Abstract
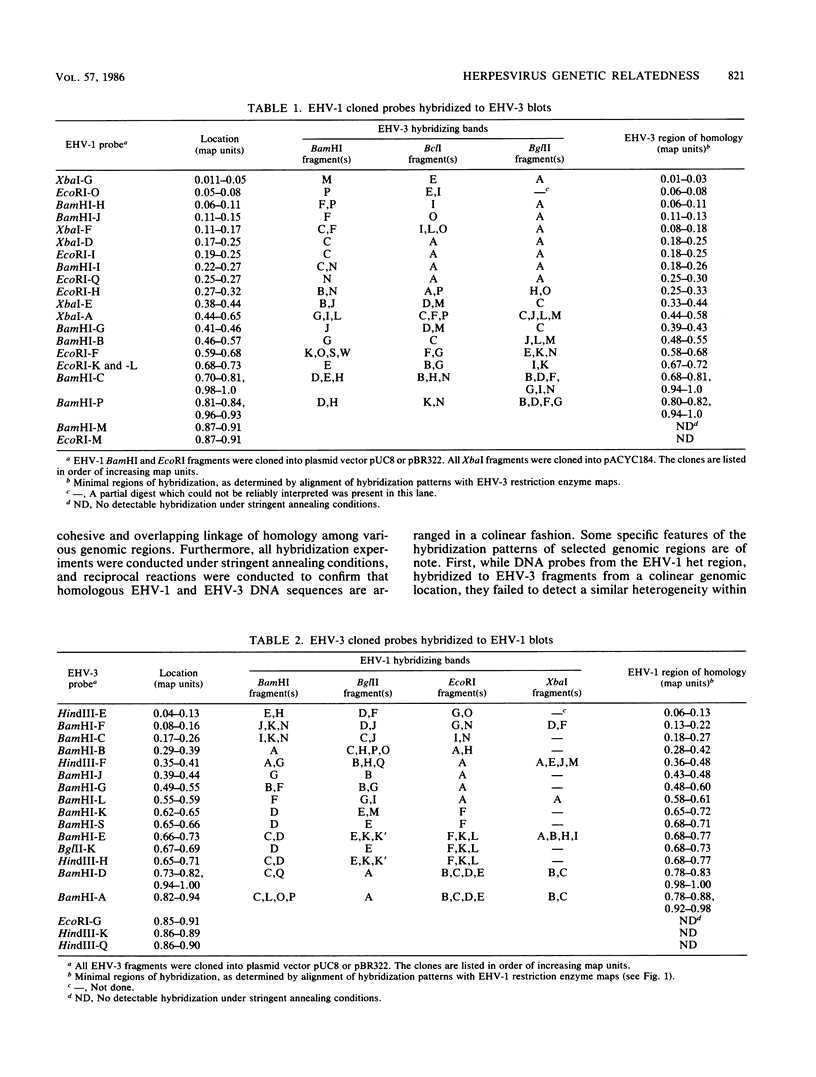
The arrangement and location of homologous DNA sequences within the genomes of equine herpesvirus type 1 (EHV-1) and EHV-3 were investigated by using Southern blot hybridization analyses conducted under stringent conditions. Recombinant plasmid libraries comprising 95 and 84% of the EHV-1 and EHV-3 genomes, respectively, were labeled with 32P-deoxynucleotides by nick translation and were used as probes in filter hybridization studies. The DNA homology between the EHV-1 and EHV-3 genomes was dispersed throughout the genomes in a colinear arrangement. Significant hybridization was detected between the EHV-1 short region inverted repeat sequences, which are known to encode immediate early transcripts, and the corresponding EHV-3 inverted repeat sequences. Interestingly, probes derived from the EHV-1 heterogeneous region, which is adjacent to the EHV-1 short region, hybridized strongly to EHV-3 DNA sequences within a similar genomic location, but did not reveal any corresponding heterogeneity within the EHV-3 genome. Our results demonstrated that there is a highly conserved evolutionary relationship between EHV-1 and EHV-3 and provided the foundation for further investigations to determine whether similarities in protein function underpin the genetic relatedness between these two herpesviruses.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atherton S. S., Sullivan D. C., Dauenhauer S. A., Ruyechan W. T., O'Callaghan D. J. Properties of the genome of equine herpesvirus type 3. Virology. 1982 Jul 15;120(1):18–32. doi: 10.1016/0042-6822(82)90003-4. [DOI] [PubMed] [Google Scholar]
- Baumann R. P., Dauenhauer S. A., Caughman G. B., Staczek J., O'Callaghan D. J. Structure and genetic complexity of the genomes of herpesvirus defective-interfering particles associated with oncogenic transformation and persistent infection. J Virol. 1984 Apr;50(1):13–21. doi: 10.1128/jvi.50.1.13-21.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell D. E., Kemp M. C., Perdue M. L., Randall C. C., Gentry G. A. Equine herpesvirus in vivo: cyclic production of a DNA density variant with repetitive sequences. Virology. 1976 Feb;69(2):737–750. doi: 10.1016/0042-6822(76)90502-x. [DOI] [PubMed] [Google Scholar]
- Caughman G. B., Staczek J., O'Callaghan D. J. Equine herpesvirus type 1 infected cell polypeptides: evidence for immediate early/early/late regulation of viral gene expression. Virology. 1985 Aug;145(1):49–61. doi: 10.1016/0042-6822(85)90200-4. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauenhauer S. A., Robinson R. A., O'Callaghan D. J. Chronic production of defective-interfering particles by hamster embryo cultures of herpesvirus persistently infected and oncogenically transformed cells. J Gen Virol. 1982 May;60(Pt 1):1–14. doi: 10.1099/0022-1317-60-1-1. [DOI] [PubMed] [Google Scholar]
- Davison A. J., Wilkie N. M. Nucleotide sequences of the joint between the L and S segments of herpes simplex virus types 1 and 2. J Gen Virol. 1981 Aug;55(Pt 2):315–331. doi: 10.1099/0022-1317-55-2-315. [DOI] [PubMed] [Google Scholar]
