Abstract
CARM1/PRMT4 (for COACTIVATOR-ASSOCIATED ARGININE METHYLTRANSFERASE1/PROTEIN ARGININE METHYLTRANSFERASE4) catalyzes asymmetric dimethylation on arginine (Arg), and its functions in gene regulation is understood only in animal systems. Here, we describe AtPRMT4a and AtPRMT4b as a pair of Arabidopsis (Arabidopsis thaliana) homologs of mammalian CARM1/PRMT4. Recombinant AtPRMT4a and AtPRMT4b could asymmetrically dimethylate histone H3 at Arg-2, Arg-17, Arg-26, and myelin basic protein in vitro. Both AtPRMT4a and AtPRMT4b exhibited nuclear as well as cytoplasmic distribution and were expressed ubiquitously in all tissues throughout development. Glutathione S-transferase pull-down assays revealed that AtPRMT4a and AtPRMT4b could form homodimers and heterodimers in vitro, and formation of the heterodimer was further confirmed by bimolecular fluorescence complementation. Simultaneous lesions in AtPRMT4a and AtPRMT4b genes led to delayed flowering, whereas single mutations in either AtPRMT4a or AtPRMT4b did not cause major developmental defects, indicating the redundancy of AtPRMT4a and AtPRMT4b. Genetic analysis also indicated that atprmt4a atprmt4b double mutants phenocopied autonomous pathway mutants. Finally, we found that asymmetric methylation at Arg-17 of histone H3 was greatly reduced in atprmt4a atprmt4b double mutants. Taken together, our results demonstrate that AtPRMT4a and AtPRMT4b are required for proper regulation of flowering time mainly through the FLOWERING LOCUS C-dependent pathway.
In animals, Arg methylation is catalyzed by a gene family called PROTEIN ARGININE METHYLTRANSFERASE (PRMT). Arg methylation has been proposed to participate in multiple cellular processes, including transcriptional regulation, RNA metabolism, nuclear trafficking, DNA repair, and signal transduction (Bedford and Richard, 2005; D.Y. Lee et al., 2005). To date, nine PRMTs have been reported in humans, and it has been shown that except for PRMT2, all of them have Arg methyltransferase activities. CARM1 (for COACTIVATOR-ASSOCIATED ARGININE METHYLTRANSFERASE1)/PRMT4 is one of the best characterized type I PRMTs. It catalyzes the formation of asymmetric dimethyl-Arg and was originally identified as the secondary coactivator of the p160 family members (Chen et al., 1999). It is involved in transcriptional activation of the nuclear receptor-responsive genes (Chen et al., 1999, 2000; Koh et al., 2001; Ma et al., 2001; Bauer et al., 2002; Daujat et al., 2002; Stallcup et al., 2003; An et al., 2004; Ananthanarayanan et al., 2004; Y.H. Lee et al., 2005; Teyssier et al., 2006) and nonnuclear receptor-associated genes (Koh et al., 2002; Covic et al., 2005; Zika et al., 2005; El Messaoudi et al., 2006; Krones-Herzig et al., 2006; Miao et al., 2006) through interactions with transcription cofactors, such as CREB-binding protein (CBP)/p300, the p160 family of coactivators, and other transcriptional coactivators at the promoter region of specific target genes. This activity is methyltransferase dependent (Ma et al., 2001; Bauer et al., 2002; Daujat et al., 2002; An et al., 2004; El Messaoudi et al., 2006). Moreover, CARM1 also negatively impacts transcription by disassembling the coactivator complex or enhancing coactivator degradation (Feng et al., 2006; Naeem et al., 2007) or by blocking the domains' interaction (Xu et al., 2001). In addition, RNA-binding proteins, such as HUR, HUD, PABP1, and TARPP (Lee and Bedford, 2002; Fujiwara et al., 2006), and splicing factors, such as CA150, SAP49, SmB, and U1C (Cheng et al., 2007), can also be methylated by CARM1. Although mammalian cells lacking CARM1 are viable, carm1 deficient mouse embryos are small in size and die during late embryonic development or immediately after birth, indicating that CARM1 is essential for developmental processes (Yadav et al., 2003). Recent studies have also revealed pleiotropic functions of CARM1 in cell differentiation (Chen et al., 2002; J. Kim et al., 2004), cell fate determination (Torres-Padilla et al., 2007), proliferation (Fujiwara et al., 2006), apoptosis (Cakouros et al., 2004), and oncogenesis (El Messaoudi et al., 2006; Majumder et al., 2006).
