Abstract
Geminiviruses are small DNA viruses that use plant replication machinery to amplify their genomes. Microarray analysis of the Arabidopsis (Arabidopsis thaliana) transcriptome in response to cabbage leaf curl virus (CaLCuV) infection uncovered 5,365 genes (false discovery rate <0.005) differentially expressed in infected rosette leaves at 12 d postinoculation. Data mining revealed that CaLCuV triggers a pathogen response via the salicylic acid pathway and induces expression of genes involved in programmed cell death, genotoxic stress, and DNA repair. CaLCuV also altered expression of cell cycle-associated genes, preferentially activating genes expressed during S and G2 and inhibiting genes active in G1 and M. A limited set of core cell cycle genes associated with cell cycle reentry, late G1, S, and early G2 had increased RNA levels, while core cell cycle genes linked to early G1 and late G2 had reduced transcripts. Fluorescence-activated cell sorting of nuclei from infected leaves revealed a depletion of the 4C population and an increase in 8C, 16C, and 32C nuclei. Infectivity studies of transgenic Arabidopsis showed that overexpression of CYCD3;1 or E2FB, both of which promote the mitotic cell cycle, strongly impaired CaLCuV infection. In contrast, overexpression of E2FA or E2FC, which can facilitate the endocycle, had no apparent effect. These results showed that geminiviruses and RNA viruses interface with the host pathogen response via a common mechanism, and that geminiviruses modulate plant cell cycle status by differentially impacting the CYCD/retinoblastoma-related protein/E2F regulatory network and facilitating progression into the endocycle.
Geminiviruses are a large, diverse family of plant-infecting viruses that cause serious crop losses worldwide (Rojas et al., 2005). They have singled-stranded DNA genomes that are transcribed, replicated, and encapsidated in the nuclei of infected cells. They also traffic within and between cells and move systemically through infected plants. Geminiviruses usurp plant enzymes and metabolites for these processes, while their hosts mount defense responses to limit infection. Studies of plant RNA viruses showed that viral infection is accompanied by many changes in the plant transcriptome (Whitham et al., 2003; Marathe et al., 2004; Yang et al., 2007). Earlier studies showed that geminiviruses induce the accumulation of a host replication protein (Nagar et al., 1995) and a protein kinase (Kong and Hanley-Bowdoin, 2002), indicating that plant DNA viruses also alter host gene expression. However, unlike RNA viruses that encode replicases to amplify their genomes, geminiviruses represent a unique opportunity to study changes in the expression of host genes involved in cell cycle regulation and DNA replication as well as the induction of the pathogen response.
The Geminiviridae is classified into four genera based on genome structure, insect vector, and host range (Fauquet et al., 2003). Cabbage leaf curl virus (CaLCuV), a member of the Begomovirus genus (Hill et al., 1998), encodes seven proteins, including two viral replication proteins designated as AL1 (AC1, C1, or Rep) and AL3 (AC3, C3, or REn), and does not specify a DNA polymerase. Instead, it depends on host DNA replication machinery to amplify its small, circular genome via a combination of rolling circle replication (RCR) and recombination-mediated replication (RDR). This dependence on host machinery constitutes a barrier to infection of mature plant cells, which have exited the cell cycle and no longer support DNA replication (for review, see Hanley-Bowdoin et al., 2004). To overcome this constraint, the geminivirus AL1 protein binds to the host retinoblastoma-related protein (RBR) and relieves repression of E2F transcription factors. This, in turn, allows activation of genes required for transition into S phase and establishment of a DNA replication-competent environment (Egelkrout et al., 2001, 2002; Desvoyes et al., 2006).
Interactions between geminivirus proteins and other host factors are also likely to impact plant gene expression mechanisms. The binding of AL3 to a NAC transcription factor enhances viral DNA replication (Selth et al., 2005), while interactions between AL1 and a putative mitotic kinesin or histone H3 might contribute to the altered chromosomal structure characteristic of infected cells and indirectly influence host gene expression (Bass et al., 2000; Kong and Hanley-Bowdoin, 2002). The viral AL2 protein binds to adenosine kinase to suppress host gene silencing (Wang et al., 2005), while interactions between the viral nuclear shuttle protein BR1 and the nuclear acetyltransferase AtNSI may prevent DNA modifications that interfere with replication and transcription (Carvalho et al., 2006). In addition, both AL1 and AL2 function as transcriptional regulators of viral genes and might impact the activities of yet-to-be-identified host genes (Eagle et al., 1994; Sunter and Bisaro, 1997).
Geminiviruses could also influence host gene expression by altering signal transduction pathways through interactions with host protein kinases. Reduced activity of a SNF1-related kinase (SnRK1) in response to AL2 binding has been implicated in host susceptibility to infection (Hao et al., 2003). The BR1 protein binds to NIK1, NIK2, and NIK3, members of the Leu-rich repeat-receptor-like kinase (RLK) family, and inhibits their phosphorylation and antiviral activities (Fontes et al., 2004; for a list of gene acronyms and descriptions, see Supplemental Table S1). In contrast, interaction and phosphorylation of BR1 by a PERK-like RLK, NsAK, is necessary for efficient infection and full symptom development (Florentino et al., 2006). AL1 binding to GRIK1 and GRIK2, which accumulate in infected cells (Kong and Hanley-Bowdoin, 2002), may modulate their proposed dual roles in controlling precursor and energy resources needed for DNA replication and activation of SnRK1 and the pathogen response (Shen and Hanley-Bowdoin, 2006). The divergent AL4 and C4 proteins may alter cell signaling through their interactions with two members of the shaggy protein kinase-related family involved in brassinosteroid signaling (Piroux et al., 2007).
The diverse interactions and activities of the viral proteins suggest that geminiviruses modulate a variety of plant processes by altering host gene expression. Serial analysis of gene expression of cassava mosaic disease annotated 30 differentially expressed genes encoding proteins associated with systemic acquired resistance, a β-tubulin, and a WRKY transcription factor among 12,786 sequenced tags (Fregene et al., 2004). Differential display analysis of Capsicum annuum response to Pepper huasteco yellow vein virus identified two clones encoding a methyltransferase and an NADP-malic enzyme (Anaya-López et al., 2005). A microarray study of Arabidopsis (Arabidopsis thaliana) cultured cells expressing the AC2 proteins of Mungbean yellow mosaic virus (MYMV) or African cassava mosaic virus (ACMV) identified 139 genes that were elevated by both viral proteins (Trinks et al., 2005). All of these studies used resistant virus/host combinations or focused on differences between resistant and susceptible infections. The best-characterized example of host gene expression change during a compatible geminivirus infection is activation of the Nicotiana benthamiana PCNA gene in response to CaLCuV or tomato golden mosaic virus infection (Egelkrout et al., 2001; Egelkrout et al., 2002).
Our limited knowledge of host gene expression during geminivirus infection in planta is due in part to the absence of a well-characterized, compatible virus/host system suitable for transcriptome profiling studies. This limitation is compounded by the technical challenge of detecting changes that occur in only a small fraction of virus-positive cells. To address these constraints and to gain new insight into geminivirus/host interactions, we established a carefully controlled experimental system based on CaLCuV and its susceptible host Arabidopsis to examine global changes in host gene expression during infection. Using this system, we showed that CaLCuV infection alters the expression of a large number of plant genes involved in diverse processes ranging from the pathogen response and programmed cell death to DNA replication and cell cycle control.
RESULTS
CaLCuV Infection of Arabidopsis Ecotype Columbia
For the gene profiling studies, we implemented an infection protocol for CaLCuV in its compatible host Arabidopsis ecotype Columbia (Col-0) that minimized secondary effects due to development and environment. Plants were grown under short-day conditions (8-h/16-h photoperiod) to produce a large number of rosette leaves and prevent flowering during the course of the experiment. This approach ensured adequate leaf material for RNA extraction and minimized any developmental effects associated with the vegetative to floral transition that would have complicated the analysis. Previous experiments showed that geminivirus-mediated activation of PCNA expression was readily detected in plants inoculated by agroinfection but not by bombardment (Egelkrout et al., 2001). Hence, Arabidopsis plants at the 16- to 18-leaf stage were agroinoculated with partial tandem dimers of CaLCuV and monitored for symptom development over time.
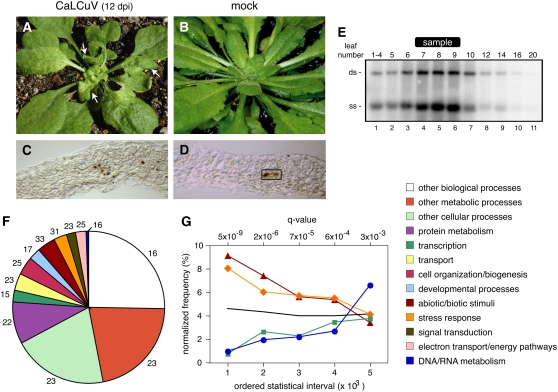
CaLCuV symptoms began to appear at 9 to 10 d postinoculation (dpi) with all plants showing symptoms by 12 dpi (Fig. 1A). A general yellowing associated with agroinfection disappeared at least 3 d before the onset of viral symptoms, which were characterized by overall stunting and chlorosis. Upper leaves were smaller, narrower, and curled and displayed a reticulated chlorosis beginning at the base and expanding through the blade as infection progressed. Lower leaves were thicker and darker and showed a moderate upward curvature and helical twisting. Mock-inoculated plants showed normal leaf development and no visible effect of agroinoculation at 12 dpi (Fig. 1B). Viral DNA accumulation was examined in leaves 5 to 20 and in a pool comprised of the four uppermost visible leaves (1–4) from a single infected Arabidopsis plant at 12 dpi. DNA gel blotting detected double- and single-stranded forms of CaLCuV DNA in all the leaves (Fig. 1E, lanes 1–11), with the highest amounts in leaves 7, 8, and 9 (lanes 4, 5, and 6). Leaves 6 and 10 contained intermediate amounts of viral DNA (Fig. 1E, lanes 3 and 7), while leaves 1 to 4, 5, 12, and 14 had lower levels (lanes 1, 2, 8, and 9). Leaves 16 and 20 had trace amounts of CaLCuV DNA (Fig. 1E, lanes 10 and 11). Similar distributions of viral DNA were seen in two other plants (data not shown).
Figure 1.
CaLCuV infection differentially impacts Arabidopsis gene expression across functional categories. CaLCuV symptoms in Arabidopsis Col-0 rosette leaves at 12 dpi are shown in A. The arrows mark the size of leaves harvested for RNA extraction. The mock-inoculated plant at 12 dpi is shown in B. Immunolocalization of the AL1 protein in leaf 9 of a CaLCuV-infected plant at 12 dpi can be observed in C. D, Control section from an equivalent, mock-inoculated leaf. Endogenous peroxidase activity is seen in vascular tissue (rectangle). Total DNA (0.5 μg) from a CaLCuV-infected plant at 12 dpi was analyzed by DNA gel blotting using a 32P-labeled probe corresponding to full-length CaLCuV A DNA. In E, the DNA samples were from a pool of leaves 1 to 4 (lane 1) or individual leaves (lanes 2–11). The leaves are numbered according to their positions relative to the shoot apex from the youngest (1–4, lane 1) to the oldest (20, lane 11). The bands corresponding to double-stranded (ds) and single-stranded (ss) forms of CaLCuV A are identified on the left. The leaves used for the microarray experiments are marked at the top (sample). A pie chart showing the distribution of the differentially expressed genes across functional categories defined by the TAIR7 GO is depicted in F. The numbers are the percentage of genes in each category differentially expressed during infection. Interval analysis of selected functional groups can be seen in G. Genes were ordered according to their q values and grouped into intervals of 1,000. The frequencies of selected functional classes normalized to their overall representation in the GO database were plotted for five intervals corresponding to the 5,000 genes with the lowest P values. The abiotic/biotic stimulus is represented by triangles, the stress response by diamonds, DNA/RNA metabolism by circles, and transcription by squares. The colors of the lines are the same as in F. The black line corresponds to the mean representation of all GO categories for each interval.
Expression Profiling of CaLCuV-Infected Arabidopsis Leaves
We used the Affymetrix ATH1 GeneChip to compare the expression profiles of mock-inoculated and CaLCuV-infected plants at 12 dpi, the earliest time after inoculation at which we could reliably identify infected leaves. At this time, leaves 7, 8, and 9 contained the highest amounts of viral DNA and were larger than 1 cm in length (Fig. 1E). Leaves greater than 1 cm in length contain few cells undergoing DNA replication, while leaves smaller than 1 cm contain significant numbers of replicating cells (Donnelly et al., 1999). Based on these observations, we hypothesized that viral effects on host gene expression would be maximal in leaves 7, 8, and 9, while leaf developmental effects related to DNA replication and the cell division cycle would be minimal. We also took into account that at a given thermal time, the expanded leaves are equivalent to the leaves described by Donnelly et al. (1999); therefore, leaves bigger than 1 cm will be suitable for sampling (Granier et al., 2002).
