Abstract
Monoclonal antibodies raised against axonemal proteins of sea urchin spermatozoa have been used to study regulatory mechanisms involved in flagellar motility. Here, we report that one of these antibodies, monoclonal antibody D-316, has an unusual perturbating effect on the motility of sea urchin sperm models; it does not affect the beat frequency, the amplitude of beating or the percentage of motile sperm models, but instead promotes a marked transformation of the flagellar beating pattern which changes from a two-dimensional to a three-dimensional type of movement. On immunoblots of axonemal proteins separated by SDS-PAGE, D-316 recognized a single polypeptide of 90 kDa. This protein was purified following its extraction by exposure of axonemes to a brief heat treatment at 40°C. The protein copurified and coimmunoprecipitated with proteins of 43 and 34 kDa, suggesting that it exists as a complex in its native form. Using D-316 as a probe, a full-length cDNA clone encoding the 90-kDa protein was obtained from a sea urchin cDNA library. The sequence predicts a highly acidic (pI = 4.0) protein of 552 amino acids with a mass of 62,720 Da (p63). Comparison with protein sequences in databases indicated that the protein is related to radial spoke proteins 4 and 6 (RSP4 and RSP6) of Chlamydomonas reinhardtii, which share 37% and 25% similarity, respectively, with p63. However, the sea urchin protein possesses structural features distinct from RSP4 and RSP6, such as the presence of three major acidic stretches which contains 25, 17, and 12 aspartate and glutamate residues of 34-, 22-, and 14-amino acid long stretches, respectively, that are predicted to form α-helical coiled-coil secondary structures. These results suggest a major role for p63 in the maintenance of a planar form of sperm flagellar beating and provide new tools to study the function of radial spoke heads in more evolved species.
INTRODUCTION
Eukaryotic flagella are complex organelles comprising over 200 distinct polypeptides assembled onto a framework of microtubules (Luck, 1984; Dutcher, 1995). The main components of the axoneme are nine outer doublet microtubules which slide relative to one another; dynein arms, which generate the force for this sliding; central pair microtubules, and radial spokes (Luck, 1984; Dutcher, 1995).
Radial spokes are protein complexes composed of a stalk that attaches to each doublet microtubule and a globular structure (spoke head) that projects toward the central pair of microtubules (Curry and Rosenbaum, 1993). The structure and function of the radial spoke complex has been analyzed using motility mutants of Chlamydomonas with paralyzed flagella (pf). Two-dimensional PAGE analysis of mutants pf14 (lacking the entire spoke), pf1 and pf17 (lacking the spoke head) and wild-type axonemes revealed that the radial spoke complex is formed by the assembly of 17 distinct polypeptides, 5 of which compose the radial spoke head (Piperno et al., 1981). These polypeptides, RSP 1, 4, 6, 9, and 10, are encoded by separate genes but a single mutation in RSP4 (pf1), RSP9 (pf17), or RSP6 (pf26) results in the absence of the entire spoke head structure, suggesting that these proteins form complexes in the axoneme. Mutations that result in disrupted assembly of radial spokes also result in flagellar paralysis but extragenic suppressor mutations that bypassed the requirement for the radial spoke and/or the central pair have been isolated (Huang et al., 1982). These genes encode seven proteins known as the dynein regulatory complex (Piperno et al., 1992). Recent data suggest that radial spokes, along with the central pair and the dynein regulatory complex, are involved in the regulation of dynein-driven microtubule sliding (Smith and Sale, 1992), possibly by phosphorylation of a regulatory inner arm component (Howard et al., 1994; King and Dutcher, 1997; Habermacher and Sale, 1997).
We have generated a panel of monoclonal antibodies (mAbs) against axonemal proteins from sea urchin sperm flagella. These antibodies were screened for their disturbing effects on flagellar motility as well as by their recognition of specific polypeptides on immunoblots. Using one of these antibodies, we have recently identified and cloned a dynein light chain and shown that this protein may play a dynamic role in the motility process (Gingras et al., 1996). In this article, we report that D-316 changes the planar movement of sea urchin spermatozoa to a helical type of movement. The purification and cloning of the protein recognized by this antibody reveal that it is related to a radial spoke head protein and thus strengthen the notion that spoke head proteins regulate the flagellar beating pattern.
EXPERIMENTAL PROCEDURES
Preparation of Sea Urchin Axonemes and mAb Production
Isolation of axonemes from the sea urchin Lytechinus pictus and Strongylocentrotus purpuratus spermatozoa was performed as previously described (Gingras et al., 1996). The production of hydridomas producing mAbs against sea urchin sperm axonemes from L. pictus, as well as the screening and cloning of mAb D-316, a mouse IgG1, were achieved as reported by Gagnon et al. (1994).
Analysis of Motility Parameters from Sea Urchin Sperm Models
The percentage of motile sperm models from Paracentrotus lividus, L. pictus, and S. purpuratus and the flagellar beat frequency of freely motile sperm models was measured by dark field microscopy with a 40× immersion objective and a stroboscopic flash illumination of variable frequency (Chadwick-Helmuth, El Monte, CA) as described by Gagnon et al. (1994). Recordings of video frames were obtained at 280–300 Hz while the microscope stage was translated. This allowed the visualization of multiple well-defined successive images of individual spermatozoa within a single video frame.
