Abstract
Every cyanobacterial species possesses multiple genes encoding AbrB-like transcriptional regulators (cyAbrBs) distinct from those conserved among other bacterial species. In this study, two genes encoding cyAbrBs in Synechocystis sp. PCC 6803, sll0359 and sll0822, were insertionally disrupted in order to examine their physiological roles. A fully segregated disrupted mutant of sll0822 (Δsll0822 mutant) but not of sll0359 was obtained, although both mutants exhibited similar phenotypes (i.e. decreases in growth rate and pigment content). The growth rate of the Δsll0822 mutant was low under any condition, but the low pigment content could be partially recovered by nitrate supplementation of the medium. DNA microarray and RNA-blot analyses revealed that the level of expression of a part of the NtcA regulon, such as urtA, amt1, glnB, sigE, and the nrt operon, was significantly decreased in the Δsll0822 mutant, although the induction of these genes upon nitrogen depletion was still observed to some extent. Sll0822 seems to work in parallel with NtcA to achieve flexible regulation of the nitrogen uptake system. The Sll0822 protein exists mainly in a dimeric form in vivo, and the amount of the protein was not affected by nitrogen availability. This observation, together with the low binding specificity of the purified histidine-tagged Sll0822 protein, implies that the activity of Sll0822 may be posttranslationally modulated in Synechocystis cells.
Following changes in environmental conditions such as photon flux density, temperature, or nutrient availability, photosynthetic organisms must modulate photosynthetic activity and various metabolic processes in order to acclimate to the new environment. The acclimation responses of the cyanobacterium Synechocystis sp. PCC 6803 are well characterized with regard to transcriptional regulation. Genome-wide investigations of gene expression using DNA microarray techniques have allowed the identification of a set of genes induced or repressed upon environmental change (Los et al., 2007). In order to understand the signal transduction mechanisms responsible for the changes in the gene expression profile, identification of transcriptional regulators working in response to environmental changes is essential. We have been characterizing putative transcriptional regulators that are highly conserved in cyanobacterial genomes but not in other bacterial genomes, since such regulators must play an important role in photosynthetic organisms. For example, PedR, a small LuxR-type regulator specific to cyanobacterial species, was shown to regulate gene expression, depending on the availability of reducing equivalents supplied from the photosynthetic electron transport chain (Nakamura and Hihara, 2006).
In this study, we focused on AbrB-like transcriptional regulators in cyanobacteria. Genes encoding AbrB-like transcriptional regulators have been discovered in a wide variety of gram-positive, gram-negative, and archaeal species (Vaughn et al., 2000; Coles et al., 2005). In most cases, the N-terminal DNA-binding domains of these proteins are conserved, whereas the C-terminal domains are significantly divergent in sequence and in size. To date, the most widely studied member of the AbrB family has been the Bacillus subtilis AbrB protein. AbrB, a homotetramer of 10.5-kD subunits (Vaughn et al., 2000; Benson et al., 2002; Bobay et al., 2004), is a pivotal regulator of transition state gene expression upon entry into stationary phase and sporulation (Phillips and Strauch, 2002). The structure of the N-terminal DNA-binding domain of AbrB is quite different from the known structures for DNA binding. It forms a swapped-hairpin β-barrel consisting of four β-strands and an α-helix (ββαββ) involved in both dimerization and DNA binding (Bobay et al., 2005; Coles et al., 2005). No DNA consensus sequence recognized by AbrB was found upstream of the target genes, and it is believed that AbrB-DNA binding interactions entail the recognition of specific stretches of three-dimensional DNA helix configurations (Bobay et al., 2004). The C-terminal domain of AbrB is an independent dimerization module required for the formation of the functional tetramer (Yao and Strauch, 2005).
Cyanobacterial AbrB proteins (hereafter called cyAbrBs) are unique in that they have an AbrB-like DNA-binding domain in the C terminal region. Multiple copies of genes encoding cyAbrBs are found in every cyanobacterial genomic sequence now available, whereas AbrB-like regulators having DNA-binding domains at their C termini are not conserved in other bacterial species. Very recently, Oliveira and Lindblad (2008) reported the isolation of the Sll0359 protein, one of the cyAbrBs in Synechocystis sp. PCC 6803, as a binding factor to the promoter region of the hox operon encoding the bidirectional NiFe hydrogenase. Similarly, by DNA affinity assay, Shalev-Malul et al. (2008) identified specific binding of a cyAbrB to the intergenic region between aoaA and aoaC encoding biosynthetic enzymes for the hepatotoxin cylindrospermopsin in Aphanizomenon ovalisporum. These reports provide the first evidence that cyAbrBs are functional in cyanobacterial cells. However, their precise functions and physiological significance are still unclear.
Here, we report that Sll0822, one of the cyAbrBs in Synechocystis sp. PCC 6803, plays an important role in the activation of nitrogen-regulated genes. The low growth rate and pigment content of the gene-disrupted mutant revealed the physiological significance of transcriptional regulation by Sll0822.
RESULTS
Phylogenetic Analysis of cyAbrBs
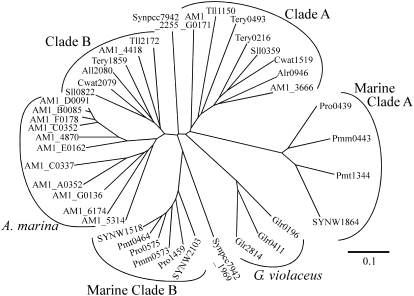
Multiple genes encode cyAbrBs in every cyanobacterial genome. Among 32 cyanobacterial species whose genomic sequences are available (January 2008), 23 species have two copies, and six species possess three copies, of genes encoding cyAbrB. Synechococcus sp. CC 9902 and Synechococcus sp. CC 9605 have four and five copies, respectively. It is notable that there are 14 copies on the chromosome and plasmids of Acaryochloris marina. Figure 1 shows the phylogenetic tree of cyAbrBs from 12 representative species. In most cases, multiple copies in a single organism are classified into two different clades, clades A and B. The marine unicellular non-N2-fixing species also have a set of cyAbrBs categorized into two different clades, marine clades A and B. These marine regulators are distinct from other cyAbrBs in that their N-terminal end is about 10 amino acids shorter (Supplemental Fig. S1). In the case of A. marina, two copies are classified into clades A and B and the remaining 12 copies form a distinct clade. CyAbrBs of Gloeobacter violaceus PCC 7421 and Synechococcus elongatus PCC 7942 do not belong to any of the groups mentioned above.
Figure 1.