In Arabidopsis (Arabidopsis thaliana), nine PRMTs are present in the genome (Niu et al., 2007). AtPRMT1a and AtPRMT1b, the pair of Arabidopsis homologs of human PRMT1, can methylate histone H4R3 and fibrillarin 2, an RNA methyltransferase in vitro (Yan et al., 2007). AtPRMT5, a type II methyltransferase, and AtPRMT10, a type I methyltransferase, can dimethylate histone H4R3 symmetrically (H4R3me2s) and asymmetrically (H4R3me2a), respectively (Niu et al., 2007; Pei et al., 2007). AtPRMT5 is involved in the promotion of vegetative growth and flowering time, since atprmt5 mutants cause pleiotropic developmental defects, including growth retardation, dark green and curled leaves, and FLOWERING LOCUS C (FLC)-dependent delayed flowering (Pei et al., 2007; Wang et al., 2007; Schmitz et al., 2008). AtPRMT10 is a plant-specific Arg methyltransferase that is also required for the repression of FLC to promote flowering (Niu et al., 2007). In addition, lesions in AtPRMT5 and AtPRMT10 cause a delayed-flowering phenotype in a nonredundant manner, reflecting the complexity of posttranslational modifications in flowering time regulation in Arabidopsis.
Physiological and genetic analyses of flowering time using the model plant Arabidopsis have shown that the floral transition is controlled by multiple environmental cues and endogenous signals. Such complex regulation ensures that Arabidopsis flowers at the optimal time for successful reproduction. An increasing number of genes are involved in this regulation. There are four main pathways that regulate Arabidopsis flowering: the photoperiod, vernalization, gibberellin (GA), and autonomous pathways (Simpson et al., 1999; Reeves and Coupland, 2000; Mouradov et al., 2002; Komeda, 2004; Amasino, 2005). FLC, a MADS box transcription factor (Michaels and Amasino, 1999; Sheldon et al., 1999), represses flowering largely by suppressing the expression of the flowering integrators FT and SOC1 (for SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1).
A central role of the autonomous pathway is to negatively regulate FLC expression; thus, lesions in autonomous pathway genes display a late-flowering phenotype associated with elevation of FLC expression (Michaels and Amasino, 2001). Increasing lines of evidence suggest that many components of the autonomous pathway are involved in chromatin remodeling and RNA processing. FVE (AtMSI4) is the plant homolog of yeast MSI (for MULTICOPY SUPPRESSOR OF IRA1) and the mammalian retinoblastoma-associated proteins RbAp46 and RbAp48, which are found in histone deacetylase complexes involved in transcriptional repression (Kenzior and Folk, 1998; Ausin et al., 2004; H.J. Kim et al., 2004). With an enhanced response of cold-induced gene expression and late flowering, fve mutants exhibit an increase in acetylation state of histone H4 at FLC chromatin (Ausin et al., 2004). FLOWERING LOCUS D (FLD), a homolog of human LYSINE DEMETHYLASE1 (LSD1), and its two relatives, LSD1-LIKE1 (LDL1) and LDL2, redundantly repress FLC expression by down-regulating acetylation levels of FLC chromatin (He et al., 2003; Jiang et al., 2007). RELATIVE OF EARLY FLOWERING6 (REF6) is a jumonji domain-containing protein that contains the conserved amino acids for histone demethylase activity (Lu et al., 2008). Mutations in REF6 show derepressed expression of FLC accompanied by hyperacetylation of histone H4 (Noh et al., 2004). In addition, some chromatin modifiers are also important for the repression of FLC expression, although chromatin status at FLC in the mutant plants may not be influenced significantly. AtHAC1 encodes a histone acetyltransferase, and loss-of-function athac1 mutants display elevated FLC expression without detectable changes of FLC chromatin modifications (Deng et al., 2007; Han et al., 2007). All of the above factors regulate FLC at the transcriptional level through direct and most likely indirect biochemical modifications at FLC chromatin.
