Abstract
Protein ubiquitination is a posttranslational regulatory process essential for plant growth and interaction with the environment. E3 ligases, to which the seven in absentia (SINA) proteins belong, determine the specificity by selecting the target proteins for ubiquitination. SINA proteins are found in animals as well as in plants, and a small gene family with highly related members has been identified in the genome of rice (Oryza sativa), Arabidopsis (Arabidopsis thaliana), Medicago truncatula, and poplar (Populus trichocarpa). To acquire insight into the function of SINA proteins in nodulation, a dominant negative form of the Arabidopsis SINAT5 was ectopically expressed in the model legume M. truncatula. After rhizobial inoculation of the 35S:SINAT5DN transgenic plants, fewer nodules were formed than in control plants, and most nodules remained small and white, a sign of impaired symbiosis. Defects in rhizobial infection and symbiosome formation were observed by extensive microscopic analysis. Besides the nodulation phenotype, transgenic plants were affected in shoot growth, leaf size, and lateral root number. This work illustrates a function for SINA E3 ligases in a broad spectrum of plant developmental processes, including nodulation.
Ubiquitination of regulatory proteins plays an important role in plant development and responses to the environment (Ellis et al., 2002; Devoto et al., 2003; Hare et al., 2003; Zeng et al., 2006). Polyubiquitination is mediated by repeated sequential action of three enzymes that activate (E1), conjugate (E2), and finally ligate (E3) ubiquitin (Ub) to target proteins that are subsequently degraded via the 26S proteasome (Glickman and Ciechanover, 2002). The E3 ligases confer the degradation specificity by selecting the target proteins. Several types have been identified, such as the anaphase-promoting complex, the Skp-Cullin-F-box complex, the E6-associated protein carboxyl terminus (HECT) domain E3 ligases, the RING E3 ligases, and the U-box proteins (Callis and Vierstra, 2000; Joazeiro and Weissman, 2000; Estelle, 2001; Stone et al., 2005). A RING finger motif or HECT domain is common among all E3 ligases and mediates the interaction with E2 enzymes to facilitate the transfer of Ub to a Lys residue of the target protein (Freemont, 1993; Hershko and Ciechanover, 1998; Pickart, 2001). Proteins may also become mono- or multi-ubiquinated. This Ub tag controls membrane protein internalization, protein sorting as well as transcription, DNA repair, and viral budding (d'Azzo et al., 2005; Piper and Luzio, 2007).
Seven in absentia (SINA) proteins are E3 ligases containing an N-terminally located RING finger domain, followed by the conserved SINA domain that is involved in substrate binding and dimerization (Hu and Fearon, 1999). The name of the protein family is derived from the first member that was characterized, the Drosophila melanogaster SINA that regulates photoreceptor differentiation by targeting the transcription factor Tramtrack for proteasomal degradation (Carthew and Rubin, 1990; Li et al., 1997; Cooper, 2007). The human SINA homologs Siah1 and Siah2 are involved in synaptic transmission, apoptosis, tumor suppression, and stress and hypoxia responses among others (Wheeler et al., 2002; Franck et al., 2006; Khurana et al., 2006; Fukuba et al., 2007).
SINA proteins are often active as dimers, although the human Siah1 is also part of an Skp-Cullin-F-box-like ubiquitination complex (Santelli et al., 2005). Several substrates of the Siah/SINA proteins have been identified and most have been shown to be degraded in an Ub-dependent fashion. Among the substrates are key transcription factors but also cytoplasmic and membrane proteins (Susini et al., 2001; Habelhah et al., 2004; Kim et al., 2004).
SINA proteins also occur in plants but have been poorly studied. So far, only the SINAT5 gene of Arabidopsis (Arabidopsis thaliana) has been functionally characterized. It attenuates the auxin-induced lateral root (LR) formation, because ectopic expression results in a lower LR number (Xie et al., 2002), while ectopic expression of a dominant-negative Cys-49→Ser RING domain mutant of SINAT5 (SINAT5DN) causes more LRs than in wild-type plants. RING domain-independent dimerization of the Arabidopsis SINAT5 dimer is essential for Ub E3 ligase activity, and the dominant-negative effect of SINAT5DN is due to the formation of inactive heterodimers between SINAT5 and SINAT5DN (Xie et al., 2002). SINAT5 affects LR formation by targeting the NAC1 transcription factor (a member of the NO APICAL MERISTEM/CUP-SHAPED COTYLEDON family) for degradation (Xie et al., 2000, 2002). Recently, the SINAT2 gene of Arabidopsis has been shown to interact with the transcription factor AtRAP2.2 involved in carotenogenesis (Welsch et al., 2007).
Besides LRs, legume plants develop another type of secondary root organ, the nodule, whose formation is triggered by compatible soil bacteria, referred to as rhizobia (Jones et al., 2007). Bacterial nodulation (Nod) factors trigger nodule development that consists of two processes, rhizobial infection and organ development. In the model legume Medicago truncatula, Sinorhizobium meliloti enters via curling of root hairs. Within the curl, wall hydrolysis and membrane invagination result in an infection thread (IT) that guides the bacteria into the cortex. Meanwhile, a nodule primordium is formed through cell division in cortex and pericycle (Timmers et al., 1999). The ITs reach the nodule primordium where bacteria enter the plant cells and differentiate into nitrogen-fixing bacteroids. At the same time, an apical meristem is installed and an indeterminate nodule develops, consisting of a meristem, an infection zone where bacteria are taken up by meristem-derived nodule cells, a nitrogen fixation zone, and the senescence zone where cells of both partners are degraded (Vasse et al., 1990).
To investigate the role of SINA proteins in nodule formation, a small SINA gene family was identified in the M. truncatula genome. All MtSINA proteins were analyzed by yeast two-hybrid assays and were capable of forming homodimers and heterodimers with each other as well as with the Arabidopsis SINAT5DN and SINAT5 proteins. Ectopic expression of SINAT5DN and SINAT5 in M. truncatula provoked effects on LR formation similar to those in Arabidopsis (Xie et al., 2002). Plants that ectopically expressed SINAT5DN showed a reduced number of nodules with a defective nitrogen fixation (Fix−) phenotype. Light and electron microscopy revealed impaired IT and defective symbiosome development, resulting in early senescence.
RESULTS
The M. truncatula SINA Gene Family
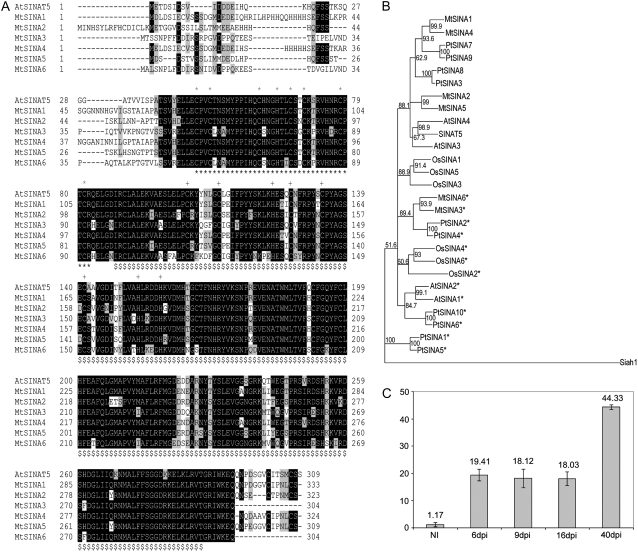
To investigate the role of SINA proteins in nodule development, M. truncatula SINAT5 homologs were isolated by TBLASTX analyses with the M. truncatula Gene Index (MTGI) of the expressed sequence tag database from The Institute for Genomic Research (http://tigrblast.tigr.org/tgi/) and the available genomic data. Six genes, designated MtSINA1 to MtSINA6, coding for SINA domain-containing proteins were identified. The derived amino acid sequences conferred 75% to 89% similarity to SINAT5.
MtSINA1 and MtSINA4 were highly similar and displayed the closest homology to SINAT5. All the protein sequences had the conserved RING finger and SINA domain, whereas only the N-terminal region was highly variable (Fig. 1A). The SINA domain of MtSINA3 and MtSINA6 was 15 amino acids shorter at the C-terminal end than that of other MtSINA proteins (Fig. 1A).
Figure 1.