Arabidopsis plants were agroinoculated with CaLCuV or mock inoculated with a control strain to eliminate effects due to Agrobacterium infection. For each sample, leaves 7, 8, and 9 from at least six plants at 12 dpi were pooled to minimize plant-to-plant variation. Three independent sets of plants were sampled to generate three biological replicas. Total RNA from the leaf samples was converted to cRNA for use as target in microarray experiments. The six target samples were hybridized, processed, and scanned in parallel, and the consistency of the perfect match data was inspected by MA plots (data not shown). LOESS normalization using the mean across the chips as the baseline (Dudoit and Fridlyand, 2002) was applied directly to the probe data because of its higher reproducibility (Chu et al., 2006). After normalization, the correlation coefficients of perfect match probes between chips ranged from 0.89 to 0.98, with a mean value of 0.93 (data not shown). The expression data was analyzed by an ANOVA model (Chu et al., 2002) using JMP Genomics (http://www.jmp.com/software/genomics/index.shtml), and the resulting P values were used to determine a false discovery rate (Storey and Tibshirani, 2003). Using a q value <0.005 (P value <0.002), we identified 5,365 RNAs that are differentially expressed in CaLCuV-infected versus mock-inoculated Arabidopsis in our experimental system. Of these, 3,004 RNAs were elevated and 2,361 were reduced in infected leaves (Supplemental Table S2). The microarray results were validated for 30 of the differentially expressed genes (q values from 0.0031–4.0 × 10−29) and for six genes with q values >0.005. The ATH1 results were confirmed for 34 of the 36 genes examined (Supplemental Fig. S1).
Functional Categorization of Differentially Expressed Genes
As a first step toward characterization of the 5,365 differentially expressed genes, we used the TAIR7 Gene Ontology (GO) descriptions to categorize the genes by biological process. All GO biological process categories were represented among the significant genes, with the number ranging from 66 to 2,552 across the various processes (Fig. 1F). Categories corresponding to abiotic/biotic stimuli (33%) and stress response (31%) were overrepresented, while the categories for DNA/RNA metabolism (16%) and transcription (15%) were underrepresented.
To gain insight into the differences between categories, the genes in order of their q values were grouped into bins of 1,000. The bins were classified by GO biological processes and normalized relative to the total number of annotations in each category. The results for the bins with the 5,000 lowest q values are shown for selected categories in Figure 1G. This analysis revealed that genes annotated in the abiotic/biotic stimulus (dark red) and stress response (orange) categories are highly represented in the first three bins, while genes annotated in the DNA/RNA metabolism (blue) and transcription (green) categories are poorly represented in the same bins. In both cases, the representations diverge from the overall mean of all categories in each bin (black line). The normalized frequencies for the four categories are more similar in bins 4 and 5 and converge on the overall mean (black line). Our analysis is consistent with more robust and, hence, more readily detected expression changes for genes in the overrepresented versus the underrepresented categories. The uneven distribution of the different categories may reflect different aspects of the infection process, with the overrepresented categories including a large fraction of systemic events and the underrepresented categories containing a large proportion of events that are cell autonomous with CaLCuV presence. This idea is supported by studies showing that the host factors PCNA and GRIK, which are expressed in young tissues of healthy plants, accumulate specifically in virus-positive cells of mature leaves during infection (Nagar et al., 1995; Kong and Hanley-Bowdoin, 2002).
Earlier studies showed that the AL1 protein is an early marker for geminivirus infection and can be detected prior to viral DNA at the cellular level (Nagar et al., 2002). To determine the relative fraction of CaLCuV-positive cells under our infection condition, we used a polyclonal antiserum to immunolocalize AL1 in infected Arabidopsis leaves at 12 dpi. The AL1 protein was readily detected in cross sections of leaf 9 (Fig. 1C), consistent with high viral DNA level in this leaf (Fig. 1E). The viral replication protein was seen most often in cells adjacent to vascular tissue but was also found in the occasional mesophyll cell and the rare epidermal cell. In all cases, AL1 was detected in nuclei, which showed quenching of 4′,6-diamidino-2-phenylinodole (DAPI) staining by the peroxidase precipitate (data not shown). Mock-inoculated leaves had background staining in the vasculature, but no nuclear signals were seen in any leaf cell types (Fig. 1D). The number of infected cells varied across sections but was no more than 10% of the total number of cells. Similar results were obtained when in situ hybridization was used to detect viral DNA (data not shown). Based on these results, we reasoned that changes in gene expression associated with cell-autonomous events would be diluted at least 10-fold, making them more difficult to detect than systemic changes that are not confined to virus-positive cells.
CaLCuV and RNA Viruses Alter the Expression of a Common Set of Genes
Although plant DNA and RNA viruses use different replication and expression strategies, many aspects of their infection cycles are related and may lead to similar changes in host gene expression. We compared the CaLCuV infection profile with the profiles of plants inoculated with Turnip mosaic virus (TuMV; Yang et al., 2007) or Cucumber mosaic virus-Y (CMV-Y; Marathe et al., 2004) in Figure 2A. The TuMV study was in the susceptible Arabidopsis Col-0 ecotype, while the CMV-Y study was in the resistant C24 ecotype. Comparison of the CaLCuV profile uncovered greater overlap with the TuMV infection profile (267/556) than the CMV-Y resistome (107/444), but both comparisons also identified many genes with opposite expression (TuMV, 72/556; CMV-Y, 77/444). In contrast, comparison to a common set of genes differentially expressed in response to five different RNA viruses (including TuMV) representing four families (Whitham et al., 2003) uncovered 63 of 102 similarly expressed genes and only three with opposite expression during CaLCuV infection (Supplemental Table S3). Together, these comparisons indicated that a subset of the host genes differentially regulated during virus infection is common to both DNA and RNA viruses (Fig. 2A).
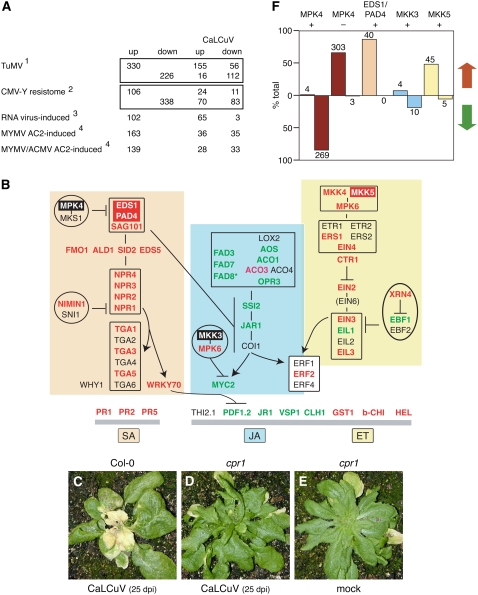
Figure 2.
Pathogen response during CaLCuV infection. Genes differentially expressed during CaLCuV infection were compared to the expression profiles of Arabidopsis genes that change in response to other plant pathogens or pathogen proteins. A, The comparisons include: (1) genes differentially expressed during the compatible TuMV infection (Yang et al., 2007); (2) genes differentially expressed during the resistance response to CMV-Y (Marathe et al., 2004); (3) transcripts elevated by five RNA viruses in planta (Whitham et al., 2003); and (4) transcripts increased in cultured cells expressing MYMV or ACMV AC2 protein (Trinks et al., 2005). The numbers of shared genes that are up- or down-regulated in CaLCuV-infected leaves are indicated for each comparison. The totals for each treatment were adjusted to include only genes represented on the ATH1 array. B, A model of the interactions between key genes involved in the SA (orange), JA (blue), and ET (yellow) pathogen response pathways. The genes are shown in their proposed order in the regulatory network or order of expression. Genes with elevated mRNA levels are in red typeface, while genes with reduced mRNA are in green typeface. The genes with red or black backgrounds are in F. The corresponding Arabidopsis gene numbers are in Supplemental Table S1. C, Infected Col-0 plant showing signs of senescence and cell death at 25 dpi is depicted. CaLCuV-inoculated cpr1 mutant (Bowling et al., 1994) at 25 dpi is shown in D, followed by a mock-inoculated cpr1 mutant at 25 dpi (E). Graph in F shows the percentage of overlap of mRNAs that are increased (red arrow) or decreased (green arrow) during CaLCuV infection to genes with reduced (MPK4 +) or elevated (MPK4 −) transcripts in an mpk4 mutant (Andreasson et al., 2005), with reduced RNA in an eds1/pad4 mutant (EDS1/PAD4 +; Bartsch et al., 2006), or with increased RNA in MKK3 (+)- or MKK5 (+)-overexpressing lines (Takahashi et al., 2007). The number of genes is indicated for each bar.
The only published microarray study for geminiviruses examined the impact of expressing individual viral proteins in suspension cells (Trinks et al., 2005). The RNA profiles of Arabidopsis cells transiently expressing MYMV or ACMV AC2 at 8 h posttransfection (Trinks et al., 2005) did not show a strong relationship to the expression profile of CaLCuV-infected leaves, with only 18% to 20% of the RNAs showing similar trends and 21% to 24% showing opposite trends. It was also reported that overexpression of MYMV AC1 altered expression of 259 RNAs, but we could not compare these to our results, because the elevated mRNAs were not identified. However, the reported lack of overlap between the AC1 and AC2 datasets is consistent with the observation that many more RNAs are differentially expressed in infected leaves than in cells expressing individual viral proteins. The low correlation between the two experimental systems may also reflect the use of different virus/host combinations (natural host versus nonhost), timing of analysis (12 dpi versus 8 h), and greater dilution of cell-autonomous changes in intact plants versus cultured cells.
Pathogen Response Genes
Plants respond to pathogens via the salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) pathways. RNA viruses typically activate the SA pathway (Whitham et al., 2006), but equivalent information is not available for geminiviruses. The gene profiling data showed that PR1, PR2, and PR5 transcripts, which are markers for the SA response (Pieterse and Van Loon, 2004), were elevated during CaLCuV infection (Fig. 2B). Genes specifying SA biosynthetic enzymes and signaling components (EDS1, PAD4, SAG101, FMO1, ALD1, SID2, EDS5, NPR1, NPR2, NPR3, and NPR4) as well as downstream transcription factors (WRKY70, TGA1, TGA3, and TGA5) also had increased RNA levels (Pieterse and Van Loon, 2004; Wiermer et al., 2005). In contrast, transcripts for some JA/ET markers (JR1, VSP1, PDF1.2, and CLH1) were reduced, whereas others (GST1, b-CHI, and HEL) were up in infected leaves (Lorenzo and Solano, 2005). RNAs encoding components of the JA pathway were down (FAD3, FAD7, AOS, ACO1, OPR3, SSI2, and MYC2) or showed no change on the arrays (COI1 and JAR1, although JAR1 transcripts were reduced in reverse transcription [RT]-PCR reactions; Lorenzo and Solano, 2005; Beckers and Spoel, 2006). The mRNAs for EIN2 and EIN5, which are associated with the ET pathway (Etheridge et al., 2006), were up, possibly leading to the downstream activation of JA late response genes.
We used semiquantitative RT-PCR to verify induction of the SA pathway and down-regulation of the ET/JA pathways. These experiments confirmed the microarray data for selected RNAs (Supplemental Fig. S1A; EDS1, PAD4, NPR2, FAD3, PR1, and PDF1.2). The microarray experiments failed to detect a significant change in JAR1 transcripts even though RT-PCR indicated that they were reduced in four independent RNA samples from infected leaves. Based on these results, we included JAR1 among the down-regulated genes. FAD8, which was not represented on the array, was also down in infected versus mock-inoculated tissues (Supplemental Fig. S1A), and COI1-induced genes were reduced in CaLCuV microarray data (data not shown), lending further support for repression of the JA pathway. In addition, we examined the expression of a general set of defense response genes involved in oxidative stress and cell wall metabolism by quantitative-RT PCR using a commercial kit (Supplemental Fig. S1C). These results paralleled the microarray data but showed a greater dynamic range. Among the analyzed RNAs, only PDF1.2 transcripts were reduced during CaLCuV infection. Transcripts specifying thioredoxins, glutathione S-transferases, and β-glucanases were represented among the elevated transcripts in CaLCuV-infected leaves. The catalase and invertase RNAs, which showed the greatest changes in the microarray experiments, also displayed strong differences in mock and infected leaves by semiquantitative RT-PCR.
We tested a number of Arabidopsis lines carrying mutations in various pathogen response genes in CaLCuV infectivity assays. These experiments did not uncover any differences with respect to timing or severity of symptoms between SA (pad4, mpk4, sid2-2, npr1, and wrky70), JA (coi1-16), or ET (ein2) pathway mutants and wild-type Col-0 plants (data not shown). Although the aggressive nature of CaLCuV infection may have masked subtle differences, similar results have also been reported for RNA viruses, suggesting that individual knockout mutations do not dramatically impact plant virus infection (Huang et al., 2005). In contrast, Arabidopsis cpr1 plants, which constitutively express PR1 (Bowling et al., 1994), were less susceptible to CaLCuV infection. CaLCuV-inoculated cpr1 plants did not display symptoms at 25 dpi (Fig. 2D), while Col-0 plants were highly symptomatic at this time (Fig. 2C). The cpr1 plants eventually developed weak symptoms (data not shown), indicating that up-regulation of the downstream components of the SA pathway impair but do not prevent CaLCuV infection. A recent study also reported a delayed symptom phenotype for cpr1 plants infected with Cauliflower mosaic virus (CaMV), which has a double-stranded DNA genome (Love et al., 2007). Interestingly, CaMV induces Arabidopsis PR1, PR2, and PR5 expression during infection (Love et al., 2005) like CaLCuV. However, CaMV up-regulates PDF1;2 expression while CaLCuV down-regulates its expression, suggesting that are differences as well as parallels in the host responses to these two DNA virus families.