Extraction of Axonemal Proteins and Mono Q Chromatography
Axonemes (5 mg/ml) were salt-extracted at 4°C by a 15-min incubation in 10 mM Tris-Cl (pH 8.0), 1 mM EDTA, and 1 mM dithiothreitol (DTT) (TED buffer) containing 0.6 M NaCl. The pellet was washed twice with TED buffer, resuspended in 1 mM Tris-Cl (pH 8.0), 0.1 mM EDTA, and 1 mM DTT and incubated at 40°C for 5 min. The extracted material (heat extract) was separated from the remaining axonemes by ultracentrifugation at 100,000 × g for 1 h at 4°C. The pellet was resuspended in TED buffer containing 0.5% sodium lauryl sarcosinate (Sarkosyl), and the solubilized material was separated by ultracentrifugation. Under these conditions, the majority of the protein recognized by D-316 was present in the heat-extracted material (see Figure 2).
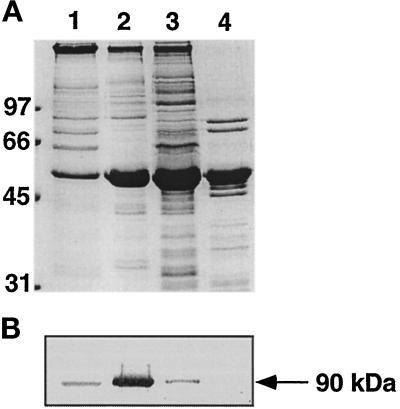
Figure 2.
Fractionation of sea urchin axonemes. Axonemes (5 mg/ml) were sequentially incubated with 0.6 M NaCl (lane 1) for 15 min, at 40°C (lane 2) for 5 min, and with 0.5% Sarkosyl (lane 3) for 60 min. Lane 4 represents the final pellet of the extraction scheme. Aliquots (10 μl) of fractions collected at each step were subjected to 11% SDS-polyacrylamide gels and the proteins were stained with Coomassie blue (A) or immunoblotted with D-316 (B).
The heat-extracted proteins (1 mg/ml) were adjusted to 20 mM Tris-Cl (pH 8.0) and applied at a flow rate of 0.5 ml/min onto a 1-ml Mono Q column previously equilibrated with the same buffer. The proteins were eluted using a linear NaCl gradient and fractions of 1 ml were collected. The presence of the protein recognized by the antibody was monitored by immunoblotting, and its relative amount was estimated by densitometric scanning.
Characterization of a 90-kDa Protein Recognized by D-316
For partial amino acid sequence analysis, the immunoreactive fractions from the Mono Q column containing the 90-kDa protein were subjected to SDS-PAGE, and the resolved proteins were electroblotted onto a polyvinylidene difluoride (PVDF) membrane for N-terminal amino acid sequencing. Endoproteinase Lys-C and CNBr proteolysis of the proteins were also used to produce internal peptides which were sequenced as previously reported (Gingras et al., 1996). The BLAST server (Altschul et al., 1990) was used to search for homologous sequences in PDB, Swiss-Prot, PIR, and Genpept databases.
For immunoprecipitation experiments, the crude heat extract (200 μg) was incubated for 4 h in the absence or presence of 25 μg of specific antibody in 150 mM NaCl and 1% Nonidet P-40. A 50% suspension (25 μl) of protein G-Sepharose beads was then added and the mixture was further incubated for 2 h. The beads were collected by low-speed centrifugation, washed extensively, and resuspended in 20 μl of Laemmli sample buffer (Laemmli, 1970). The immunoprecipitates were separated by SDS-PAGE (Laemmli, 1970).
Cloning, Sequencing, and Expression of the Sea Urchin 90-kDa Protein
A S. purpuratus testis cDNA library made in the λZAP vector (kindly provided by Dr. V. Vacquier, University of California at San Diego, San Diego, CA) was screened using mAb D316 (Sambrook et al., 1989). Up to 4 × 105 plaques were screened and three positive clones were purified. The cDNA inserts were isolated by helper phage excision and insert sizes were determined by digesting the plasmids with EcoRI. The clone containing the largest insert (2.4 kb) was sequenced on both strands using an automated sequencing unit (Sheldon Biotechnology Center, McGill University, Montreal, Quebec, Canada).
For the expression of the protein, a DNA fragment containing the initiator methionine through an internal EcoRI site was prepared by the polymerase chain reaction using Vent polymerase (New England BioLabs, Beverly, MA) and 100 pmol of the primers 5′-CGGGATCCATGATGGAAGAACCTCAA-3′ and 5′-AGAATTCTGCTAAG-GTGATCATATAA-3′. Cycling parameters were 4 min at 94°C, followed by 28 cycles of 1.5 min at 94°C, 1.5 min at 50°C, and 2 min at 72°C. The 200-bp product was gel purified, digested with BamHI and EcoRI, and ligated to a 1.8-kb EcoRI–HindIII fragment of the cDNA clone. The ligation product was inserted in a pTrchis vector (Invitrogen, San Diego, CA) digested with BamHI and HindIII and sequenced. The polyhistidine-tagged protein was purified by Mono Q and metal chelate-affinity chromatographic steps (Pharmacia Biotech, Montreal, Quebec, Canada).
