Abstract
Prenylation is a posttranslational protein modification essential for developmental processes and response to abscisic acid. Following prenylation, the three C-terminal residues are proteoliticaly removed and in turn the free carboxyl group of the isoprenyl cysteine is methylated. The proteolysis and methylation, collectively referred to as CaaX processing, are catalyzed by Ste24 endoprotease or Rce1 endoprotease and by an isoprenyl cysteine methyltransferase (ICMT). Arabidopsis (Arabidopsis thaliana) contains single STE24 and RCE1 and two ICMT homologs. Here we show that in yeast (Saccharomyces cerevisiae) AtRCE1 promoted a-mating factor secretion and membrane localization of a ROP GTPase. Furthermore, green fluorescent protein fusion proteins of AtSTE24, AtRCE1, AtICMTA, and AtICMTB are colocalized in the endoplasmic reticulum, indicating that prenylated proteins reach this compartment and that CaaX processing is likely required for subcellular targeting. AtICMTB can process yeast a-factor more efficiently than AtICMTA. Sequence and mutational analyses revealed that the higher activity AtICMTB is conferred by five residues, which are conserved between yeast Ste14p, human ICMT, and AtICMTB but not in AtICMTA. Quantitative real-time reverse transcription-polymerase chain reaction and microarray data show that AtICMTA expression is significantly lower compared to AtICMTB. AtICMTA null mutants have a wild-type phenotype, indicating that its function is redundant. However, AtICMT RNAi lines had fasciated inflorescence stems, altered phylotaxis, and developed multiple buds without stem elongation. The phenotype of the ICMT RNAi lines is similar to farnesyltransferase β-subunit mutant enhanced response to abscisic acid2 but is more subtle. Collectively, the data suggest that AtICMTB is likely the major ICMT and that methylation modulates activity of prenylated proteins.
Prenylation is a posttranslational protein modification essential for the function of diverse proteins. Plant mutants lacking prenyltransferase function have pleotropic phenotypes including enlargement of the shoot apical meristem, hypersensitivity to abscisic acid (ABA), retarded growth rate, and delayed flowering (Cutler et al., 1996; Pei et al., 1998; Running et al., 1998, 2004; Bonetta et al., 2000; Yalovsky et al., 2000; Ziegelhoffer et al., 2000; Allen et al., 2002; Johnson et al., 2005).
Prenylation involves the attachment of either the 15-carbon farnesyl or the 20-carbon geranygeranylgeranyl isoprenoid moieties to conserved CaaX box or double Cys C-terminal sequence motifs. Protein farnesyltransferase (PFT) attaches the farnesyl group from farnesyldiphosphate prenyl group donor via a thioether linkage to the Cys of a CaaX box sequence motif (C, Cys; a, usually an aliphatic amino acid; X can be any amino acid but commonly Ser, Met, Cys, Ala, or Gln). A second protein prenyltransferase, geranylgeranyltransferase type I (PGGT-I), uses geranylgeranyldiphosphate to modify CaaX box containing proteins in which X residue is usually a Leu. A third protein prenyltransferase, Rab geranylgeranyltransferase, uses geranylgeranyldiphosphate to modify C-terminal double Cys motifs in Rab proteins (Zhang and Casey, 1996; Yalovsky et al., 1999; Galichet and Gruissem, 2003; Maurer-Stroh et al., 2003).
Prenylation is followed by the proteolytic cleavage of the last three amino acids of the protein (aaX), leaving an isoprenyl carboxylate group at the C-terminal end of the protein. The prenylated isoprenyl Cys is in turn methylated (Supplemental Fig. S1). The proteolytic cleavage and methylation of prenylated proteins are collectively referred to as CaaX processing (Young et al., 2000). The prenyl-dependent endoproteolysis is catalyzed by either of the two endoproteases, designated Rce1 and Ste24 (Afc1; Boyartchuk et al., 1997; Young et al., 2000). The methylation of the newly exposed isoprenyl carboxylate group is catalyzed by an isoprenyl carboxy methyltransferase (Icmt) also known as Ste14 in yeast (Saccharomyces cerevisiae; Clarke, 1992; Romano et al., 1998; Young et al., 2000). In yeast and animal cells, the CaaX proteases and methyltransferase are localized in the endoplasmic reticulum (ER) membranes, unlike the prenyltransferases, which are soluble enzymes (Dai et al., 1998; Romano et al., 1998; Schmidt et al., 1998). This indicates that CaaX proteins are prenylated in the cytoplasm and then processed in the endomembrane system. In animals, CaaX processing affects fundamental processes. In fibroblasts derived from Rce1 or Icmt knockout mice, farnesylated Ras but not Rho proteins were mislocalized from the plasma membrane to the endomembrane (Kim et al., 1999; Bergo et al., 2001; Michaelson et al., 2005). Rce1 and Icmt knockout mice die during embryogenesis (Kim et al., 1999; Bergo et al., 2001) and the mutations affect oncogenic Ras transforming ability (Bergo et al., 2002, 2004). Disruption of Ste24 induces premature aging, a condition known as Hutchinson-Gilford progeria syndrome, resulting from accumulation of prenylated unprocessed prelamin A (Bergo et al., 2002; Pendas et al., 2002; Young et al., 2005).
In Arabidopsis (Arabidopsis thaliana), CaaX proteolysis is catalyzed by conserved AtSTE24 and AtRCE1/AtFACE-2 (Bracha et al., 2002; Cadinanos et al., 2003). AtSTE24 was demonstrated to be localized in the ER (Bracha et al., 2002). However, the subcellular localization of AtRCE1/AtFACE-2 has not been determined, nor has its effect on subcellular localization of prenylated plant proteins. Plant protein extracts have been shown to contain ICMT activity (Crowell et al., 1998; Rodriguez-Concepcion et al., 2000), and two genes encoding ICMT have been cloned (Rodriguez-Concepcion et al., 2000; Crowell and Kennedy, 2001; Narasimha Chary et al., 2002). Following the identification of the second ICMT gene in Arabidopsis, the two genes were designated AtICMTA/AtSTE14A and AtICMTB/AtSTE14B (Narasimha Chary et al., 2002). It was shown that AtICMTA and AtICMTB are differentially expressed and that in vitro AtICMTB had higher catalytic activity (Narasimha Chary et al., 2002). Treatment of plants with high concentration of the methyltransferase inhibiter acetyl farnesyl Cys (AFC) induced ABA hypersensitivity (Narasimha Chary et al., 2002), reminiscent of the ABA hypersensitivity of enhanced response to abscisic acid1 (era1) PFT β-subunit, PGGT-I β-subunit, and pluripetala (plp) PFT and PGGT-I α-subunit mutants (Cutler et al., 1996; Pei et al., 1998; Running et al., 2004; Johnson et al., 2005).
CaaX processing has been suggested to further increase protein affinity to cell membranes (Hancock et al., 1991; Kato et al., 1992; Ghomashchi et al., 1995; Parish et al., 1995), protect proteins from degradation (Backlund, 1997), and facilitate functional interactions with other proteins (Higgins and Casey, 1994; Rosenberg et al., 1998). More recently it has been shown that CaaX processing is required for subcellular targeting of prenylated proteins. Consistent with the ER localization of CaaX processing enzymes (Dai et al., 1998; Romano et al., 1998; Schmidt et al., 1998), Ras proteins accumulate in the endomembrane system in Rce1 and Icmt mutant cells (Michaelson et al., 2005).
In plants treated with the ICMT inhibitor AFC, targeting of the prenylated calmodulin CaM53 to the plasma membrane was compromised and it accumulated in endomembranes (Rodriguez-Concepcion et al., 2000).
This work highlights the conservation and uniqueness of CaaX processing in plants. We show that all the CaaX processing enzymes are localized in the ER and can complement respective yeast mutants. Uniquely, Arabidopsis has two ICMT horologes, ICMTA and ICMTB. In yeast cells ICMTA is less active than ICMTB. Sequence and mutational analyses revealed the residues responsible for this activity difference. Real-time reverse transcription (RT)-PCR and whole-genome expression analysis show that ICMTA expression levels are significantly lower than ICMTB. A T-DNA mutant analysis shows that ICMTA function is redundant but that ICMT RNA interference (RNAi) plants display subtle phenotypes reminiscent of era1 PFT β-subunit and plp PFT and PGGT-I shared α-subunit mutants.