Signaling and the Pathogen Response
EDS1 and PAD4 are key activators of the SA pathway (Wiermer et al., 2005), while the mitogen-activated protein kinase MPK4 is a negative regulator of the EDS1/PAD4 pathway (Brodersen et al., 2006). During CaLCuV infection, EDS1 and PAD4 transcripts were elevated in both the microarray and RT-PCR analyses (Supplemental Fig. S1A). MPK4 RNA levels were also elevated in the microarray experiment, but this was not confirmed by semiquantitative RT-PCR of four independent RNA samples (Supplemental Fig. S1A). Comparison of our microarray data to expression profiles of Arabidopsis lines carrying knockout mutations in these regulators revealed that 87% of the genes displaying EDS1/PAD4-dependent expression (Bartsch et al., 2006) and 67% of MPK4-repressed genes (Andreasson et al., 2005) showed enhanced expression in CaLCuV-infected leaves (Fig. 2F). In contrast, 84% of the genes showing MPK4-dependent expression (Andreasson et al., 2005) were down. For both the eds1/pad4 and mpk4 mutant studies, many of the affected genes are known components of the SA pathway and among the most significant genes differentially expressed during CaLCuV infection (Supplemental Table S4). Together, these results showed that CaLCuV infection activates the SA pathway through induction of EDS1 and PAD4 and that MPK4 activity is suppressed posttranscriptionally during infection.
The mitogen-activated protein kinases MKK4, MKK5, and MPK6 are activated in response to pathogen infection as part of the ET pathway, and MKK3 and MPK6 regulate transcription in the JA pathway (Takahashi et al., 2007). MKK4, MKK5, and MPK6 transcripts were elevated in CaLCuV-infected leaves, while MKK3 RNA levels did not change. Comparison of our microarray data to expression profiles of Arabidopsis lines that inducibly express MKK3 or MKK5 (Takahashi et al., 2007) revealed that 55% of the genes activated by MKK5 overexpression were differentially expressed in CaLCuV-infected leaves, with the majority showing up-regulation (Fig. 2F). There was less overlap between the CaLCuV profile and MKK3-induced genes and the trend was generally opposite. These results are consistent with the activation of the MKK5 cascade and not the MKK3 cascade during CaLCuV infection.
Senescence
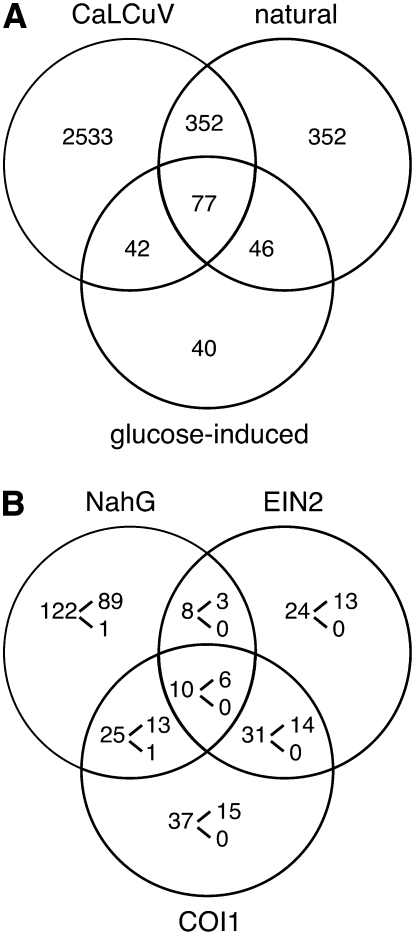
Arabidopsis plants infected with CaLCuV at the 16- to 18-leaf stage display strong chlorosis and signs of desiccation at 25 dpi (Fig. 2C) and die approximately 6 weeks postinoculation. Mock-inoculated plants do not show signs of tissue death (data not shown) and flower during the same time frame. Because of its extent and delay, the cell death cannot be attributed to induction of a hypersensitive response and, instead, may reflect premature senescence or some other form of programmed cell death. To address this possibility, we identified several genes associated with senescence that were differentially expressed during CaLCuV infection (see Supplemental Table S5). We also compared the expression profiles of CaLCuV infection, natural leaf senescence (Buchanan-Wollaston et al., 2005), and Glc-induced senescence (Pourtau et al., 2006). The Venn diagram in Figure 3A shows that the profiles of the three treatments overlap and share 77 common genes. However, 82% (349/424) of the genes shared between CaLCuV and natural senescence were absent in the Glc treatment, while only 36% (42/117) in common between CaLCuV and Glc-induced senescence were not included in the natural senescence profile. There was little overlap between the expression profiles during infection and nitrogen starvation-induced senescence (Pourtau et al., 2006; data not shown).
Figure 3.
CaLCuV infection induces senescence. A, Genes differentially expressed during CaLCuV infection at 12 dpi compared to genes that are up-regulated during natural senescence (Buchanan-Wollaston et al., 2005) and Glc-induced senescence (Pourtau et al., 2006). B, Genes differentially expressed during infection compared to genes (first number in each section) that are expressed in response to SA (NahG), ET (ein2), or JA (coi1) during senescence (Buchanan-Wollaston et al., 2005). The numbers of mRNAs elevated during infection are given at the top of the bracket, while the numbers of depressed transcripts are given below.
Hallmarks of leaf senescence include a decline in photosynthetic capacity, nitrogen recycling, and protein degradation (Lim and Nam, 2005). During CaLCuV infection, there was a general reduction in mRNAs encoding parts of the photosynthetic apparatus, including both photosystems, light-harvesting complexes, and dark reaction enzymes, as well as enzymes involved in chlorophyll synthesis (see Supplemental Table S6). Genes (GDH2, GAD1, GLN1;3, and GLN1;4) specifying enzymes involved in nitrogen recycling via Gln were more highly expressed in infected tissues. Genes encoding components of the ubiquitin-proteasome pathway also showed increased transcript accumulation (Smalle and Vierstra, 2004). A total of 32 genes specifying 11 core and 13 regulatory subunits of the 26S proteasome complex had elevated transcripts. There were also increases in the mRNAs specifying two of two E1 ubiquitin-activating enzymes, eight of 37 E2 ubiquitin-conjugating enzymes, and 149 of 1,570 E3 ubiquitin ligases. The elevated E3 transcripts represented eight of the 10 classes, with the majority in the F-box, RING, and PHD groups (http://plantsubq.genomics.purdue.edu/plantsubq/html/master.html). Only 23 E3 ligase genes showed reduced mRNA levels.
We examined the involvement of the SA, JA, and ET signaling pathways in induction of senescence-associated gene expression during CaLCuV infection. The Venn diagram in Figure 3B compares senescence-associated genes that are not activated in NahG-expressing plants or in ein2 or coi1 mutants (Buchanan-Wollaston et al., 2005). Of the 122 genes that are specifically activated through the SA pathway (down in NahG plants), 72% had elevated transcripts in CaLCuV-infected leaves. The correlation was less for genes specifically activated through the ET (ein2 mutant) and JA (coi1 mutant) pathways, which display 54% and 40% overlap, respectively, with the CaLCuV profile. When the intersecting regions of the Venn diagram were included in the comparisons, the SA pathway included 73% of the total number of senescence-associated genes induced in CaLCuV plants, and the SA and ET pathways together comprised 90%. Transcripts for several senescence markers (PAP1, STP13, GLN1;4, and GPT2) not associated with the SA or ET pathways were also enhanced during CaLCuV infection, indicating that induction of programmed cell death was not simply a consequence of activating the pathogen response. In addition, mRNAs for some SA-responsive genes specifically associated with senescence like WRKY53 were increased during CaLCuV infection. In contrast, the WRKY53 repressor ESR, which is regulated through the JA pathway, was down during infection.
Genotoxic Stress and DNA Repair
The single-stranded and nicked viral DNA forms that accumulate in infected nuclei may be perceived as damaged DNA and induce a genotoxic stress response (Weitzman et al., 2004). To address this possibility, we compared the CaLCuV infection profile with the profiles induced by four treatments known to cause genotoxic stress (Chen et al., 2003; Molinier et al., 2005). UV-C radiation and mitomycin C induce the formation of nucleotide dimers, bleomycin (Blm) causes double-stranded DNA breaks (DSBs), and xylanase (Xyl) is a general fungal elicitor that induces an oxidative burst and the defense response. Comparison of genes represented on the two array platforms used for CaLCuV and genotoxic stress profiling revealed limited overlap (UV-C, 201/732; Blm, 54/584; Xyl, 210/758 genes) as well as opposite effects on expression (UV-C, 51/732; Blm, 79/584; Xyl, 71/758). In contrast, comparison to a common set of genes differentially expressed in response to all three treatments uncovered 105 of 198 similarly expressed genes and only three with opposite expression. A higher correlation was also seen with a combined Blm/mitomycin C treatment, with 68 of 130 genes showing similar expression and seven displaying opposite expression. Thus, CaLCuV infection alters the expression of a core set of genes involved in response to genotoxic stress. However, the CaLCuV profile only shows weak overlap (13 up and eight down) with a set of 67 transcripts that are increased by nine different types of stress (Swindell, 2006), indicating that infection does not activate a general stress response.
Like all eukaryotes, plants have evolved multiple DNA repair pathways (Kimura and Sakaguchi, 2006). Base excision repair (BER), nucleotide excision repair (NER), and photoreactivation repair DNA cross-links, while nonhomologous end-joining and homologous recombination repair DSBs. To gain insight into the impact of geminivirus infection on the various repair pathways and DNA damage tolerance, we analyzed the expression profiles of Arabidopsis genes involved in these processes (Table I). CaLCuV infection and genotoxic stress altered the expression of components of BER, NER, and DSB pathways but did not alter expression of genes encoding the replication-associated mismatch repair machinery or the photolyases involved in photoreactivation.
Table I.
Differentially expressed DNA repair genes
Bold type designates similar regulation during CaLCuV infection and genotoxic stress. No results are shown for genes not on the array. *, Plastid protein. nc, No change.
| Gene Identifier | Gene Name | CaLCuV | Genotoxic Stressa |
|---|---|---|---|
| Damage tolerance | |||
| At3g04880 | DRT102 | up | nc |
| At1g23260 | MMS1 | up | |
| At1g70660 | MMS2 | up | |
| At2g39100 | RAD18 | nc | up/down |
| At5g44740 | RAD30 | nc | down |
| At4g35740 | RECQ3 | nc | up |
| At1g10930 | RECQ4A | nc | down |
| At1g67500 | REV3 | nc | up |
| At1g52410 | TSA1 | down | |
| At5g13930 | CHS (TT4) | down | down |
| At3g55120 | CFI (TT5) | down | up/down |
| At3g51240 | F3H (TT6) | down | |
| At3g12610 | *DRT100 | down | |
| At1g30480 | *DRT111 | nc | up |
| At1g20340 | *DRT112 | down | nc |
| BER | |||
| At4t02390 | PARP1 | up | up/down |
| At2g31320 | PARP2 | up | up |
| At1g80420 | XRCC1 | up | |
| At2g41460 | ARP | nc | up |
| At3g10010 | DML2 (DNA glycosylase) | up | |
| At1g75090 | DNA glycosylase | down | |
| At3g12710 | DNA glycosylase | down | |
| At4g12740 | DNA glycosylase | down | |
| At5g44680 | DNA glycosylase | down | |
| At3g50880 | *Base excision | up | |
| At1g21710 | *OGG1 | up | nc |
| NER | |||
| At3g50360 | CEN2 | up | |
| At4g37010 | CEN2-like | up | up |
| At1g03750 | CSB-like | nc | up |
| At2g18760 | ERCC6/CSB | up | up |
| At1g79650 | RAD23 | up | |
| At3g02540 | RAD23-3 | down | |
| At5g38470 | RAD23-4 | up | |
| At1g55750 | TFB1-1 | nc | up |
| At1g55680 | TFB1-2 | up | nc |
| At1g12400 | TFB5 | up | |
| DSB repair | |||
| At4g21070 | BRCA1 | nc | up |
| At3g22880 | DMC1 | up | nc |
| At1g64750 | DSS1(1) | up | nc |
| At5g45010 | DSS1(V) | up | |
| At3g57300 | INO80 | up | |
| At1g48050 | KU80 | up | |
| At5g61460 | MIM | nc | up/down |
| At5g66130 | RAD17 | up | |
| At2g31970 | RAD50 | up | nc |
| At2g28460 | RAD51B | nc | up |
| At2g45280 | RAD51C | nc | up |
| At1g10930 | REC4QA | down | nc |
| At3g26680 | SMN1 | nc | up |
Expression of genes involved in DSB repair during CaLCuV infection is of particular interest because of the role of recombination in the geminivirus DNA replication process (Jeske et al., 2001). Increased RAD50 and KU80 mRNA levels in infected leaves (Table I) suggested that both the homologous recombination and nonhomologous end-joining pathways are activated during infection (Schuermann et al., 2005). Increased expression of DMC1, DSS1(1), and DSS1(V), which encode proteins that interact with the RAD51 recombinase, further supported activation of recombination during infection. None of the transcripts for the four Arabidopsis RAD51 genes was elevated during infection, even though RAD51B and RAD51C are activated by genotoxic stress. However, activation of DNA repair pathways may only occur in cells that replicate the viral DNA and, as such, may be difficult to detect.