RESULTS
Effect of mAb D-316 on the Motility of Sea Urchin Spermatozoa
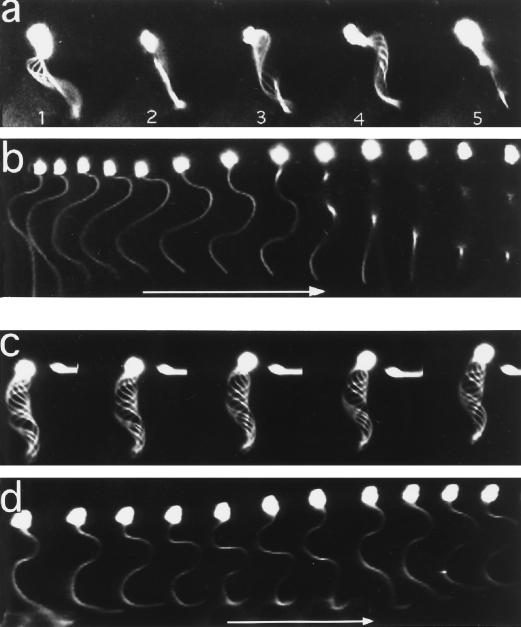
Addition of mAb D-316 to demembranated-reactivated sea urchin spermatozoa (sperm models) from P. lividus or L. pictus had no significant effect on the beat frequency, amplitude of beating, and percentage of motile sperm models within the first 10 min of incubation (our unpublished observations). However, as the time of contact progressed, the flagellar beating pattern changed from a two-dimensional beating into a three-dimensional movement. As shown in Figure 1, the average beating plane of the sperm model is rotating around its trajectory axis: the main curvature of the axoneme alternates from the left (Figure 1a, image 1) of the main axis, then from the top (or bottom) in the second image, then on the right (images 3 and 4), and then from the bottom (or top). In this example, where the sperm model was exposed to D-316 for 3 min, a 360° rotation of the average beating plane took 120 ms, indicating an average rotation frequency of 8 Hz. Whereas the overall sperm trajectory is helicoidal, a more detailed analysis of individual images taken 3 ms apart clearly demonstrate the three-dimensional beating of the spermatozoon (Figure 1b). In contrast, flagellar beating of spermatozoa incubated without D-316 or with control antibodies remained planar during the entire 20-min incubation period (Figure 1, c and d).
Figure 1.
Video images of demembranated/reactivated sperm models. Sperm models were incubated with (c and d) or without (a and b) D-316 and analyzed by dark-field microscopy using 300-Hz stroboscopic illumination and a translation of the microscope stage (b and d). (a and b) control sperm models without antibody at 171 and 178 s after reactivation; (c and d) Sperm models with D-316 (17 μg/ml) at 176 and 179 s; (a and c) overlapping images; (b and d) individual images taken 3 ms apart. Arrow, 15 μm.
Extraction and Purification of the 90-kDa Protein Recognized by mAb D-316 from Sea Urchin Axonemes and Association with Axonemal Components
Sequential extraction of the sea urchin axoneme by high-salt solutions, heat, and detergent were used to solubilize the protein recognized by mAb D316. As shown in Figure 2, the majority (85%) of an immunoreactive protein of 90 kDa was extracted by a brief exposure of the axoneme to 40°C, whereas small amounts of the protein were solubilized with high-salt solutions and detergent. This heat extract was thus used as the starting material for the purification of the 90-kDa protein.
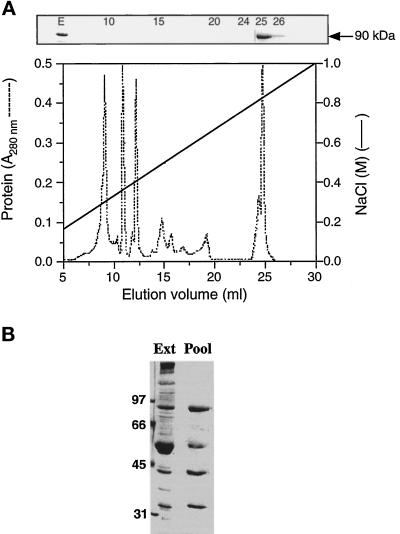
Mono Q anion exchange chromatography of the heat extract resulted in the separation of the 90-kDa protein from the bulk of extracted proteins (Figure 3A) and in its elution at very high NaCl concentrations. SDS-PAGE analysis of the immunoreactive fractions showed that the 90-kDa protein coeluted with proteins of 55 kDa, 43 kDa, and 35 kDa (Figure 3B). These proteins still coeluted along with the 90-kDa polypeptide when elution was performed with 0.6% of the detergent CHAPS, 1 mM DTT, and even upon reverse-phase high-pressure liquid chromatography with a trifluoroacetic acid/acetonitrile mobile phase (our unpublished observations), suggesting that they are tightly associated or of very similar properties.