RESULTS
Arabidopsis AtRCE1 Is Functionally Conserved
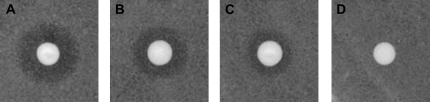
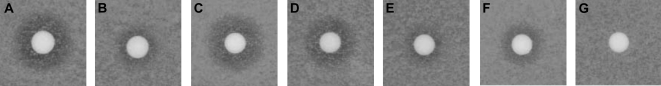
RCE1 is considered as the major CaaX protease. The function of AtRCE1 was tested by complementation in yeast using pheromone diffusion halo assays (Supplemental Fig. S2; Bracha et al., 2002). This assay can be used to quantitatively evaluate CaaX processing efficiency in vivo since the size of the growth inhibition halos correspond to the efficiency of a-factor processing (Tam et al., 1998; Schmidt et al., 2000; Trueblood et al., 2000). AtRCE1 complemented a-factor secretion in ste24Δ rce1Δ double-mutant yeast strain (JRY6959) when expressed from a 2μ high-copy-number plasmid (Fig. 1). Unlike Ste24, Rce1 can only catalyze CaaX proteolysis but not the additional proteolytic step required for maturation and of a-factor and its efficient secretion (Supplemental Fig. S2; Boyartchuk and Rine, 1998; Tam et al., 1998). The halos that formed around the AtRCE1-expressing ste24Δ rce1Δ cells (SYY563) were smaller compared to the halos that formed around the SYY500 AtSTE24-expressing ste24Δ rce1Δ cells (Fig. 1, B and C), indicating that a-factor secretion was indeed less efficient. The halos that formed around Ste24 Rce1 wild-type cells (JRY6958) were bigger than either AtSTE24- or AtRCE1-complemented cells (Fig. 1A), indicating that a-factor processing by neither plant CaaX proteases was as efficient as by yeast Ste24. These data corroborated functional analysis of AtRCE1 function in vitro (Cadinanos et al., 2003).
Figure 1.
Arabidopsis AtSTE24 and AtRCE1 facilitate yeast a-factor processing. Growth inhibition halos of a-factor hypersensitive sst2α cells formed around wild-type Ste24 Rce1 cells (A), ste24Δ rce1Δ expressing AtSTE24 (B), or ste24Δ rce1Δ cells expressing AtRCE1 (C), but not around ste24Δ rce1Δ cells transformed with empty vector control (D).
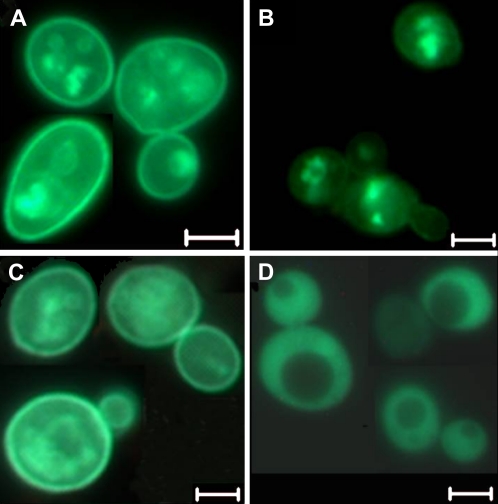
To determine whether AtRCE1 can process plant target proteins, we created a yeast strain in which AtRCE1 and GFP-AtROP9 (GFP-AtRAC7) were coexpressed in ste24Δ rce1Δ mutant background (SYY569; Fig. 2). AtROP9 (AtRAC7) is a type-II ROP/RAC GTPase that is farnesylated in yeast (Lavy et al., 2002). In wild-type Ste24 Rce1 background (SYY538), GFP-AtROP9 was localized in the plasma membrane and endomembrane (Fig. 2A). In contrast, in ste24Δ rce1Δ double-mutant background (SYY542), GFP-AtROP9 was only localized in the endomembrane fraction (Fig. 2B). Coexpression of AtRCE1 (SYY569) restored plasma membrane localization of GFP-ROP9, confirming that AtRCE1 processed AtROP9 (Fig. 2C). The nonprenylated GFP:Atrop9mS mutant in which the prenyl acceptor Cys was mutated to Ser was dispersed in the cytoplasm following expression in Ste24 Rce1 wild-type cells (SYY539; Fig. 2D). These data show that prenylation promotes association of proteins with the endomembrane system, while CaaX processing is required for subcellular targeting.
Figure 2.
AtRCE1 promotes plasma membrane localization of GFP:AtROP9 in ste24Δ rce1Δ double-mutant yeast cells. A, GFP:AtROP9 detected in both plasma membranes and endomembranes following expression in wild-type Ste24 Rce1 cells. B, GFP:AtROP9 detected only in the endomembranes in ste24Δ rce1Δ double-mutant cells. C, AtRCE1 restored plasma membrane localization of GFP-AtROP9 in ste24Δ rce1Δ cells. D, Nonprenylated GFP:Atrop9mS mutant protein dispersed in the cytoplasm following expression in wild-type Ste24 Rce1 cells. Bars are 5 μm. [See online article for color version of this figure.]
Subcellular Localization of CaaX Processing Enzymes
We have previously demonstrated that AtSTE24 is localized in the ER (Bracha et al., 2002) and that ICMTA is localized in endomembranes (Rodriguez-Concepcion et al., 2000). If in plants too prenylated proteins reach the ER, where they undergo CaaX processing, AtRCE1, ICMTA, and ICMTB should also be localized in the ER. To test this assumption, the subcellular localization of AtRCE1, AtICMTA, and AtICMTB was determined.
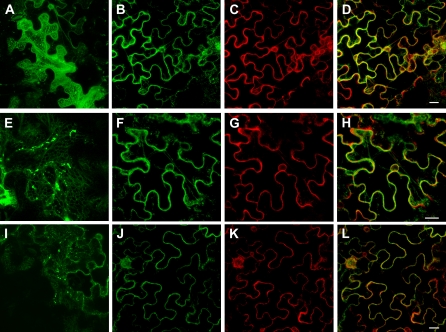
Following transient expression, AtICMTA, AtICMTB, and AtRCE1 GFP fusion proteins had typical to ER, reticulate, and perinuclear subcellular distribution patterns (Fig. 3, A, E, and I; Supplemental Fig. S3, B, D, E, and G). Colocalization analysis of AtICMTA, AtICMTB, and AtRCE1 together with the ER marker ER-mCherry (Nelson et al., 2007) were carried out to confirm the ER localization. The results (Fig. 3, B–D, F–H, and J–L) show that fusion proteins of all three CaaX processing enzymes were colocalized with the marker confirming their ER localization. Colocalization of cyan fluorescent protein (CFP)-AtICMTA with yellow fluorescent protein (YFP)-AtSTE24 and of YFP-ICMTA with CFP-AtRCE1 was used to further confirm the ER colocalization of the CaaX processing enzymes in the ER (Supplemental Fig. S3, A–F).
Figure 3.
Subcellular localization of AtICMTA, AtICMTB, and AtRCE1 GFP fusion proteins following transient expression in N. benthamiana leaf epidermal cells. GFP-AtICMTA (A–D), GFP-AtICMTB (E–H), and GFP-AtRCE1 (I–L) were expressed alone (A, E, and I) or coexpressed with the ER-mCherry (Nelson et al., 2007; B–D, F–H, and J–L). Shown are GFP channel (B, F, and J), mCherry channel (C, G, and K), and GFP/mCherry overlay (D, H, and L). Bars are 10 μm.