SINA E3 ligase family. A, Alignment of the M. truncatula SINA protein family and Arabidopsis SINAT5. The RING finger motif and the SINA domain are marked by asterisks and dollar signs, respectively. Conserved Cys and His residues in the RING (*) and Zn finger (+) motifs are marked on top of the sequence. MtSINA1 (TC102374, EU189945); MtSINA2 (TC102369, EU189946); MtSINA3 (TC109024, EU189947); MtSINA4 (TC102612, EU189948); MtSINA5 (TC104350, EU189949); and MtSINA6 (TC109632, EU189950). B, Phylogram of M. truncatula (Mt), Arabidopsis (At), rice (Os), and poplar (Pt) conserved SINA nucleotide sequence regions guided by amino acid alignment. The human Siah1 sequence was used to root the tree. Asterisks indicate the SINA genes encoding a protein with a shorter C-terminal SINA domain. C, Transcript level of MtSINA4 in roots (NI) and in developing nodules at 6 (6dpi), 9 (9dpi), 16 (16dpi), and 40 (40dpi) dpi as measured by qRT-PCR.
The SINA proteins of M. truncatula, Arabidopsis, rice (Oryza sativa), and poplar (Populus trichocarpa) were compared by phylogenetic analysis. Database searches revealed six sequences in M. truncatula, five in Arabidopsis, six in rice, and 10 in poplar. Except for a short N-terminal region, all the plant SINA proteins were highly conserved (Supplemental Fig. S1). Like M. truncatula, the other plants also contained SINA proteins with shorter SINA domains (Supplemental Fig. S1). Because of this high conservation, a phylogram was made with the conserved nucleotide sequence regions guided by amino acid alignment to identify orthologous genes (Fig. 1B; Supplemental Figs. S1 and S2). Neighbor-joining analysis resulted in a phylogram that divided the SINA sequences into six branches (Fig. 1B).
The SINA sequences from Arabidopsis, M. truncatula, and poplar, but not from rice, which contain a long SINA domain, clustered together in one branch (Fig. 1B). Within that branch, no orthologous relationships could be established. On the other hand, the SINA sequences that lacked the C-terminal end clustered in four separate branches (Fig. 1B, asterisks). The sixth branch of the tree grouped the rice proteins with long SINA domains (Fig. 1B). The phylogram demonstrates that most SINA proteins, including the six from M. truncatula, cluster in pairs, suggesting genome duplications for possible neofunctionalization.
To identify possible amino acid motifs that would be specific for each branch, a MEME motif analysis was done (http://meme.sdsc.edu/meme/meme.html; data not shown). However, no motifs were found that were specific for a certain branch.
The temporal expression of the six MtSINA genes during nodule formation was analyzed by quantitative reverse transcription (qRT)-PCR. Because M. truncatula nodule formation is not synchronized, tissue from uninoculated roots and from roots 6, 9, 16, and 40 d postinoculation (dpi) was carefully selected with fluorescence stereomicroscopy. The developmental stages that were specifically harvested at the different time points are presented in Supplemental Figure S3. The transcript levels of MtSINA1, MtSINA2, MtSINA3, MtSINA5, and MtSINA6, although detectable, did not change significantly during nodule development (data not shown). However, for MtSINA4, an increase in transcript level was observed already at 6 dpi, a stage at which bacteria invade the cortex and nodule primordia develop (Fig. 1C). The expression level remained the same throughout nodule development and increased again in mature elongated nodules.
MtSINA Proteins Interact with Each Other and with SINAT5 and SINAT5DN
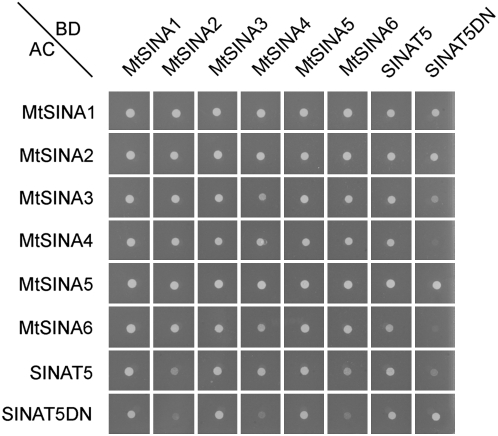
Several studies have shown that SINA proteins can form homodimers as well as heterodimers (Hu and Fearon, 1999; Depaux et al., 2006). To unravel whether the interaction between the different MtSINA proteins was specific, a pairwise yeast two-hybrid analysis was done. The MtSINA clones were fused to the GAL4 activation domain (AD) or DNA-binding domain (BD) and tested in one-to-one combinations (Fig. 2; see “Materials and Methods”). Likewise, every interaction between two SINA proteins was tested in both directions. When selected on −Trp/Leu/His medium in the presence of 10 mm 3-amino 1,2,4-triazole (3-AT), yeast growth was observed for most interactions. Weak colony growth was considered only as a sign of a weak interaction when it occurred in the two reciprocal combinations. As this was not the case for any of the MtSINA-MtSINA interactions in the presence of 10 mm 3-AT (Fig. 2) and 80 mm 3-AT (data not shown), all interactions were considered strong.
Figure 2.
Pairwise yeast two-hybrid interaction of MtSINA proteins, SINAT5, and SINAT5DN. The clones were fused to the GAL4 BD (horizontal) or GAL4 AD (vertical) and tested by a pairwise yeast two-hybrid analysis. Yeast colonies were spotted with equal density on −Trp/Leu/His medium in the presence of 10 mm 3-AT.
Furthermore, all six MtSINA proteins interacted with SINAT5 and SINAT5DN in the presence of 5 mm 3-AT (data not shown). The interactions of MtSINA4 and MtSINA6 with SINAT5DN and of MtSINA6 with SINAT5 were weak, as illustrated by the faint, or absence of, growth on 10 mm 3-AT (Fig. 2). The other combinations resulted in yeast growth even in the presence of 40 to 80 mm 3-AT (data not shown).
To verify these results, co-immunoprecipitation of the six MtSINA proteins with SINAT5DN was analyzed (see Supplemental Materials and Methods S1). SINAT5DN and the MtSINA proteins were tagged with a 3-hemagglutinin (3-HA) and 3-MYC epitope, respectively, and used to transiently cotransform in Arabidopsis protoplasts. After cotransformation with 3-HA-SINAT5DN, only MYC-MtSINA1 and MYC-MtSINA3 accumulated in amounts high enough for co-immunoprecipitations. The anti-HA/3-HA-SINAT5DN complexes from protoplasts coexpressing 3-HA-SINAT5DN and MYC-MtSINA1 or MYC-MtSINA3 were immunoprecipitated. As negative control, extracts were used from protoplasts expressing only MYC-MtSINA1 or MYC-MtSINA3. SDS-PAGE of the precipitated fractions followed by immunoblotting with the MYC antibody revealed the presence of MtSINA1 and MtSINA3 in the anti-HA/3-HA-SINAT5DN complexes (Supplemental Fig. S4, arrows).
Heterologous Expression of SINAT5 and SINAT5DN in M. truncatula
To unravel the role of SINA proteins in nodulation, the dominant-negative form, SINAT5DN, was ectopically expressed in M. truncatula. This approach was chosen because not all MtSINA sequences were available at the onset of the experiments. Moreover, and importantly, the yeast two-hybrid analysis revealed that SINAT5DN interacted with all the endogenous MtSINA proteins. Based on these observations and on the results in Arabidopsis (Xie et al., 2002), we anticipated that the Arabidopsis SINAT5DN would inhibit endogenous MtSINA proteins by producing nonfunctional heterodimers. The M. truncatula ‘R108’ was selected for Agrobacterium tumefaciens-mediated transformation of the 35S:SINAT5DN and, as a control, 35S:SINAT5 and 35S:GUS constructs because of the shorter generation time and the more efficient transformation protocol compared to those of the model cultivar Jemalong J5 (Cook, 1999; Frugoli and Harris, 2001; see “Materials and Methods”).
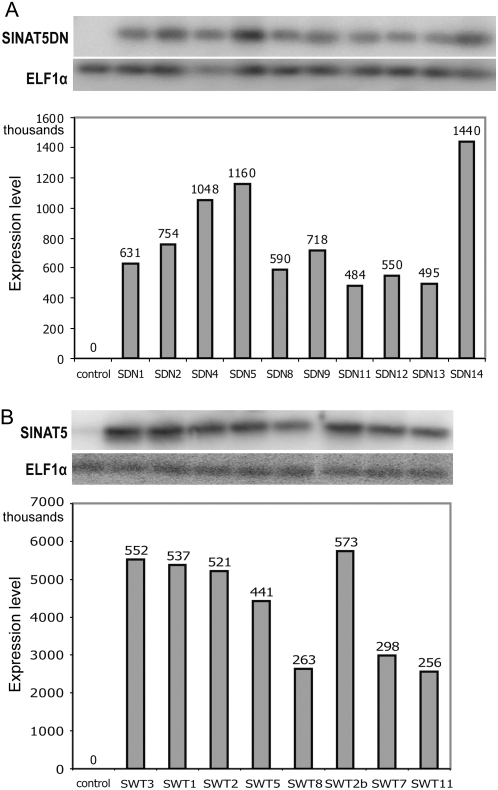
In nine independent lines transformed with 35S:SINAT5DN, different levels of SINAT5DN transcripts were observed in leaves (Fig. 3). For the 35S:SINAT5 construct, DNA gel-blot hybridization indicated 27 different lines (data not shown) with various expression levels (Fig. 3).