RNA processing also plays an important role in flowering time control. In the autonomous pathway, there are now three genes encoding RNA-binding proteins: the plant-specific RNA recognition motif-containing proteins FCA (Macknight et al., 1997) and FPA (Schomburg et al., 2001) and the K homology domain-containing protein FLK (Lim et al., 2004; Mockler et al., 2004). Lesions in these genes cause late flowering. A recent study showed that FCA requires FLD for FLC repression, which is associated with decreased H3K4 dimethylation (Liu et al., 2007). In addition, FY is an mRNA 3′ end processing factor that can physically interact with FCA, and this interaction is required for FCA autoregulation and flowering regulation (Simpson et al., 2003).
Despite recent advances in our understanding of CARM1 functions in Drosophila, mouse, and human, the roles of CARM1 homologs in Arabidopsis have not been addressed. In this study, we show that AtPRMT4a and AtPRMT4b, a pair of Arg methyltransferases in Arabidopsis sharing the highest homology with CARM1, methylate histone H3 and myelin basic protein (MBP) in vitro. Both single mutant plants did not exhibit obvious developmental defects, whereas lesions in both AtPRMT4a and AtPRMT4b caused FLC-dependent late flowering. The global level of asymmetric dimethylation on histone H3 Arg-17 was decreased in atprmt4a atprmt4b double mutants. Thus, we demonstrate that, in addition to AtPRMT5 and AtPRMT10, AtPRMT4a and AtPRMT4b are two new AtPRMTs redundantly required for proper flowering time regulation in Arabidopsis.
RESULTS
AtPRMT4a and AtPRMT4b Are Homologs of Animal CARM1
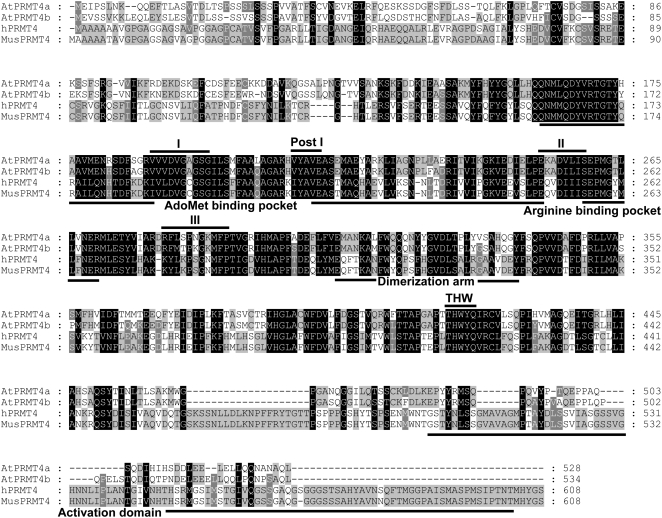
There are two CARM1/PRMT4 homologs in the Arabidopsis genome: AtPRMT4a (At5g49020) and AtPRMT4b (At3g06930; Niu et al., 2007). Protein sequence analysis showed that AtPRMT4a and AtPRMT4b share 43% and 45% identity to human CARM1, respectively. AtPRMT4a and AtPRMT4b are 79% identical at the amino acid level and exhibit amino acid conservation within the characteristic S-adenosyl-Met (AdoMet)-dependent methyltransferase motifs I, post-I, II, and III as well as in amino acids outside these regions, including the THW loop, the AdoMet-binding pocket, and the Arg-binding pocket common to all PRMT family members (Matsuda et al., 2007; Fig. 1), which suggested that AtPRMT4a and AtPRMT4b might have protein Arg methyltransferase activities. In addition, the dimerization arms residing in the central region were also well conserved, indicating that both AtPRMT4a and AtPRMT4b had the potential to form homodimers or heterodimers. Notably, unlike mammalian CARM1, AtPRMT4a and AtPRMT4b did not contain the typical activation domain at the C-terminal ends, suggesting that a minimal set of amino acids might be sufficient to compose the activation domain in Arabidopsis.