ICMTB Is Likely the Major Carboxymethyl Isoprenyl Transferase in Arabidopsis
Based on its reversibility, the isoprenyl carboxy methylation step has been suggested to play a regulatory role (Young et al., 2000). Recently, a prenyl Cys methylesterase has been identified in Arabidopsis (Deem et al., 2006), suggesting that methylation/demethylation cycles of prenylated proteins may regulate their function. This has prompted us to study the role of prenyl Cys methytransferases and their function in Arabidopsis
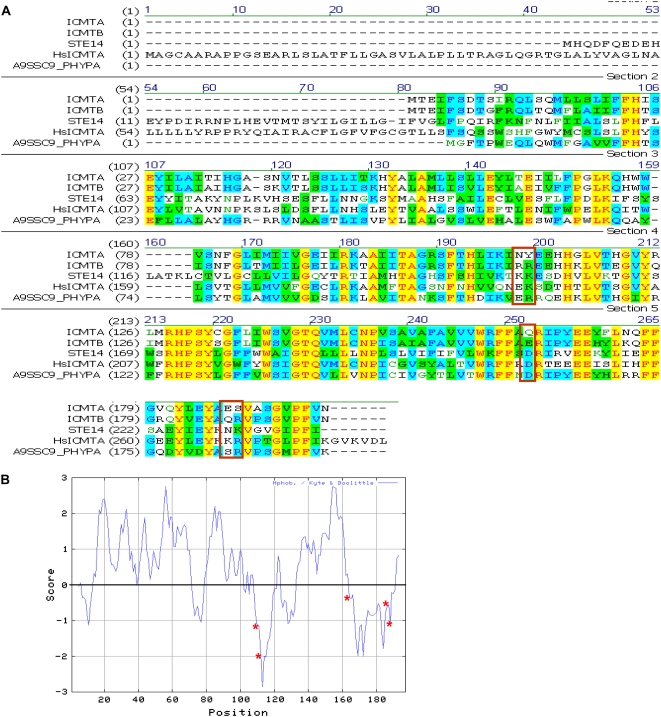
In vitro, AtICMTB/AtSTE14B (At5g08335) was shown to be more active than AtICMTA/AtSTE14A (At5g23320; Narasimha Chary et al., 2002). Sequence comparison highlighted five amino acids that are conserved between AtICMTB and yeast Ste14p and differ in AtICMTA (Fig. 4A, red rectangles). These residues include: K154 and K155 in Ste14p and R111 and R112 in AtICMTB, D208 in Ste14p and E165 in AtICMTB, N230 and K231 in Ste14p, and Q187 and R188 in AtICMTB. Interestingly, these residues are also structurally conserved in human (HsICMT) and moss Physcomitrella patens (PpICMT) ICMT proteins (Fig. 4A). Hydropathy plot analysis showed that AtICMTB and AtICMTA have six to eight transmembrane helices and that their overall topology is similar to Ste14p (Fig. 4B). Interestingly, all the five residues that are conserved between AtICMTB and Ste14p, HsICMT and PpICMT, and differ in AtICMTA are located on hydrophylic domains (Fig. 4B, asterisks), which were shown to be essential for Ste14p activity (Romano and Michaelis, 2001).
Figure 4.
Sequence alignment of AtICMTA, AtICMTB, yeast Ste14p, and human and P. patens ICMT. A, Identical residues in all five proteins are highlighted in yellow. Identical residues between two of the proteins are highlighted in blue. Similar residues are highlighted in green. Red rectangles denote five residues that are structurally conserved between AtICMTB and Ste14p human and moss ICMTs and differ in AtICMTA. B, Kyte and Doolittle hydropathy plot of ICMTB with a nine-amino acid window size. Hydrophobic residues appear above the midline and hydrophilic residues below the midline. Asterisks (*) mark the positions of the AtICMTB Ste14, HsICMT, PhyPaICMT conserved, and AtICMTA unique residues.
Pheromone diffusion halo assays were carried out to compare the activities of AtICMTA and AtICMTB in yeast and whether differences in their ability to process a-factor correspond to the structural conservation between AtICMTB and Ste14p (Fig. 5). The halos that formed around ste14Δ mutant cells that expressed AtICMTB from a 2μ high-copy-number plasmid (SYY564) were larger than the halos that formed around ste14Δ cells that expressed AtICMTA (SYY526) that are comparable in size to the halos that formed around wild-type Ste14 cells (Fig. 5, A–C). When AtICMTB was expressed from a single-copy CEN plasmid (SYY565), the halo size was smaller, indicating that methylation of a-factor by AtICMTB was less efficient than by yeast Ste14. An AtICMTA mutant, AticmtAmR111R112E165Q187R188, harboring point mutations that converted five amino acids to their corresponding residues in AtICMTB (N111/R, Y112/R, Q165/E, E187/Q, and S188/R) was created. The halos that formed around ste14Δ cells that expressed AticmtAmR111R112E165Q187R188 (SYY579 and SYY578) were comparable to the halos that formed around AtICMTB-expressing cells (Fig. 5, D and F). Halo sizes were calculated relative to wild-type Ste14 cells, average size of which was taken as 1. The average halo size around AtICMTA-complemented cells (when expressed from a 2μ plasmid) was 0.57 of wild-type Ste14 cells with a se of ±0.09. The average halo sizes around AtICMTB- and AticmtAmR111R112E165Q187R188-complemented cells (expressed from 2μ plasmids) were 1.05 ± 0.07 and 0.96 ± 0.06, respectively. These results indicated that the a-factor was processed at similar efficiencies by AtICMTB and the mutant AticmtAmR111R112E165Q187R188 and confirmed that the reduced activity of AtICMTA results from changes in structurally conserved residues rather than technical problems of the experimental setup. Interestingly, activity of the AticmtAmE165 single mutant and the AticmtAmR111R112E165 triple mutant was similar to wild-type AtICMTA (data not shown), suggesting that all five residues are critical for efficient ICMT activity.
Figure 5.
Pheromone diffusion halo assay identifies amino acids responsible for reduced efficiency of a-factor processing by AtICMTA. Growth inhibition halos produced by a-factor secretion on a mat of sst2α cells. A, Wild-type Ste14 cells. B to G, ste14Δ mutants cells expressing: AtICMTA (B), AtICMTB (C), AticmtAmR154R155E165Q187R188 (D), AtICMTB (E), AticmtAmR154R155E165Q187R188 (F), and vector control (G). B, C, D, and G cells were transformed with 2μ high-copy-number plasmids. E and F cells were transformed with CEN single-copy plasmids.
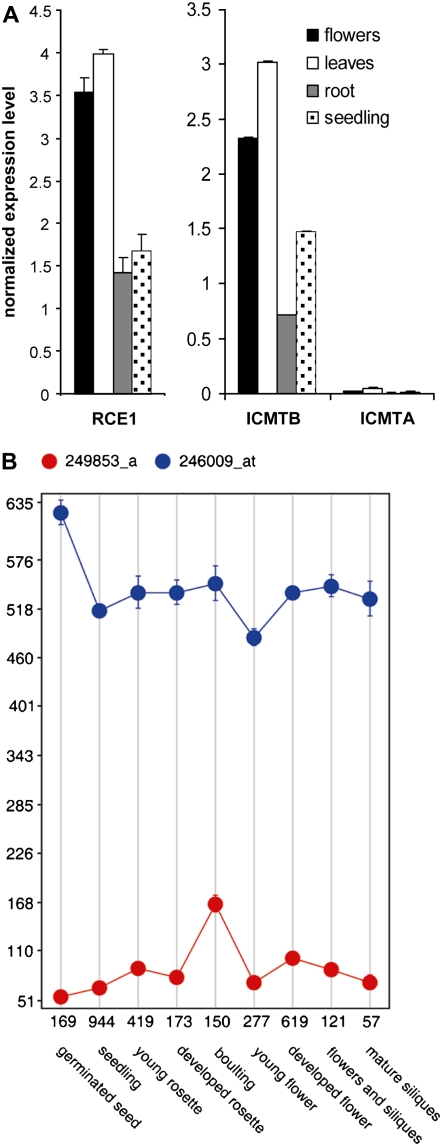
Real-time quantitative RT-PCR (Q-PCR) experiments showed that AtICMTB was expressed at significantly higher levels compared to AtICMTA (Fig. 6A). The Q-PCR data were corroborated by examining microarray expression data using Genevestigator (Zimmermann et al., 2004; Laule et al., 2006; Fig. 6B; Supplemental Fig. S4). The microarray data show that expression of ICMTA is low in most tissues and throughout development and that in all cases the expression of ICMTB is higher. The Q-PCR analyses showed that AtRCE1 is expressed at similar levels to AtICMTB (Fig. 6A). Unfortunately, AtRCE1 (At2G36305) is currently not represented on the ATH1 Affymetrix microarray chips and thus whole-genome analysis of its expression is currently unavailable. The high sensitivity of the Q-PCR enabled detection of slightly higher relative expression levels of AtRCE1 and AtICMTB in flowers and leaves relative to roots and seedlings.
Figure 6.
Expression levels of AtICMTA and AtICMTB and AtRCE1. A, Q-PCR determined relative and normalized expression levels of AtRCE1, AtICMTA, and AtICMTB. ACTIN was use as internal control in the Q-PCR. Average was calculated using three replicates. The error bars represent sd. B, Microarray data taken from Genevestigator (Zimmermann et al., 2004; Laule et al., 2006) show the expression levels of AtICMTA (red) and AtICMTB (blue) during development of Arabidopsis.