DNA repair and replication depend on adequate dNTP pools and share many enzymes and factors (Kimura and Sakaguchi, 2006). During CaLCuV infection, nine Arabidopsis genes encoding dNTP biosynthetic enzymes or DNA replication machinery (Shultz et al., 2007) were differentially expressed, while 10 were differentially expressed during genotoxic stress (Table II). The datasets share four genes: one encoding the POLD3 subunit, two encoding RPA1 subunits, and one encoding the TSO small subunit of ribonucleotide reductase. RPA, a trimeric single-stranded DNA binding complex, has been implicated in all four eukaryotic repair pathways as well as in DNA damage checkpoint control (Zou et al., 2006). POLD, the four-subunit replicative DNA polymerase, is involved in NER and mismatch repair (Hubscher et al., 2002). POLD expression during infection was also supported by up-regulation of the POLD4 gene specifying the smallest subunit of the polymerase complex. Based on earlier studies showing that CaLCuV infection induces transcription of the PCNA gene in N. benthamiana (Egelkrout et al., 2002), we anticipated that expression of the POLD4 clamp would also be elevated during infection. However, the gene profiling results indicated that transcripts for the PCNA (At1g07370) gene were reduced, while RNA corresponding to the PCNA-like gene (At2g29570) did not change in infected leaves. Semiquantitative RT-PCR confirmed these results (Supplemental Fig. S1B). Down-regulation of the PCNA gene is consistent with its expression only in embryonic tissues (https://www.genevestigator.ethz.ch/). The lack of detectable up-regulation of the PCNA-like gene may reflect the difficulty in detecting cell autonomous events during infection, especially if such genes are expressed in a limited time period during the process.
Table II.
Differentially expressed DNA replication and core cell cycle genes
Bold type indicates similar regulation during CaLCuV infection and genotoxic stress. No results are shown for genes not on the array. nc, No change.
| Gene Identifier | Gene Name | CaLCuV | Genotoxic Stressa |
|---|---|---|---|
| DNA replication | |||
| At1g44900 | MCM2 | nc | up/down |
| At5g46280 | MCM3 | nc | up |
| At5g44635 | MCM5 | down | |
| At4g02060 | MCM7 | down | nc |
| At2g37560 | ORC2 | nc | up |
| At4g29910 | ORC5 | nc | up |
| At1g07370 | PCNA | down | |
| At1g78650 | POLD3 | up | up |
| At1g09815 | POLD4 | up | |
| At2g27120 | POLE1 | nc | up |
| At1g19080 | PSF3 | up | |
| At2g06510 | RPA1 | up | up |
| At4g19130 | RPA1 | up | up |
| At5g61000 | RPA1 | down | |
| At3g07800 | TK | up | |
| At3g27060 | TSO2 | up | up |
| Core cell cycle | |||
| At1g76540 | CDKB2;1 | down | |
| At1g66750 | CDKD;2 | up | |
| At5g63370 | CDKG:1 | up | |
| At5g44290 | CKL5 | up | nc |
| At1g03740 | CKL6 | up | up |
| At1g70210 | CYCD1;1 | down | down |
| At4g34160 | CYCD3;1 | nc | down |
| At5g67260 | CYCD3;2 | down | |
| At5g65420 | CYCD4;1 | nc | up |
| At5g10440 | CYCD4;2 | up | |
| At1g77390 | CYCA1;2 | nc | up |
| At5g25380 | CYCA2;1 | nc | down |
| At4g37490 | CYCB1;1 | up | up |
| At5g06150 | CYCB1;2 | down | |
| At2g26760 | CYCB1;4 | nc | down |
| At2g17620 | CYCB2;1 | nc | down |
| At4g35620 | CYCB2;2 | nc | up |
| At1g16330 | CYCB3;1 | nc | down |
| At1g27630 | CYCT;1 | nc | down |
| At4g34090 | CYL1 | down | up/down |
| At3g12280 | RBR | nc | |
| At2g36010 | E2Fa | nc | up |
| At1g47870 | E2Fc | up | |
| At3g48160 | E2Fe | down | |
| At5g03455 | CDC25 | up | |
| At1g02970 | WEE1 | nc | up |
| At2g26760 | KRP1 | nc | down |
Compiled from Chen et al. (2003) and Molinier et al. (2005).
Because many of the plant genes encoding dNTP biosynthetic enzymes and replication proteins are under cell cycle control, we compared the expression patterns of Arabidopsis core cell cycle genes (Vandepoele et al., 2002; Menges et al., 2005) during infection and genotoxic stress (Table II). This comparison uncovered similar expression patterns for four core cell cycle genes among the 12 and 18 genes that showed differential expression during infection and DNA damage, respectively. CKL6 and CYCB1;1 mRNAs were up, while CYCD1;1 and CYL1 transcripts were down in both cases. Up-regulation of CYCB1;1 expression during infection is associated with a q value of 7.5 × 10−8, making it the most significant core cell cycle gene in our dataset. CYCB1;1 RNA levels are highly induced by a variety of genotoxic stresses, suggesting that activation of CYCB1;1 expression is a general feature of genotoxic stress (De Veylder et al., 2007). The CaLCuV profile did not show a change in expression for WEE1 or its upstream activators ATR and ATM, which have been implicated in activation of Arabidopsis DNA checkpoints in response to DNA damage (De Schutter et al., 2007).
A limitation of the above comparisons is that the genotoxic stress studies used array formats with fewer features and, in some instances, different genes than those represented on the ATH1 array. As a consequence, it was only possible to compare expression changes for a subset of the DNA repair, DNA replication, and core cell cycle genes shared by the array formats. Even with this constraint, the results indicated that while there was overlap between the expression profiles, not all of the changes in the repair and cell cycle pathways during infection can be attributed to genotoxic stress.
Cell Cycle Regulation
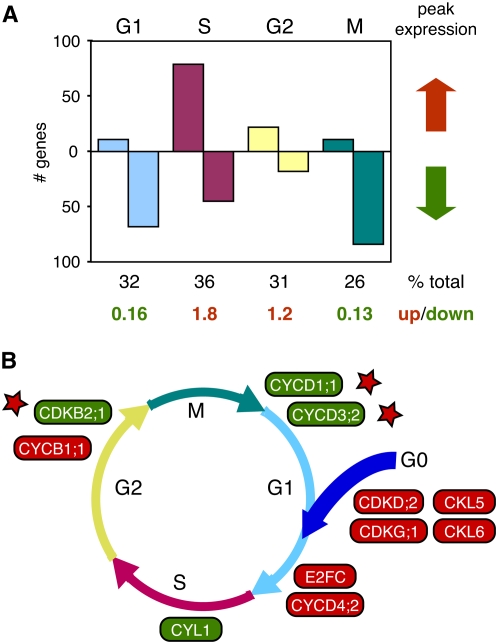
To gain further insight into modulation of host gene expression leading to viral DNA replication, we compared the CaLCuV profile to those generated from cultured Arabidopsis cells synchronized with aphidicolin (Menges et al., 2003). Of the 1,081 genes that showed cell cycle regulation, 120 were up and 214 were down during CaLCuV infection. Classification of the oscillating cell cycle-associated genes by peak expression phase revealed that a higher proportion of S phase (35%) than M phase (26%) genes were differentially expressed during infection, while the percentages of G1 (32%) and G2 (31%) phase genes were intermediate (Fig. 4A). Genes with peak expression in G1 and M were primarily down, while S and G2 phase genes tended to be up. The highest fraction of elevated transcripts was associated with S phase, while the greatest proportion of reduced RNAs was associated with M phase. These distributions are not compatible with a general activation of the cell division cycle by geminivirus infection and, instead, indicated that infection specifically activates genes needed to establish a replication-competent environment and prevents expression of genes necessary for mitosis. This idea is supported by an earlier study showing that both viral and plant chromosomal DNA replicate in infected cells in the absence of proliferation (Nagar et al., 2002).
Figure 4.
CaLCuV infection alters expression of cell cycle genes. Cell cycle-associated genes that are up-regulated (red arrow) or down-regulated (green arrow) in CaLCuV-infected leaves are grouped according to their peak expression phase (top) during the cell division cycle (Menges et al., 2003) can be seen in A. The total number of genes associated with each phase that are differentially expressed during infection and the ratio of increased versus reduced transcripts are shown at the bottom. B, A model showing the core cell cycle transcripts that are higher (red) or lower (green) during CaLCuV infection is proposed. Each gene is positioned where it is thought to act or shows peak expression during the cell division cycle or during reentry (Menges et al., 2005). The stars mark three genes with reduced transcripts in infected leaves and function at the indicated cell cycle stage. The Arabidopsis gene numbers for the genes in A and C are in Supplemental Table S1.
We identified transcripts for 12 core cell cycle genes that differentially accumulate during infection in the microarray experiments and confirmed their expression patterns by semiquantitative RT-PCR (Supplemental Fig. S1B). The same expression pattern was also detected at 9 dpi, which is prior to CaLCuV symptom appearance, by quantitative RT-PCR (Supplemental Table S8). Ten of the differentially expressed core cell cycle genes were grouped in Figure 4B according to when they are thought to act or are maximally expressed during the cell cycle or reentry (Menges et al., 2005; De Veylder et al., 2007). For this analysis, we did not consider CDC25, which has an uncertain role in the plant cell division cycle, or CDKD;2, which functions primarily during transcription. It is also not known if CYL1 is involved in cell cycle control, although it is most highly expressed during S phase (Menges et al., 2005). All three mRNAs were increased during infection.
Three core cell cycle genes that are enhanced upon cell cycle reentry (CDKG;1, CKL5, and CKL6) had elevated transcripts in infected tissue (Fig. 4B), suggesting that CaLCuV induces quiescent cells to reenter the cell division cycle. Because there is a general down-regulation of G1-associated genes during infection (Fig. 4A), it is likely that infected cells only transit through late G1. This idea is supported by down-regulation of CYCD1;1 and CYCD3;2, which encode early activators of G1 (Masubelele et al., 2005; Dewitte et al., 2007). In contrast, transcripts corresponding to the late G1 cyclin CYCD4;2 (Menges et al., 2005) were increased by infection. E2FC mRNA was also elevated in infected leaves. We did not detect changes in the expression of other components of the RBR/E2F network, which regulates transcription at the G1/S boundary (Gutierrez et al., 2002).
CaLCuV infection has opposite effects on the expression of CYCB1;1 and CDKB2;1, both of which promote mitosis and growth (De Veylder et al., 2007). Expression of CYCB1;1, which is induced upon cell cycle reentry and remains high throughout the cell cycle, is increased by infection. It has been proposed that elevated CYCB1;1 expression leads to sequestering factors necessary for M phase and G2 arrest (De Veylder et al., 2007). Consistent with this idea, CDKB2;1 expression, which peaks at the G2/M boundary, is reduced by infection (Fig. 4B). The CYCB1;1 and CDKB2;1 expression profiles resulting in G2 block correlate with the absence of dividing cells and enations and shut down of the meristem during infection (Fig. 1; J.T. Ascencio-Ibáñez and L. Hanley-Bowdoin, unpublished data).
CaLCuV Infection and Endoreduplication
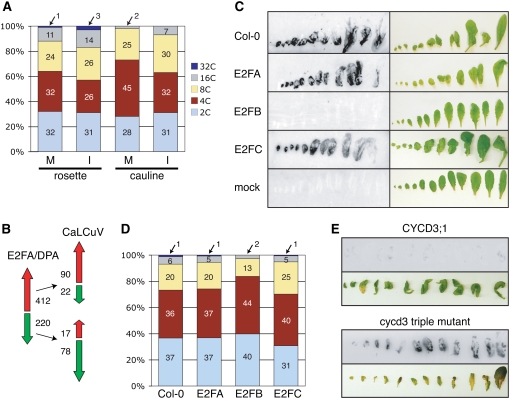
The asymmetric expression patterns observed for cell cycle-associated and core cell cycle genes suggested that CaLCuV infection specifically activates S phase and inhibits M phase. This could be accomplished by blocking transit into M phase or by bypassing M phase as part of an endocycle. To distinguish between these possibilities, we asked if infected leaves are enriched for 4C cells indicative of a G2 block or contain a larger fraction of ≥8C cells indicative of the endocycle. Efforts to isolate intact nuclei from plants inoculated at the 16- to18-leaf stage were not successful, most likely because of the strong cell death phenotype. To minimize this effect, we inoculated older plants, which are less severely impacted by CaLCuV. These plants produced an inflorescence stem, allowing us to compare rosette and cauline leaves. FACS analysis of nuclei isolated from both leaf types of infected and mock-inoculated plants at 28 dpi showed a small increase in ploidy levels in infected versus control nuclei, with a 7% increase in 8C, 16C, and 32C nuclei in rosette leaves and a 10% increase in cauline leaves (Fig. 5A). In both cases, the increase in 8C, 16C, and 32C nuclei was accompanied by a reduction in 4C nuclei. In two independent replicates, rosette and cauline leaves showed 9% and 12% increases in 8C, 16C, and 32C nuclei, respectively, and a concomitant reduction in 4C nuclei (data not shown). The numbers of 4C (P value = 0.024) and ≥8C (P value = 0.0041) nuclei in mock and infected leaves were significantly different in a two-tailed Student's t test, but there was no significant difference for the 2C nuclei (P value = 0.68; Supplemental Table S8). The 7% to 12% increase in 8C, 16C, and 32C nuclei in infected leaves is reminiscent of the small proportion of virus-positive cells in CaLCuV infected leaves (Fig. 1C), suggesting that the increase in ploidy is cell autonomous with viral presence.