Figure 3.
Mono Q anion exchange chromatography of heat-extracted axonemal proteins. (A) The heat extract (2 mg of protein) was applied to a 1-ml Mono Q column previously equilibrated with 20 mM Tris-Cl (pH 8.0). The proteins were eluted at a flow rate of 0.5 ml/min using the indicated linear salt gradient, and 1-ml fractions were collected. The presence of p90 in the indicated fractions (upper panel) and in the starting extract (E) was monitored by immunoblotting with mAb D-316. (B) SDS-PAGE analysis of the immunoreactive fractions. Fractions 25 and 26 from the Mono Q column were pooled and an aliquot (10 μl) was analyzed by SDS-PAGE on a 11% gel.
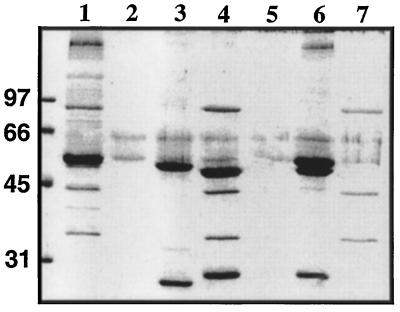
The possible association between these polypeptides was further examined by immunoprecipitation experiments of the crude heat extract using mAb D-316 (Figure 4): in the presence of D316, the 90-kDa, 55-kDa, 50-kDa, 43-kDa, and 34-kDa proteins were all precipitated by the antibody. Controls performed in the absence of primary antibody or using an unrelated mAb, D405–14, which recognizes a dynein light chain (Gingras et al., 1996), showed the presence of some tubulins that bound nonspecifically to the beads (Figure 4, lanes 2 and 3). Immunoprecipitation using antitubulin antibodies resulted in a large amount of tubulins and a number of other proteins but was completely devoided of the 90-kDa, 43-kDa, and 34-kDa proteins. These results thus suggest that the 90-kDa protein recognized by mAb D-316 is tightly associated with proteins of 43 kDa and 34 kDa in the sea urchin axoneme, whereas association of these proteins with tubulin remains elusive. Interestingly, the three proteins (90 kDa, 43 kDa, and 34 kDa) were also found to cosediment upon sucrose gradient centrifugation of the heat extract (our unpublished observations).
Figure 4.
Immunoprecipitation of p90 and associated proteins from axonemal crude extracts. The crude heat extract (200 μg of protein, lane 1) was incubated in the absence (lane 2) or in the presence of the unrelated mAb D405–14 (lane 3), mAb D-316 (lanes 4 and 5), and the antitubulin D-66 produced in our laboratory (lane 6). The immune complexes were precipitated with protein G Sepharose beads, except for lane 5 where unconjugated Sepharose beads were used. The samples were subjected to SDS-PAGE and the resulting gel was stained with Coomassie blue. Lane 7 represents the protein composition of the Mono Q-active fraction pool.
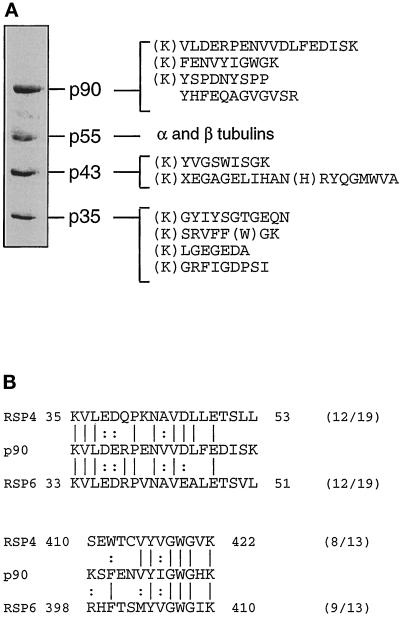
Partial amino acid sequence analysis of these proteins was performed (Figure 5A). As a first step, N-terminal sequencing of the proteins was attempted but failed to yield any information except for the 55-kDa protein where the sequences revealed the presence of a mixture of α- and β-tubulin only. Immunoblot analysis of the fraction using a panel of anti-α- and β-tubulin antibodies (Gagnon et al., 1996) confirmed this result (our unpublished observations). Four internal peptides were obtained for the 90-kDa protein following LysC-endoproteinase and CNBr digestions. Two of these peptides showed significant homologies to radial spoke head proteins RSP4 and RSP6 from Chlamydomonas axoneme (Figure 5B). In the case of the 43-kDa and 35-kDa proteins, internal fragmentation yielded two and four peptides, respectively. Comparison of their sequences with those in the databases failed to reveal any significant similarity with known proteins, suggesting that they may represent novel axonemal components.
Figure 5.