Figure 3.
Quantification of SINAT5 expression levels in T0 leaf RNA of different 35S:SINAT5DN (SDN) and 35S:SINAT5 (SWT) lines as measured by RT-PCR. A, RNA levels of T0 35S:SINAT5DN. Lines SDN1 and SDN8 are clonal with comparable expression levels of the transgene as a result. B, RNA levels of T0 35S:SINAT5. ELF1α indicates the levels of the constitutively expressed ELF1α gene. Control corresponds to SINAT5 and SINAT5DN expression levels in nontransformed control M. truncatula lines.
The different lines were grown for seed setting and for segregation analysis of T1 plants with glufosinate ammonium or phosphinothricin (PPT; see “Materials and Methods”). Lines with a Mendelian segregation pattern were selected for further experiments.
Shoot and Root Phenotype in 35S:SINAT5DN and 35S:SINAT5 Transgenic M. truncatula Plants
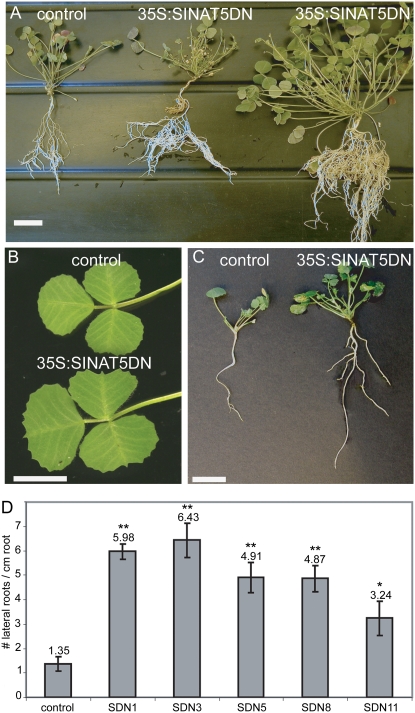
Growth of 35S:SINAT5DN plants was more vigorous than that of control plants. After 8 weeks under greenhouse conditions in rich soils, several 35S:SINAT5DN lines had grown to approximately 150% of the size of the control plants (Fig. 4A) and had a larger leaf area (Fig. 4B) and more shoots. Moreover, 20-d-old in vitro-grown plants had more LRs (Fig. 4C). Wild-type and 35S:GUS plants had consistently similar phenotypes in all experiments and are referred to as “control” or “control plants.”
Figure 4.
M. truncatula 35S:SINAT5DN shoot and root phenotype. A, Eight-week-old T0 35S:SINAT5DN plants grown in rich soils, demonstrating vigorous growth and more roots compared to the T0 control plant. B, Trifoliate leaf of control and 35S:SINAT5DN T2 plants. C, LR phenotype of 35S:SINAT5DN and control plants after 20 d of in vitro growth. D, Average number of LRs/cm of 35S:SINAT5DN T1 M. truncatula lines (SDN1, SDN3, SDN5, SDN8, and SDN11) and control plants after 6 d of NAA treatment on 12-d-old plants. Error bars, se measurements. Significantly different values are labeled with a different number of asterisks as defined by a two-tailed t test (* = P < 0.05; ** <0.001 for the transgenic line compared to the control). Bars = 3 cm (A) and 1 cm (B and C).
Plants with a high transgene expression level (as measured in leaf tissue) were grown in vitro, and the number of LRs/cm was determined (Fig. 4D). Control and 35S:SINAT5DN transgenic plants had an average of 2.16 ± 0.51 LRs/cm and 4.12 ± 0.57 LRs/cm, respectively (data not shown). Although these results are indicative of a negative effect of SINAT5 on LR formation due to variation within each plant population, the difference between control roots and several transgenic lines was statistically not significant (0.3 > P > 0.05). Moreover, the SINAT5 effect on LR formation has been shown to be an auxin-dependent mechanism (Xie et al., 2002). Therefore, the effect of 35S:SINAT5DN on auxin-induced LR formation was tested by growing plants in the presence of 1-α-naphthalene acetic acid (NAA). Indeed, when 6-d-old 35S:SINAT5DN transgenic plants were transferred to NAA-containing medium for another 6 d, an average of 4.95 ± 0.33 LRs/cm was observed versus 1.35 ± 0.29 LRs/cm in control lines (Fig. 4D). A two-tailed t test showed that the number of LR/cm root differed significantly between control roots and different transgenic lines (Fig. 4D, asterisks).
Ten PPT-selected plants from six independent M. truncatula 35S:SINAT5 T1 lines were grown in vitro on nitrogen-rich medium for 40 d to analyze root and shoot growth (“Materials and Methods”). No obvious shoot phenotype was observed, but the roots had fewer LRs than the control plants (Fig. 5A). Ectopic expression of SINAT5 resulted in a significantly lower number of LRs/cm (Fig. 5B). The average number was 1.32 ± 0.018 and 2.16 ± 0.15 LRs/cm in 35S:SINAT5 and control plants, respectively. A two-tailed t test revealed that the difference in number of LR/cm root was significantly different between control and most transgenic 35S:SINAT5 (Fig. 5B, asterisks).
Figure 5.
M. truncatula 35S:SINAT5 LR phenotype. A, Root phenotype of T1 35S:SINAT5 and control plants after 40 d of in vitro growth. Bar = 1 cm. B, Average number of LRs/cm on 10 T1 plants of 35S:SINAT5 lines (SWT1–SWT6) compared to control (control). Error bars, se measurements. Significantly different values are labeled with a different number of asterisks as defined by a two-tailed t test (* = P < 0.05; **= P < 0.01 for the transgenic line compared to the control).
Effect of 35S:SINAT5DN and 35S:SINAT5 on Nodule Number in M. truncatula
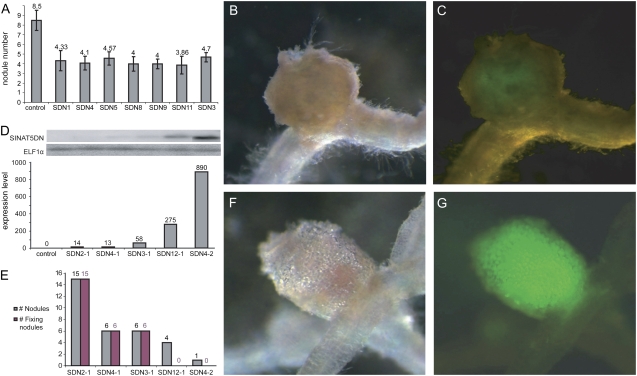
The effect on LR growth indicated that SINAT5 was active in M. truncatula and that SINAT5DN exerted a dominant-negative effect on endogenous MtSINA proteins. To investigate the effect of 35S:SINAT5DN on nodule formation, 10 plants of seven transgenic lines were grown under nitrogen-poor conditions and inoculated with S. meliloti 41 (pHC60-gfp; Cheng and Walker, 1998). An average of 4.22 ± 0.29 nodules/plant was obtained for the 35S:SINAT5DN lines, whereas control plants had 8.53 ± 1.02 nodules/plant at 22 dpi (Fig. 6A; P = 5.19E-04 < 0.001). Of the nodules that appeared on 35S:SINAT5DN plants, 40% were white, reflecting a lack of active leghemoglobin (compare Fig. 6, B and F), a sign for Fix−. Fluorescence microscopy analysis of these nodules revealed that the central tissue was less fluorescent than that of the wild-type nodules (Fig. 6, C and G), indicating that fewer bacteria had invaded the nodule. Whereas control nodules of M. truncatula are cylindrical because of the presence of a persistent apical meristem, the Fix− nodules of 35S:SINAT5DN were round-shaped, implying a precocious arrest of meristematic activity (Fig. 6B). By RT-PCR on root tissue of the different T1 lines, a correlation was found between the level of transgene expression and the phenotype (Fig. 6, D and E). Compared to control lines, low-expression lines carried fewer, but functional, nodules, whereas the nodules in high-expression lines were Fix− (Fig. 6, D and E).
Figure 6.
Nodule numbers of M. truncatula 35S:SINAT5DN plants. A, Average number of nodules formed on 10 T1 plants of seven different 35S:SINAT5DN R108 lines (SDN) compared to the number formed on control plants. B, C, F, and G, Stereomicroscopic images of 35S:SINAT5 Fix− (B and C) and wild-type (F and G) nodules 22 dpi with S. meliloti Rm41 (pHC60-gfp). The white (B) and pinkish (F) colors are indicative for the absence and presence of leghemoglobin, respectively. C and G, Fluorescent microscopic images showing the weak (C) and strong (G) bacterial GFP activity. D, RT-PCR hybridized with probes specific for SINAT5DN and ELF1α on root RNA of the transgenic lines showing the relative expression level of the transgene in different lines. E, Total and Fix+ nodules per plant of different 35S:SINAT5DN lines at 25 dpi.