The activity and expression data suggested that AtICMTA has a minor or restricted function and that AtICMTB is likely the major isoprenyl methyltransferase in Arabidopsis. Mutant analyses were carried out to test this assumption.
The Function of ICMTA and ICMTB in Arabidopsis
An Arabidopsis line (P65H9) with a T-DNA insertion three nucleotides upstream of the termination TGA codon of AtICMTA (Supplemental Figs. S5 and S6) was identified in screens of the University of Wisconsin Biotechnology Center T-DNA collection (www.biotech.wisc.edu). No RNA was detected in AticmtA−/− homozygote mutant plants by RT-PCR with AtICMTA gene-specific primers (Supplemental Figs. S5 and S7; Supplemental Table S1), indicating the mutant is a null. An RT-PCR reaction on the same RNA template using AtICMTB-specific primers (Supplemental Figs. S5 and S7; Supplemental Table S1) showed that the mutation had no effect on its expression (Supplemental Fig. S5). Cloning and sequencing showed that the weak band that was amplified by the AtICMTA-specific primers was AtICMTB. The AticmtA−/− plants had no visible phenotype including increased sensitivity to ABA, indicating that AtICMTA function is redundant with ICMTB.
Unfortunately, no mutant with compromised AtICMTB expression has been identified. To further explore the function of isoprenyl Cys methylation in plants we generated ICMT RNAi plants (Aticmtsil). These plants expressed two 380-bp fragments of the ICMTA coding region in reverse orientation. ICMTA and ICMTB show approximately 80% identity at the nucleotide level (Supplemental Fig. S7), making it highly likely that the RNAi construct would silence both ICMT genes. One hundred and ten independent transgenic lines were isolated, of which a total of 63 (70%) had a similar abnormal phenotype (Fig. 7). Six independent T1 lines were selected for further analysis.
Figure 7.
Phenotype of ICMT RNAi plants. A to C, Altered phylotaxis of flower arrangement on inflorescence stems. D, Development of multiple axillary buds. E, A fasciated inflorescence stem. F, Adjacent siliques on a stem tip results from development of multiple buds. G, Wild-type Col-0 inflorescence stem. H, era1-2 inflorescence stem. Note the altered phylotaxis (arrowheads). [See online article for color version of this figure.]
Aticmtsil plants had altered phyllotaxis (Fig. 7, A–C, E, and F). Flowers developed from the same region of the inflorescence stem with minimal or no elongation of the internodes (Fig. 7, A–C and F). Occasionally, secondary axillary flowers developed within flowers, indicating partial conversion of flowers into inflorescence (Fig. 7C). Siliques developed without internode elongation (Fig. 7F). Multiple buds developed in the axil of cauline leaves (Fig. 7D). All the plants developed fasciated bifurcated stems (Fig. 7E). Interestingly, altered phyllotaxis, fasciated stems, and development of axillary flowers were also observed in the era1 PFT β-subunit mutant plants (Fig. 7H), suggesting functional relatedness of the two genes. Unlike in era1 and plp plants (Running et al., 1998; Yalovsky et al., 2000; Running et al., 2004), Aticmtsil flowers did not have extra flower organs or additional rosette leaves and were not late flowering and partially male sterile.
It was difficult to observe consistent reduction in mRNA levels in the Aticmtsil plants, suggesting a posttranscriptional silencing mechanism. To prove that the phenotype of the plants was associated with the ICMT RNAi we monitored the cosegregation of the mutant phenotype and the kanamycin-resistant selection marker. To this end, approximately 100 seedlings from each of three independent transgenic lines were grown on soil without selection and their phenotypes were scored. Plants were allowed to self, their seeds were harvested, germinated on a kanamycin-containing selection medium, and the ratios between resistant and sensitive seedlings were determined. In turn, plants were transferred from the selection plates to soil and their phenotypes were scored again. In nonsegregating populations in which all the plants had a mutant phenotype 100% of the progeny were kanamycin resistant. The progeny of plants without a mutant phenotype were all kanamycin sensitive. The progeny of plants heterozygote to the RNAi segregated in a 3 to 1 ratio with respect to the selection marker. All the kanamycin-resistant plants showed a mutant phenotype after transferring to soil. In contrast, rescued kanamycin-sensitive plants that were transferred to soil did not show the mutant phenotype. The segregation analysis confirmed that the abnormal phenotype of the Aticmtsil plants were associated with the AtICMT RNAi transgene.
DISCUSSION
In this and a previous work (Bracha et al., 2002), we showed that the plant CaaX processing machinery is evolutionarily conserved. The Arabidopsis CaaX proteases AtSTE24 and AtRCE1 can complement corresponding yeast mutants promoting a-factor processing (Fig. 1; Bracha et al., 2002). Proteolysis of plant prenylated proteins by either AtRCE1 or AtSTE24 was sufficient to promote their plasma membrane localization in yeast cells (Fig. 2; Bracha et al., 2002). Both AtRCE1 and AtSTE24 are expressed in most tissues (Fig. 6; Supplemental Fig. S4; Bracha et al., 2002). All CaaX processing enzymes are localized in the ER (Fig. 3), like their homologs in yeast and mammalian cells. These findings strongly suggest that following prenylation, proteins are targeted to the ER where they undergo CaaX processing, which in turn is likely required for their subcellular targeting. Our expression, activity, and mutant analyses suggests that AtICMTB is likely the major prenyl-dependent carboxy methyltransferase in Arabidopsis and that AtICMTA function is most likely redundant (Figs. 4–7). The similarity of the Aticmtsil lines to era1 and plp suggests that isoprenyl Cys methylation modulates the function of prenylated proteins.
Evaluation of CaaX Processing Activity by Plant CaaX Proteases in Yeast
Both Ste24 and Rce1 can carry out the CaaX processing cleavage of prenylated a-factor. The processed a-factor is then methylated and in turn cleaved again by Ste24 at the internal N-terminal site and then by Ste23 to yield the mature secreted a-factor (Supplemental Fig. S2; Boyartchuk and Rine, 1998; Tam et al., 1998). The Arabidopsis AtRCE1 homolog has been previously identified and characterized biochemically (Cadinanos et al., 2003). When expressed in rce1Δ ste24Δ mutant cells, AtRCE1 was able to restore a-factor production (Fig. 1), demonstrating its functionality as a CaaX protease in vivo in yeast. The yeast Ste24 and Rce1 enzymes are known to have partially overlapping substrate specificity (Trueblood et al., 2000). It was not known whether this specificity pattern extends to other organisms. Our results indicate that this may be the case. We found that the substrate specificity of AtRCE1 is partially overlapping with that of AtSTE24 as demonstrated by their ability to process both the Arabidopsis AtROP9 (Fig. 2C; Bracha et al., 2002) and the yeast a-factor. The halos that formed around the AtRCE1-complemented ste24Δ rce1Δ cells were smaller than the halos formed around cells that were complemented with AtSTE24. The differences in halo sizes indicate a-factor processing by AtRCE1 was less efficient than by AtSTE24, likely owing the dual role of AtSTE24 in a-factor processing.
The subcellular localization of the farnesylated GFP-AtROP9 in wild-type, rce1Δ ste24Δ, and rce1Δ ste24Δ cells complemented with AtRCE1 revealed a role for CaaX proteolysis in targeting of prenylated protein to the plasma membrane. Consistently, in fibroblasts derived from rce1 or icmt knockout mice, farnesylated Ras accumulated in the endomembrane (Michaelson et al., 2005). Likewise, the geranylgeranylated petunia (Petunia hybrida) calmodulin CaM53 (Rodriguez-Concepcion et al., 1999; Caldelari et al., 2001) accumulated in the endomembranes following transient expression of Nicotiana benthamiana leaf epidermal cells treated with the carboxymethylation inhibitor AFC (Rodriguez-Concepcion et al., 2000).
Subcellular Targeting of Prenylated Proteins
Our results suggest the following targeting pathway for CaaX proteins in plant cells. Nascent CaaX proteins are synthesized in the cytosol, where they encounter one of two protein prenyltransferases that modify the CaaX Cys by thioether linkage of a C-15 farnesyl or C-20 geranylgeranyl lipid. Prenylated CaaX proteins have high affinity for the ER, where they encounter a prenyl-CaaX-specific protease, either AtSTE24 or AtRCE1 that removes the -aaX amino acids. The newly exposed C-terminal isoprenyl Cys then becomes a substrate for ER-localized ICMT that methylesterifies the α-carboxyl group of the isoprenyl Cys. The end product of this three-step posttranslational modification process contains a hydrophobic domain at the C terminus of an otherwise hydrophilic protein. It remains to be established whether the fully modified proteins in plant cells arrive at their final location via the secretory pathway or through a different route.