Partial amino acid sequencing of the active fraction from the Mono Q chromatography. (A) The immunoreactive fraction from the Mono Q column was subjected to SDS-PAGE and electroblotted to a PVDF membrane (same lane pool as in Figure 3B). Each separated band was digested with endoproteinase Lys-C and/or CNBr digestions and peptides were separated by high-pressure liquid chromatography were sequenced. The lysine residues in parentheses are deduced from the known specificity of endoproteinase Lys-C. Other residues in parentheses represent amino acids for which identification was carried out with a lower degree of confidence. (B) Search in databases indicated a significant similarity between two peptides from p90 with two peptides from radial spoke head proteins RSP4 and RSP6. Identities are indicated by vertical lines and similarities by dots.
Cloning, Sequencing, and Expression of the 90-kDa Protein
Using D-316 as a probe, a sea urchin λZAP cDNA testis library was screened, leading to the identification of three positive clones, one of which contained a full-length cDNA encoding the protein. Assuming that translation initiation occurs at the first in-frame ATG triplet (nucleotide position 234), the isolated cDNA clone encodes a protein of 552 amino acids with a predicted molecular mass of 62,720 Da (p63) and a very acidic isoelectric point of 4.0 (Figure 6). The four internal peptide sequences that were determined from the purified protein all matched perfectly with the deduced amino acid sequence (Figure 6, underlined). Since these peptides were not used to obtain the clone, this confirms that the isolated cDNA encodes the protein that we have purified as a 90-kDa protein.
Figure 6.
Sequence of the axonemal protein recognized by mAb D-316. Nucleotide and deduced amino acid sequence of the cDNA clone encoding the p90 from sea urchin axoneme, whose real molecular weight is 62,720 Da. Numbers on the right refer to amino acid residues, whereas those on the left refer to nucleotides. The stop codon is indicated by an asterisk. The underlined residues represent matches for peptide sequences that were obtained from the purified protein. This sequence will be available under GenBank accession number U73123.
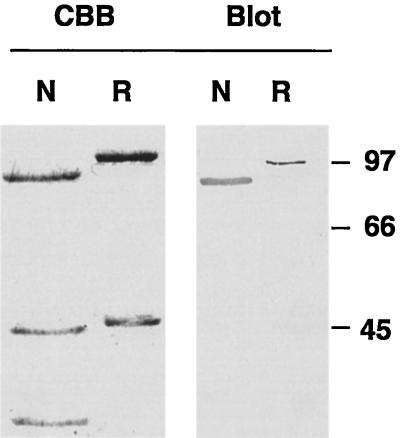
The identity of the cloned protein with the native one was further studied by expression in Escherichia coli of the cloned protein as a polyhistidine-tagged protein. As shown in Figure 7, the recombinant protein has an immunoreactivity similar to that of the native counterpart toward D-316. In these conditions, the cloned protein migrates with a slightly reduced mobility in SDS-PAGE, compared with the isolated native p63, probably due to the presence of a 30-amino acid residues stretch encoded by the expression vector upstream of the polylinker region. The similarity in the electrophoretic migration of the native and bacterially expressed proteins on SDS-PAGE strongly suggests that some intrinsic structural features of the protein promote a large increase in apparent molecular mass from 63 to 90 kDa.
Figure 7.
Expression of the cloned p63 as a polyhistidine-tagged protein. Purified native p63 and its associated proteins (Mono Q pool, N) and recombinant p63 (R) (2 μg each) were separated by SDS-PAGE and the resulting gel was stained with Coomassie blue (CBB) or immunoblotted with mAb D-316 (Blot). Molecular mass standards are indicated on the right.
The protein is rich in proline (10%) and acidic residues (25% glutamate and aspartate). The acidic residues are particularly clustered in three major stretches from residues 207 to 240, 403 to 424, and 539 to 552, which have a strong probability of forming α-helical coiled-coil structures. These acidic domains are separated by less acidic regions containing a high proportion of proline residues and show a strong propensity to form several β-turns.