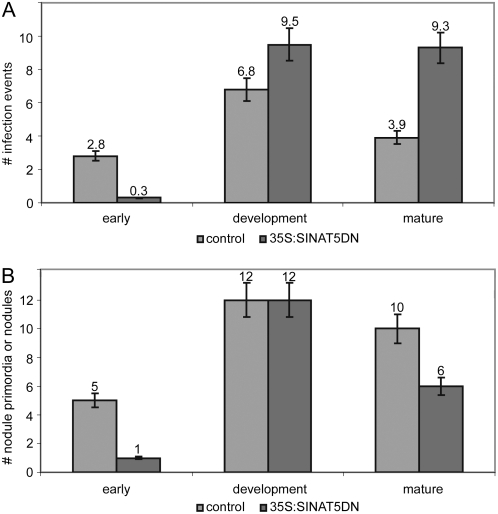
To discriminate between an effect on infection or nodule primordium formation, 10 plants of two different 35S:SINAT5DN T2 lines were inoculated with Rm41(pHC60-gfp), and the infected root hairs in addition to the infections that occurred on the nodule primordia were counted at 10, 20, and 29 dpi (Fig. 7A). Moreover, the developing nodules were counted at these time points (Fig. 7B), corresponding more precisely to the number of incipient primordia at 10 dpi (Fig. 7, early), developing primordia at 20 dpi (Fig. 7, development), and mature functional nodules at 29 dpi (Fig. 7, mature). At the early developmental stage, 35S:SINAT5DN lines had fewer additional infection events than the control plants (on average 0.3 versus 2.8 per plant; Fig. 7A) and fewer nodule primordia (on average one versus five per plant; Fig. 7B). However, later on during development, the number of nodule primordia on 35S:SINAT5DN plants became equal to that on control plants (on average 12 per plant). When the additional infection events were counted at the “development” stage, the number was considerably higher than that in control plants (on average 9.5 versus 6.8; Fig. 7A). Finally, on average, 10 mature nodules were observed on control plants and only six on the transgenic lines (Fig. 7B). Interestingly, whereas the number of infection events in control roots had decreased (on average 3.9), at the mature stage it remained equal (on average 9.3) to that of the “development” stage in transgenic 35S:SINAT5DN plants (Fig. 7A). These observations show that nodule initiation (infection and primordium formation) is delayed but not inhibited. The high number of infection events observed at later stages in 35S:SINAT5DN nodulation and the relatively lower number of primordia that developed into functional nodules are indicative of interference with infection or symbiosome development rather than with primordium development.
Figure 7.
Infection events and nodule primordia or mature nodules during nodulation of T2 35S:SINAT5DN and control M. truncatula plants. A, Number of infection events referring to the number of root hairs with curls entrapping a bacterial microcolony or an IT in addition to the infected root hairs that are linked with nodule primordia. B, Number of nodule primordia at incipient (early) and developing (development) nodulation stages, and number of nodules at the mature nodulation stage. Error bars, se measurements.
The nodulation capacity of the transgenic 35S:SINAT5 lines was also analyzed. Plants of the 27 different 35S:SINAT5 R108 lines were germinated and inoculated. Nodule development did not differ from that in control lines (data not shown).
Microscopic Analysis of 35S:SINAT5DN Nodule Development
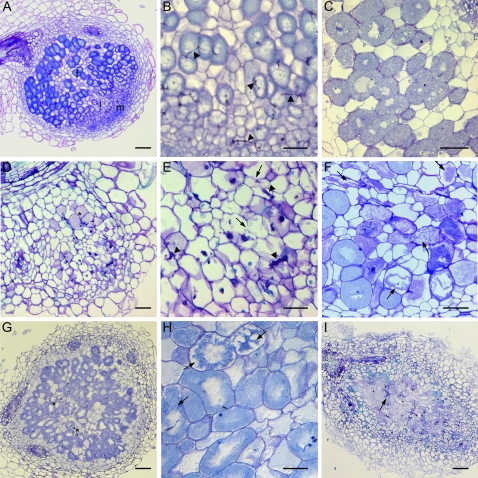
To unravel the stages during which nodule development is blocked in the 35S:SINAT5DN transgenic plants, nodules at different time points were analyzed by light microscopy. At 16 dpi, small round nodules were observed on control roots, with an apical meristem, infection zone, and small fixation zone (Fig. 8A). In sections of 35S:SINAT5DN nodules at the same time point, the central tissue of the incipient nodule was completely disordered, and the typically zonated structure was not visible (Fig. 8D). Serial sectioning through such nodules revealed that the meristem was absent. The ITs had reached the primordium cells but had a thick swollen appearance (compare Fig. 8, D and E with A and B). Some infected cells were observed (Fig. 8D, asterisks), but the staining was more pinkish than that in infected cells of control plants (Fig. 8, C and D), indicating an early symbiosome senescence (compare Fig. 8, D and F; Van de Velde et al., 2006). Plant cells were also senescent, because cell walls were disrupted (Fig. 8E, arrows).
Figure 8.
Bright-field microscopy on 35S:SINAT5DN and control M. truncatula nodules. A, Young wild-type nodule containing a nodule meristem (m), infection zone (i), and fixation (f) zone. B, Detail of infection zone of young control nodule. Arrowheads indicate ITs. C, Wild-type fixation zone (25 dpi) with infected cells filled with symbiosomes. D, Medial section through a young 35S:SINAT5DN nodule with a disordered central tissue and few infected cells (asterisks). E, Detail of infected region in D. Thicker ITs (arrowheads) appear and cells show signs of degradation (arrows). F, Senescent nodule tissue of a wild-type M. truncatula nodule (61 dpi). Arrows mark cells with senescent features. G, Medial section through a 35S:SINAT5DN nodule more developed than the 35S:SINAT5DN nodule in D. Asterisks show senescing regions. H, Detail of G with arrows indicating cells with signs of senescence. I, Cylindrical 35S:SINAT5DN nodule demonstrating early senescence of the nodule central tissue. Bars = 100 μm (B–F and H), 200 μm (A and G), and 400 μm (I).
In some cases, at 25 dpi, the development of the nodule had proceeded and a fixation zone was visible (Fig. 8G), but again with signs of early nodule senescence (Fig. 8, G and H, asterisks and arrows, respectively). Eventually, some cylindrical nodules were present (Fig. 8I). Sectioning through such nodules often revealed a central senescing tissue (Fig. 8I) and only a residual or no apical meristem.
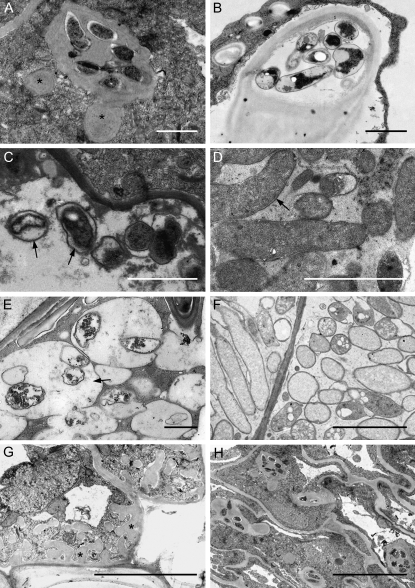
Electron microscopical analysis on young and mature control and 35S:SINAT5DN nodules revealed that the IT matrix in 35S:SINAT5DN lines was more dense than that of control nodules (Fig. 9, A and B). The bacteria within the ITs looked healthy, but fewer bacteria and more matrix were seen (Fig. 9A). Moreover, the ITs were not regular and bulge-like structures were observed (Fig. 9A, asterisks). Once the ITs had reached the nodule primordium, bacterial uptake took place, but the symbiosome development was hampered (Fig. 9, C and D). Whereas normally the symbiosome membrane tightly encloses a single bacteroid and only a narrow symbiosome space is present (as seen by the thin transparent line between the bacteroid and symbiosome membrane; Fig. 9D, arrow), in the 35S:SINATDN nodules, the symbiosome membrane only loosely surrounded the bacteroid, resulting in a clearly visible symbiosome space (Fig. 9, C and E, arrows). At later stages, large symbiosomes could be detected with several degrading bacteroids (Fig. 9E). Senescence of the plant cells also occurred, visible by cell wall appositions (Fig. 9G) and loss of cell integrity (Fig. 9H).
Figure 9.
