950 [DOI] [PubMed] [Google Scholar]
- Maleck K, Dietrich RA (1999) Defense on multiple fronts: How do plants cope with diverse enemies? Trends Plant Sci 4 215–219 [DOI] [PubMed] [Google Scholar]
- Manes CL, Beeckman T, Ritsema T, Montagu VV, Goethals K, Holsters M (2004) Phenotypic alterations in Arabidopsis thaliana plants caused by Rhodococcus fascians infection. J Plant Res 117 139–145 [DOI] [PubMed] [Google Scholar]
- Mayrose M, Ekengren SK, Melech-Bonfil S, Martin GB, Sessa G (2006) A novel link between tomato GRAS genes, plant disease resistance and mechanical stress response. Mol Plant Pathol 7 593–604 [DOI] [PubMed] [Google Scholar]
- Ning SB, Wang L, Song YC (2002) Identification of programmed cell death in situ in individual plant cells in vivo using a chromosome preparation technique. J Exp Bot 53 651–658 [DOI] [PubMed] [Google Scholar]
- Nomura K, Melotto M, He SY (2005) Suppression of host defense in compatible plant-Pseudomonas syringae interactions. Curr Opin Plant Biol 8 361–368 [DOI] [PubMed] [Google Scholar]
- Ober D, Hartmann T (1999) Deoxyhypusine synthase from tobacco. J Biol Chem 274 32040–32047 [DOI] [PubMed] [Google Scholar]
- Orozco-Cardenas M, Ryan CA (1999) Hydrogen peroxide is generated systemically in plant leaves by wounding and systemin via the octadecanoid pathway. Proc Natl Acad Sci USA 96 6553–6557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Aravind L, Wolff EC, Kaevel J, Kim YS, Park MH (2006) Molecular cloning, expression, and structural prediction of deoxyhypusine hydroxylase: a HEAT-repeat-containing metalloenxyme. Proc Natl Acad Sci USA 103 51–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH, Cooper HL, Folk JE (1981) Identification of hypusine, an unusual amino acid, in a protein from human lymphocytes and of spermidine as its biosynthetic precursor. Proc Natl Acad Sci USA 78 2869–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilloff RK, Devadas SK, Enyedi A, Raina R (2002) The Arabidopsis gain-of-function mutant dll1 spontaneously develops lesions mimicking cell death associated with disease. Plant J 30 61–70 [DOI] [PubMed] [Google Scholar]
- Pozo OD, Pedley KF, Martin GB (2004) MAPKKKα is a positive regulator of cell death associated with both plant immunity and disease. EMBO J 23 3072–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosorius O, Reichart B, Kratzer F, Heger P, Dabauvalle MC, Hauber J (1999) Nuclear pore localization and nucleocytoplasmic transport of eIF-5A: evidence for direct interaction with the export receptor CRM1. J Cell Sci 112 2369–2380 [DOI] [PubMed] [Google Scholar]
- Schardl CL, Byrd AD, Benzion G, Altschuler MA, Hildebrand DF, Hunt AG (1987) Design and construction of a versatile system for the expression of foreign genes in plants. Gene 61 1–11 [DOI] [PubMed] [Google Scholar]
- Senda K, Ogawa K (2004) Induction of PR-1 accumulation accompanied by runaway cell death in the lsd1 mutant of Arabidopsis is dependent on glutathione levels but independent of the redox state of glutathione. Plant Cell Physiol 45 1578–1585 [DOI] [PubMed] [Google Scholar]
- Shi XP, Yin KC, Zimolo ZA, Stern AM, Waxman L (1996) The subcellular distribution of eukaryotic translation initiation factor, eIF-5A, in cultured cells. Exp Cell Res 225 348–356 [DOI] [PubMed] [Google Scholar]
- Tao Y, Xie Z, Chen W, Glazebrook J, Chang HS, Han B, Zhu T, Zou G, Katagiri F (2003) Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CA, Sun Z, Cliche DO, Ming H, Eshaque B, Jin S, Hopkins MT, Thai B, Thompson JE (2007) Eukaryotic translation initiation factor 5A induces apoptosis in colon cancer cells and associates with the nucleus in response to tumour necrosis factor alpha signaling. Exp Cell Res 313 437–449 [DOI] [PubMed] [Google Scholar]
- Thompson JE, Hopkins MT, Taylor C, Wang TW (2004) Regulation of senescence by eukaryotic translation initiation factor 5A: implications for plant growth and development. Trends Plant Sci 9 174–179 [DOI] [PubMed] [Google Scholar]
- Tome ME, Fiser SM, Payne CM, Gerner EW (1997) Excess putrescine accumulation inhibits the formation of modified eukaryotic initiation factor 5A (eIF-5A) and induces apoptosis. Biochem J 328 847–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tome ME, Gerner EW (1997) Cellular eukaryotic initiation factor 5A content as a mediator of polyamine effects on growth and apoptosis. Biol Signals 6 150–156 [DOI] [PubMed] [Google Scholar]
- Wang H, Li J, Bostock RM, Gilchrist DG (1996) Apoptosis: a functional paradigm for programmed plant cell death induced by a host-selective phytotoxin and invoked during development. Plant Cell 8 375–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TW, Lu L, Wang D, Thompson JE (2001) Isolation and characterization of senescence-induced cDNAs encoding deoxyhypusine synthase and eucaryotic translation initiation factor 5A from tomato. J Biol Chem 276 17541–17549 [DOI] [PubMed] [Google Scholar]
- Wang TW, Lu L, Zhang CG, Taylor CA, Thompson JE (2003) Pleiotropic effects of suppressing deoxyhypusine synthase expression in Arabidopsis thaliana. Plant Mol Biol 52 1223–1235 [DOI] [PubMed] [Google Scholar]
- Wolff EC, Folk JE, Park MH (1997) Enzyme-substrate intermediate formation at lysine 329 of human deoxyhypusine synthase. J Biol Chem 272 15865–15871 [DOI] [PubMed] [Google Scholar]
- Yao N, Imai S, Tada Y, Nakayashiki H, Tosa Y, Park P (2002) Apoptotic cell death is a common response to pathogen attack in oats. Mol Plant Microbe Interact 15 1000–1007 [DOI] [PubMed] [Google Scholar]
- Zhang C, Gutsche AT, Shapiro AD (2004) Feedback control of the Arabidopsis hypersensitive response. Mol Plant Microbe Interact 17 357–365 [DOI] [PubMed] [Google Scholar]