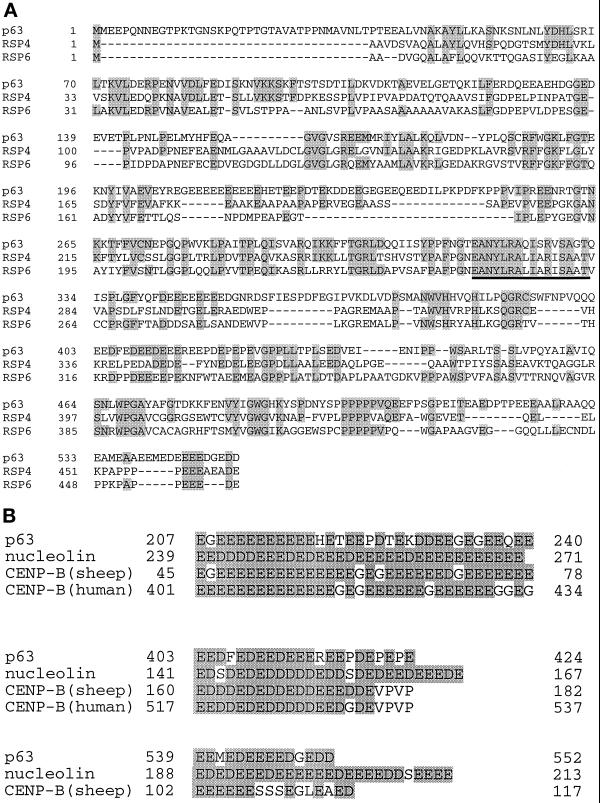
Sequence comparisons indicate that p63 shares similarities with radial spoke head proteins RSP4 and RSP6 of Chlamydomonas axoneme (Figure 8A). p63 show 28% identity (37% similarity) with RSP4 whereas it is 20% identical (25% similar) to RSP6. The relationship between the three proteins is the highest in region 250–334 of p63 with 60% and 55% similarity to RSP4 and RSP6, respectively. Interestingly, this region is also the most conserved between RSP4 and RSP6 and corresponds to the most basic domain (pI = 10) in all three proteins. However, the three acidic repeats present in p63 were not found to the same extent in the Chlamydomonas radial spoke head proteins. In fact, searching through protein sequence databases, we observed that this feature has been documented only in the case of nucleolin and for the major centromere autoantigen CENP-B (Figure 8B). Nucleolin is an ubiquitous protein located in the nucleolus of eukaryotic cells and is thought to play a role in RNA transcription and ribosome assembly (Lapeyre et al., 1987), whereas CENP-B interacts with centromeric heterochromatin in chromosomes (Earnshaw et al., 1987).
Figure 8.
Sequence relationship of sea urchin p63 with Chlamydomonas radial spoke head proteins RSP4 and RSP6 and to proteins containing acidic domains. (A) Sequence comparison between p63 and RSP 4 and 6. The alignment was generated using the program GeneJockey. Gaps introduced by the program to maximize alignment are indicated by dashes and shaded residues represent regions of identity. The underlined region represents the sequence with the highest similarity between the three proteins. (B) Sequence comparison between acidic domains of p63 and those of nucleolin and CENP-B. Shaded areas represent acidic (aspartate or glutamate) residues that are present in the three proteins.
DISCUSSION
In this article, we report the properties of D-316, a mAb that induces a very unusual perturbating effect on the beating of sea urchin sperm axoneme. Whereas most mAbs characterized so far affect flagellar motility by interfering with beat amplitude, symmetry of beating and shear angle (Asai and Brokaw, 1980; Okuno et al., 1981; Asai et al., 1982; Gagnon et al., 1994, 1996), or flagellar beat frequency (Cosson et al., 1996), D-316 modified the beating pattern causing the sequential and repetitive disappearence of the proximal and distal portions from the focal plane of the microscopic field. This unusual behavior is apparently explained by the transformation of the usual planar (two-dimensional) type of movement characteristic of sea urchin sperm flagella beating at the surface or the bottom of a drop of medium on a microscope slide into a three-dimensional type of movement.
Molecular cloning of the cDNA encoding the protein recognized by D-316 revealed a very acidic protein of about 63 kDa with significant homologies to Chlamydomonas axonemal radial spoke head proteins RSP4 and RSP6, suggesting that the protein is a sea urchin sperm homologue of these proteins. The isolated cDNA was surprisingly shorter than expected given the electrophoretic mobility of the native protein (apparent molecular mass of 90 kDa). However, the cloned protein expressed in E. coli had similar mobility and immunoreactivity to those of the native protein, thereby suggesting that the protein has intrinsic structural features that slow its mobility in polyacrylamide gels. Interestingly, this feature has also been observed for RSP4 (49.8 kDa) and RSP6 (48.8 kDa) which migrate like 76-kDa and 67-kDa polypeptides on SDS-PAGE, respectively (Piperno et al., 1981). In those cases, it has been suggested that the high proline content of the proteins may promote the formation of flexible lateral domains that extend toward the central pair or adjacent spokes (Curry et al., 1992).
Sequence comparison of the sea urchin protein with RSP4 and RSP6 indicates that a region of the three proteins has been particularly conserved throughout evolution, suggesting that this region may be essential to the function of the radial spoke heads. However, the sea urchin spoke head protein showed distinct structural features. The most notable of these features is the presence of acidic stretches throughout its sequence that alternate with more basic regions containing proline-rich sequences. These repeats showed a strong probability of forming α-helical coiled-coil structures which may play important roles for the function of the protein. The only known proteins that possess similar acidic stretches are nucleolin, the major nucleolus protein and CENP-B, the major centromere autoantigen. In the case of nucleolin, these regions are postulated to be involved in the binding of the protein to histones (Lapeyre et al., 1987). In the case of CENP-B, the acidic clusters are, in combination with hydrophobic stretches, within the C-terminal 20 kDa of the protein thought to be involved in its dimerization (Yoda et al., 1992). Although it remains to be established, it is tempting to speculate that these regions in p63 are involved in protein–protein interactions.