Transmission electron microscopy of 35S:SINAT5DN and control M. truncatula young and mature nodules. A and B, Cross sections through an IT of a 35S:SINAT5DN nodule (A), with fewer bacteria and a denser matrix than a wild-type IT (B). Asterisks indicate bulge-like structures. C, Deformed symbiosomes (arrows) in 35S:SINAT5DN-infected cells. D, Wild-type symbiosomes. The symbiosome membrane tightly encloses a single bacteroid. Arrow marks symbiosome space. E, Hampered 35S:SINAT5DN symbiosome development leading to large symbiosomes containing several degrading bacteroids. F, Section through infected cells of a control nodule. Each bacteroid is surrounded by a symbiosome membrane. G, Cell wall appositions (asterisks) as a sign of plant cell defense in 35S:SINAT5DN nodule tissue. H, Loss of plant cell integrity in the 35S:SINAT5DN nodules displaying the initiation of senescence. Bars = 1 μm (A–G) and 2 μm (H).
DISCUSSION
Nodule formation is a tightly regulated process that integrates specific signal exchange and the coordinated activation of developmental mechanisms to synchronize bacterial invasion and organ development. Ub-mediated proteolysis is a common regulatory system and, hence, presumably involved in nodulation.
The central components of the Ub-mediated degradation machinery are E3 ligases that specifically select the target proteins. Until now, only two E3 ligases have been shown to play a role in nodule development (Vinardell et al., 2003; Shimomura et al., 2006). The anaphase-promoting complex, APCCCS52A, degrades mitotic cyclins in the infected cells to induce endoreduplication, resulting in polyploidy needed to assure cell enlargement to host the huge numbers of nitrogen-fixing bacteroids (Vinardell et al., 2003; Kondorosi et al., 2005). In Lotus japonicus, a RING-H2 finger domain protein has been identified that might have a function in IT formation (Shimomura et al., 2006). We provide evidence that another family of E3 ligases, the SINA proteins, is important for IT growth and symbiosome differentiation.
SINA domain proteins exist in animals as well as in plants. They are mostly active as homo- or heterodimers and consist of a RING finger domain and a SINA domain that controls substrate recognition and dimerization (Hu and Fearon, 1999). BLAST searches against expressed sequence tags and genomic databases revealed that Arabidopsis, rice, poplar, and M. truncatula contain a small family of conserved sequences. In human and mouse, only two and three SINA genes have been found, respectively. Hence, plants might have extended the use of E3 ligases for posttranslational control of various processes.
Because of the strong amino acid conservation within the RING and SINA domain, the corresponding nucleotide sequence regions were used to investigate the phylogenetic relation among the SINA sequences. A phylogram separated the plant SINA proteins into six different groups. In contrast to Wang et al. (2008), we could not identify protein domains that specify each group of proteins. However, each plant species contained two groups of SINA proteins that were separated by the presence or absence of a C-terminal extension of approximately 15 amino acids. Strikingly, each phylogenetic branch contained only one kind of protein. The SINA proteins without the C-terminal stretch split in four branches. The other SINA proteins were divided into two groups. SINAT5 belonged to a branch with all long SINA proteins from the tested dicots. The M. truncatula ortholog of SINAT5 could not be identified within that cluster. Whether the phylogenic separation reflects different biological functions has to be resolved. The high conservation within the RING finger and SINA domains hints at critical functions during plant development.
Two-hybrid analysis revealed a low specificity to form SINA protein dimers, because all MtSINA proteins and the Arabidopsis SINAT5 protein could interact. Most probably, in vivo, the formation of heterodimers depends on spatial and temporal expression together with subcellular localization. qRT-PCR analysis indicated that only the level of MtSINA4 transcripts increased during nodule formation. The expression level of the other MtSINA genes did not vary significantly between nodules and roots. Thus, MtSINA4 is probably important for nodule development, but heterodimerization with other MtSINA proteins might be critical for function. Indeed, the other five MtSINA genes were transcribed in roots and nodules. ESTs have been identified from root and nodule cDNA libraries, and the corresponding SINA proteins have been characterized by yeast two-hybrid analysis as interactors of SINAT5 and SINAT5DN (data not shown). It would be interesting to know where the MtSINA proteins meet in the nodule at cellular and subcellular levels.
SINAT5 Is Active in M. truncatula
The only characterized SINA protein in plants is SINAT5 from Arabidopsis. The protein is involved in LR formation because its ectopic production results in fewer LRs, whereas that of a dominant-negative form produces more LRs (Xie et al., 2002). The dominant-negative protein has an amino acid substitution in the RING domain with the loss of the Ub ligase activity as a consequence, while the binding with the target AtNAC1 remains intact (Xie et al., 2002). Upon heterodimerization, the activity of the interacting wild-type SINAT5 protein is inhibited (Xie et al., 2002).
Transgenic M. truncatula lines that ectopically express SINAT5 or SINAT5DN have several features reminiscent of phenotypes of transgenic Arabidopsis plants. The 35S:SINAT5DN plants grew more vigorously and had larger leaves than the control plants. This phenotype has not been described in Arabidopsis but has been reported for the transgenic plants that ectopically express the NAC1 gene, coding for a NAC transcription factor that is a target of SINAT5 (Xie et al., 2000, 2002). Also the number of LRs was modulated by introducing SINAT5 and SINAT5DN into M. truncatula, similarly to what was observed for Arabidopsis. 35S:SINAT5 and 35S:SINAT5DN plants produced fewer and more LRs/cm, respectively. The effect on LR formation was even more pronounced when roots were treated with auxin, indicating that not the increased shoot growth was responsible for the increase in LR number but that the SINA proteins were involved directly in the process of auxin-induced LR formation.
Together, these experiments suggest that SINAT5 can target M. truncatula proteins for degradation and that SINAT5DN is able to inhibit endogenous SINA proteins. The M. truncatula ortholog of SINAT5 is still unknown, although MtSINA1 is a good candidate. The clone has been found in cDNA libraries from developing roots (MTGI on www.tigr.org), and the deduced amino acid sequence is highly similar to that of SINAT5.
MtSINA Proteins Are Involved in the Formation of ITs and in Symbiosome Development
Ectopic expression of SINAT5DN had a drastic effect on nodulation, more precisely on IT formation and symbiosome development. Bacterial infection was delayed and eventually ITs were formed, but the infection did not proceed well, resulting in abundant initiation of new infections at later time points. This observation is typical for mutants with a defective IT growth when the mechanism that restricts the number of infection events is not switched on (e.g. Tsyganov et al., 2002; Tansengco et al., 2003; Veereshlingam et al., 2004). Light and electron microscopy confirmed that the ITs were deformed: they were broader than those of the wild type, had a denser matrix with fewer bacteria, and the walls were irregular with bulge-like structures. Eventually, some ITs reached the nodule primordia and penetrated between the primordium cells, releasing bacteria into the plant cell cytoplasm. However, the formation of symbiosomes was affected. The bacterial uptake, via unwalled infection droplets, occurred, but the freshly formed symbiosomes did not look normal. In wild-type plants, the symbiosome membrane closely surrounds the bacteroid, whereas a broad symbiosome space between the bacteroid and the symbiosome membrane was observed in 35S:SINAT5DN nodules. Large symbiosomes were seen with several degenerating bacteroids, leading to early nodule senescence and small, round Fix− nodules. In addition, these defects were often accompanied by a general plant defense, because wall appositions and loss of cell integrity were observed within the cells of the central tissue.
As the two symbiotic partners meet closely within the ITs and symbiosomes, continuous mutual recognition is needed for proper nodule functioning (Jones et al., 2007). The bacterial lipopolysaccharide (LPS) molecules play an important role for sustained infection and bacteroid survival within the symbiosomes (Mathis et al., 2005; Jones et al., 2007). Irregular ITs and hampered symbiosome development are often seen in nodules that had been elicited by LPS-defective symbionts (Priefer, 1989; Putnoky et al., 1990; Glazebrook et al., 1993; Perotto et al., 1994; Niehaus et al., 1998; Campbell et al., 2002; Laus et al., 2004). Moreover, infection with surface polysaccharide-defective bacteria mostly results in the elicitation of the plant's defense (Tellström et al., 2007). Thus, SINA proteins could plausibly contribute to LPS recognition for symbiosome development. Nevertheless, the phenotype is not fully identical: in contrast to what was reported for inoculation with LPS mutant bacteria, we never observed large hypertrophied ITs on 35S:SINAT5DN plants.