Unrooted phylogenetic tree for cyAbrBs present in the 12 cyanobacterial genomes generated using the neighbor-joining algorithm. Cyanobacterial species used for the analysis and accession numbers of genes encoding cyAbrBs are as follows. Synechocystis sp. PCC 6803: sll0822 (gi 16331736) and sll0359 (gi 16331120); Crocosphaera watsonii WH 8501: Cwat2079 (gi 67923953) and Cwat1519 (gi 67925462); Synechococcus elongatus PCC 7942: Synpcc7942_2255 (gi 81301064) and Synpcc7942_1969 (gi 81300778); Thermosynechococcus elongatus BP-1: tll2172 (gi 22299715) and tll1150 (gi 22298693); Gloeobacter violaceus PCC 7421: glr0411 (gi 37519980), glr0196 (gi 37519765), and glr2814 (gi 37522383); Anabaena sp. PCC 7120: all2080 (gi 17131171) and alr0946 (gi 17228441); Synechococcus sp. WH 8102: SYNW2103 (gi 33866635), SYNW1518 (gi 33866052), and SYNW1864 (gi 33866396); Prochlorococcus marinus SS120: Pro-0575 (gi 33240026), Pro-1459 (gi 33240908), and Pro-0439 (gi 33239891); P. marinus MED4: PMM0573 (gi 33861130) and PMM0443 (gi 33861000); P. marinus MIT9313: PMT0464 (gi 33862737) and PMT1344 (gi 33863611); Trichodesmium erythraeum IMS101: tery1859 (gi 113475527), tery0493 (gi 113474361), and tery0216 (gi 113474117); Acaryochloris marina MBIC 11017: AM1_4418 (gi 158337537), AM1_5314 (gi158338414), AM1_4870 (gi158337983), AM1_E0162 (gi1158341078), AM1_A0352 (gi 158339613), AM1_B0085 (gi 158339955), AM1_C0337 (gi 158340409), AM1_C0352 (gi 158340424), AM1_D0091 (gi 158340732), AM1_6174 (gi 158339248), AM1_F0178 (gi 158341155), AM1_3666 (gi 158340732), AM1_G0171 (gi 158341500), and AM1_G0136 (gi 158341465). The unrooted phylogenetic tree was generated using the ClustalW program (version 1.83) at the DNA Data Bank of Japan (http://clustalw.ddbj.nig.ac.jp/top-j.html) and displayed using TreeView (version 1.6.6).
Supplemental Figure S1 shows a comparison of the amino acid sequences of cyAbrBs used for the generation of the phylogenetic tree. The N-terminal regions of cyAbrBs are highly conserved but show no significant homology to known protein motifs. In the C-terminal region, the overall structure of the ββαββ motif characteristic of AbrB-like DNA-binding domains (Coles et al., 2005) is well conserved among cyAbrBs, although sequence similarity to AbrB itself is rather low. The sequence identity of cyAbrBs between clades A and B is approximately 50%.
Characterization of Synechocystis sp. PCC 6803 Mutants Lacking cyAbrBs
Synechocystis sp. PCC 6803 possesses two genes for cyAbrB, sll0359 (clade A) and sll0822 (clade B), as well as three genes for the AbrB-like regulators, slr0724, ssl1300, and ssr7040, which have N-terminal DNA-binding domains. To elucidate the function of the cyAbrBs and the physiological significance of a set of cyAbrBs belonging to two different clades, we individually inactivated both sll0359 and sll0822 by insertion of an antibiotic resistance cassette (Supplemental Fig. S2A). The Δsll0822 mutant replaced all wild-type copies with the mutant allele, whereas considerable numbers of wild-type copies remained in the genome of the Δsll0359 mutant (Supplemental Fig. S2B). Despite the differences in the extent of segregation, these mutants exhibited similar phenotypes (Table I). When grown under normal growth conditions (20 μmol photons m−2 s−1), the doubling time of the mutants was about 1.8 times longer than that of the wild type. Decreases in the amount of chlorophyll and phycocyanin were also observed in both mutants. Long-term cultivation was unfavorable for these mutants, since their growth rates and pigment contents tended to recover, probably due to suppressor mutations. Hereafter, we proceed with the characterization of the completely segregated Δsll0822 mutant.
Table I.
Growth rates and pigment contents of wild-type, Δsll0359, and Δsll0822 strains grown under photoautotrophic conditions
| Strain | Doubling Time | Chlorophyll Content | Phycocyanin Content |
|---|---|---|---|
| h | fg/cell | ||
| Wild type | 20.0 ± 1.4 | 48.1 ± 3.6 | 393.3 ± 34.7 |
| Δsll0359 | 35.0 ± 1.2 | 41.5 ± 2.4 | 316.7 ± 21.4 |
| Δsll0822 | 37.6 ± 5.3 | 38.4 ± 5.7 | 295.0 ± 37.5 |
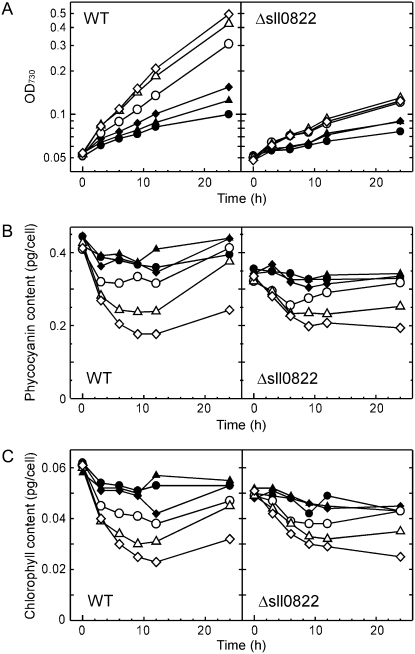
Figure 2 shows the growth properties and pigment contents of wild-type and Δsll0822 mutant cells under different photon flux densities. The upshift of photon flux density did not improve the growth of the mutant. The growth of the Δsll0822 mutant was always slower than that of the wild type under a given light condition (Fig. 2A). The pigment content of the mutant decreased normally upon the upshift of photon flux density (Fig. 2, B and C), indicating that the capacity for light acclimation was not abolished by inactivation of sll0822.
Figure 2.
Effects of photon flux density on growth properties and pigment content of the wild type (WT) and the Δsll0822 mutant. At time 0, cells grown under 5 μmol photons m−2 s−1 were transferred to conditions with a photon flux density of 5 (black circles), 10 (black triangles), 20 (black diamonds), 50 (white circles), 100 (white triangles), or 200 (white diamonds) μmol photons m−2 s−1. Changes in OD730 (A), phycocyanin content (B), and chlorophyll content (C) were monitored.
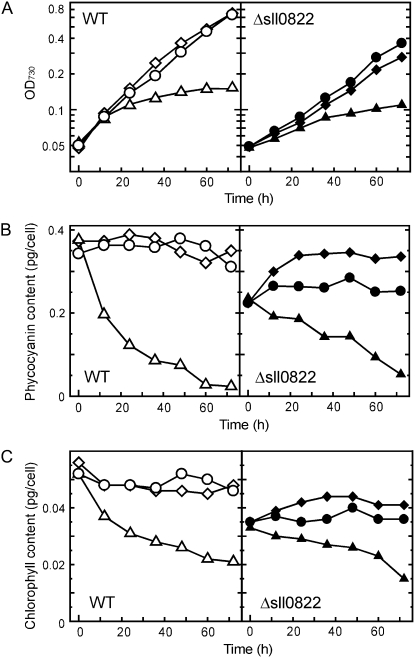
Nitrogen availability significantly affects cellular pigment content in cyanobacterial cells (Allen and Smith, 1969; Collier and Grossman, 1992). Thus, we examined the effect of nitrogen availability on the growth properties and pigment content of the Δsll0822 mutant. When cells were transferred from normal BG11 medium to nitrogen-free medium, growth inhibition (Fig. 3A) and a decrease in pigment content (Fig. 3, B and C) were similar in both the wild type and the Δsll0822 mutant. Supplementation of normal BG11 medium with a 4-fold excess of nitrate (final concentration = 70.6 mm) did not affect the growth properties of either strain (Fig. 3A). However, the pigment content of the mutant, especially phycocyanin content, was significantly recovered by supplementation with nitrate (Fig. 3, B and C).