In this respect, the analysis of the native p63 revealed that the protein is likely to be associated with axonemal proteins of 43 kDa and 35 kDa since these proteins copurify with p63 under both native and denaturing conditions, they coimmunoprecipitate with p63, and they cosediment with the protein upon sucrose gradient centrifugation. In fact, we have not been able to date to resolve these proteins from each other. Although we have not studied in details the p63-associated proteins, partial amino acid analysis suggest that they may represent novel axonemal components. It will be interesting to determine whether these proteins are radial spoke head components or other axonemal proteins that associate with spoke heads. It should be noted, however, that radial spoke head proteins may have a propensity to form macromolecular complexes since, in Chlamydomonas, radial spoke head proteins RSP4, 6, 9, and 10 form a complex in the cell body shortly after their synthesis (Luck et al., 1977).
Mutants of Chlamydomonas defective for radial spokes have little or no flagellar activity (Huang et al., 1981). However, a series of extragenic mutations that suppress the flagellar paralysis of such mutants without restoring their ultrastructural defects have been isolated (Huang et al., 1982), allowing the study of flagellar movement that is generated in the absence of radial spoke heads. For example, cells carrying the pf17 mutation, which produce nonmotile flagella characterized by the absence of radial spoke heads, became motile when the mutation was suppressed as in mutants suppf1 or suppf3 (Brokaw et al., 1982). However, the restored motility is very distinct from that of wild-type cells with a symmetric and large amplitude pattern (Brokaw et al., 1982). Beside indicating that radial spokes are not necessary for bend initiation and bend propagation, these results suggest that the function of the radial spoke system may be to convert a symmetric bending pattern into the asymmetric pattern required for efficient swimming (Brokaw et al., 1982). In these studies, the mutants always show a typical disappearence of an entire axonemal structure, which may complicate the identification of the molecules that are directly responsible for the altered flagellar beating pattern.
The present study shows that the single radial spoke head protein p63, targeted with the specific mAb D-316, is involved in the regulation of the tridimensional characteristics of the sea urchin sperm beat pattern. This observation agrees very well with previous data obtained using naturally occurring axonemes that lack the entire spoke system. For example, the Asian horseshoe crab sperm flagella lack these structures and beat with a three-dimensional helical wave (Ishijima et al., 1988), whereas its closely related American counterpart possesses radial spokes and show a typical two-dimensional flagellar bending pattern. In addition, the eel flagellum, which lacks both outer arms and the spoke-central pair system, generates helicoidal rather than planar waves (Gibbons et al., 1983). In light of our results, these observations support a model where the radial spokes, and especially some spoke head proteins, play a crucial role in the determination of the tridimensional pattern of flagellar beating.
ACKNOWLEDGMENTS
We are grateful to Dr. V. Vacquier for his kind gift of the sea urchin testis library and to S. Audebert for stimulating discussions. This work was supported by grants from the Medical Research Council, Canada (to C.G. and H.Z.), by the Centre National de la Recherche Scientifique, France (to J.C., P.H., and C.C.), and by the Commissariat à l’énergie Atomique, France (to J.G.). The support of a France-Quebec exchange program is also gratefully acknowledged.
Footnotes
The nucleotide sequence reported in this article has been submitted to the GenBank/EMBL data bank with accession number U73123.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Asai DJ, Brokaw CJ. Effects of antibodies against tubulin on the movement of reactivated sea urchin sperm flagella. J Cell Biol. 1980;87:114–123. doi: 10.1083/jcb.87.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai DJ, Brokaw CJ, Thompson WC, Wilson L. Two different monoclonal antibodies to alpha-tubulin inhibit the bending of reactivated sea urchin spermatozoa. Cell Motil Cytoskeleton. 1982;2:599–614. doi: 10.1002/cm.970020608. [DOI] [PubMed] [Google Scholar]
- Brokaw CJ, Luck DJL, Huang B. Analysis of the movement of Chlamydomonas flagella: the function of the radial-spoke system is revealed by comparison of wild-type and mutant flagella. J Cell Biol. 1982;92:722–732. doi: 10.1083/jcb.92.3.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson J, White D, Huitorel P, Edde B, Cibert C, Audebert S, Gagnon C. Inhibition of flagellar beat frequency by a new anti-beta-tubulin antibody. Cell Motil Cytoskeleton. 1996;35:100–112. doi: 10.1002/(SICI)1097-0169(1996)35:2<100::AID-CM3>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Curry AM, Williams BD, Rosenbaum JL. Sequence analysis reveals homology between two proteins of the flagellar radial spoke. Mol Cell Biol. 1992;12:3967–3977. doi: 10.1128/mcb.12.9.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry AM, Rosenbaum JL. Flagellar radial spoke: A model molecular genetic system for studying organelle assembly. Cell Motil Cytoskeleton. 1993;24:224–232. doi: 10.1002/cm.970240403. [DOI] [PubMed] [Google Scholar]