The Function of ICMTA and ICMTB
AtICMTB/AtSTE14B had lower KM and a higher catalytic activity compared to AtICMTA/AtSTE14A in carboxy methylation assays in vitro. Expression analysis demonstrated that AtICMTB/AtSTE14B is more widely expressed than AtICMTA/AtSTE14. Based on these observations, it has been suggested that AtICMTB/STE14B is the major prenyl-dependent carboxy methyltransferase in Arabidopsis (Narasimha Chary et al., 2002). In an extension to this work, we show that AtICMTB also has higher activity in yeast. Analysis of the sequence differences between AtICMTA and AtICMTB revealed five amino acids conserved between AtICMTB, yeast Ste14p, human, and moss ICMT proteins that differ in ICMTA (Fig. 4). Our results suggest that these five amino acids are responsible for the different activities of AtICMTA and AtICMTB (Fig. 5).
Based on a topology model, Ste14p contains six membrane spans, two of which form a helical hairpin. According to this model, most of the Ste14p hydrophilic regions are located in the cytosol (Romano and Michaelis, 2001). Hydropathy analysis suggests that the overall topology of Ste14p is preserved among all members of the ICMT family (Romano and Michaelis, 2001). AtICMTB and AtICMTA have similar topology and contain between six to eight transmembrane helices (Fig. 4B). In contrast to their differing N-terminal topologies, ICMT proteins from different organisms share similar C-terminal hydropathy profiles (Romano and Michaelis, 2001).
Sequence analysis revealed that four of the five amino acids unique to AtICMTA (N111, Y112, Q165, and S188) introduce neutral or hydrophobic residues into cytosolic spans, which may disrupt the topology or structure of AtICMTA. It is noteworthy that Q165 is located in the region close to the end of the transmembrane span and the beginning of the cytosolic part (Fig. 4B). The charged amino acid E187 in AtICMTA is represented by the neutral residues Q187 and N230 in AtICMTB and Ste14p, respectively. Our findings that activity of AtICMTA is reduced compared to AtICMTB and that substituting all five residues to AtICMTB-like restored activity in yeast (Fig. 5) are consistent with the hypothesis that residues in the C-terminal hydrophilic domains play a critical role in the function of ICMT (Romano and Michaelis, 2001).
Interestingly, querying of plant sequence databases revealed small families of ICMT proteins in monocot and dicot of plant species. For example, we identified four putative ICMT proteins in rice (Oryza sativa; Supplemental Fig. S8) and in grape vine (Vitis vinifera; data not shown). The sequence alignment of ICMT protein from Arabidopsis, poplar (Populus spp.), rice, Physomitrella, yeast, and human shows that while AtICMTA contains a Ser residue at position 188, all the ICMT proteins have positively charged Lys or Arg residues at this position (Supplemental Fig. S8). As noted above, activity of AticmtAmE165 single mutant and the AticmtAmR111R112E165 triple mutant was similar to wild-type AtICMTA (data not shown). Taken together, these data suggest that a positively charged residue at position 188 is critical for catalytic activity of ICMT. In contrast, the charged residues at positions 111 and 112 are less conserved (Supplemental Fig. S8). The existence of small ICMT protein families in different plant species and identity between AtICMTA and poplar ICMT at positions 111 (N) and 165 (Q) and between AtICMTA and rice ICMT protein at position 112 (Y) may indicate that the ICMT proteins have diverged early during the evolution of higher plants.
The Q-PCR analysis showed that AtICMTB is expressed at significantly higher levels compared to AtICMTA (Fig. 6), confirming an earlier finding (Narasimha Chary et al., 2002). We further extended the expression analysis using microarray data available on the web (Zimmermann et al., 2004; Laule et al., 2006). This analysis confirmed the Q-PCR experiments and demonstrated that in all tissues, developmental stages, and under any stimuli, the expression of AtICMTB is higher (Fig. 6; Supplemental Fig. S4).
The activity and expression analysis strongly suggested that AtICMTB is the major isoprenyl carboxymehtyltransferase in Arabidopsis. The lack of noticeable developmental abnormalities and ABA hypersensitivity in the AtICMTA knockout mutant supports this conclusion. In contrast, Aticmtsil plants had distinguishable developmental alterations, implicating that likely function of both ICMTA and ICMTB genes has been compromised. While unique AtICMTA activities cannot be ruled out at this stage, it is questionable whether it has any essential function.
The Phenotype of Aticmtsil Plants
The fasciation and bifurcation of the stem and the altered phyllotaxis of the Aticmtsil plants suggested that organization of the shoot apical meristem was compromised. A similar phenotype was observed in the era1-2 PFTB and plp PFTA/PGGTA mutants (Yalovsky et al., 2000; Running et al., 2004). Thus, prenylation and CaaX processing are required for proper function of the shoot apical meristem. Interestingly, the Aticmtsil plants were not ABA hypersensitive (data not shown).
The relatively mild phenotype of the Aticmtsil plants could have been a result of incomplete silencing of AtICMTA and AtICMTB expression. Alternatively, the mild phenotype could be due the partial function of prenylated but nonmethylated proteins. Although it was difficult to observe consistent reduction in RNA levels of AtICMTA and AtICMTB in the RNAi lines, the presence of the mutant phenotype in 70% of the T1 lines and the strict cosegregation of the abnormal phenotype with the RNAi construct indicate that the expression of the ICMT genes was very likely affected. The absence of similar phenotypes in the icmtA−/− T-DNA knockout plants and the 80% sequence identity at the nucleotide level between AtICMTA and AtICMTB further support the notion that the RNAi affected expression of both genes.
It is noteworthy that the relatively subtle phenotype of the Aticmtsil plants is consistent with findings on the function of ICMT yeast and mammals. Yeast rce1Δ ste24Δ and ste14Δ mutants cannot mate but their growth rate is normal. In contrast, ram1Δ (PFT β-subunit mutant) cells display a retarded growth rate due to malfunction of Ras1p and Ras2p and cdc43Δ (PGGT-I β-subunit) mutant cells are nonviable, primarily due to the malfunction of Cdc42p (Trueblood et al., 1993). Thus, in yeast prenylated nonmethylated Ras and Cdc42 proteins are still functional but the a-factor is not processed and secreted. Rce1 and Icmt knockout mice die during embryogenesis but fibroblasts obtained from these embryos can be maintained in culture (Michaelson et al., 2005), indicating that cell division was not completely compromised. Future analysis will be required to determine whether in plants many prenylated nonmethylated proteins are still functional.
The importance of α-carboxyl methylation of prenylated proteins is illustrated by the fact that this is the final and the only potentially reversible step during prenylation and CaaX processing. Consequently, the targeting and function of prenylated plant proteins may be modulated by the methylation/demethylation status of the proteins. Consistent with this hypothesis, specific prenyl Cys methyl ester hydrolase activities have been described in mammals and more recently also in plants (Perez-Sala et al., 1991; Tan and Rando, 1992; Valentijn and Jamieson, 1998; Van Dessel et al., 2001; Deem et al., 2006).
MATERIALS AND METHODS
Molecular Cloning
All plasmids used in this study are listed in Supplemental Table S2. Plasmids were generated using standard protocols. Oligonucleotide primers used in PCR reaction to generate appropriate restriction sites, site-directed mutagenesis, and sequencing are listed in Supplemental Table S1. All fragments and construct were sequenced to verify that no PCR-generated errors were introduced and that relevant fragments are cloned in the correct frame. Escherichia coli DH5α was used for all recombinant plasmid constructions.
Plasmid production for routine DNA manipulations and sequencing was prepared with QIAGENE mini prep purification kit according to the manufacturer's protocol.
Yeast (Saccharomyces cerevisiae) expression vectors were generated by cloning the corresponding cDNAs into the polycloning sites of high-copy (2μ) or low-copy (CEN) yeast vectors. The yeast vectors contain a glyceraldehyde-3-P dehydrogenase gene promoter and a phosphoglycerate kinase gene terminator, separated by restriction sites.
Plant expression vectors were generated by cloning a SphI, HindIII, or NotI cassette, containing the cauliflower mosaic virus 35S promoter, gene of interest, and NOS 3′ end, into the plant binary vector pCAMBIA2300 (SphI, HindIII).