Figure 3.
Effects of nitrogen availability on growth properties and pigment content of the wild type (WT) and the Δsll0822 mutant. At time 0, cells grown in normal BG11 medium were transferred to nitrogen-free medium (triangles), normal BG11 medium (circles; nitrate concentration, 17.7 mm), or nitrate-excess medium (diamonds; nitrate concentration, 70.6 mm). Changes in OD730 (A), phycocyanin content (B), and chlorophyll content (C) were monitored.
Identification of Target Genes of Sll0822
To identify the target genes of Sll0822, we performed DNA microarray analysis of wild-type and Δsll0822 mutant cells grown under normal growth conditions (20 μmol photons m−2 s−1; normal BG11 medium). Comprehensive information on the effects of sll0822 disruption of the gene expression profile is presented as Supplemental Table S1. Table II shows the list of genes whose expression levels were significantly affected by the disruption of sll0822. Genes whose mean induction rate (Δsll0822 mutant/wild type) minus sd was greater than 2.0 or whose mean induction rate plus sd was less than 0.5 are listed. It is notable that the expression level of nitrogen-controlled genes was lowered by the disruption of sll0822. Among them, sigE encodes a group 2 sigma factor that is involved in the activation of sugar catabolic genes (Osanai et al., 2005a) and is induced upon nitrogen starvation (Muro-Pastor et al., 2001). The gene product of glnB is the PII protein, which works as both a sensor and a regulator of carbon and nitrogen balance (Forchhammer, 2004). In addition to these genes encoding regulatory proteins, genes related to nitrogen uptake systems were significantly affected. nrtABCD encodes subunits of the ATP-binding cassette (ABC)-type nitrate/nitrite transporter (Aichi et al., 2001), and urtA encodes a subunit of the ABC-type urea transporter (Valladares et al., 2002). In the mutant, expression levels of genes encoding the other subunits of the urea transporter, urtB, urtC, urtD, and urtE, as well as genes for the monocomponent ammonium permeases, amt1, amt2, and amt3 (Montesinos et al., 1998), were also lowered to 50% to 70% of wild-type levels (Supplemental Table S2). It is also of note that expression levels of many other genes encoding channels or transporters were lowered in the mutant. These include glcP encoding a Glc transporter, aqpZ encoding a water channel protein, slr0646 and sll1270 encoding subunits of amino acid transporters, and mscL encoding a mechanosensitive channel (Table II). Genes encoding the subunits of PSI, psaK1 and psaF, are also listed. As for the other PSI subunit genes, expression levels of psaJ, psaC, and psaL in the mutant were less than 70% of the wild-type level, whereas those of psaAB and psaE were only slightly affected in the mutant (Supplemental Table S1). Among genes whose expression level increased upon the disruption of sll0822, special attention should be paid to hoxU and hoxF, encoding subunits of the bidirectional hydrogenase. Not only these two genes but also other members of the hoxEFUYH operon (sll1220–sll1226) showed higher expression in the Δsll0822 mutant (Supplemental Table S1). The hoxEFUYH operon was identified as positively regulated target genes of Sll0359 (Oliveira and Lindblad, 2008).
Table II.
List of open reading frames whose expression levels were affected by disruption of sll0822
Values are means ± sd obtained from three independent duplicate results of DNA microarray experiments (n = 6). Genes with an induction rate minus sd greater than 2.0 or those with an induction rate plus sd less than 0.5 are listed.
| Open Reading Frame No. | Description | Induction by Disruption of sll0822 |
|---|---|---|
| fold | ||
| Genes whose expression levels were decreased by disruption of sll0822 | ||
| sll1009 | 0.16 ± 0.09 | |
| sll0771 | Glc transporter (glcP) | 0.17 ± 0.14 |
| slr2057 | Water channel protein (apqZ) | 0.19 ± 0.04 |
| ssr0390 | PSI subunit X (psaK1) | 0.21 ± 0.13 |
| sll0064 | Substrate-binding protein of putative polar amino acid transport system | 0.24 ± 0.12 |
| slr0646 | Probable d-alanyl-d-Ala carboxypeptidase | 0.24 ± 0.15 |
| sll1689 | Group 2 RNA polymerase sigma factor SigE (sigE, rpoD2-V) | 0.26 ± 0.04 |
| slr0447 | ABC-type urea transport system substrate-binding protein (urtA) | 0.26 ± 0.06 |
| sll1641 | Glu decarboxylase | 0.27 ± 0.20 |
| sll1890 | CobN-like protein | 0.28 ± 0.08 |
| sll0051 | 0.29 ± 0.10 | |
| sll0444 | 0.31 ± 0.07 | |
| sll1451 | Nitrate/nitrite transport system permease protein (nrtB) | 0.31 ± 0.08 |
| slr1746 | Glu racemase | 0.32 ± 0.10 |
| sll1486 | 0.32 ± 0.10 | |
| sll1453 | Nitrate/nitrite transport system ATP-binding protein (nrtD) | 0.32 ± 0.15 |
| sll0024 | 0.32 ± 0.15 | |
| sll1270 | Substrate-binding protein of putative polar amino acid transport system (bgtB) | 0.32 ± 0.16 |
| sll0872 | 0.33 ± 0.08 | |
| sll1452 | Nitrate/nitrite transport system ATP-binding protein (nrtC) | 0.33 ± 0.10 |
| slr1275 | 0.35 ± 0.14 | |
| sll1450 | Nitrate/nitrite transport system substrate-binding protein (nrtA) | 0.35 ± 0.15 |
| slr0875 | Large-conductance mechanosensitive channel (mscL) | 0.37 ± 0.04 |
| sll0576 | Putative sugar-nucleotide epimerase/dehydratease | 0.37 ± 0.10 |
| sll1484 | Type 2 NADH dehydrogenase (ndbC) | 0.37 ± 0.12 |
| ssl0707 | Nitrogen regulatory protein PII (glnB) | 0.38 ± 0.06 |
| sll0819 | PSI subunit III (psaF) | 0.39 ± 0.10 |
| sll1665 | 0.42 ± 0.06 | |
| sll1077 | Agmatinase (speB2) | 0.42 ± 0.08 |
| Genes whose expression levels were enhanced by disruption of sll0822 | ||
| slr0303 | 29.38 ± 15.82 | |