Figure 5.
Ploidy and infectivity studies of wild-type Arabidopsis and plants altered in the CYCD/RBR/E2F pathway. A, Ploidy distribution of nuclei from rosette and cauline leaves of mock-inoculated and infected Arabidopsis plants. The percentage of 2C, 4C, 8C, 16C, or 32C nuclei in each FACS profile is indicated. The experiment was repeated twice with similar results (data not shown). B, A comparison of CaLCuV expression profile to the RNA profile of transgenic Arabidopsis overexpressing E2FA and its DPA partner is presented (Vandepoele et al., 2005). The red and green arrows indicate increased and reduced transcripts, respectively. The number of genes represented by each arrow is indicated. C, Viral DNA in leaves from mock and CaLCuV-inoculated Col-0, E2FA-, E2FB-, or E2FC-overexpressing plants was detected by tissue printing at 12 d after bombardment. The leaves were photographed prior to rubbing onto nylon membrane. D, Ploidy distribution of nuclei from untreated, mature rosette leaves of Col-0, E2FA-, E2FB-, or E2FC-overexpressing plants is presented. The percentage of 2C, 4C, 8C, 16C, or 32C nuclei from FACS profile is indicated. The experiment was repeated twice with similar results (data not shown). E, Viral DNA in leaves from mock and CaLCuV-inoculated plants carrying a triple knockout in CYCD3;1, 2, and 3 or overexpressing CYCD3;1 was detected by tissue printing at 22 d after bombardment.
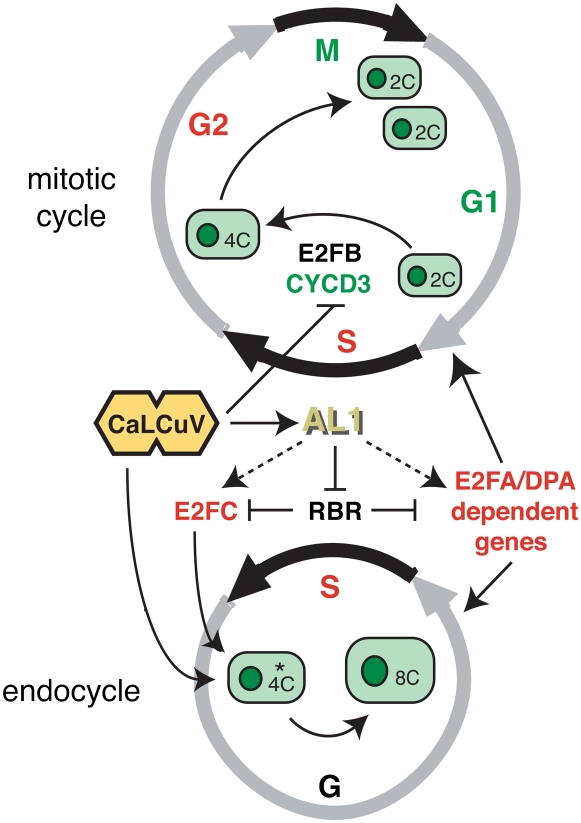
Several plant genes, including CDKB1;1, CDKA;1, CYCD2;1, CYCD3;1, CYCD3;2, CYCD3;3, CYCA2;3, E2FC, E2FE, KRP2, SIM, ILP1, FAS1, and CCS52A, influence the balance of mitotic and endoreduplicating cells or the number of endocycles (Boudolf et al., 2004; Verkest et al., 2005; Churchman et al., 2006; Qi and John, 2007; Ramírez-Parra and Gutiérrez, 2007). Of those represented on the ATH1 array, E2FC and ILP1 were elevated in infected leaves and CYCD3;2 was reduced. ILP1 negatively regulates CYCA2 transcription (Yoshizumi et al., 2006), but we did not detect expression changes for any of the four CYCA2 genes encoded by the Arabidopsis genome. Interestingly, E2FC and CYCD3;2 are part of the RBR/E2F regulatory network, which is modulated by geminivirus infection (Hanley-Bowdoin et al., 2004).
CaLCuV Infection Is Differentially Impacted by E2F Transcription Factors
To gain insight into the impact of geminivirus infection on the RBR/E2F network, we compared genes up- or down-regulated by ectopic expression of E2FA and its DPA partner in Arabidopsis (Vlieghe et al., 2003). This comparison uncovered 107 genes (28%) that showed similar trends and 39 (6%) that showed opposite trends in infected leaves (Fig. 5B). We then examined the expression of Arabidopsis genes with predicted E2F sites in their promoters (Vandepoele et al., 2005). This analysis revealed that 24% of the 5,861 putative E2F target genes were differentially expressed in CaLCuV-infected leaves (data not shown). Together, these results suggested that geminivirus infection impacts the expression of a subset of E2F-regulated genes, including those controlled by E2FA/DPA.
We also asked if overexpression of E2FA, E2FB, or E2FC, the three Arabidopsis E2F family members that bind to RBR (Mariconti et al., 2002), impacts CaLCuV infection. Plants ectopically expressing E2FA or E2FC developed severe symptoms at the same time as wild-type plants and did not flower (data not shown). In contrast, E2FB-overexpressing plants showed very mild symptoms in floral tissues after a significant delay, while none of the E2FA and E2FC plants underwent the vegetative to floral transition. Viral DNA accumulation was readily detected by tissue printing of rosette leaves of E2FA and E2FC plants but not in the leaves of E2FB plants (Fig. 5C). The E2FB resistance phenotype was observed when plants were inoculated by bombardment or agroinfection, ruling out any effect due to the inoculation procedure.
We examined the ploidy distributions of uninfected rosette leaves from the E2FA-, E2FB-, and E2FC-overexpressing lines. FACS analysis was performed on nuclei from mature leaves of plants with 16 to 18 true leaves. The plants were at the same developmental stage and grown under the same conditions as those used for the microarray experiments. The FACS profiles detected similar numbers of 8C, 16C, and 32C nuclei in wild type (27%), E2FA (26%), and E2FC (24%) leaves (Fig. 5D). In contrast, only 12% of the nuclei isolated from E2FB leaves had ploidy values ≥8C. Based on these results, one possibility is that E2FB plants are resistant to CaLCuV infection because of a reduced capacity to undergo endocycling and support viral replication.
The cyclin D family plays a key role in regulating the E2F/RBR pathway by activating cyclin-dependent kinase (CDK)-mediated phosphorylation of RBR and disrupting its interactions with E2F (De Veylder et al., 2007). Hence, we asked if overexpression of CYCD3;1 (Dewitte et al., 2003) or loss of CYCD3 function (DeWitte et al., 2007) impacts CaLCuV infection. Plants carrying mutations in CYCD3;1, CYCD3;2, and CYCD3;3 genes developed severe symptoms by 12 dpi, while plants overexpressing CYCD3;1 showed no evidence of disease even at later times (data not shown). High levels of viral DNA were detected in rosette leaves of cycd3 mutants at 22 dpi by tissue printing (Fig. 5E). In contrast, only trace amounts of viral DNA were observed in leaves from CYCD3;1-overexpressing plants. Quantitative RT-PCR showed that the ratios of PR1 transcripts in E2FB- and CYCD3;1-overexpressing plants to wild-type plants were 0.93 and 0.76, respectively, ruling out that the resistance phenotypes reflected induction of pathogen response genes (Supplemental Table S8). Together, these results are consistent with CaLCuV replicating in endocycling cells and the proposed role of the CYCD3 subfamily in promoting the mitotic cycle and inhibiting the endocycle (Dewitte et al., 2007).
DISCUSSION
Host-pathogen interactions involve a complex set of events that depend on the nature of the interacting partners, developmental stage, and environmental signals. In plants, pathosystems share a common set of responses that are intertwined with unique responses characteristic of the specific host/pathogen combination. Analysis of the Arabidopsis transcriptome in response to CaLCuV infection showed that like RNA viruses (Whitham et al., 2006), plant DNA viruses activate the SA pathway leading to the expression of PR genes and the induction of programmed cell death. The general pathogen response is superimposed on expression changes in core cell cycle and DNA replication/repair machinery due to the unique dependence of geminiviruses on host enzymes to amplify their genomes. The extent and complexity of the response also reflect the asynchronous nature of the CaLCuV infection in intact plants and contributions by both systemic and cell-autonomous events.
A recurring theme in the CaLCuV expression data is cross talk between the various pathways. Increased expression of the EDS1/PAD4/SAG101 complex and downstream genes encoding biosynthetic enzymes, transcriptional regulators, and PR proteins (Bartsch et al., 2006) and suppression of the MPK4 signaling pathway (Andreasson et al., 2005) provided strong evidence for the activation of a pathogen response through the SA pathway (Wiermer et al., 2005). This conclusion was further supported by the observation that CaLCuV symptoms are strongly delayed and attenuated in cpr1 plants that constitutively express PR1 (Bowling et al., 1994). The overlap between the CaLCuV expression profile and senescence-associated genes regulated by SA also implicated SA signaling in induction of programmed cell death (Buchanan-Wollaston et al., 2005). However, the CaLCuV expression profile suggested that regulation of the pathogen response and senescence during infection is more complex. Although the expression of genes encoding JA biosynthetic enzymes was reduced, consistent with the ability of the SA pathway to suppress JA signaling (Beckers and Spoel, 2006), selected JA marker transcripts were elevated. This observation is indicative of cross talk between the JA and ET pathways, which share the ERF transcription factors, (Lorenzo and Solano, 2005), and is compatible with activation of MKK5 but not the MKK3 cascade (Takahashi et al., 2007) during infection. As observed for the SA pathway, there was significant overlap between the CaLCuV expression profile and senescence-associated genes regulated by EIN2 (Buchanan-Wollaston et al., 2005), implicating ET signaling in programmed cell death as well as the pathogen response.
There was also cross talk between the genotoxic response, the cell cycle changes, and the programmed cell death phenotype associated with CaLCuV infection. CYCB1;1 is expressed in proliferating cell populations of developing leaves (Beemster et al., 2005), in response to DNA damage (Chen et al., 2003), and in CaLCuV-infected leaves. RAD17, which increased transcript levels during infection, has been implicated in checkpoint control and shown to be necessary for DSB repair in Arabidopsis (Heitzeberg et al., 2004). Recent animal studies linked activation of the DNA damage checkpoint, aberrant DNA replication, and induction of senescence (Bartkova et al., 2006; Di Micco et al., 2006). A similar relationship in plants is supported by the observation that silencing of the core cell cycle gene encoding the RBR protein causes necrosis in mature leaves (Jordan et al., 2007). Thus, inhibition of RBR by CaLCuV AL1 binding might contribute to the activation of programmed cell death as well as to the induction of host genes encoding DNA replication factors (Egelkrout et al., 2002; Arguello-Astorga et al., 2004).
Activation of both core cell cycle and DNA repair genes during CaLCuV infection indicated that geminiviruses might rely on both host DNA synthesis pathways for their replication, consistent with their use of RCR and RDR mechanisms (Jeske et al., 2001). However, because of the low frequency of homologous recombination, which is required for RDR, versus nonhomologous end joining in plants (Kimura and Sakaguchi, 2006), the relative contributions of RCR and RDR to viral DNA accumulation may differ significantly. A very early event in infection is likely to be AL1/RBR binding, leading to reprogramming of cell cycle controls, the accumulation of host replication machinery, and the onset of RCR. The accumulation of viral DNA replication products and intermediates could then trigger a genotoxic response and the synthesis of host repair proteins and potentially a switch to RDR. A model in which RCR precedes RDR is congruent with the absolute dependence of geminivirus infection on a functional origin that is recognized by its cognate AL1 protein (Fontes et al., 1994), analogous to bacteriophage T4, which replicates by sequential RCR to RDR (Cox, 2001).