- Ellis R. W., Keller P. M., Lowe R. S., Zivin R. A. Use of a bacterial expression vector to map the varicella-zoster virus major glycoprotein gene, gC. J Virol. 1985 Jan;53(1):81–88. doi: 10.1128/jvi.53.1.81-88.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry B. E., Newcomb W. W., O'Callaghan D. J. Alterations in virus protein synthesis and capsid production in infection with DI particles of herpesvirus. J Gen Virol. 1980 Apr;47(2):343–353. doi: 10.1099/0022-1317-47-2-343. [DOI] [PubMed] [Google Scholar]
- Henry B. E., Newcomb W. W., O'Callaghan D. J. Biological and biochemical properties of defective interfering particles of equine herpesvirus type 1. Virology. 1979 Jan 30;92(2):495–506. doi: 10.1016/0042-6822(79)90152-1. [DOI] [PubMed] [Google Scholar]
- Henry B. E., Robinson R. A., Dauenhauer S. A., Atherton S. S., Hayward G. S., O'Callaghan D. J. Structure of the genome of equine herpesvirus type 1. Virology. 1981 Nov;115(1):97–114. doi: 10.1016/0042-6822(81)90092-1. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Howley P. M., Israel M. A., Law M. F., Martin M. A. A rapid method for detecting and mapping homology between heterologous DNAs. Evaluation of polyomavirus genomes. J Biol Chem. 1979 Jun 10;254(11):4876–4883. [PubMed] [Google Scholar]
- Locker H., Frenkel N. BamI, KpnI, and SalI restriction enzyme maps of the DNAs of herpes simplex virus strains Justin and F: occurrence of heterogeneities in defined regions of the viral DNA. J Virol. 1979 Nov;32(2):429–441. doi: 10.1128/jvi.32.2.429-441.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettenleiter T. C., Lukacs N., Rziha H. J. Mapping of the structural gene of pseudorabies virus glycoprotein A and identification of two non-glycosylated precursor polypeptides. J Virol. 1985 Jan;53(1):52–57. doi: 10.1128/jvi.53.1.52-57.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G., Heston L., Countryman J. P3HR-1 Epstein-Barr virus with heterogeneous DNA is an independent replicon maintained by cell-to-cell spread. J Virol. 1985 Apr;54(1):45–52. doi: 10.1128/jvi.54.1.45-52.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Site-specific inversion sequence of the herpes simplex virus genome: domain and structural features. Proc Natl Acad Sci U S A. 1981 Nov;78(11):7047–7051. doi: 10.1073/pnas.78.11.7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Structure and role of the herpes simplex virus DNA termini in inversion, circularization and generation of virion DNA. Cell. 1982 Nov;31(1):89–97. doi: 10.1016/0092-8674(82)90408-1. [DOI] [PubMed] [Google Scholar]
- O'Callaghan D. J., Cheevers W. P., Gentry G. A., Randall C. C. Kinetics of cellular and viral DNA synthesis in equine abortion (herpes) virus infection of L-M cells. Virology. 1968 Sep;36(1):104–114. doi: 10.1016/0042-6822(68)90120-7. [DOI] [PubMed] [Google Scholar]
- Perdue M. L., Kemp M. C., Randall C. C., O'Callaghan D. J. Studies of the molecular anatomy of the L-M cell strain of equine herpes virus type 1: proteins of the nucleocapsid and intact virion. Virology. 1974 May;59(1):201–216. doi: 10.1016/0042-6822(74)90216-5. [DOI] [PubMed] [Google Scholar]