Fusion Protein to the C Terminus
A fragment containing a full-length cDNA sequence of the gene of interest, without the ATG and with a stop codon, was cloned in frame with the C terminus of plasmids containing GFP, CFP, and YFP.
Fusion Protein to the N Terminus
A fragment containing a full-length cDNA sequence of the gene of interest, with the ATG and without a stop codon, was cloned in frame with the N terminus of plasmids containing GFP, CFP, and YFP.
Generation of AticmtAmR111R112E165Q187R188 Mutant
pSY60 was used as the template together with primers SYP50 and SYP51 to mutate AtICMTA Q165 to E165 using a QuikChange site-directed mutagenesis kit (Stratagene) to create pSY915. AticmtAmE165 was sequenced to verify that no PCR-generated errors were introduced. pSY915 was used as the template together with primers SYP54 and SYP55 to mutate AticmtAmE165 N111 and Y112 to R111 and R112, respectively, using a QuikChange (Stratagene) to create pSY902. pSY902 was used as the template together with primers SYP52 and SYP53 to mutate AticmtAmE165Q187R188 E187 and S188 to Q187 and R188 using a QuikChange (Stratagene) to create pSY904. AticmtAmR111R112E165Q187R188 was sequenced to verify that no PCR-generated errors were introduced.
Construction of AtICMTA-RNAi Plasmid
A fragment from position 1 to 380 of AtICMTA was amplified with oligonucleotide primers SYP304 and SYP305. The resulting fragment was purified, digested with NcoI and XhoI, and cloned into pGFP-MRC to generate pSY302. An identical fragment was amplified with oligonucleotide primers SYP306 and SYP307, purified, digested with XbaI and SacI, and cloned into pSY302 to generate pSY303. Cloning was such that both fragments were flanking GFP in a reverse orientation to one another. Next, pSY303 was digested with HindIII to isolate a cassette containing the 35S promoter, reverse-oriented AtICMTA fragments flanking GFP, and NOS transcriptional terminator. This cassette was subcloned into pCAMBIA2300 to generate pSY304.
PCR
PCR was used for gene amplification, gene detection, and cloning. Unless mentioned, reaction conditions were as follows: 5 to 100 ng of DNA, 0.05 mm dNTPs, 7.5 μm of each primer, 1× reaction buffer (10 mm Tris, pH 8.8, 50 mm KCl, 0.08% Nonidet P-40), 1.5 mm MgCl2, 0.1 mg/mL bovine serum albumin, and 0.125 to 0.25 μL of Taq DNA polymerase or Pfu DNA polymerase (Promega) that was added in a hot-start fashion following the annealing step of the first cycle template in a final volume of 25 μL. The reaction conditions were: 2 min at 94°C, followed by 30 repeats of 30 s at 94°C, 30 s at 55°C, 1 min per 1 kb at 72°C, and an additional step of 3 min at 72°C.
Yeast Work
All yeast strains used in this study are listed in Supplemental Table S3. Yeast transformation was carried out with a modified lithium acetate transformation protocol (Ito et al., 1983; Yalovsky et al., 1997).
Subcellular Localization of GFP-AtROP/AtRAC7 in Yeast
AtROP9 and Atrop9mS were subcloned in yeast expression vector pJR1138-GFP to obtain plasmids pSY32 and pSY31, respectively (Supplemental Table S2; Lavy et al., 2002). Yeast strains JRY6959, JRY6958, and SYY500 (Supplemental Table S3) were transformed with the resulting plasmids to obtain yeast strains SYY535, SYY538, SYY539, and SYY542. Subcellular localization of the GFP fusion proteins was determined using a confocal laser-scanning microscope following overnight growth of cells at 30°C on rich media plates. To prevent cell motion and obtain sharp images, cells were loaded onto slides in 0.1% low melting agarose.
Growth Arrest Pheromone Diffusion (Halo) Assays
Assays were carried out essentially as described previously (Trueblood et al., 2000), with the following modifications. Three microliters of MATa cell slurry (approximately 107 cells) were spotted onto a solid, rich medium (yeast peptone dextrose) plate containing 0.01% Triton X-100 that had been spread with a lawn (approximately 2 × 106 cells) of the MATα sst2α cells (JRY3443). After 1 d of growth at 28°C, the relative amounts of a-factor produced by each MATa strain were evident from the size of the growth inhibition zone (halo) surrounding the MATa cells. The relative halo sizes were directly proportional to the amount of a-factor exported from the MATa cells (Ito et al., 1983). Experiments were repeated three times with four to five replicates.
Plant Material
Nicotiana benthamiana and Arabidopsis (Arabidopsis thaliana; Columbia-0 and Wassilewskija) plants were grown as described previously (Bracha et al., 2002; Lavy et al., 2002).
Transient Expression Assays
Young leaves of N. benthamiana were injected on the abaxial side with Agrobacterium tumefaciens GV3101/pMP90 strains harboring appropriate plasmids as described previously (Bracha et al., 2002; Lavy et al., 2002). ER localization was determined by coexpression with ER-mCherry, which is composed of the N-terminal signal peptide of Arabidopsis WALL-ASSOCIATED KINASE2 and a C-terminal ER-retention signal (His-Asp-Glu-Leu; Nelson et al., 2007), or with GFP-AtSTE24 (Bracha et al., 2002). Leaves were observed for GFP and mCherry fluorescence at 24 to 48 h postinjection.
Arabidopsis Transformation
Stable Arabidopsis transgenic plants were obtained using the floral-dip method (Clough and Bent, 1998). Transgenic plants were selected by either growth on kanamycin containing 0.5× Murashige and Skoog medium or bar spraying soil-grown plants with BASTA. AtICMTA plants were also analyzed by PCR to reveal transgene existence.
Plant DNA and RNA Isolation and Q-PCR
DNA Isolation
Leaves were ground with a mortar and pestle and 1 to 2 g were used for DNA isolation using GenElute plant genomic DNA miniprep kit (Sigma) according to the manufacturer's protocol.
RNA Isolation
Leaves were ground with a mortar and pestle and 100 to 200 mg were used for total RNA isolation using SV total RNA isolation kit (Promega) according to the manufacturer's protocol.
Q-PCR
Total RNA was isolated from leaves, flowers, seedlings, and roots using SV total isolation RNA kit (Promega). For RT, 1 μg of total RNA was denatured at 70°C for 5 min in the presence of 0.5 μg of oligo(dT)15 primers. The tubes were immediately chilled on ice and reverse transcribed with 200 units of Moloney murine leukemia virus reverse transcriptase according to the manufacturer's instructions (Promega) in a total volume of 25 μL at 42°C for 60 min. Quantification of AtRCE1, AtICMTA, and AtICMTB RNA levels was carried out by Q-PCR using an ABI Prism 7700 StepOnePlus instrument (Applied Biosystems). Study samples were run in triplicate on eight-well optical PCR strips (Applied Biosystems) in a final volume of 10 μL. Primers were designed according to Roche Universal ProbeLibrary (https://www.roche-applied-science.com/sis/rtpcr/upl/index.jsp). The following primers pairs were used: AtRCE1, 500 nmol L−1 of each SYP1008 + SYP1009 to amplify a 106-bp fragment from positions 265 to 371; AtICMTA, 125 nmol L−1 of each SYP1006 + SYP1007 to amplify a 59-bp fragment from positions 252 to 311; and AtICMTB, 500 nmol L−1 of each SYP1004 + SYP 1005 to amplify a 120-bp fragment from positions 179 to 299. The PCR cycles were run as follows: 10 min initial denaturation at 95°C, followed by 40 subsequent cycles of 15 s denaturation at 95°C followed by 1 min primer annealing and elongation at 60°C. The specificity of the unique amplification product was determined by melting curve analysis according to the manufacturer's instructions (Applied Biosystems). Relative quantities of RNA were calculated by the standard curve method (Applied Biosystems Incorporated 2001 User Bulletin #2: ABI PRISM 7700 sequence detection system [http://www.appliedbiosystems.com]). DNA dilution series were prepared to calculate amplification efficiency coefficient for each gene. The relative levels of AtRCE1, AtICMTA, and AtICMTB RNA were calculated according to the amplification efficiency coefficient and normalized against ACTIN8 gene standard, level of which was taken as 1. The analysis was repeated with three independent biological replicates.