| sll0330 | Sepiapterine reductase | 21.00 ± 14.19 |
| slr1704 | 14.18 ± 8.60 | |
| slr0304 | 13.15 ± 5.92 | |
| sll1236 | 11.57 ± 3.68 | |
| sll1722 | 11.34 ± 7.34 | |
| ssr2194 | 10.25 ± 2.18 | |
| slr1185 | Cytochrome b6-f complex Rieske iron sulfur protein (petC2) | 8.03 ± 2.65 |
| sll1723 | Probable glycosyltransferase | 6.70 ± 4.21 |
| slr1851 | 6.28 ± 2.53 | |
| slr0112 | 5.78 ± 3.22 | |
| slr1237 | Cytosine deaminase | 4.58 ± 1.06 |
| slr0229 | 3-Hydroxyisobutyrate dehydrogenase | 4.41 ± 0.80 |
| slr1811 | 4.28 ± 1.87 | |
| sll0910 | 4.27 ± 1.96 | |
| slr1812 | 4.19 ± 1.68 | |
| slr0581 | 4.15 ± 0.66 | |
| sll0274 | 4.14 ± 2.02 | |
| slr2076 | 60-kD chaperonin (groEL1) | 4.05 ± 1.99 |
| sll1130 | 3.96 ± 0.22 | |
| slr2075 | 10-kD chaperonin (groES) | 3.92 ± 1.34 |
| sll0470 | 3.90 ± 0.68 | |
| slr1852 | 3.79 ± 0.89 | |
| sll0947 | Light-repressed protein A homolog (lrtA) | 3.77 ± 1.65 |
| sll1566 | Glucosylglycerolphosphate synthase (ggpS) | 3.66 ± 0.87 |
| sll1221 | Diaphorase subunit of the bidirectional hydrogenase (hoxF) | 3.54 ± 1.02 |
| slr0551 | 3.36 ± 1.21 | |
| slr1814 | 3.33 ± 1.22 | |
| slr1853 | Carboxymuconolactone decarboxylase | 3.31 ± 1.20 |
| sll0502 | Arginyl-tRNA synthetase (argS) | 3.30 ± 1.17 |
| sll0563 | 3.24 ± 0.83 | |
| sll0314 | Periplasmic protein, function unknown | 3.23 ± 1.00 |
| ssr1528 | 3.08 ± 0.59 | |
| slr1854 | 2.98 ± 0.58 | |
| slr1855 | 2.89 ± 0.60 | |
| ssl2245 | 2.86 ± 0.71 | |
| sll0740 | 2.85 ± 0.74 | |
| sll1223 | Diaphorase subunit of the bidirectional hydrogenase (hoxU) | 2.77 ± 0.68 |
| sll1426 | 2.73 ± 0.70 | |
| sll0668 | Putative transposase | 2.40 ± 0.37 |
| slr1339 | 2.38 ± 0.23 | |
| sll1891 | 2.35 ± 0.32 | |
| ssl2501 | 2.34 ± 0.31 | |
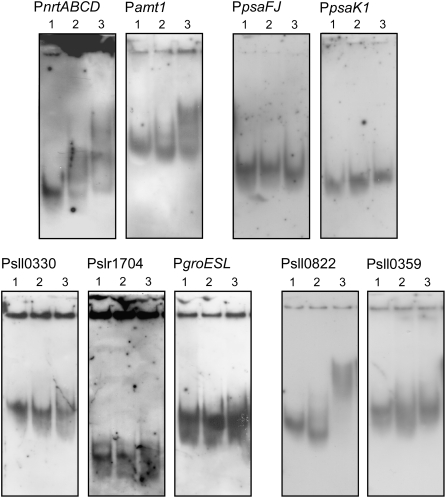
Next, we investigated whether Sll0822 can bind to the upstream regions of the above listed putative target genes. Recombinant His-tagged Sll0822 was overexpressed in Escherichia coli, purified to near homogeneity (Supplemental Fig. S3), and used for gel mobility shift assays with upstream DNA fragments of putative target genes. The binding specificity of the His-Sll0822 protein appeared to be low, since nonspecific band shifts with unrelated DNA sequences were observed upon the addition of 40 ng of His-Sll0822 protein (data not shown). However, when 20 ng of His-Sll0822 protein was added, the interaction with the specific promoters was reproducibly observed (Fig. 4). His-Sll0822 bound to the upstream regions of nrtABCD and amt1, whose expression levels were lowered by disruption of sll0822. On the other hand, no band shifts were observed using the upstream regions of psaFJ and psaK1, although expression of these genes was also lowered in the Δsll0822 mutant. As for urtA, glnB, and sigE, a slightly shifted smear was observed upon the addition of 20 ng of His-Sll0822 (data not shown). The upstream regions of sll0330, slr1704, and groESL, whose expression levels were increased in the Δsll0822 mutant, did not show an interaction with His-Sll0822.
Figure 4.
Gel mobility shift assays of the promoter segments of the putative target genes with purified His-Sll0822. DIG-labeled promoter segments of the putative target genes of Sll0822 were incubated with 0 (lane 1), 10 (lane 2), and 20 (lane 3) ng of purified His-Sll0822.
Many AbrB-like regulators having N-terminal DNA-binding domains are negatively autoregulated (Strauch et al., 1989; Bagyan et al., 1996). Thus, we tested the binding of His-Sll0822 to its own promoter region (Fig. 4). Mobility of the sll0822 upstream DNA fragment was clearly altered by incubation with purified His-Sll0822. On the other hand, no band shift was observed with the upstream region of sll0359.
Transcriptional Regulation by Sll0822 under Different Nitrogen Conditions
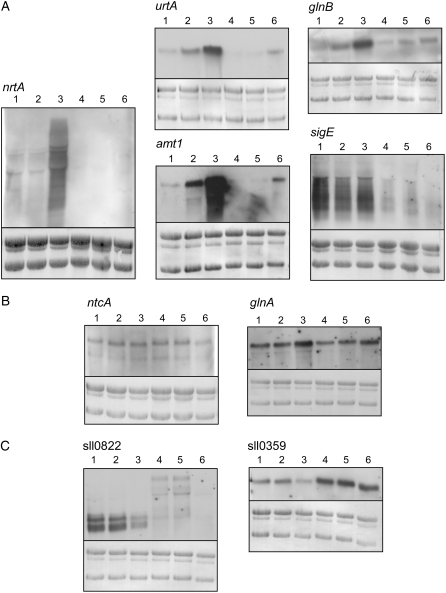
In cyanobacteria, most of the genes encoding nitrogen assimilation enzymes or transporters are repressed by ammonium, and their transcription is activated by the transcriptional factor NtcA in the absence of ammonium (Herrero et al., 2001). The nitrogen-related genes shown to be regulated by Sll0822 in this study are also under the control of NtcA. Thus, to clarify the role of Sll0822 in nitrogen regulation, we examined the effect of disruption of sll0822 on gene expression under different nitrogen conditions (Fig. 5A). In the Δsll0822 mutant, transcript levels of urtA, glnB, nrtA, amt1, and sigE were much lower than those of wild-type cells irrespective of nitrogen availability. This observation indicates that Sll0822 as well as NtcA are essential for the expression of these genes. However, not all NtcA target genes require Sll0822 for their expression (Supplemental Table S2). As shown in Figure 5B, transcript levels of ntcA and glnA were only slightly affected by disruption of sll0822. Figure 5C shows the transcript levels of sll0822 and sll0359 under different nitrogen conditions. A single transcript of 530 bp was detected for sll0359, whereas two transcripts of 650 and 510 bp were detected for sll0822. Both sll0359 and sll0822 are likely to be transcribed monocistronically in our study, which is different from the observation by Oliveira and Lindblad (2008) detecting a single long transcript of 1.4 kb for sll0359. In wild-type cells, down-regulation of sll0359 and sll0822 was observed upon nitrogen depletion (Fig. 5C). Transcript levels continued to decrease during incubation in nitrogen-free medium and became undetectable after 12 h (data not shown). In the Δsll0822 mutant, the expression level of sll0359 was much higher than that in wild-type cells irrespective of nitrogen conditions.