- Dutcher SK. Flagellar assembly in two hundred and fifty easy-to-follow steps. Trends Genet. 1995;11:398–404. doi: 10.1016/s0168-9525(00)89123-4. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Sullivan KF, Machlin PS, Cooke CA, Kaiser DA, Pollard TD, Rothfield NF, Cleveland DW. Molecular cloning of cDNA for CENP-B, the major human centromere autoantigen. J Cell Biol. 1987;104:817–829. doi: 10.1083/jcb.104.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon C, White D, Cosson J, Huitorel P, Eddé B, Desbruyeres E, Paturle-Lafenachère L, Multigner L, Job D, Cibert C. The polyglutamylated lateral chain of alpha-tubulin plays a key role in flagellar motility. J Cell Sci. 1996;109:1545–1553. doi: 10.1242/jcs.109.6.1545. [DOI] [PubMed] [Google Scholar]
- Gagnon C, White D, Huitorel P, Cosson J. A monoclonal antibody against the dynein IC1 peptide of sea urchin spermatozoa inhibits the motility of sea urchin, dinoflagellate, and human flagellar axonemes. Mol Biol Cell. 1994;5:1051–1063. doi: 10.1091/mbc.5.9.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons BH, Gibbons IR, Baccetti B. Structure and motility of the 9 + 0 flagellum of eel spermatozoa. J Submicrosc Cytol. 1983;15:15–20. [PubMed] [Google Scholar]
- Gingras D, White D, Garin J, Multigner L, Job D, Cosson J, Huitorel P, Zingg H, Dumas F, Gagnon C. Purification, cloning and sequence analysis of a Mr = 30,000 protein from sea urchin axonemes that is important for sperm motility. Relationship of the protein to a dynein light chain. J Biol Chem. 1996;271:12807–12813. doi: 10.1074/jbc.271.22.12807. [DOI] [PubMed] [Google Scholar]
- Habermacher G, Sale WS. Regulation of flagellar dynein by phosphorylation of a 138-kD inner arm dynein intermediate chain. J Cell Biol. 1997;136:167–176. doi: 10.1083/jcb.136.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard DR, Habermacher G, Glass DB, Smith EF, Sale WS. Regulation of Chlamydomonas flagellar dynein by an axonemal protein kinase. J Cell Biol. 1994;127:1683–1692. doi: 10.1083/jcb.127.6.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Piperno G, Ramanis Z, Luck DJL. Radial spokes of Chlamydomonas flagella: genetic analysis of assembly and function. J Cell Biol. 1981;88:80–88. doi: 10.1083/jcb.88.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Ramanis Z, Luck DJL. Suppressor mutations in Chlamydomonas reveal a regulatory mechanism for flagellar function. Cell. 1982;28:115–124. doi: 10.1016/0092-8674(82)90381-6. [DOI] [PubMed] [Google Scholar]
- Ishijima SK, Sekiguchi S, Hiramoto Y. Comparative study of the beat patterns of American and Asian horseshoe crab sperm: evidence for a role of the central pair complex in forming planar wave forms in flagella. Cell Motil Cytoskeleton. 1988;9:264–270. [Google Scholar]
- King SJ, Dutcher SK. Phosphoregulation of an inner dynein arm complex in Chlamydomonas reinhardtii is altered in phototactic strains. J Cell Biol. 1997;136:177–191. doi: 10.1083/jcb.136.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lapeyre B, Bourbon H, Amalric F. Nucleolin, the major nucleolar protein of growing eukaryotic cells: an unusual protein structure revealed by the nucleotide sequence. Proc Natl Acad USA. 1987;84:1472–1476. doi: 10.1073/pnas.84.6.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck DJL. Genetic and biochemical dissection of the eucaryotic flagellum. J Cell Biol. 1984;98:789–794. doi: 10.1083/jcb.98.3.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck DJL, Piperno G, Ramanis Z, Huang B. Flagellar mutants of Chlamydomonas: studies of radial spoke-defective strains by dikaryon and revertant analysis. Proc Natl Acad Sci USA. 1977;74:3456–3460. doi: 10.1073/pnas.74.8.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Huang B, Ramanis Z, Luck DJL. Radial spokes of Chlamydomonas flagella: polypeptide composition and phosphorylation of stalk components. J Cell Biol. 1981;88:73–79. doi: 10.1083/jcb.88.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Mead K, Shestak W. The inner dynein arms I2 interact with a “dynein regulatory complex” in Chlamydomonas flagella. J Cell Biol. 1992;118:1455–1463. doi: 10.1083/jcb.118.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Smith EF, Sale WS. Regulation of dynein-driven microtubule sliding by the radial spokes in flagella. Science. 1992;257:1557–1559. doi: 10.1126/science.1387971. [DOI] [PubMed] [Google Scholar]
- Yoda K, Kitagawa K, Masumoto H, Muro Y, Okazaki T. A human centromere protein, CENP-B, has a DNA binding domain containing four potential alpha helices at the NH2 terminus, which is separable from dimerizing activity. J Cell Biol. 1992;119:1413–1427. doi: 10.1083/jcb.119.6.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]