Identification and Analysis of AtICMTA T-DNA Mutant
DNA pulls of the University of Wisconsin Biotechnology Center T-DNA mutant collection were screened with oligonucleotide primers SYP300 and JL202 or SYP301 and JL202. Southern blot of the amplified PCR fragments with an AtICMTA gene-specific probe was used to verify identification of potential T-DNA insertion in AtICMTA. In turn, the relevant seed stock, P65H9, was ordered and the T-DNA insertion site was determined by sequencing. Expression level of AtICMTA and AtICMTB in the mutant homozygote plants was determined by RT-PCR with nucleotide primers SYP304 and SYP305, and SYP306 and SYP307, respectively.
Plant Imaging and Confocal Laser Fluorescence Imaging
Images of plants were obtained with Nikon Coolpix CCD camera or with Zeiss Axiocam CCD camera mounted on a Zeiss SV11 stereomicroscope.
Confocal Imaging
Imaging was carried out on Leica TCS-SL confocal laser-scanning microscope using 20× multi-immersion or 63× water-immersion objectives with numerical apertures of 0.7 and 1.2, respectively. GFP excitation was carried out with argon laser at 488 nm together with a 500 nm beam splitter and spectral detector set between 510 to 535 nm. mCherry was excited with argon laser at 514 nm together with 458/514 dichroic beam splitter and spectral detector set between 580 to 630. Dual imaging of GFP and mCherry was carried out in a sequential mode to avoid channel bleeding. The reticulate ER images are of single confocal sections and the colocalization images are maximal projection stacks of multiple confocal sections. Image processing was carried out with Leica LCS, Zeiss LSM browser, and Adobe Photoshop.
ABA Germination Assays
ABA germination assays were carried out as described previously (Running et al., 2004).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At2g36305, At5g23320, At5g08335, A9P_B59POPTR, Os04g0602900, Q01SH5_ORYSA, OSI016666, OsJ_015369, and A9SSC9_PHYPA.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Prenylation and CaaX processing of proteins.
Supplemental Figure S2. Processing of the a-mating factor precursor and pheromone diffusion halo assay.
Supplemental Figure S3. Colocalization of AtSTE24, AtICMTA, AtRCE1, and AtICMTB in the ER following transient expression in N. benthamiana leaf epidermal cells.
Supplemental Figure S4. Expression pattern of AtICMTA and AtICMTB.
Supplemental Figure S5. Identification and analysis of an AtICMTA T-DNA insertion mutant.
Supplemental Figure S6. Nucleotide sequence of AtICMTA highlighting the insertion site of the T-DNA.
Supplemental Figure S7. Nucleotide sequence alignment of AtICMTA and AtICMTB.
Supplemental Figure S8. Amino acid sequence analysis of plant ICMT proteins.
Supplemental Table S1. Oligonucleotide primers used in this study.
Supplemental Table S2. Plasmids used in this study.
Supplemental Table S3. Yeast strains used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Nir Ohad for access to the Q-PCR machine, Dr. Tali Yahalom for technical advice, and members of the Yalovsky lab for their help during this project.
This work was supported by the Israel Science Foundation (grant no. ISF 312/07 to S.Y.), the U.S.-Israel Binational Agricultural Research and Development Fund (grant no. BARD 4032/07 to S.Y.), and the German Israel Foundation (grant no. GIF 834/2005 to S.Y.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Shaul Yalovsky (shauly@tauex.tau.ac.il).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Allen GJ, Murata Y, Chu SP, Nafisi M, Schroeder JI (2002) Hypersensitivity of abscisic acid-induced cytosolic calcium increases in the Arabidopsis farnesyltransferase mutant era1-2. Plant Cell 14 1649–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backlund PS Jr (1997) Post-translational processing of RhoA: carboxyl methylation of the carboxyl-terminal prenylcysteine increases the half-life of Rhoa. J Biol Chem 272 33175–33180 [DOI] [PubMed] [Google Scholar]
- Bergo MO, Ambroziak P, Gregory C, George A, Otto JC, Kim E, Nagase H, Casey PJ, Balmain A, Young SG (2002) Absence of the CAAX endoprotease Rce1: effects on cell growth and transformation. Mol Cell Biol 22 171–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergo MO, Gavino B, Ross J, Schmidt WK, Hong C, Kendall LV, Mohr A, Meta M, Genant H, Jiang Y et al (2002) Zmpste24 deficiency in mice causes spontaneous bone fractures, muscle weakness, and a prelamin A processing defect. Proc Natl Acad Sci USA 99 13049–13054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergo MO, Gavino BJ, Hong C, Beigneux AP, McMahon M, Casey PJ, Young SG (2004) Inactivation of Icmt inhibits transformation by oncogenic K-Ras and B-Raf. J Clin Invest 113 539–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergo MO, Leung GK, Ambroziak P, Otto JC, Casey PJ, Gomes AQ, Seabra MC, Young SG (2001) Isoprenylcysteine carboxyl methyltransferase deficiency in mice. J Biol Chem 276 5841–5845 [DOI] [PubMed] [Google Scholar]
- Bonetta D, Bayliss P, Sun S, Sage T, McCourt P (2000) Farnesylation is involved in meristem organization in Arabidopsis. Planta 211 182–190 [DOI] [PubMed] [Google Scholar]
- Boyartchuk VL, Ashby MN, Rine J (1997) Modulation of Ras and a-factor function by carboxyl-terminal proteolysis. Science 275 1796–1800 [DOI] [PubMed] [Google Scholar]
- Boyartchuk VL, Rine J (1998) Roles of prenyl protein proteases in maturation of Saccharomyces cerevisiae a-factor. Genetics 150 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha K, Lavy M, Yalovsky S (2002) The Arabidopsis AtSTE24 is a CaaX protease with broad substrate specificity. J Biol Chem 277 29856–29864 [DOI] [PubMed] [Google Scholar]
- Cadinanos J, Varela I, Mandel DA, Schmidt WK, Diaz-Perales A, Lopez-Otin C, Freije JM (2003) AtFACE-2, a functional prenylated protein protease from Arabidopsis thaliana related to mammalian Ras-converting enzymes. J Biol Chem 278 42091–42097 [DOI] [PubMed] [Google Scholar]
- Caldelari D, Sternberg H, Rodriguez-Concepcion M, Gruissem W, Yalovsky S (2001) Efficient prenylation by a plant geranylgeranyltransferase-I requires a functional CaaL box motif and a proximal polybasic domain. Plant Physiol 126 1416–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke S (1992) Protein isoprenylation and methylation at carboxyl-terminal cysteine residues. Annu Rev Biochem 61 355–386 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Crowell DN, Kennedy M (2001) Identification and functional expression in yeast of a prenylcysteine alpha-carboxyl methyltransferase gene from Arabidopsis thaliana. Plant Mol Biol 45 469–476 [DOI] [PubMed] [Google Scholar]
- Crowell DN, Sen SE, Randall SK (1998) Prenylcysteine alpha-carboxyl methyltransferse in suspension-cultured tobacco cells. Plant Physiol 118 115–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S, Ghassemian M, Bonetta D, Cooney S, McCourt P (1996) A protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis. Science 273 1239–1241 [DOI] [PubMed] [Google Scholar]
- Dai Q, Choy E, Chiu V, Romano J, Slivka SR, Steitz SA, Michaelis S, Philips MR (1998) Mammalian prenylcysteine carboxyl methyltransferase is in the endoplasmic reticulum. J Biol Chem 273 15030–15034 [DOI] [PubMed] [Google Scholar]
- Deem AK, Bultema RL, Crowell DN (2006) Prenylcysteine methylesterase in Arabidopsis thaliana. Gene 380 159–166 [DOI] [PubMed] [Google Scholar]
- Galichet A, Gruissem W (2003) Protein farnesylation in plants—conserved mechanisms but different targets. Curr Opin Plant Biol 6 530–535 [DOI] [PubMed] [Google Scholar]
- Ghomashchi F, Zhang X, Liu L, Gelb MH (1995) Binding of prenylated and polybasic peptides to membranes: affinities and intervesicle exchange. Biochemistry 34 11910–11918 [DOI] [PubMed] [Google Scholar]