Besides a role in microsymbiont recognition, MtSINA proteins might also be involved in a common mechanism that underlies both IT and symbiosome formation. For instance, endomembrane vesicle traffic is needed for both growth and differentiation of ITs and symbiosomes (Brewin, 2004; Jones et al., 2007). The pea (Pisum sativum) sym31 mutant shows an aberrant targeting of vesicles toward the symbiosomes with a phenotype similar to that of the 35S:SINAT5DN transgenic plants (Borisov et al., 1992; Dahiya et al., 1998). Interestingly, but still preliminary, two-hybrid analysis revealed two candidate SINA protein interactors that code for genes that are involved in vesicle trafficking (G. Den Herder, M. Holsters, and S. Goormachtig, unpublished data). Further investigation on their role during nodule formation might reveal insights into how the endomembrane system supports IT as well as symbiosome development.
Besides sym31, many other legume Fix− mutants have related symbiosome defects as observed here (Tsyganov et al., 1998; Borisov et al., 1992; Morzhina et al., 2000; Suganuma et al., 2003). However, the phenotype of most of these mutants does not fully correspond to that of 35S:SINAT5DN. To our knowledge, the most similar mutant is sym33/SGRfix-2 that makes fewer nodules with thick ITs, which are often closed and in which bacteroids do not develop properly after sporadic release into the plant cells (Tsyganov et al., 1998). Unfortunately, the interrupted gene is still unknown.
In summary, we show that SINA proteins are important for nodule formation, although a specific MtSINA protein for nodule formation cannot be appointed yet. Certainly, MtSINA4 is a good candidate, because it was the only up-regulated gene during nodule formation. However, heterodimerization might complicate the picture. Future experiments, including protein localizations and RNA interference to knock down each MtSINA protein separately or in combination, will give us further insight into the SINA protein complexes that rule nodule formation.
MATERIALS AND METHODS
Plant Material, Bacterial Strains, and Growth Conditions
Medicago truncatula ‘R108’ plants were grown and inoculated as described (Mergaert et al., 2003; www.isv.cnrs-gif.fr/embo01/index.html). Sinorhizobium meliloti Sm1021, Sm1021 (pHC60-GFP), and Rm41 (pHC60-GFP; Cheng and Walker, 1998) were grown at 28°C in yeast extract broth medium (Vervliet et al., 1975) supplemented with 10 mg/L tetracycline for the pHC60-GFP strain.
In vitro growth occurred in square petri dishes (12 × 12 cm) as described (www.isv.cnrs-gif.fr/embo01/index.html) supplemented with 1 mm NH4NO3. For the 35S:SINAT5 and 35S:SINAT5DN lines, glufosinate ammonium (Pestanal, Sigma-Aldrich) was added to the medium.
LRs were counted and the main root length was measured (5-mm accuracy) after 20 or 40 d of growth for 35S:SINAT5DN and 35:SINAT5 lines, respectively. For counting of LRs and measurement of the main root length, in vitro-grown 6-d-old 35S:SINAT5DN lines and wild-type R108 were transferred to plates containing 2 μm NAA (Duchefa) for another 6 d.
For nodulation experiments, 35S:SINAT5 and 35S:SINAT5DN lines, grown in vitro for 2 weeks on medium supplemented with PPT, were transferred to perlite and inoculated with Rm41 (pHC60-GFP). Nodule morphology was analyzed with light and electron microscopy on T1 and T2 plants at the time points indicated.
For qRT-PCR analysis, M. truncatula ‘J5’ plants were grown for 7 d in perlite and inoculated with Sm1021 (pHC60-GFP). Tissue was collected by visualizing the green fluorescent bacteria under a stereomicroscope MZFLII (Leica) equipped with a blue-light source and a Leica GFP Plus filter set (λex = 480/40; λem = 510 nm LP barrier filter). Uninoculated root sections were isolated at the same developmental stage as the 6-dpi stage and contained the root elongation zone (Supplemental Fig. S3).
For seed setting of the wild-type and transgenic progeny, seedlings were transferred to vermiculite:sand (2:1) mixtures and grown for 4 weeks, after which they were transferred to a soil:sand (2:1) mixture and further grown in the greenhouse under nitrogen-rich conditions (www.isv.cnrs-gif.fr/embo01/index.html).
The analysis of shoot and leaf growth in 35S:GUS control, 35S:SINAT5DN and 35S:SINAT5 transgenic, and R108 wild-type plants was carried out after 8 weeks of growth in soil under greenhouse conditions. For shoot measurements, the control and transgenic plants were removed from the soil and measured from the shoot tip to the initiation of the root, with approximately 5-mm accuracy). This experiment was done for several plants of at least two different transgenic lines.
Agrobacterium tumefaciens-Mediated Transformation of M. truncatula
Young leaves from 4-week-old M. truncatula ‘R108’ plants were transformed with A. tumefaciens AGL0 containing the plasmids pBA002(35S:SINAT5WT) and pBA002(35S:SINAT5DN; kindly provided by N.-H. Chua, Rockefeller University, New York) as described (www.isv.cnrs-gif.fr/embo01/index.html). Transgenic tissue was selected on the appropriate medium (www.isv.cnrs-gif.fr/embo01/index.html) containing 3 mg L−1 PPT. Regenerated transgenic shoots were grown on plates for 1 month and then transferred to vermiculite in the growth chamber. Seed pods were harvested and T1 seedlings were selected on nitrogen-rich medium supplemented with 3 mg L−1 PPT (see above) to segregate the transgenic plants and to obtain seed for the T2 generation. M. truncatula ‘J5’ was transformed with pBA002(35S:SINAT5DN) according to the same procedure and had a phenotype similar to that of 35S:SINAT5DN R108 transgenic lines (data not shown).
Bioinformatics Analyses
The coding sequences of SINAT5 (GenBank accession no. AF480944) were analyzed by TBLASTX on the MTGI database (release 8.0; www.tigr.org/tdb/tgi) and on the genomic bacterial artificial chromosome sequences available in the GenBank. For DNA sequence data, percentages of identity and similarity between amino acid sequences were determined with the GCG package (Accelrys), including the GAP program. ClustalW was used for the alignments.
For the phylogenetic analysis of the SINA homologs, sequences from Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), and poplar (Populus trichocarpa) were retrieved from GenBank and the Joint Genome Institute (http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html). To calculate the SINA nucleotide phylogenetic tree, the SINA amino acid sequences were aligned with ClustalX 1.8 (Thompson et al., 1997). The conserved regions were defined as those with at least 70% identity among the aligned sequences, as identified with BioEdit 7.0.9 (Hall, 1999). The corresponding nucleotide sequences of the conserved regions were aligned guided by the protein sequence alignment with MacClade 4.0 (Maddison and Maddison, 1989). A neighbor-joining bootstrap analysis with 1,000 replicates was carried out to generate the phylogram with PHYLIP 3.6 (Felsenstein, 2005). Branches with bootstrap support values <50% were collapsed. Siah1 was used to root the tree.
Statistical Analysis of LR and Nodule Numbers
For each line, the number of LRs/cm was counted for n plants. Standard errors were calculated as the sd divided by the square root of the n plants. Control versus total transgenic population as well as versus separate transgenic line populations were statistically analyzed by a two-tailed t test.
In two experiments, 10 control and 10 T1 plants per 35S:SINAT5 line were taken after 40 d of growth. For the effect of auxin-induced LR growth, five to eight plants of five 35S:SINAT5DN lines and controls were studied after 12 d of growth of which 6 d after auxin treatment.
Nodules were counted on control (n = 19) and T1 35S:SINAT5DN plants (n = 51).
Microscopy
For bright-field microscopy (D'Haeze et al., 1998), sections (5 μm) of embedded nodules 16 and 25 dpi were stained with 0.5% toluidine blue, and images were taken with an Axiocam digital camera (Zeiss). Electron microscopy was as described (Van de Velde et al., 2006).
Expression Analysis
RNA was prepared from harvested tissue homogenized in TRIzol Reagent (Invitrogen) and phase-separated with chloroform by precipitation with isopropanol. First-strand cDNA was synthesized and amplified by RT-PCR according to Van de Velde et al. (2006) and Corich et al. (1998), respectively. The probes were generated from the purified PCR product with SINAT5 or Elongation factor 1α (ELF1α) primers (Supplemental Table S1) and pBA002(35S:SINAT5) plasmid DNA or M. truncatula cDNA as template sequence, respectively, and radioactively labeled with the Rediprime II Random Prime Labeling system (GE-Heathcare). After hybridization and exposure, radioactive signals were quantified for each line by phosphorimaging.
qRT-PCR was done as described by Vlieghe et al. (2005). For the sequences of the primers used, see Supplemental Table S1. MtELF1α gene-specific primers were used for normalization of each gene and MtENOD40 gene-specific primers (Supplemental Table S1) as control for early nodulation stages (data not shown; Supplemental Fig. S1). For all stages, biological repeats were done in triplicate.