Figure 5.
Effects of disruption of sll0822 on gene expression under different nitrogen conditions. Ammonium-grown wild-type and Δsll0822 mutant cells were harvested, washed, and inoculated into fresh medium of different nitrogen conditions. After 1 h, total RNA was collected from wild-type (lanes 1–3) and Δsll0822 mutant (lanes 4–6) cells incubated in ammonium-containing medium (lanes 1 and 4), nitrate-containing medium (lanes 2 and 5), or nitrogen-free medium (lanes 3 and 6). RNA-blot analysis was performed as described in “Materials and Methods.” The amount of total RNA loaded per lane was as follows: 2 μg for urtA, glnB, glnA, sll0822, and sll0359, 4 μg for amt1 and ntcA, 6 μg for sigE, and 10 μg for nrtA. For comparison within each RNA blot, total RNA was stained with methylene blue.
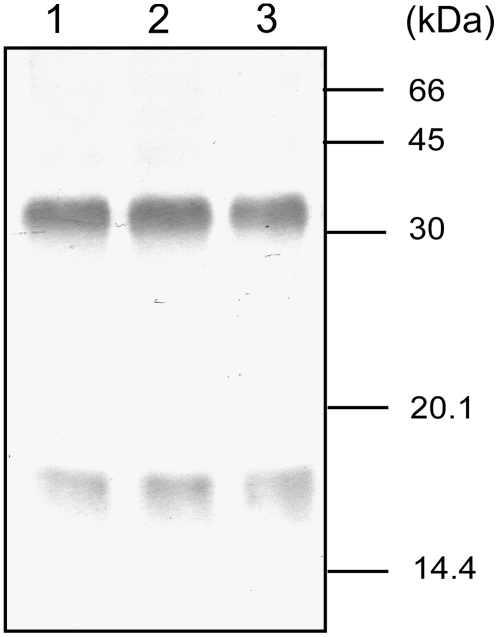
To examine the amount and oligomerization state of the Sll0822 protein in vivo, crude extracts of wild-type cells under different nitrogen conditions were subjected to immunoblot analysis using polyclonal antibodies raised against purified Sll0822. No band was detected when a crude extract from the Δsll0822 mutant was used, demonstrating that the Sll0822 antibody did not cross-react with the Sll0359 protein (data not shown). When a crude extract of wild-type cells was separated by SDS-PAGE, Sll0822 protein was detected mainly as a monomer of 17 kD (data not shown). To detect the native oligomerization state of Sll0822, we treated the crude extract with the cross-linker 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC) prior to separation by SDS-PAGE. Most of the Sll0822 protein was detected as a dimeric form of 34 kD by this method (Fig. 6). This observation suggests that the oligomeric structure of cyAbrB is different from that of AbrB in B. subtilis, since AbrB was shown to exist as a tetramer in vivo. We observed that the banding pattern of Sll0822 remained unchanged after both 1 h (data not shown) and 12 h (Fig. 6) of incubation under different nitrogen conditions. It is of note that neither the amount nor the oligomerization state of Sll0822 was affected by the different nitrogen conditions.
Figure 6.
The amount and oligomerization state of Sll0822 protein in vivo. Ammonium-grown wild-type cells were harvested, washed, and inoculated into fresh medium of different nitrogen conditions. After 12 h, crude extracts were prepared from cells incubated in ammonium-containing medium (lane 1), nitrate-containing medium (lane 2), or nitrogen-free medium (lane 3) and treated with the cross-linker EDC. Total protein equivalent to 1 × 107 cells per lane was separated by 15% (w/v) SDS-PAGE, and Sll0822 was detected by its specific antibody.
DISCUSSION
Physiological Significance of Sll0822
In this study, we identified Sll0822 as a novel regulator essential for the high-level expression of nitrogen-controlled genes. Upon disruption of the sll0822 gene, the expression levels of several members of the NtcA regulon, such as the nrt operon, urtA-E, amt1-3, glnB, and sigE, decreased significantly irrespective of nitrogen conditions, although the induction upon nitrogen depletion was still observed to some extent (Fig. 5). The nrt, urt, and amt genes encode components of the nitrate/nitrite transporter, urea transporter, and ammonium permease, respectively. It is likely that Sll0822 is important for the normal expression of nitrogen uptake systems. The growth rates and pigment contents of Δsll0822 mutants were much lower than those of wild-type plants when grown in normal BG11 medium (Table I; Figs. 2 and 3). The low pigment content of the mutant could be partially recovered by an increase in nitrate concentration of the medium (Fig. 3, B and C). Cyanobacterial cells faced with nutrient depletion down-regulate pigment contents to recycle the limiting nutrients and to decrease the capacity for light harvesting (Allen and Smith, 1969; Collier and Grossman, 1992). The defect in the activation of genes related to nitrogen uptake may cause nitrogen limitation within the mutant cells, leading to a decline in pigment content even under nitrate-replete conditions. In the mutant, a decrease in the expression levels of psaK1 and psaFJ, encoding subunits of PSI, was observed (Table II). However, the His-Sll0822 protein did not bind to their upstream regions (Fig. 4). This indicates that the low expression level of PSI genes could be due to a secondary effect of nitrogen limitation within the mutant cells. We also observed that the expression levels of various genes encoding transporters or channels were lowered in the mutant (Table II). The low growth rate of the mutant could be attributed to defects in some transportation systems.
Relationship between Sll0822 and NtcA
The partial overlap of the target genes of Sll0822 with those of NtcA raises a question concerning the relationship between these two regulators. Sll0822 is not located upstream of NtcA in the signal transduction cascade, considering that transcript levels of ntcA were not affected by the disruption of sll0822 (Fig. 5). Similarly, it is not likely that NtcA regulates the transcript level of sll0822, since no NtcA-binding site was found in the upstream region of sll0822. As for the possibility of protein-protein interactions, neither the cross-linking experiment conducted in this study (Fig. 6) nor a yeast two-hybrid assay performed at the Kazusa DNA Research Institute (Sato et al., 2007) detected an interaction between Sll0822 and NtcA. Thus, at present, we assume that Sll0822 and NtcA work independently on shared target promoters.
To date, several studies have identified additional factors required for the transcriptional regulation of NtcA-regulated genes in Synechocystis sp. PCC 6803. For example, the nrtA operon and the nirA operon involved in nitrate assimilation require the LysR family regulator NtcB for high-level expression and for nitrite-responsive activation (Aichi et al., 2001). In the case of ammonium assimilation-related genes, such as amt1, glnA, glnN, and gifB, the PII protein seems to be involved in their negative regulation (Takatani and Omata, 2006). Under nitrogen-depleted conditions, induction of several NtcA-regulated genes, such as the nrtA operon, nblA, and sigE, is dependent on PamA, a PII-binding transmembrane protein (Osanai et al., 2005b). The NtcA regulon appears to include subgroups of genes subject to different transcriptional regulation, and the above-mentioned regulators may provide fine-tuning of NtcA-dependent mechanisms. It is possible that Sll0822 also works in parallel with NtcA to achieve flexible regulation of nitrogen uptake.