A longstanding question is the extent to which geminivirus infection impacts host DNA replication (Accotto et al., 1993; Nagar et al., 2002). Up-regulation of S phase-associated genes during CaLCuV infection is consistent with the establishment of a replication-competent environment, while increased ploidy in infected leaves demonstrated that infection induces cells to endocycle and fully replicate their genomes. Increased ploidy levels have also been reported in Arabidopsis cauline leaves infected with the nanovirus Fava bean necrotic yellow virus (Lageix et al., 2007). Like geminiviruses, nanoviruses rely on host DNA replication enzymes to amplify their single-stranded DNA genomes and encode a protein (CLINK) that binds to RBR and induces the expression of host replication machinery. These parallels suggest that geminiviruses and nanoviruses interact with their hosts through conserved mechanisms that ultimately lead to the establishment of an endocycle. However, hyperplasia and enations have been observed for a few geminivirus/host combinations, indicating that some plant DNA viruses can induce mitosis (Esau and Hoefert, 1978; Briddon, 2003). The capacity to induce cell division is associated with the viral C4 protein (Latham et al., 1997), but C4 is not essential for infection (Stanley and Latham, 1992). Hence, geminiviruses that induce cell division do not depend on the mitotic cell cycle to replicate their genomes, underscoring the universal involvement of the endocycle in plant DNA virus replication. This idea is supported further by the ability of maize streak virus RepA, the RBR-binding protein of monocot-infecting geminiviruses, to induce endoreduplication (Desvoyes et al., 2006).
Mammalian DNA tumor viruses also induce chromosomal DNA replication, resulting in 2C, 4C, and >4C cell populations (Friedrich et al., 1992; Belyavskyi et al., 1996). Unlike plant DNA viruses, rereplication in animal cells is not associated with discrete C values characteristic of complete endocycles. This difference may reflect the fundamentally different ways the cell cycle and DNA replication are integrated with developmental processes in the two kingdoms. In mammals, growth generally corresponds to increased cell number, and the endocycle is confined to a few specialized cell types. In contrast, in plants, the endocycle is commonplace and linked to growth. It often represents the final stage of DNA replication during plant development and, as such, may be more readily induced by viral infection. The mechanisms leading to rereplication in mammalian cells are not known, but there is evidence that checkpoint controls are disrupted and that interactions between viral oncoproteins and retinoblastoma family members are dispensable (Wu et al., 2004; Cherubini et al., 2006). In contrast, the RBR/E2F pathway has been implicated in endoreduplication in plants and insects, which also undergo extensive endocycling during development (De Veylder et al., 2007; Swanhart et al., 2007). However, a recent report showed that mammalian primary cells depleted for Rb can undergo full rounds of genome amplication, suggesting that Rb suppresses the endocycle in nontransformed cells (Srinivasan et al., 2007). In contrast, another recent report indicated that depletion of Arabidopsis RBR is not sufficient to induce endoreduplication in cultured plant cells (Hirano et al., 2008), suggesting that the mechanisms that govern induction of the endocycle may differ in plant and animal systems.
Like mammalian DNA viruses (Felsani et al., 2006), geminiviruses bypass G1 cell cycle controls by binding to RBR and altering the expression E2F target genes. Several lines of evidence indicate that geminiviruses selectively impact E2F activity related to endocycling. First, E2FC expression was elevated during CaLCuV infection. Arabidopsis plants that overexpress E2FC and its DPB partner have increased DNA content, and E2FC RNAi lines undergo hyperplasia (del Pozo et al., 2006). Both of these phenotypes are indicative of E2FC promoting the endocycle. Second, although we did not observe increased E2FA expression, many genes differentially expressed in plants ectopically expressing E2FA and DPA showed similar trends in CaLCuV-infected leaves (Vlieghe et al., 2003; Vandepoele et al., 2005), suggesting that geminivirus infection alters expression of a subset of E2FA/DPA-regulated genes. Third, like CaLCuV infection, inactivation of RBR by binding to maize streak virus RepA resulted in up-regulation of E2FA and E2FC expression and an increased number of endocycling cells in older developing leaves (Desvoyes et al., 2006). Strikingly, E2FA overexpression and RBR inactivation enhances cell division early in leaf development and the endocycle later in development (De Veylder et al., 2002; Desvoyes et al., 2006), indicating that geminivirus infection distinguishes between the mitotic and endocycling activities of RBR and E2FA and selectively activates endoreduplication.
During CaLCuV infection, the increase in ploidy was accompanied by a reduction in 4C but not 2C cells, indicating that the virus preferentially targets 4C cells. A recent study proposed that the CYCD3 family triggers a commitment to the mitotic cell cycle (Dewitte et al., 2007). This model predicts the existence of two 4C populations, one programmed to complete mitosis and the other destined to enter the endocycle (Fig. 6). Several observations suggested that geminiviruses specifically target the second population. First, CYCD3;2 RNA levels were reduced in infected leaves. Down-regulation of CYCD3 expression would favor accumulation of the endocycle 4C population and facilitate infection. A triple CYCD3 mutant line did not show enhanced susceptibility to CaLCuV, most likely because of the strong infection phenotype of the virus. However, plants ectopically expressing CYCD3;1 were highly resistant to infection, indicating that although these plants contain a larger proportion of 4C cells (Dewitte et al., 2003), they are not suitable targets for viral infection, because they are preprogrammed to undergo mitosis and not to enter the endocycle. Similarly, the resistance phenotype of E2FB-overexpressing plants but not E2FA or E2FC lines indicated that reducing the number of endocycle-competent cells strongly impairs infection. Up-regulation of CYCD4;2 in infected leaves was consistent with its expression outside of meristematic regions (Kono et al., 2007) and demonstrated that viral infection does not result in a general reduction of CYCD expression.
Figure 6.
Model for CaLCuV induction of the endocycle. During infection, CaLCuV selectively infects the 4C cell population (marked by the asterisk) predisposed to undergo endoreduplication but not 2C or 4C cells in the mitotic cycle. The viral AL1 protein, which is produced very early in the infection process, binds to RBR and relieves repression of E2FC and E2FA, leading to activation of S phase genes and an increase in endocycling cells. Up-regulation of E2FC expression also promotes the endocycle during infection, while down-regulation of CYCD3 expression prevents the mitotic cycle possibly through suppression of E2FB activity. Genes with elevated transcripts in CaLCuV-infected leaves are in red, while genes with reduced mRNAs are in green.
At first glance, geminivirus targeting of 4C cells and their dependence on the RBR/E2F pathway may seem incompatible. However, the endocycle consists of only two phases, S and a gap phase that prepares the cell to replicate its genome. In the absence of CYCD3-CDK activity, RBR and E2F may play a role in inducing 4C cells to transition to the endocycle early in infection. Alternatively, if the 4C cells are already in the endocycle gap phase, the RBR/E2F pathway may facilitate transition into S phase of the first endocycle. In both cases, AL1 binding to RBR would modulate E2FA and E2FC activities, leading to changes in host gene expression and induction of the endocycle (Fig. 6). These mechanisms are not mutually exclusive and may involve atypical E2F family members that do not interact with RBR (Vlieghe et al., 2005) as well as the canonical E2Fs. Future studies that examine the transcript and protein accumulation patterns of core cell cycle and DNA replication genes in infected cells, as well as the efficiency and tissue specificity of infection of various core cell cycle mutant and overexpressing lines, will provide insight into requirements for the establishment of a DNA replication-competent environment during geminivirus infection.
MATERIALS AND METHODS
Plant Growth and Inoculation Conditions
Arabidopsis (Arabidopsis thaliana) Col-0 plants were grown and inoculated with CaLCuV as described previously (Shen and Hanley-Bowdoin, 2006). Control plants were mock inoculated using the same Agrobacterium strain transformed with pNSB690, which contains a 35S promoter uidA expression cassette. Typically, 12 plants were inoculated with CaLCuV, while six were mock inoculated per tray. The tray was covered with a clear plastic lid for 4 d and then uncovered for the remainder of the growth period. Symptoms were monitored from the first dpi until the CaLCuV-infected plants were dead at 6 weeks postinfection.
Transgenic Arabidopsis plants carrying expression cassettes for E2FA, E2FB, E2FC, or CYCD3;1, with mutations in CYCD3;1, CYCD3;2, and CYCD3;3 or carrying the cpr1 mutation (Bowling et al., 1994; del Pozo et al., 2006; Sozzani et al., 2006; Dewitte et al., 2003, 2007), were grown under the same conditions. Overexpression of the transgenes was verified for each homozygous line by RT-PCR (data not shown). The plants were agroinoculated as described above or inoculated by bombardment using 1-μm gold microprojectiles coated with plasmid DNA (mock) or a mixture of CaLCuV A (pCPCBLCVA.003) and B (pCPCBLCVB.002) replicon plasmids (Turnage et al., 2002).
Viral DNA Hybridization
Visible rosette leaves were collected at 12 dpi. Leaves 1 to 4 (with leaf 1 being the youngest) were pooled, while leaves 5 to 20 were harvested individually. DNA (0.5 μg) isolated from each leaf sample was digested with SacI, resolved by agarose gel electrophoresis, and visualized by DNA gel blotting using a 32P-labeled, unit-length CaLCuV A probe (Ascencio-Ibañez and Settlage, 2007). Viral DNA accumulation was monitored independently in three plants. Tissue printing of viral DNA was performed by rubbing leaves onto nylon membranes followed by UV crosslinking and hybridization with a 32P-labeled, unit-length CaLCuV A probe.
AL1 Immunolocalization
The coding region for CaLCuV AL1 was released from pNSB958 (Arguello-Astorga et al., 2004) as a 1.2-kb NdeI/XhoI fragment and subcloned into the Escherichia coli expression vector pET-16b (Novagen). The resulting pNSB1044 clone encodes an AL1 protein fused to a 31-amino acid N-terminal extension with 11 His residues and designated as His-AL1. The expression cassette was transformed into the E. coli BL21 strain and His-AL1 was produced by isopropylthio-β-galactoside induction and purified by nickel-nitrilotriacetic acid agarose agarose affinity chromatography according to the manufacturer's instructions (Qiagen) with the following modifications to the recommended growth conditions. A 5-mL aliquot from a fresh overnight culture was diluted into 100 mL LB supplemented with 100 μg/mL ampicillin in a 250-mL baffle flask and grown at 28°C to an OD of 0.8. Recombinant protein expression was induced by the addition of 1 mm isopropylthio-β-galactoside, followed by incubation at 37°C for 2.5 h. Antibodies were produced by Cocalico by immunizing a rabbit with 0.25 mg purified His-AL1 and boosting with 0.25 mg three times at 14, 21, and 49 d. The antiserum was verified by immunoblotting total protein extracts from CaLCuV-infected and mock inoculated leaves (Shen and Hanley-Bowdoin, 2006).
Leaf 9 from CaLCuV-infected plants at 12 dpi and equivalent leaves from mock-inoculated plants were fixed and processed as previously reported (Shen and Hanley-Bowdoin, 2006) with the following modifications. After a 10-min exposure to AEC reagents (Vector Laboratories), sections were washed for 5 s in ethanol to reduce background. Sections were immediately washed with phosphate-buffered saline, pH 7.4, counter-stained with 1 μg/mL DAPI for 10 min, and mounted onto glass slides in 90% (v/v) glycerol in phosphate-buffered saline. Sections were observed with a Nikon Eclipse E800 microscope.
RNA Isolation, Labeling, and Hybridization
Leaves 7 to 9 were harvested, pooled, flash-frozen, ground in liquid nitrogen, and stored at −80°C. Three biological replicas corresponding to infected and mock-inoculated samples were collected and processed independently as described below. Total RNA was extracted from each sample using the Plant RNeasy Mini kit (Qiagen). Double-stranded cDNA was synthesized using total RNA (5 μg) template and an oligo(dT)-T7 primer (Qiagen) in 20-μL reactions using the Superscript II system (Invitrogen) according to Affymetrix protocols. Biotinylated cRNA was synthesized using 10 μL of the resulting cDNA using the BioArray High Yield Transcript Labeling kit (Enzo) and cleaned with RNeasy columns (Qiagen) according to the manufacturer's protocol except for a double passage through the column to increase yield. The cRNA (15 μg) was hybridized to an ATH1 GeneChip (Affymetrix P/N 510690) for 16 h according to the Affymetrix GeneChip protocol. The arrays were washed and stained using the EukGe-Wsv4 protocol in a GeneChip Fluidic Station 450 and scanned using a GeneChip Scanner (Affymetrix). Array quality was assessed following the Affymetrix recommended parameters (GeneChip Expression Analysis, Technical Manual, 701021 rev 1). The hybridization, washing, and scanning protocols were performed at the NCSU Genome Research Laboratory. The microarray data have been submitted to ArrayExpress (www.ebi.ac.uk/arrayexpress; accession no. E-ATMX-34).
Microarray Data Analysis
The GeneChip scanning data were applied at probe-level to directly extract the intensities from.CEL files. The data were first transformed by logarithm base 2 and normalized across the chips by LOESS normalization (Dudoit and Fridlyand, 2002) with the probe-wise mean intensities using the mean across the arrays for each probe as baseline. Normalization was performed using SAS/Proc LOESS (SAS Institute).
The following mixed ANOVA model (Chu et al., 2002) was applied for further statistical analysis on the normalized data:
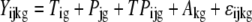 |
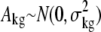 |
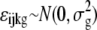 |
The indexes i, j, k, and g indicate the treatment group, probe, chip, and probe set (gene), respectively. The response variable, Y, represents the normalized intensity. T and P represent the treatment and probe main effects, respectively. TP represents the interaction effect of the two main effects. A represents the random chip effect assumed to be normal distributed, while ɛ corresponds to the stochastic error assumed to be normal distributed and independent to random chip effect. The P values from the ANOVA were converted to q values using the R program (version 2.5.1; Storey and Tibshirani, 2003).