Yeast Two-Hybrid Techniques
The coding sequences of SINAT5, SINAT5DN, and the six MtSINA genes were fused to the GAL4 BD and the GAL4 AD of the pBDGAL4 vector and pADGAL4 vector, respectively (Agilent), used in pairwise yeast two-hybrid analyses according to the manufacturer's procedure, and plated with equal yeast density on triple selection medium. Auto-activation was analyzed in the PJ694A yeast strain (James et al., 1996). The strength of the interactions was tested by addition of 0, 5, 10, 20, 40, and 80 mm of 3-AT to this medium. Growth was analyzed after 3 d.
Accession Numbers
Sequence data from this article are deposited in the GenBank/EMBL databases with the following accession numbers: EU189945 (MtSINA1), EU189946 (MtSINA2), EU189947 (MtSINA3), EU189948 (MtSINA4), EU189949 (MtSINA5), and EU189950 (MtSINA6).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of SINA protein sequences of M. truncatula, Arabidopsis, rice, poplar, and human Siah1.
Supplemental Figure S2. Nucleotide sequence alignment (690-bp assembled conserved regions) used to design the phylogenetic tree shown in Fig. 1B.
Supplemental Figure S3. Stereomicroscopic images representing the different stages that were harvested for qRT-PCR analysis.
Supplemental Figure S4. Co-immunoprecipitation of SINAT5DN with MtSINA1 and MtSINA3.
Supplemental Table S1. Primers used to amplify the open reading frames corresponding to the MtSINA genes and primers used for qRT-PCR analysis.
Supplemental Materials and Methods S1. Transient expression in Arabidopsis protoplasts and immunoprecipitation.
Supplementary Material
Acknowledgments
The authors thank Professor Nam-Hai Chua for providing the SINAT5 clones, Wilson Ardiles for sequencing, Martine De Cock for help in preparing the manuscript, Stephanie De Bodt for helpful comments for the phylogenetic analysis, and Marnik Vuylsteke for help with the statistical analysis.
This work was supported by the Research Foundation-Flanders (“Krediet aan Navorsers” grant nos. 1.5.088.99N and 1.5.192.01N), by the Institute for the Promotion of Innovation by Science and Technology in Flanders (predoctoral fellowship to G.D.H.), and by the Organisation for Economic Co-operation and Development (postdoctoral fellowship to M.R.C.).
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) are: Marcelle Holsters (marcelle.holsters@psb.ugent.be) and Sofie Goormachtig (sofie.goormachtig@psb.ugent.be).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Borisov AY, Morzhina EV, Kulikova OA, Tchetkova SA, Lebsky VK, Tikhonovich IA (1992) New symbiotic mutants of pea (Pisum sativum L.) affecting either nodule initiation or symbiosome development. Symbiosis 14 297–313 [Google Scholar]
- Brewin NJ (2004) Plant cell wall remodelling in the Rhizobium–legume symbiosis. Crit Rev Plant Sci 23 293–316 [Google Scholar]
- Callis J, Vierstra RD (2000) Protein degradation in signaling. Curr Opin Plant Biol 3 381–386 [DOI] [PubMed] [Google Scholar]
- Campbell GRO, Reuhs BL, Walker GC (2002) Chronic intracellular infection of alfalfa nodules by Sinorhizobium meliloti requires correct lipopolysaccharide core. Proc Natl Acad Sci USA 99 3938–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Rubin GM (1990) seven in absentia, a gene required for specification of R7 cell fate in the Drosophila eye. Cell 63 561–577 [DOI] [PubMed] [Google Scholar]
- Cheng H-P, Walker GC (1998) Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J Bacteriol 180 5183–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DR (1999) Medicago truncatula—a model in the making! Curr Opin Plant Biol 2 301–304 [DOI] [PubMed] [Google Scholar]
- Cooper SE (2007) In vivo function of a novel Siah protein in Drosophila. Mech Dev 124 584–591 [DOI] [PubMed] [Google Scholar]
- Corich V, Goormachtig S, Lievens S, Van Montagu M, Holsters M (1998) Patterns of ENOD40 gene expression in stem-borne nodules of Sesbania rostrata. Plant Mol Biol 37 67–76 [DOI] [PubMed] [Google Scholar]
- d'Azzo A, Bongiovanni A, Nastasi T (2005) E3 ubiquitin ligases as regulators of membrane protein trafficking and degradation. Traffic 6 429–441 [DOI] [PubMed] [Google Scholar]
- Dahiya P, Sherrier DJ, Kardailsky IV, Borisov AY, Brewin NJ (1998) Symbiotic gene sym31 controls the presence of a lectinlike glycoprotein in the symbiosome compartment of nitrogen-fixing pea nodules. Mol Plant Microbe Interact 11 915–923 [Google Scholar]
- Depaux A, Regnier-Ricard F, Germani A, Varin-Blank N (2006) Dimerization of hSiah proteins regulates their stability. Biochem Biophys Res Commun 348 857–863 [DOI] [PubMed] [Google Scholar]
- Devoto A, Muskett PR, Shirasu K (2003) Role of ubiquitination in the regulation of plant defence against pathogens. Curr Opin Plant Biol 6 307–311 [DOI] [PubMed] [Google Scholar]
- D'Haeze W, Gao M, De Rycke R, Van Montagu M, Engler G, Holsters M (1998) Roles for azorhizobial Nod factors and surface polysaccharides in intercellular invasion and nodule penetration, respectively. Mol Plant Microbe Interact 11 999–1008 [Google Scholar]
- Ellis C, Turner JG, Devoto A (2002) Protein complexes mediate signalling in plant responses to hormones, light, sucrose and pathogens. Plant Mol Biol 50 971–980 [DOI] [PubMed] [Google Scholar]
- Estelle M (2001) Proteases and cellular regulation in plants. Curr Opin Plant Biol 4 254–260 [DOI] [PubMed] [Google Scholar]
- Felsenstein J (2005) PHYLIP (Phylogeny Inference Package) version 3.6. Department of Genetics, University of Washington, Seattle. http://evolution.genetics.washington.edu/phylip.html (June 18, 2007)
- Franck T, Krueger R, Woitalla D, Müller T, Engelender S, Riess O (2006) Mutation analysis of the seven in absentia homolog 1 (SIAH1) gene in Parkinson's disease. J Neural Transm 113 1903–1908 [DOI] [PubMed] [Google Scholar]
- Freemont PS (1993) The RING finger. A novel protein sequence motif related to the zincfinger. Ann N Y Acad Sci 684 174–192 [DOI] [PubMed] [Google Scholar]
- Frugoli J, Harris J (2001) Medicago truncatula on the move! Plant Cell 13 458–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuba H, Yamashita H, Nagano Y, Jin HG, Hiji M, Ohtsuki T, Takahashi T, Kohriyama T, Matsumoto M (2007) Siah-1 facilitates ubiquitination and degradation of factor inhibiting HIF-1α (FIH). Biochem Biophys Res Commun 353 324–329 [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Ichige A, Walker GC (1993) A Rhizobium meliloti homolog of the Escherichia coli peptide-antibiotic transport protein Sbm1 is essential for bacteroid development. Genes Dev 7 1485–1497 [DOI] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A (2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82 373–428 [DOI] [PubMed] [Google Scholar]
- Habelhah H, Laine A, Erdjument-Bromage H, Tempst P, Gershwin ME, Bowtell DDL, Ronai Z (2004) Regulation of 2-oxoglutarate (α-ketoglutarate) dehydrogenase stability by the RING finger ubiquitin ligase Siah. J Biol Chem 279 53782–53788 [DOI] [PubMed] [Google Scholar]
- Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41 95–98 [Google Scholar]
- Hare PD, Seo HS, Yang J-Y, Chua N-H (2003) Modulation of sensitivity and selectivity in plant signaling by proteasomal destabilization. Curr Opin Plant Biol 6 453–462 [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67 425–479 [DOI] [PubMed] [Google Scholar]
- Hu G, Fearon ER (1999) Siah-1 N-terminal RING domain is required for proteolysis function, and C-terminal sequences regulate oligomerization and binding to target proteins. Mol Cell Biol 19 724–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro CAP, Weissman AM (2000) RING finger proteins: mediators of ubiquitin ligase activity. Cell 102 549–552 [DOI] [PubMed] [Google Scholar]
- Jones KM, Kabayashi H, Davies BW, Taga ME, Walker GC (2007) How rhizobial symbionts invade plants: the Sinorhizobium–Medicago model. Nat Rev Microbiol 5 619–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana A, Nakayama K, Williams S, Davis RJ, Mustelin T, Ronai Z (2006) Regulation of the Ring finger E3 ligase Siah2 by p38 MAPK. J Biol Chem 281 35316–35326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Jeong W, Ahn K, Ahn C, Kang S (2004) Siah-1 interacts with the intracellular region of polycystin-1 and affects its stability via the ubiquitin-proteasome pathway. J Am Soc Nephrol 15 2042–2049 [DOI] [PubMed] [Google Scholar]