Figure 1.
Amino acid sequence alignment of PRMT4 from Arabidopsis, human, and mouse. Amino acids that match all and a majority consensus are boxed in black and gray, respectively. The signature methyltransferase motifs I, post-I, II, III, and THW loop are indicated at top with black lines. The AdoMet binding pocket, Arg-binding pocket, dimerization arm, and activation domain are indicated at bottom. The accession numbers of the used protein sequences are as follows: NP_199713 for AtPRMT4a; NP_187349 for AtPRMT4b; NP_954592 for hPRMT4; and NP_067506 for MusPRMT4.
AtPRMT4a and AtPRMT4b Methylate Core Histone H3 and Nonhistone Proteins in Vitro
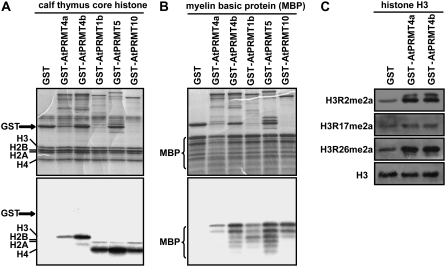
To test the methyltransferase activities, full-length AtPRMT4a and AtPRMT4b proteins were expressed as glutathione S-transferase (GST) fusion proteins in Escherichia coli. The purified recombinant proteins were evaluated in vitro for methyltransferase activities with two different substrates: calf thymus core histones and MBP. In contrast to AtPRMT1b, AtPRMT5, and AtPRMT10, which mainly methylate histone H4 and H2A, both AtPRMT4a and AtPRMT4b preferentially methylate H3 (Fig. 2A). In addition to histone H3, the nonhistone protein MBP was also methylated by AtPRMT4a and AtPRMT4b, similar to AtPRMT1b, AtPRMT5, and AtPRMT10 (Fig. 2B). Thus, methyltransferase activity assay results show that AtPRMT4a and AtPRMT4b are bona fide protein Arg methyltransferases.
Figure 2.
Methyltransferase activities of AtPRMT4a and AtPRMT4b in vitro. A and B, GST fusion AtPRMT proteins were incubated with the indicated substrates: calf thymus core histones (A) and MBP in the presence of the methyl donor S-adenosyl-l-[methyl-3H] (B). Coomassie Brilliant Blue-stained gels are shown at top, and the autoradiographs of the gels are shown at bottom. C, Equal amounts of histone H3 methylated by AtPRMT4a, AtPRMT4b, or control GST were immunoblotted with antibodies against H3R2me2a, H3R17me2a, and H3R26me2, respectively. For the loading control, the membrane used for anti-H3R17me2a was stripped and immunoblotted with anti-H3 antibody.
AtPRMT4a and AtPRMT4b Asymmetrically Dimethylate Histone H3 at Arg-2, Arg-17, and Arg-26 in Vitro
The human CARM1/PRMT4 can asymmetrically dimethylate histone H3 at Arg-2, Arg-17, and Arg-26 (Schurter et al., 2001). To further investigate the biochemical characteristics of AtPRMT4a and AtPRMT4b, calf thymus histone H3 incubated with AtPRMT4a and AtPRMT4b was probed with antibodies against asymmetrically dimethylated histone H3 at Arg-2 (H3R2me2a), Arg-17 (H3R17me2a), and Arg-26 (H3R26me2a). As shown in Figure 2C, the modification levels of H3R2me2a, H3R17me2a, and H3R26me2a were increased when histone H3 was incubated with AtPRMT4a and AtPRMT4b compared with GST protein, which was used as a negative control. This result indicates that both AtPRMT4a and AtPRMT4b can asymmetrically dimethylate histone H3 at Arg-2, Arg-17, and Arg-26 in vitro.