- Hancock JF, Cadwallader K, Marshall CJ (1991) Methylation and proteolysis are essential for efficient membrane binding of prenylated p21K-ras(B). EMBO J 10 641–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JB, Casey PJ (1994) In vitro processing of recombinant G protein gamma subunits: requirements for assembly of an active beta gamma complex. J Biol Chem 269 9067–9073 [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A (1983) Transformation of intact yeast cells treated with alkali cations. J Bacteriol 153 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CD, Chary SN, Chernoff EA, Zeng Q, Running MP, Crowell DN (2005) Protein geranylgeranyltransferase I is involved in specific aspects of abscisic acid and auxin signaling in Arabidopsis. Plant Physiol 139 722–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Cox AD, Hisaka MM, Graham SM, Buss JE, Der CJ (1992) Isoprenoid addition to ras protein is the critical modiification for its membrane association and transforming activity. Proc Natl Acad Sci USA 89 6403–6407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Ambroziak P, Otto JC, Taylor B, Ashby M, Shannon K, Casey PJ, Young SG (1999) Disruption of the mouse Rce1 gene results in defective Ras processing and mislocalization of Ras within cells. J Biol Chem 274 8383–8390 [DOI] [PubMed] [Google Scholar]
- Laule O, Hirsch-Hoffmann M, Hruz T, Gruissem W, Zimmermann P (2006) Web-based analysis of the mouse transcriptome using Genevestigator. BMC Bioinformatics 7 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavy M, Bracha-Drori K, Sternberg H, Yalovsky S (2002) Cell-specific prenylation independent mechanism regulates targeting of type-II RACs. Plant Cell 14 2431–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer-Stroh S, Washietl S, Eisenhaber F (2003) Protein prenyltransferases. Genome Biol 4 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson D, Ali W, Chiu VK, Bergo M, Silletti J, Wright L, Young SG, Philips M (2005) Postprenylation CAAX processing is required for proper localization of Ras but not Rho GTPases. Mol Biol Cell 16 1606–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimha Chary S, Bultema RL, Packard CE, Crowell DN (2002) Prenylcysteine alpha-carboxyl methyltransferase expression and function in Arabidopsis thaliana. Plant J 32 735–747 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenfuhr A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51 1126–1136 [DOI] [PubMed] [Google Scholar]
- Parish CA, Smrcka AV, Rando RR (1995) Functional significance of beta gamma-subunit carboxymethylation for the activation of phospholipase C and phosphoinositide 3-kinase. Biochemistry 34 7722–7727 [DOI] [PubMed] [Google Scholar]
- Pei ZM, Ghassemian M, Kwak CM, McCourt P, Schroeder JI (1998) Role of farnesyltransferase in ABA regulation of guard cell anion channels and plant water loss. Science 282 287–290 [DOI] [PubMed] [Google Scholar]
- Pendas AM, Zhou Z, Cadinanos J, Freije JM, Wang J, Hultenby K, Astudillo A, Wernerson A, Rodriguez F, Tryggvason K, et al (2002) Defective prelamin A processing and muscular and adipocyte alterations in Zmpste24 metalloproteinase-deficient mice. Nat Genet 31 94–99 [DOI] [PubMed] [Google Scholar]
- Perez-Sala D, Tan EW, Canada FJ, Rando RR (1991) Methylation and demethylation reactions of guanine nucleotide-binding proteins of retinal rod outer segments. Proc Natl Acad Sci USA 88 3043–3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Concepcion M, Toledo-Ortiz G, Yalovsky S, Caldelari D, Gruissem W (2000) Carboxyl-methylation of prenylated calmodulin CaM53 is required for efficient plasma membrane targeting of the protein. Plant J 24 775–784 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Concepcion M, Yalovsky S, Zik M, Fromm H, Gruissem W (1999) The prenylation status of a novel plant calmodulin directs plasma membrane or nuclear localization of the protein. EMBO J 18 1996–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano JD, Michaelis S (2001) Topological and mutational analysis of Saccharomyces cerevisiae Ste14p, founding member of the isoprenylcysteine carboxyl methyltransferase family. Mol Biol Cell 12 1957–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano JD, Schmidt WK, Michaelis S (1998) The Saccharomyces cerevisiae prenylcysteine carboxyl methyltransferase Ste14p is in the endoplasmic reticulum membrane. Mol Biol Cell 9 2231–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg SJ, Rane MJ, Dean WL, Corpier CL, Hoffman JL, McLeish KR (1998) Effect of gamma subunit carboxyl methylation on the interaction of G protein alpha subunits with beta gamma subunits of defined composition. Cell Signal 10 131–136 [DOI] [PubMed] [Google Scholar]
- Running MP, Fletcher JC, Meyerowitz EM (1998) The WIGGUM gene is required for proper regulation of floral meristem size in Arabidopsis. Development 125 2545–2553 [DOI] [PubMed] [Google Scholar]
- Running MP, Lavy M, Sternberg H, Galichet A, Gruissem W, Hake S, Ori N, Yalovsky S (2004) Enlarged meristems and delayed growth in plp mutants result from lack of CaaX prenyltransferases. Proc Natl Acad Sci USA 101 7815–7820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt WK, Tam A, Fujimura-Kamada K, Michaelis S (1998) Endoplasmic reticulum membrane localization of Rce1p and Ste24p, yeast proteases involved in carboxyl-terminal CAAX protein processing and amino-terminal a-factor cleavage. Proc Natl Acad Sci USA 95 11175–11180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt WK, Tam A, Michaelis S (2000) Reconstitution of the Ste24p-dependent N-terminal proteolytic step in yeast a-factor biogenesis. J Biol Chem 275 6227–6233 [DOI] [PubMed] [Google Scholar]
- Tam A, Nouvet FJ, Fujimura-Kamada K, Slunt H, Sisodia SS, Michaelis S (1998) Dual roles for Ste24p in yeast a-factor maturation: NH2-terminal proteolysis and COOH-terminal CAAX processing. J Cell Biol 142 635–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan EW, Rando RR (1992) Identification of an isoprenylated cysteine methyl ester hydrolase activity in bovine rod outer segment membranes. Biochemistry 31 5572–5578 [DOI] [PubMed] [Google Scholar]
- Trueblood CE, Boyartchuk VL, Picologlou EA, Rozema D, Poulter CD, Rine J (2000) The CaaX proteases, Afc1p and Rce1p, have overlapping but distinct substrate specificities. Mol Cell Biol 20 4381–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueblood CE, Ohya Y, Rine J (1993) Genetic evidence for in vivo cross-specificity of the CaaX-box protein prenyltransferases farnesyltransferase and geranylgeranyltransferase-I in Saccharomyces cerevisiae. Mol Cell Biol 13 4260–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentijn JA, Jamieson JD (1998) Carboxyl methylation of rab3D is developmentally regulated in the rat pancreas: correlation with exocrine function. Eur J Cell Biol 76 204–211 [DOI] [PubMed] [Google Scholar]
- Van Dessel GA, De Busser HM, Lagrou AR (2001) On the occurrence of multiple isoprenylated cysteine methyl ester hydrolase activities in bovine adrenal medulla. Biochem Biophys Res Commun 284 50–56 [DOI] [PubMed] [Google Scholar]
- Yalovsky S, Kulukian A, Rodriguez-Concepcion M, Young CA, Gruissem W (2000) Functional requirement of plant farnesyltransferase during development in Arabidopsis. Plant Cell 12 1267–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalovsky S, Rodr Guez-Concepcion M, Gruissem W (1999) Lipid modifications of proteins—slipping in and out of membranes. Trends Plant Sci 4 439–445 [DOI] [PubMed] [Google Scholar]
- Yalovsky S, Trueblood CE, Callan KL, Narita JO, Jenkins SM, Rine J, Gruissem W (1997) Plant farnesyltransferase can restore yeast Ras signaling and mating. Mol Cell Biol 17 1986–1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SG, Ambroziak P, Kim E, Clarke S (2000) Postprenylation protein processing: CXXX (CaaX) endoproteases and isoprenylcyteine carboxyl methyltransferase. In F Tamanoi, DS Sigman, eds, Protein Lipidation, Ed 3, Vol 21. Academic Press, San Diego, pp 156–213
- Young SG, Fong LG, Michaelis S (2005) Prelamin A, Zmpste24, misshapen cell nuclei, and progeria—new evidence suggesting that protein farnesylation could be important for disease pathogenesis. J Lipid Res 46 2531–2558 [DOI] [PubMed] [Google Scholar]
- Zhang FL, Casey PJ (1996) Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem 65 241–269 [DOI] [PubMed] [Google Scholar]
- Ziegelhoffer EC, Medrano LJ, Meyerowitz EM (2000) Cloning of the Arabidopsis WIGGUM gene identifies a role for farnesylation in meristem development. Proc Natl Acad Sci USA 97 7633–7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.