- Kondorosi E, Redondo-Nieto M, Kondorosi A (2005) Ubiquitin-mediated proteolysis. To be in the right place at the right moment during nodule development. Plant Physiol 137 1197–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laus MC, Logman TJ, van Brussel AAN, Carlson RW, Azadi P, Gao M-Y, Kijne JW (2004) Involvement of exo5 in production of surface polysaccharides in Rhizobium leguminosarum and its role in nodulation of Vicia sativa subsp. nigra. J Bacteriol 186 6617–6625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Carthew RW, Lai Z-C (1997) Photoreceptor cell differentiation requires regulated proteolysis of the transcriptional repressor Tramtrack. Cell 90 469–478 [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR (1989) Interactive analysis of phylogeny and character evolution using the computer program MacClade. Folia Primatol (Basel) 53 190–202 [DOI] [PubMed] [Google Scholar]
- Mathis R, Van Gijsegem F, De Rycke R, D'Haeze W, Van Maelsaeke E, Anthonio E, Van Montagu M, Holsters M, Vereecke D (2005) Lipopolysaccharides as a communication signal for progression of legume endosymbiosis. Proc Natl Acad Sci USA 102 2655–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergaert P, Nikovics K, Kelemen Z, Maunoury N, Vaubert D, Kondorosi A, Kondorosi E (2003) A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol 132 161–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morzhina EV, Tsyganov VE, Borisov AY, Lebsky VK, Tikhonovich IA (2000) Four developmental stages identified by genetic dissection of pea (Pisum sativum L.) root nodule morphogenesis. Plant Sci 155 75–83 [DOI] [PubMed] [Google Scholar]
- Niehaus K, Lagares A, Pühler A (1998) A Sinorhizobium meliloti lipopolysaccharide mutant induces effective nodules on the host plant Medicago sativa (alfalfa) but fails to establish a symbiosis with Medicago truncatula. Mol Plant Microbe Interact 11 906–914 [Google Scholar]
- Perotto S, Brewin NJ, Kannenberg EL (1994) Cytological evidence for a host defense response that reduces cell and tissue invasion in pea nodules by lipopolysaccharide-defective mutants of Rhizobium leguminosarum strain 3841. Mol Plant Microbe Interact 7 99–112 [Google Scholar]
- Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70 503–533 [DOI] [PubMed] [Google Scholar]
- Piper RC, Luzio JP (2007) Ubiquitin-dependent sorting of integral membrane proteins for degradation in lysosomes. Curr Opin Cell Biol 19 459–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priefer UB (1989) Genes involved in lipopolysaccharide production and symbiosis are clustered on the chromosome of Rhizobium leguminosarum biovar viciae VF39. J Bacteriol 171 6161–6168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnoky P, Petrovics G, Kereszt A, Grosskopf E, Cam Ha DT, Bánfalvi Z, Kondorosi Á (1990) Rhizobium meliloti lipopolysaccharide and exopolysaccharide can have the same function in the plant–bacterium interaction. J Bacteriol 172 5450–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santelli E, Leone M, Li C, Fukushima T, Preece NE, Olson AJ, Ely KR, Reed JC, Pellecchia M, Liddington RC, et al (2005) Structural analysis of Siah1-Siah-interacting protein interactions and insights into the assembly of an E3 ligase multiprotein complex. J Biol Chem 280 34278–34287 [DOI] [PubMed] [Google Scholar]
- Shimomura K, Nomura M, Tajima S, Kouchi H (2006) LjnsRING, a novel RING finger protein, is required for symbiotic interactions between Mesorhizobium loti and Lotus japonicus. Plant Cell Physiol 47 1572–1581 [DOI] [PubMed] [Google Scholar]
- Stone SL, Hauksdóttir H, Troy A, Herschleb J, Kraft E, Callis J (2005) Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol 137 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma N, Nakamura Y, Yamamoto M, Ohta T, Koiwa T, Koiwa H, Akao S, Kawaguchi M (2003) The Lotus japonicus Sen1 gene controls rhizobial differentiation into nitrogen-fixing bacteroids in nodules. Mol Genet Genomics 269 312–320 [DOI] [PubMed] [Google Scholar]
- Susini L, Passer BJ, Amzallag-Elbaz N, Juven-Gershon T, Prieur S, Privat N, Tuynder M, Gendron M-C, Israël A, Amson R, et al (2001) Siah-1 binds and regulates the function of Numb. Proc Natl Acad Sci USA 98 15067–15072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tansengco ML, Hayashi M, Kawaguchi M, Imaizumi-Anraku H, Murooka Y (2003) crinkle, a novel symbiotic mutant that affects the infection thread growth and alters the root hair, trichome, and seed development in Lotus japonicus. Plant Physiol 131 1054–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellström V, Usadel B, Thimm O, Stitt M, Küster H, Niehaus K (2007) The lipopolysaccharide of Sinorhizobium meliloti suppresses defense-associated gene expression in cell cultures of the host plant Medicago truncatula. Plant Physiol 143 825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers ACJ, Auriac M-C, Truchet G (1999) Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126 3617–3628 [DOI] [PubMed] [Google Scholar]
- Tsyganov VE, Morzhina EV, Stefanov SY, Borisov AY, Lebsky VK, Tikhonovich IA (1998) The pea (Pisum sativum L.) genes sym33 and sym40 control infection thread formation and root nodule function. Mol Gen Genet 259 491–503 [DOI] [PubMed] [Google Scholar]
- Tsyganov VE, Voroshilova VA, Priefer UB, Borisov AY, Tikhonovich IA (2002) Genetic dissection of the initiation of the infection process and nodule tissue development in the Rhizobium-pea (Pisum sativum L.) symbiosis. Ann Bot (Lond) 89 357–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Velde W, Pérez Guerra JC, De Keyser A, De Rycke R, Rombauts S, Maunoury N, Mergaert P, Kondorosi E, Holsters M, Goormachtig S (2006) Aging in legume symbiosis. A molecular view on nodule senescence in Medicago truncatula. Plant Physiol 141 711–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasse J, de Billy F, Camut S, Truchet G (1990) Correlation between ultrastructural differentiation of bacteroids and nitrogen fixation in alfalfa nodules. J Bacteriol 172 4295–4306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veereshlingam H, Haynes JG, Penmetsa RV, Cook DR, Sherrier DJ, Dickstein R (2004) nip, a symbiotic Medicago truncatula mutant that forms root nodules with aberrant infection threads and plant defense-like response. Plant Physiol 136 3692–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vervliet G, Holsters M, Teuchy H, Van Montagu M, Schell J (1975) Characterization of different plaque-forming and defective temperate phages in Agrobacterium strains. J Gen Virol 26 33–48 [DOI] [PubMed] [Google Scholar]
- Vinardell JM, Fedorova E, Cebolla A, Kevei Z, Horvath G, Kelemen Z, Tarayre S, Roudier F, Mergaert P, Kondorosi A, et al (2003) Endoreduplication mediated by the anaphase-promoting complex activator CCS52A is required for symbiotic cell differentiation in Medicago truncatula nodules. Plant Cell 15 2093–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieghe K, Boudolf V, Beemster GTS, Maes S, Magyar Z, Atanassova A, de Almeida Engler J, De Groodt R, Inzé D, De Veylder L (2005) The DP-E2F-like DEL1 gene controls the endocycle in Arabidopsis thaliana. Curr Biol 15 59–63 [DOI] [PubMed] [Google Scholar]
- Wang M, Jin Y, Fu J, Zhu Y, Zheng J, Hu J, Wang G (2008) Genome-wide analysis of SINA family in plants and their phylogenetic relationships. DNA Seq 19 206–216 [DOI] [PubMed] [Google Scholar]
- Welsch R, Maass D, Voegel T, DellaPenna D, Beyer P (2007) Transcription factor RAP2.2. and its interacting partner SINAT2: stable elements in the carotenogenesis of Arabidopsis leaves. Plant Physiol 145 1073–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TC, Chin L-S, Li Y, Roudabush FL, Li L (2002) Regulation of synaptophysin degradation by mammalian homologues of Seven in Absentia. J Biol Chem 277 10273–10282 [DOI] [PubMed] [Google Scholar]
- Xie Q, Guo HS, Dallman G, Fang S, Weissman AM, Chua NH (2002) SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419 167–170 [DOI] [PubMed] [Google Scholar]
- Xie Q, Frugis G, Colgan D, Chua N-H (2000) Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev 14 3024–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L-R, Vega-Sánchez ME, Zhu T, Wang G-L (2006) Ubiquitination-mediated protein degradation and modification: an emerging theme in plant-microbe interactions. Cell Res 16 413–426 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.