Regulatory Mechanism of the Activity of Sll0822
Transcript levels of sll0822 decreased under nitrogen-depleted conditions (Fig. 5), which may be a result of negative autoregulation. After 12 h of incubation, the sll0822 transcript was barely detectable, whereas the amount of Sll0822 protein remained unchanged (Fig. 6). This indicates that Sll0822 is a highly stable protein, and transcriptional regulation may not be important for the regulation of its activity. Instead, it is possible that the regulation of Sll0822 is achieved by posttranslational modification, as inferred from the information described below. Two-dimensional gel electrophoresis of the soluble fraction of Synechocystis sp. PCC 6803 (Sazuka et al., 1999) detected two spots for Sll0822 with the same molecular mass but different pI values (7.8 and 8.6). Using tandem mass spectrometry analysis, Shalev-Malul et al. (2008) found that a cyAbrB in A. ovalisporum is posttranslationally modified by N-acetylation and methylation of specific amino acid residues. We postulated that the low binding specificity of His-Sll0822 prepared from E. coli was due to the lack of posttranslational modification. Thus, we overexpressed and purified His-Sll0822 from Synechocystis cells and used it for gel mobility shift assays. However, no improvement of the binding specificity was observed (data not shown). There must be an as yet unknown mechanism for the regulation of the binding activity of Sll0822 within cyanobacterial cells.
Relationship between Sll0822 and Sll0359
At least two genes encoding cyAbrBs are found in every cyanobacterial genome currently available. A set of genes belonging to clades A and B is conserved in most cases (Fig. 1), and this corresponds to sll0359 and sll0822 in Synechocystis sp. PCC 6803. Disruption of either sll0359 or sll0822 resulted in decreases in growth rate and pigment content (Table I). This indicates that cyAbrBs belonging to both clades A and B are required for normal cell growth and that their function is not redundant. Sll0359 may play a more important role than Sll0822, considering that complete segregation was attained in the Δsll0822 mutant but not in the Δsll0359 mutant. Increases in the level of expression of sll0359 upon deletion of sll0822 (Fig. 5) indicate the intimate relationship between these two cyAbrBs. When His-tagged Sll0822 protein was expressed in Synechocystis cells and purified using a nickel-chelating column, a small amount of Sll0359 was copurified. Similarly, when His-Sll0359 was expressed in Synechocystis cells, copurification of Sll0822 was detected (data not shown). A bidirectional interaction between Sll0822 and Sll0359 was also detected in the yeast two-hybrid assay performed at the Kazusa DNA Research Institute (Sato et al., 2007). There is likely to be a protein-protein interaction between Sll0822 and Sll0359 in vivo. However, the formation of a heterodimer is not plausible, since the amount of copurified protein was much smaller than that of the purified His-tagged protein in our experiments. In B. subtilis, the AbrB paralog, Abh, was recently shown to regulate antimicrobial gene expression (Strauch et al., 2007). The target genes of AbrB and Abh overlap, and the effects of deletion of abrB and abh differ either qualitatively or quantitatively for each of the shared target promoters. It was proposed that AbrB and Abh can form mixed multimers that have DNA-binding properties different from those of the homotetramers. We observed that the expression level of the hoxEFUYH operon, reported to be positively regulated by Sll0359 (Oliveira and Lindblad, 2008), increased significantly upon disruption of sll0822 (Table II; Supplemental Table S1). Sll0822 and Sll0359 may have a regulatory interaction similar to that proposed for AbrB and Abh, and this may be the reason why a set of cyAbrBs belonging to clades A and B is highly conserved among cyanobacterial species.
CONCLUSION
This study revealed the importance of Sll0822, one of two cyAbrBs in Synechocystis sp. PCC 6803, in the activation of nitrogen-regulated genes. In the Δsll0822 mutant, significant decreases in growth rate and cellular pigment content were observed, demonstrating the physiological significance of this protein. Sll0822 existed mainly in a dimeric form in vivo, and its amount was not affected by nitrogen availability. The other cyAbrB in Synechocystis sp. PCC 6803, Sll0359, was shown to be essential for growth, and a decrease in copy number of sll0359 resulted in a phenotype similar to that observed in the Δsll0822 mutant. Significant conservation of multiple genes encoding cyAbrBs among cyanobacterial genomes implies that these multiple genes have their own specific roles in regulatory processes. Additional studies on the interactions between Sll0822 and Sll0359 should further unravel the mechanisms of transcriptional regulation.
MATERIALS AND METHODS
Strains and Culture Conditions
A Glc-tolerant wild-type strain of Synechocystis sp. PCC 6803 was grown at 32°C in BG11 medium containing 20 mm HEPES-NaOH, pH 7.0, under continuous illumination at 20 μmol photons m−2 s−1 with bubbling of air. Mutants were grown under the same conditions, except that antibiotics were added to the medium: 20 μg mL−1 kanamycin for Δsll0822 and 20 μg mL−1 spectinomycin for Δsll0359. When ammonium was used as a nitrogen source, nitrate was replaced by 10 mm NH4Cl. Cell density was estimated by optical density at 730 nm (OD730) using a spectrophotometer (model UV-160A; Shimadzu). Cultures containing 1.0 × 108 cells mL−1 give an OD730 of 1.0 with this spectrophotometer.
Construction of Δsll0822 and Δsll0359 Mutants
A gene-flanking segment of sll0822 (900 bp; nucleotides 2,862,739–2,861,840 according to the numbering in CyanoBase) was amplified by PCR from genomic DNA of the wild-type strain using the primers 0822delF (5′-CGTCGCAGGGTAATCAAC-3′) and 0822delR (5′-GTATGAGGAAATCAACAG-3′) and cloned into the pT7Blue T-vector (Novagen). A kanamycin resistance cartridge, which had been excised from the plasmid pRL161 by digestion with HincII, was inserted into the coding region of sll0822 at the StyI site. For the construction of the Δsll0359 mutant, a gene-flanking segment (900 bp; nucleotides 2,146,600–2,145,701) was amplified by PCR using the primers 0359delF (5′-TCATTGCTCCATGGCGGC-3′) and 0359delR (5′-TACGACCCACCACCAGGG-3′) and cloned into the pT7Blue T-vector. A spectinomycin resistance cartridge, which had been excised from the plasmid pRL453 by digestion with SmaI, was inserted into the coding region of sll0359 at the NheI site. Wild-type cells were transformed with these constructs, and transformants were selected using appropriate antibiotics.
Determination of Pigment Content
In vivo absorption spectra of whole cells suspended in BG11 medium were measured at room temperature using a spectrophotometer (model 557; Hitachi) with an end-on photomultiplier. Chlorophyll content was calculated from the peak heights of absorption spectra using the equations of Arnon et al. (1974).
DNA Microarray Analysis
Isolation of total RNA using the RNeasy Midi kit (Qiagen) and subsequent DNA microarray analysis with CyanoCHIP (version 1.6; Takara) were performed as described previously (Hihara et al., 2003).