Nuclei Isolation and Ploidy Analysis
Plants grown under the same conditions used for the microarray experiments were allowed to bolt, and the first flower bolts were removed at the base. The plants were inoculated with CaLCuV DNA by bombardment. For each sample, rosette and cauline leaves were harvested and pooled from at least six plants at 24 dpi and frozen in liquid nitrogen prior to nuclei isolation. The frozen tissues were chopped on ice with a single-edge razor blade in a cold glass petri dish containing lysis buffer (15 mm Tris-HCl, pH 7.5, 2 mm Na2EDTA, 80 mm KCl, 20 mm NaCl, 15 mm β-mercaptoethanol, 0.1% Triton X-100, and 2 mg/L DAPI). The chopped leaf suspension was incubated on ice for 10 min and filtered through a three-tiered nylon mesh (100, 50, and 30 μm). The nuclei suspension was filtered through a 20-μm nylon mesh and immediately subjected to FACS analysis using an InFlux cell sorter (Cytopeia) with a 355-nm UV laser (20 mW) tuned to excitation at 460/50 nm. Debris and aggregates were excluded from the populations by gating. FlowJo software (version 6.4.7) was used for FACS analysis.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Validation of microarray results.
Supplemental Table S1. Genes cited in the text and corresponding references.
Supplemental Table S2. Significant genes changing during CaLCuV infection.
Supplemental Table S3. Genes with increased transcripts during RNA virus infection and significantly altered by CaLCuV infection.
Supplemental Table S4. Comparisons to host pathogen pathway profiles.
Supplemental Table S5. Expression of senescence-associated genes.
Supplemental Table S6. Photosynthesis-related genes affected by CaLCuV infection.
Supplemental Table S7. Primer sets used in this study.
Supplemental Table S8. Core cell cycle genes at 9 dpi, PR1 expression in E2FB and CYCB3;1 overexpressors, and T-test for ploidy analysis of infected leaves.
Supplemental Methods and Materials S1. RT-PCR methods.
Supplementary Material
Acknowledgments
We thank Dr. Crisanto Gutiérrez (Universidad Autónoma de Madrid) for providing the Arabidopsis E2FC-overexpressing line, Dr. James Murray (Cambridge University) for providing the CYCD3 lines, and Dr. Steven Spoel (Duke University) for the cpr1 mutant. We also thank Drs. José Alonso (North Carolina State University) and Dominique Robertson (North Carolina State University) for their comments on the manuscript.
This work was supported by the National Science Foundation (grant nos. MCB–0110536 and DBI−0421651 to L.H.-B.) and by MiUR (grant no. 2006057940 to R.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: José Trinidad Ascencio-Ibáñez (jtascenc@ncsu.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Accotto GP, Mullineaux PM, Brown SC, Marie D (1993) Digitaria streak geminivirus replicative forms are abundant in S-phase nuclei of infected cells. Virology 195 257–259 [DOI] [PubMed] [Google Scholar]
- Anaya-López JL, Pérez-Mora E, Torres-Pacheco I, Muñoz-Sánchez CI, Guevara-Olvera L, González-Chavira M, Ochoa-Alejo N, Rivera-Bustamante RF, Guevara-González RG (2005) Inducible gene expression by Pepper huasteco virus in Capsicum chinense plants with resistance to geminivirus infections. Can J Plant Pathol 27 276–282 [Google Scholar]
- Andreasson E, Jenkins T, Brodersen P, Thorgrimsen S, Petersen NH, Zhu S, Qiu JL, Micheelsen P, Rocher A, Petersen M, et al (2005) The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J 24 2579–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguello-Astorga G, Lopez-Ochoa L, Kong LJ, Orozco BM, Settlage SB, Hanley-Bowdoin L (2004) A novel motif in geminivirus replication proteins interacts with the plant retinoblastoma homolog RBR. J Virol 78 4817–4826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascencio-Ibañez JT, Settlage SB (2007) DNA abrasion onto plants is an effective method for geminivirus infection and virus-induced gene silencing. J Virol Methods 142 198–203 [DOI] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al (2006) Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444 633–637 [DOI] [PubMed] [Google Scholar]
- Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE (2006) Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18 1038–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass HW, Nagar S, Hanley-Bowdoin L, Robertson D (2000) Chromosome condensation induced by geminivirus infection of mature plant cells. J Cell Sci 113 1149–1160 [DOI] [PubMed] [Google Scholar]
- Beckers GJ, Spoel SH (2006) Fine-tuning plant defence signalling: salicylate versus jasmonate. Plant Biol (Stuttg) 8 1–10 [DOI] [PubMed] [Google Scholar]
- Beemster GT, De Veylder L, Vercruysse S, West G, Rombaut D, Van Hummelen P, Galichet A, Gruissem W, Inze D, Vuylsteke M (2005) Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol 138 734–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyavskyi M, Westerman M, Dimichele L, Wilson VG (1996) Perturbation of the host cell cycle and DNA replication by the bovine papillomavirus replication protein E1. Virology 219 206–219 [DOI] [PubMed] [Google Scholar]
- Boudolf V, Vlieghe K, Beemster GT, Magyar Z, Torres Acosta JA, Maes S, Van Der Schueren E, Inze D, De Veylder L (2004) The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2FA-DPA control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16 2683–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling SA, Guo A, Cao H, Gordon S, Klessig DF, Dong X (1994) A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6 1845–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briddon RW (2003) Cotton leaf curl disease, a multicomponent begomovirus complex. Mol Plant Pathol 4 427–434 [DOI] [PubMed] [Google Scholar]
- Brodersen P, Petersen M, Bjorn Nielsen H, Zhu S, Newman MA, Shokat KM, Rietz S, Parker J, Mundy J (2006) Arabidopsis MAP kinase 4 regulates salicylic acid- and jasmonic acid/ethylene-dependent responses via EDS1 and PAD4. Plant J 47 532–546 [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, et al (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42 567–585 [DOI] [PubMed] [Google Scholar]
- Carvalho MF, Turgeon R, Lazarowitz SG (2006) The geminivirus nuclear shuttle protein NSP inhibits the activity of AtNSI, a vascular-expressed Arabidopsis acetyltransferase regulated with the sink-to-source transition. Plant Physiol 140 1317–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IP, Haehnel U, Altschmied L, Schubert I, Puchta H (2003) The transcriptional response of Arabidopsis to genotoxic stress: a high-density colony array study (HDCA). Plant J 35 771–786 [DOI] [PubMed] [Google Scholar]
- Cherubini G, Petouchoff T, Grossi M, Piersanti S, Cundari E, Saggio I (2006) E1B55K-deleted adenovirus (ONYX-015) overrides G1/S and G2/M checkpoints and causes mitotic catastrophe and endoreduplication in p53-proficient normal cells. Cell Cycle 5 2244–2252 [DOI] [PubMed] [Google Scholar]
- Chu TM, Deng S, Wolfinger RD (2006) Modeling Affymetrix data at the probe level. In DB Allen, GP Page, TM Beasley, JW Edwards, eds, DNA Microarray and Statistical Genomics Techniques: Design, Analysis, and Interpretation of Experiment. Chapman & Hall/CRC, Boca Raton, FL, pp 197–222
- Chu TM, Weir B, Wolfinger R (2002) A systematic statistical linear modeling approach to oligonucleotide array experiments. Math Biosci 176 35–51 [DOI] [PubMed] [Google Scholar]
- Churchman ML, Brown ML, Kato N, Kirik V, Hulskamp M, Inze D, De Veylder L, Walker JD, Zheng Z, Oppenheimer DG, et al (2006) SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana. Plant Cell 18 3147–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MM (2001) Historical overview: searching for replication help in all of the Rec places. Proc Natl Acad Sci USA 98 8173–8180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Schutter K, Joubes J, Cools T, Verkest A, Corellou F, Babiychuk E, Van Der Schueren E, Beeckman T, Kushnir S, Inze D, et al (2007) Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell 19 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GT, de Almeida Engler J, Ormenese S, Maes S, Naudts M, Van Der Schueren E, Jacqmard A, Engler G, et al (2002) Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2FA-DPA transcription factor. EMBO J 21 1360–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Inze D (2007) The ins and outs of the plant cell cycle. Nat Rev Mol Cell Biol 8 655–665 [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Diaz-Trivino S, Cisneros N, Gutierrez C (2006) The balance between cell division and endoreplication depends on E2FC-DPB, transcription factors regulated by the ubiquitin-SCFSKP2A pathway in Arabidopsis. Plant Cell 18 2224–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvoyes B, Ramirez-Parra E, Xie Q, Chua NH, Gutierrez C (2006) Cell type-specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant Physiol 140 67–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W, Riou-Khamlichi C, Scofield S, Healy JM, Jacqmard A, Kilby NJ, Murray JA (2003) Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15 79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W, Scofield S, Alcasabas AA, Maughan SC, Menges M, Braun N, Collins C, Nieuwland J, Prinsen E, Sundaresan V, et al (2007) Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc Natl Acad Sci USA 104 14537–14542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, et al (2006) Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444 638–642 [DOI] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H (1999) Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol 215 407–419 [DOI] [PubMed] [Google Scholar]
- Dudoit S, Fridlyand J (2002) A prediction-based resampling method for estimating the number of clusters in a dataset. Genome Biol 3 research0036.1–research0036.21 [DOI] [PMC free article] [PubMed]
- Eagle PA, Orozco BM, Hanley-Bowdoin L (1994) A DNA sequence required for geminivirus replication also mediates transcriptional regulation. Plant Cell 6 1157–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelkrout E, Mariconti L, Cella R, Robertson D, Hanley-Bowdoin L (2002) The activity of the proliferating cell nuclear antigen promoter is differentially regulated by two E2F elements during plant development. Plant Cell 14 3225–3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelkrout EM, Robertson D, Hanley-Bowdoin L (2001) Proliferating cell nuclear antigen transcription is repressed through an E2F consensus element and activated by geminivirus infection in mature leaves. Plant Cell 13 1437–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau K, Hoefert LL (1978) Hyperplastic phloem in sugar beet leaves infected with the beet curly top virus. Am J Bot 65 772–783 [Google Scholar]
- Etheridge N, Hall BP, Schaller GE (2006) Progress report: ethylene signaling and responses. Planta 223 387–391 [DOI] [PubMed] [Google Scholar]
- Fauquet CM, Bisaro DM, Briddon RW, Brown JK, Harrison BD, Rybicki EP, Stenger DC, Stanley J (2003) Revision of taxonomic criteria for species demarcation in the family Geminiviridae, and an updated list of begomovirus species. Arch Virol 148 405–421 [DOI] [PubMed] [Google Scholar]
- Felsani A, Mileo AM, Paggi MG (2006) Retinoblastoma family proteins as key targets of the small DNA virus oncoproteins. Oncogene 25 5277–5285 [DOI] [PubMed] [Google Scholar]
- Florentino LH, Santos AA, Fontenelle MR, Pinheiro GL, Zerbini FM, Baracat-Pereira MC, Fontes EP (2006) A PERK-like receptor kinase interacts with the geminivirus nuclear shuttle protein and potentiates viral infection. J Virol 80 6648–6656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes EP, Santos AA, Luz DF, Waclawovsky AJ, Chory J (2004) The geminivirus nuclear shuttle protein is a virulence factor that suppresses transmembrane receptor kinase activity. Genes Dev 18 2545–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes EPB, Gladfelter HJ, Schaffer RL, Petty ITD, Hanley-Bowdoin L (1994) Geminivirus replication origins have a modular organization. Plant Cell 6 405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fregene M, Matsumura H, Akano A, Dixon A, Terauchi R (2004) Serial analysis of gene expression (SAGE) of host-plant resistance to the cassava mosaic disease (CMD). Plant Mol Biol 56 563–571 [DOI] [PubMed] [Google Scholar]
- Friedrich TD, Laffin J, Lehman JM (1992) Simian virus 40 large T-antigen function is required for induction of tetraploid DNA content during lytic infection. J Virol 66 4576–4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granier C, Massonet C, Turc O, Muller B, Chenu K, Tardieu F (2002) Individual leaf development in Arabidopsis thaliana: a stable thermal-time-based programme. Ann Bot (Lond) 89 595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez C, Ramirez-Parra E, Castellano MM, del Pozo JC (2002) G(1) to S transition: more than a cell cycle engine switch. Curr Opin Plant Biol 5 480–486 [DOI] [PubMed] [Google Scholar]
- Hanley-Bowdoin L, Settlage SB, Robertson D (2004) Reprogramming plant gene expression: a prerequisite to geminivirus DNA replication. Mol Plant Pathol 5 149–156 [DOI] [PubMed] [Google Scholar]