- Rea T. J., Timmins J. G., Long G. W., Post L. E. Mapping and sequence of the gene for the pseudorabies virus glycoprotein which accumulates in the medium of infected cells. J Virol. 1985 Apr;54(1):21–29. doi: 10.1128/jvi.54.1.21-29.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Robinson R. A., Henry B. E., Duff R. G., O'Callaghan D. J. Oncogenic transformation by equine herpesviruses (EHV). I. Properties of hamster embryo cells transformed by ultraviolet-irradiated EHV-1. Virology. 1980 Mar;101(2):335–362. doi: 10.1016/0042-6822(80)90449-3. [DOI] [PubMed] [Google Scholar]
- Robinson R. A., O'Callaghan D. J. A specific viral DNA sequence is stably integrated in herpesvirus oncogenically transformed cells. Cell. 1983 Feb;32(2):569–578. doi: 10.1016/0092-8674(83)90476-2. [DOI] [PubMed] [Google Scholar]
- Robinson R. A., Tucker P. W., Dauenhauer S. A., O'Callaghan D. J. Molecular cloning of equine herpesvirus type 1 DNA: analysis of standard and defective viral genomes and viral sequences in oncogenically transformed cells. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6684–6688. doi: 10.1073/pnas.78.11.6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson R. A., Vance R. B., O'Callaghan D. J. Oncogenic transformation by by equine herpesviruses. II. Coestablishment of persistent infection and oncogenic transformation of hamster embryo cells by equine herpesvirus type 1 preparations enriched for defective interfering particles. J Virol. 1980 Oct;36(1):204–219. doi: 10.1128/jvi.36.1.204-219.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyechan W. T., Dauenhauer S. A., O'Callaghan D. J. Electron microscopic study of equine herpesvirus type 1 DNA. J Virol. 1982 Apr;42(1):297–300. doi: 10.1128/jvi.42.1.297-300.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spaete R. R., Frenkel N. The herpes simplex virus amplicon: analyses of cis-acting replication functions. Proc Natl Acad Sci U S A. 1985 Feb;82(3):694–698. doi: 10.1073/pnas.82.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staczek J., Atherton S. S., O'Callaghan D. J. Genetic relatedness of the genomes of equine herpesvirus types 1, 2, and 3. J Virol. 1983 Feb;45(2):855–858. doi: 10.1128/jvi.45.2.855-858.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan D. C., Atherton S. S., Staczek J., O'Callaghan D. J. Structure of the genome of equine herpesvirus type 3. Virology. 1984 Jan 30;132(2):352–367. doi: 10.1016/0042-6822(84)90041-2. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Tamashiro J. C., Filpula D., Friedmann T., Spector D. H. Structure of the heterogeneous L-S junction region of human cytomegalovirus strain AD169 DNA. J Virol. 1984 Nov;52(2):541–548. doi: 10.1128/jvi.52.2.541-548.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Vlazny D. A., Frenkel N. Replication of herpes simplex virus DNA: localization of replication recognition signals within defective virus genomes. Proc Natl Acad Sci U S A. 1981 Feb;78(2):742–746. doi: 10.1073/pnas.78.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalley J. M., Robertson G. R., Davison A. J. Analysis of the genome of equine herpesvirus type 1: arrangement of cleavage sites for restriction endonucleases EcoRI, BglII and BamHI. J Gen Virol. 1981 Dec;57(Pt 2):307–323. doi: 10.1099/0022-1317-57-2-307. [DOI] [PubMed] [Google Scholar]