Overexpression and Purification of Sll0822
The sll0822 coding region was PCR amplified using the primers 0822-F (5′-AACATATGGCTAAATCAAACGCA-3′) and 0822-R (5′-AAGGATCCTTACTCTTCTTCGTCGTC-3′), cloned into the pT7Blue T-vector (Novagen), digested with NdeI and BamHI (sites underlined), and subcloned into the same restriction sites in pET28a (Novagen) to create pET0822 for the expression of a fusion protein with an N-terminal His tag. The nucleotide sequence was confirmed by DNA sequencing using the BigDye terminator method (Applied Biosystems). Escherichia coli BL21(DE3) harboring pET0822 was grown to an OD600 of 0.6 in 500 mL of 2× yeast extract-tryptone medium containing 20 μg mL−1 kanamycin, induced with 0.013% isopropyl β-d-thiogalactoside for 5 h, and harvested by centrifugation. Cells were resuspended in 15 mL of 20 mm phosphate buffer, pH 7.4, containing 0.5 m NaCl and 60 mm imidazole and disrupted by 10 rounds of sonication for 30 s each at 4°C. After the removal of whole cells and insoluble material by centrifugation, the soluble protein fraction was filtered through a 0.2-μm filter (DISMIC-25cs; ADVANTEC). His-Sll0822 was purified by nickel-affinity column chromatography using a HiTrap Chelating HP column (Amersham Biosciences). The soluble protein fraction was applied to the column equilibrated with 20 mm phosphate buffer, pH 7.4, containing 0.5 m NaCl and 60 mm imidazole, washed with the same buffer, and eluted with 20 mm phosphate buffer, pH 7.4, containing 0.5 m NaCl and 300 mm imidazole. Purified His-Sll0822 was desalted by a HiTrap Desalting column (Amersham Biosciences). Protein composition was examined by electrophoresis on a 15% (w/v) SDS-polyacrylamide gel followed by staining with Coomassie Brilliant Blue R-250.
Gel Mobility Shift Assay
For preparation of the probes for gel mobility shift assays, the following DNA fragments corresponding to the whole intergenic region upstream of each gene were obtained by PCR amplification: sll0822 (nucleotides 2,862,557–2,862,395 according to the numbering in CyanoBase; 163 bp), nrtABCD (1,010,709–1,010,575; 135 bp), psaFJ (1,688,212–1,688,054; 135 bp), psaK1 (156,259–156,390; 132 bp), sll0359 (2,146,387–2,146,586; 200 bp), slr1704 (728,490–728,659; 170 bp), and groESL (915,053–915,312; 260 bp). In the case of the following genes with long intergenic regions, a part of the intergenic region was amplified: amt1 (2,971,201–2,971,400; 200 bp) and sll0330 (2,385,028–2,384,729; 300 bp). The 3′ end of the DNA fragment for each probe was labeled with digoxigenin (DIG)-ddUTP by the terminal transferase method according to the manufacturer's instructions (DIG Gel Shift kit; Roche). Assays were performed using a DIG Gel Shift kit as described previously (Nakamura and Hihara, 2006).
RNA-Blot Analysis
Isolation of RNA by the hot phenol method and RNA-blot analyses, using a DIG RNA Labeling and Detection kit (Roche), were performed as described previously (Muramatsu and Hihara, 2003). To generate RNA probes by in vitro transcription, template DNA fragments were prepared by PCR using the following primers: urtA-F (5′-ATGACTAACCCTTTTGGA-3′) and SP6-urtA-R (5′-ATTTAGGTGACACTATAGAATACAGCAACCAATCCACCGCC-3′), PnrtA-F (5′-GCCAGCACCAATGCAGTA-3′) and SP6-nrtA-R (5′-ATTTAGGTGACACTATAGAATACAATGCCTTGGCCATTGAC-3′), PglnB-F (5′-AGAGGAAAAGTTTTTCGA-3′) and T7-glnB-R (5′-TAATACGACTCACTATAGGGCGATTAAATAGCTTCGGTATC-3′), amt1-F (5′-GCCCATTTCCAGAAGGAT-3′) and T7-amt1-R (5′-TAATACGACTCACTATAGGGCGAGCCAGGAGGTAAAAGTAG-3′), sigE-F (5′-ATGAGCGATATGTCTTCC-3′) and T7-sigE-R (5′-TAATACGACTCACTATAGGGCGAATCGCCCCTTCTTGGATC-3′), ntcA-F (5′-ATGGATCAGTCCCTAACC-3′) and SP6-ntcA-R (5′-ATTTAGGTGACACTATAGAATACTTAGGTAAACTGTTGACT-3′), glnA-F (5′-ATGGCCAGAACCCCCCAG-3′) and T7-glnA-R (5′-TAATACGACTCACTATAGGGCGATCTTCCCGACCAGTGTTC-3′), 0822-F and 0822-R, and 0359-F (5′-AACATATGCCAAACGCCTCCACC-3′) and 0359-R-Eco (5′-AAGAATTCTTATACTTCCTCTTCGTC-3′). The underlining indicates the T7 polymerase recognition sequence (TAATACGACTCACTATAGGGCGA) or the SP6 polymerase recognition sequence (ATTTAGGTGACACTATAGAATAC) added to the reverse primers in order to use the PCR products directly as templates for in vitro transcription reactions. In the cases of sll0822 and sll0359, PCR products were cloned into the pT7Blue T-vector containing a T7 polymerase recognition sequence, and the resultant plasmids were used as templates for in vitro transcription.
Immunoblot Analysis
Fifty milliliters of exponential growth phase cell culture was harvested by centrifugation. The pellet was resuspended in 200 μL of breakage buffer (20 mm Tris-HCl, pH 8.0, and 100 mm NaCl). The cell suspension was mixed with approximately 100 μL of glass beads (diameter, 0.1 mm; BioSpec Products), and cells were disrupted by four rounds of vigorous vortexing for 2 min followed by cooling on ice for 1 min. After removal of unbroken cells and debris by centrifugation at 700g for 3 min, cell extracts were mixed with an equal volume of 2× SDS sample buffer (62.5 mm Tris-HCl, pH 6.8, 4% SDS [w/v], 20% glycerol [v/v], and 0.01% bromphenol blue [w/v]). SDS-gel electrophoresis was performed according to the procedure of Laemmli (1970) using a 15% (w/v) acrylamide gel. After blotting to a polyvinylidene fluoride membrane (Immobilon-P; Millipore), samples were probed with polyclonal antibodies raised against the His-Sll0822 recombinant protein. The bound antibodies were detected using goat anti-rabbit IgG secondary antibodies conjugated to alkaline phosphatase. When required, cell extracts were treated with a cross-linker, EDC, before the addition of 2× SDS sample buffer. Samples were incubated with 25 mm EDC, 10 mm Tris-HCl, pH 8.0, 200 mm NaCl, and 20% glycerol (v/v) at 25°C for 2 h.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Multiple sequence alignment of cyAbrBs from 12 cyanobacterial genomes.
Supplemental Figure S2. Insertional inactivation of sll0359 and sll0822 genes.
Supplemental Figure S3. Overexpression and purification of His-Sll0822 from E. coli cells.
Supplemental Table S1. Comparison of gene expression levels between wild-type and Δsll0822 mutant cells grown under normal growth conditions by DNA microarray analysis.
Supplemental Table S2. Effects of disruption of sll0822 on the expression levels of the NtcA regulon.
Supplementary Material
This work was supported by a Grant-in-Aid for Young Scientists from the Japan Society for the Promotion of Science (to Y.H.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Yukako Hihara (hihara@molbiol.saitama-u.ac.jp).