- Hao L, Wang H, Sunter G, Bisaro DM (2003) Geminivirus AL2 and L2 proteins interact with and inactivate SNF1 kinase. Plant Cell 15 1034–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzeberg F, Chen IP, Hartung F, Orel N, Angelis KJ, Puchta H (2004) The Rad17 homologue of Arabidopsis is involved in the regulation of DNA damage repair and homologous recombination. Plant J 38 954–968 [DOI] [PubMed] [Google Scholar]
- Hill JE, Strandberg JO, Hiebert E, Lazarowitz SG (1998) Asymmetric infectivity of pseudorecombinants of cabbage leaf curl virus and squash leaf curl virus: Implications for bipartite geminivirus evolution and movement. Virology 250 283–292 [DOI] [PubMed] [Google Scholar]
- Hirano H, Harashima H, Shinmyo A, Sekine M (2008) Arabidopsis RETINOBLASTOMA-RELATED PROTEIN 1 is involved in G1 phase cell cycle arrest caused by sucrose starvation. Plant Mol Biol 66 259–275 [DOI] [PubMed] [Google Scholar]
- Huang Z, Yeakley JM, Garcia EW, Holdridge JD, Fan JB, Whitham S (2005) Salicylic acid-dependent expression of host genes in compatible Arabidopsis-virus interactions. Plant Physiol 137 1147–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubscher U, Maga G, Spadari S (2002) Eukaryotic DNA polymerases. Annu Rev Biochem 71 133–163 [DOI] [PubMed] [Google Scholar]
- Jeske H, Lutgemeier M, Preiss W (2001) DNA forms indicate rolling circle and recombination-dependent replication of Abutilon mosaic virus. EMBO J 20 6158–6167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan CV, Chen W, Hanley-Bowdoin L, Robertson D (2007) Geminivirus-induced gene silencing of the tobacco retinoblastoma-related gene results in cell death. Plant Mol Biol 65 163–175 [DOI] [PubMed] [Google Scholar]
- Kimura S, Sakaguchi K (2006) DNA repair in plants. Chem Rev 106 753–766 [DOI] [PubMed] [Google Scholar]
- Kong L, Hanley-Bowdoin L (2002) A geminivirus replication protein interacts with a protein kinase and a motor protein that display different expression patterns during plant development and infection. Plant Cell 14 1817–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono A, Umeda-Hara C, Adachi S, Nagata N, Konomi M, Nakagawa T, Uchimiya H, Umeda M (2007) The Arabidopsis D-type cyclin CYCD4 controls cell division in the stomatal lineage of the hypocotyl epidermis. Plant Cell 19 1265–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lageix S, Catrice O, Deragon JM, Gronenborn B, Pelissier T, Ramirez BC (2007) The nanovirus-encoded Clink protein affects plant cell cycle regulation through interaction with the retinoblastoma-related protein. J Virol 81 4177–4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham JR, Saunders K, Pinner MS, Stanley J (1997) Induction of plant cell division by beet curly top virus gene C4. Plant J 11 1273–1283 [Google Scholar]
- Lim PO, Nam HG (2005) The molecular and genetic control of leaf senescence and longevity in Arabidopsis. Curr Top Dev Biol 67 49–83 [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Solano R (2005) Molecular players regulating the jasmonate signalling network. Curr Opin Plant Biol 8 532–540 [DOI] [PubMed] [Google Scholar]
- Love AJ, Laval V, Geri C, Laird J, Deri Tomos A, Hooks MA, Milner J (2007) Components of Arabidopsis defense- and ethylene-signaling pathways regulate susceptibility to Cauliflower mosaic virus by restricting long-distance movement. Mol Plant Microbe Interact 20 659–670 [DOI] [PubMed] [Google Scholar]
- Love AJ, Wook Yun B, Laval V, Loake GJ, Milner J (2005) Cauliflower mosaic virus, a compatible pathogen of Arabidopsis, engages three distinct defnse-signaling pathways and activates rapid systemic generation of reactive oxygen species. Plant Physiol 139 935–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marathe R, Guan Z, Anandalakshmi R, Zhao H, Dinesh-Kumar SP (2004) Study of Arabidopsis thaliana resistome in response to cucumber mosaic virus infection using whole genome microarray. Plant Mol Biol 55 501–520 [DOI] [PubMed] [Google Scholar]
- Mariconti L, Pellegrini B, Cantoni R, Stevens R, Bergounioux C, Cella R, Albani D (2002) The E2F family of transcription factors from Arabidopsis thaliana: novel and conserved components of the retinoblastoma/E2F pathway in plants. J Biol Chem 277 9911–9919 [DOI] [PubMed] [Google Scholar]
- Masubelele NH, Dewitte W, Menges M, Maughan S, Collins C, Huntley R, Nieuwland J, Scofield S, Murray JA (2005) D-type cyclins activate division in the root apex to promote seed germination in Arabidopsis. Proc Natl Acad Sci USA 102 15694–15699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, de Jager SM, Gruissem W, Murray JA (2005) Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J 41 546–566 [DOI] [PubMed] [Google Scholar]
- Menges M, Hennig L, Gruissem W, Murray JAH (2003) Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol Biol 53 423–442 [DOI] [PubMed] [Google Scholar]
- Molinier J, Oakeley EJ, Niederhauser O, Kovalchuk I, Hohn B (2005) Dynamic response of plant genome to ultraviolet radiation and other genotoxic stresses. Mutat Res 571 235–247 [DOI] [PubMed] [Google Scholar]
- Nagar S, Hanley-Bowdoin L, Robertson D (2002) Host DNA replication is induced by geminivirus infection of differentiated plant cells. Plant Cell 14 2995–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagar S, Pedersen TJ, Carrick K, Hanley-Bowdoin L, Robertson D (1995) A geminivirus induces expression of a host DNA synthesis protein in terminally differentiated plant cells. Plant Cell 7 705–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CM, Van Loon LC (2004) NPR1: the spider in the web of induced resistance signaling pathways. Curr Opin Plant Biol 7 456–464 [DOI] [PubMed] [Google Scholar]
- Piroux N, Saunders K, Page A, Stanley J (2007) Geminivirus pathogenicity protein C4 interacts with Arabidopsis thaliana shaggy-related protein kinase AtSKeta, a component of the brassinosteroid signalling pathway. Virology 362 428–440 [DOI] [PubMed] [Google Scholar]
- Pourtau N, Jennings R, Pelzer E, Pallas J, Wingler A (2006) Effect of sugar-induced senescence on gene expression and implications for the regulation of senescence in Arabidopsis. Planta 224 556–568 [DOI] [PubMed] [Google Scholar]
- Qi R, John PC (2007) Expression of genomic AtCYCD2;1 in Arabidopsis induces cell division at smaller cell sizes: implications for the control of plant growth. Plant Physiol 144 1587–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Parra E, Gutiérrez C (2007) E2F regulates FASCIATA1, a chromatin assembly gene whose loss switches on the endocycle and activates gene expression by changing the epigenetic status. Plant Physiol 144 105–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas MR, Hagen C, Lucas WJ, Gilbertson RL (2005) Exploiting chinks in the plant's armor: evolution and emergence of geminiviruses. Annu Rev Phytopathol 43 361–394 [DOI] [PubMed] [Google Scholar]
- Schuermann D, Molinier J, Fritsch O, Hohn B (2005) The dual nature of homologous recombination in plants. Trends Genet 21 172–181 [DOI] [PubMed] [Google Scholar]
- Selth LA, Dogra SC, Rasheed MS, Healy H, Randles JW, Rezaian MA (2005) A NAC domain protein interacts with Tomato leaf curl virus replication accessory protein and enhances viral replication. Plant Cell 17 311–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WH, Hanley-Bowdoin L (2006) Geminivirus infection up-regulates the expression of two Arabidopsis protein kinases related to yeast SNF1 and mammalian AMPK activating kinases. Plant Physiol 142 1642–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz RW, Tatineni V, Hanley-Bowdoin L, Thompson WF (2007) Genome-wide analysis of core DNA replication machinery in Arabidopsis and rice. Plant Physiol 144 1697–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55 555–590 [DOI] [PubMed] [Google Scholar]
- Sozzani R, Maggio C, Varotto S, Canova S, Bergounioux C, Albani D, Cella R (2006) Interplay between Arabidopsis activating factors E2FB and E2FA in cell cycle progression and development. Plant Physiol 140 1355–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan SV, Mayhew CN, Schwemberger S, Zagorski W, Knudsen ES (2007) RB loss promotes aberrant ploidy by deregulating levels and activity of DNA replication factors. J Biol Chem 282 23867–23877 [DOI] [PubMed] [Google Scholar]
- Stanley J, Latham JR (1992) A symptom variant of beet curly top geminivirus produced by mutation of open reading frame C4. Virology 190 506–509 [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunter G, Bisaro DM (1997) Regulation of a geminivirus coat protein promoter by AL2 protein (TrAP): Evidence for activation and derepression mechanisms. Virology 232 269–280 [DOI] [PubMed] [Google Scholar]
- Swanhart LM, Sanders AN, Duronio RJ (2007) Normal regulation of Rbf1/E2f1 target genes in Drosophila type 1 protein phosphatase mutants. Dev Dyn 236 2567–2577 [DOI] [PubMed] [Google Scholar]
- Swindell WR (2006) The association among gene expression responses to nine abiotic stress treatments in Arabidopsis thaliana. Genetics 174 1811–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi F, Yoshida R, Ichimura K, Mizoguchi T, Seo S, Yonezawa M, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K (2007) The mitogen-activated protein kinase cascade MKK3-MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. Plant Cell 19 805–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinks D, Rajeswaran R, Shivaprasad PV, Akbergenov R, Oakeley EJ, Veluthambi K, Hohn T, Pooggin MM (2005) Suppression of RNA silencing by a geminivirus nuclear protein, AC2, correlates with transactivation of host genes. J Virol 79 2517–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnage MA, Muangsan N, Peele CG, Robertson D (2002) Geminivirus-based vectors for gene silencing in Arabidopsis. Plant J 30 107–114 [DOI] [PubMed] [Google Scholar]
- Vandepoele K, Raes J, DeVeylder L, Rouze P, Rombauts S, Inze D (2002) Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14 903–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Vlieghe K, Florquin K, Hennig L, Beemster GT, Gruissem W, Van de Peer Y, Inze D, De Veylder L (2005) Genome-wide identification of potential plant E2F target genes. Plant Physiol 139 316–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkest A, Manes CL, Vercruysse S, Maes S, Van Der Schueren E, Beeckman T, Genschik P, Kuiper M, Inze D, De Veylder L (2005) The cyclin-dependent kinase inhibitor KRP2 controls the onset of the endoreduplication cycle during Arabidopsis leaf development through inhibition of mitotic CDKA;1 kinase complexes. Plant Cell 17 1723–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieghe K, Boudolf V, Beemster GT, Maes S, Magyar Z, Atanassova A, de Almeida Engler J, De Groodt R, Inze D, De Veylder L (2005) The DP-E2F-like gene DEL1 controls the endocycle in Arabidopsis thaliana. Curr Biol 15 59–63 [DOI] [PubMed] [Google Scholar]
- Vlieghe K, Vuylsteke M, Florquin K, Rombauts S, Maes S, Ormenese S, Van Hummelen P, Van de Peer Y, Inze D, De Veylder L (2003) Microarray analysis of E2FA-DPA-overexpressing plants uncovers a cross-talking genetic network between DNA replication and nitrogen assimilation. J Cell Sci 116 4249–4259 [DOI] [PubMed] [Google Scholar]
- Wang H, Buckley KJ, Yang X, Buchmann RC, Bisaro DM (2005) Adenosine kinase inhibition and suppression of RNA silencing by geminivirus AL2 and L2 proteins. J Virol 79 7410–7418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzman MD, Carson CT, Schwartz RA, Lilley CE (2004) Interactions of viruses with the cellular DNA repair machinery. DNA Repair (Amst) 3 1165–1173 [DOI] [PubMed] [Google Scholar]
- Whitham SA, Quan S, Chang HS, Cooper B, Estes B, Zhu T, Wang X, Hou YM (2003) Diverse RNA viruses elicit the expression of common sets of genes in susceptible Arabidopsis thaliana plants. Plant J 33 271–283 [DOI] [PubMed] [Google Scholar]
- Whitham SA, Yang C, Goodin MM (2006) Global impact: elucidating plant responses to viral infection. Mol Plant Microbe Interact 19 1207–1215 [DOI] [PubMed] [Google Scholar]
- Wiermer M, Feys BJ, Parker JE (2005) Plant immunity: the EDS1 regulatory node. Curr Opin Plant Biol 8 383–389 [DOI] [PubMed] [Google Scholar]
- Wu X, Avni D, Chiba T, Yan F, Zhao Q, Lin Y, Heng H, Livingston D (2004) SV40 T antigen interacts with Nbs1 to disrupt DNA replication control. Genes Dev 18 1305–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Guo R, Jie F, Nettleton D, Peng J, Carr T, Yeakley JM, Fan JB, Whitham SA (2007) Spatial analysis of Arabidopsis thaliana gene expression in response to Turnip mosaic virus infection. Mol Plant Microbe Interact 20 358–370 [DOI] [PubMed] [Google Scholar]
- Yoshizumi T, Tsumoto Y, Takiguchi T, Nagata N, Yamamoto YY, Kawashima M, Ichikawa T, Nakazawa M, Yamamoto N, Matsui M (2006) INCREASED LEVEL OF POLYPLOIDY1, a conserved repressor of CYCLINA2 transcription, controls endoreduplication in Arabidopsis. Plant Cell 18 2452–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Liu Y, Wu X, Shell SM (2006) Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. J Cell Physiol 208 267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.