The online version of this article contains Web-only data.
References
- Aichi M, Takatani N, Omata T (2001) Role of NtcB in activation of nitrate assimilation genes in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol 183 5840–5847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MM, Smith AJ (1969) Nitrogen chlorosis in blue-green algae. Arch Mikrobiol 69 114–120 [DOI] [PubMed] [Google Scholar]
- Arnon DI, McSwain BD, Tsujimoto HY, Wada K (1974) Photochemical activity and components of membrane preparations from blue-green algae. I. Coexistence of two photosystems in relation to chlorophyll a and removal of phycocyanin. Biochim Biophys Acta 357 231–245 [DOI] [PubMed] [Google Scholar]
- Bagyan I, Hobot J, Cutting S (1996) A compartmentalized regulator of developmental gene expression in Bacillus subtilis. J Bacteriol 178 4500–4507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson LM, Vaughn JL, Strauch MA, Bobay BG, Thompson R, Naylor S, Cavanagh J (2002) Macromolecular assembly of the transition state regulator AbrB in its unbound and complexed states probed by microelectrospray ionization mass spectrometry. Anal Biochem 306 222–227 [DOI] [PubMed] [Google Scholar]
- Bobay BG, Andreeva A, Mueller GA, Cavanagh J, Murzin AG (2005) Revised structure of the AbrB N-terminal domain unifies a diverse superfamily of putative DNA-binding proteins. FEBS Lett 579 5669–5674 [DOI] [PubMed] [Google Scholar]
- Bobay BG, Benson L, Naylor S, Feeney B, Clark AC, Goshe MB, Strauch MA, Thompson R, Cavanagh J (2004) Evaluation of the DNA binding tendencies of the transition state regulator AbrB. Biochemistry 43 16106–16118 [DOI] [PubMed] [Google Scholar]
- Coles M, Djuranovic S, Söding J, Frickey T, Koretke K, Truffault V, Martin J, Lupas AN (2005) AbrB-like transcription factors assume a swapped hairpin fold that is evolutionarily related to double-psi beta barrels. Structure 13 919–928 [DOI] [PubMed] [Google Scholar]
- Collier JL, Grossman AR (1992) Chlorosis induced by nutrient deprivation in Synechococcus sp. strain PCC 7942: not all bleaching is the same. J Bacteriol 174 4718–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forchhammer K (2004) Global carbon/nitrogen control by PII signal transduction in cyanobacteria: from signals to targets. FEMS Microbiol Rev 28 319–333 [DOI] [PubMed] [Google Scholar]
- Herrero A, Muro-Pastor AM, Flores E (2001) Nitrogen control in cyanobacteria. J Bacteriol 183 411–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hihara Y, Sonoike K, Kanehisa M, Ikeuchi M (2003) DNA microarray analysis of redox-responsive genes in the genome of the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol 185 1719–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- Los DA, Suzuki I, Zinchenko VV, Murata N (2007) Stress responses in Synechocystis: regulated genes and regulatory systems. In A Herrero, E Flores, eds, Cyanobacteria: Molecular Biology, Genomics and Evolution. Horizon Scientific Press, Norfolk, UK, pp 117–157
- Montesinos ML, Muro-Pastor AM, Herrero A, Flores E (1998) Ammonium/methylammonium permeases of a cyanobacterium: identification and analysis of three nitrogen-regulated amt genes in Synechocystis sp. PCC 6803. J Biol Chem 273 31463–31470 [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Hihara Y (2003) Transcriptional regulation of genes encoding subunits of photosystem I during acclimation to high-light conditions in Synechocystis sp. PCC 6803. Planta 216 446–453 [DOI] [PubMed] [Google Scholar]
- Muro-Pastor AM, Herrero A, Flores E (2001) Nitrogen-regulated group 2 sigma factor from Synechocystis sp. strain PCC 6803 involved in survival under nitrogen stress. J Bacteriol 183 1090–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Hihara Y (2006) Photon flux density-dependent gene expression in Synechocystis sp. PCC 6803 is regulated by a small, redox-responsive, LuxR-type regulator. J Biol Chem 281 36758–36766 [DOI] [PubMed] [Google Scholar]
- Oliveira P, Lindblad P (2008) An AbrB-like protein regulates the expression of the bidirectional hydrogenase in Synechocystis sp. strain PCC 6803. J Bacteriol 190 1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osanai T, Imamura S, Asayama M, Shirai M, Suzuki I, Murata N, Tanaka K (2005. a) Nitrogen induction of sugar catabolic gene expression in Synechocystis sp. PCC 6803. DNA Res 13 185–195 [DOI] [PubMed] [Google Scholar]
- Osanai T, Sato S, Tabata S, Tanaka K (2005. b) Identification of PamA as a PII-binding membrane protein important in nitrogen-related and sugar-catabolic gene expression in Synechocystis sp. PCC 6803. J Biol Chem 280 34684–34690 [DOI] [PubMed] [Google Scholar]
- Phillips ZE, Strauch MA (2002) Bacillus subtilis sporulation and stationary phase gene expression. Cell Mol Life Sci 59 392–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Shimoda Y, Muraki A, Kohara M, Nakamura Y, Tabata S (2007) A large-scale protein protein interaction analysis in Synechocystis sp. PCC6803. DNA Res 14 207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazuka T, Yamaguchi M, Ohara O (1999) Cyano2Dbase updated: linkage of 234 protein spots to corresponding genes through N-terminal microsequencing. Electrophoresis 20 2160–2171 [DOI] [PubMed] [Google Scholar]
- Shalev-Malul G, Lieman-Hurwitz J, Viner-Mozzini Y, Sukenik A, Gaathon A, Lebendiker M, Kaplan A (2008) An AbrB-like protein might be involved in the regulation of cylindrospermopsin production by Aphanizomenon ovalisporum. Environ Microbiol 10 988–999 [DOI] [PubMed] [Google Scholar]
- Strauch MA, Bobay BG, Cavanagh J, Yao F, Wilson A, Le Breton Y (2007) Abh and AbrB control of Bacillus subtilis antimicrobial gene expression. J Bacteriol 189 7720–7732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch MA, Perego M, Burbulys D, Hoch JA (1989) The transition state transcription regulator AbrB of Bacillus subtilis is autoregulated during vegetative growth. Mol Microbiol 3 1203–1209 [DOI] [PubMed] [Google Scholar]
- Takatani N, Omata T (2006) Effects of PII deficiency on expression of the genes involved in ammonium utilization in the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Cell Physiol 47 679–688 [DOI] [PubMed] [Google Scholar]
- Valladares A, Montesinos ML, Herrero A, Flores E (2002) An ABC-type, high-affinity urea permease identified in cyanobacteria. Mol Microbiol 43 703–715 [DOI] [PubMed] [Google Scholar]
- Vaughn JL, Feher V, Naylor S, Strauch MA, Cavanagh J (2000) Novel DNA binding domain and genetic regulation model of Bacillus subtilis transition state regulator abrB. Nat Struct Biol 7 1139–1146 [DOI] [PubMed] [Google Scholar]
- Yao F, Strauch MA (2005) Independent and interchangeable multimerization domains of the AbrB, Abh, and SpoVT global regulatory proteins. J Bacteriol 187 6354–6362 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.