AtPRMT4a and AtPRMT4b Are Present in Both the Nucleus and the Cytoplasm
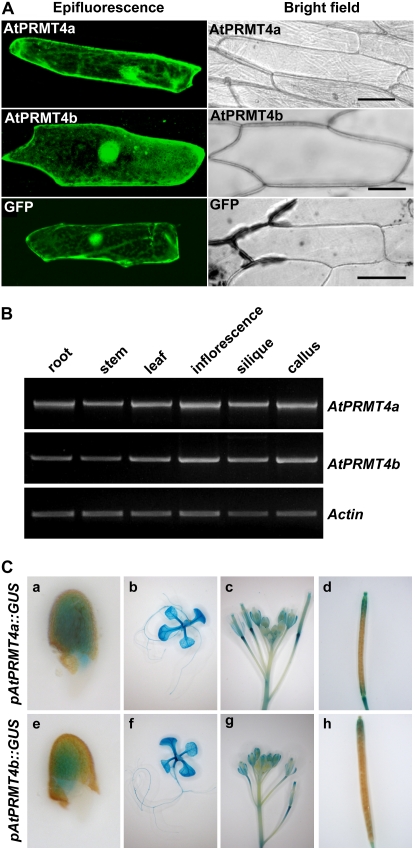
In human, different PRMTs have distinct subcellular localization patterns, reflecting their proper functions at the subcellular level (Frankel et al., 2002; J. Lee et al., 2005). To evaluate the subcellular localization of AtPRMT4a and AtPRMT4b, AtPRMT4a and AtPRMT4b cDNAs were fused in-frame with the 3′ end of GFP driven by the constitutive 35S promoter and transiently expressed in onion (Allium cepa) epidermal cells. The distribution of these fusion proteins was observed by confocal fluorescence microscopy. Although previous studies in HeLa cells (Frankel et al., 2002) and mouse embryonic fibroblast cells (Yadav et al., 2003) demonstrated the nuclear existence of CARM1, our results revealed that both GFP-AtPRMT4a and GFP-AtPRMT4b displayed cytoplasmic as well as intense nuclear localization (Fig. 3A), consistent with a recent finding of localization of CARM1 in PC12 cells (Fujiwara et al., 2006). Taken together with their different substrates, such as histone H3 in the nucleus and MBP in the cytoplasm, our results suggest that AtPRMT4a and AtPRMT4b may function both inside and outside of the nucleus. However, we cannot rule out the notion that some aspects of this localization pattern are due to overexpression in onion cells or the GFP moiety.
Figure 3.
AtPRMT4a and AtPRMT4b exhibited similar subcellular distribution and expression patterns. A, AtPRMT4a and AtPRMT4b cDNAs were fused in-frame to the C-terminal end of GFP. The resulting constructs were transiently transformed into onion epidermal cells by DNA-coated gold particle bombardment, and protein localization was observed by confocal microscopy, shown as epifluorescence and bright-field images. Bars = 75 μm. B, Semiquantitative RT-PCR was performed to examine the spatial expression of AtPRMT4a and AtPRMT4b in root, stem, leaf, inflorescence, silique, and callus. Actin2/7 was used as a constitutive positive control. C, Histochemical GUS activity analysis of pAtPRMT4a∷GUS and pAtPRMT4b∷GUS transgenic plants in the Col-0 background. a and e, Germinating seeds; b and f, 10-d-old-seedlings; c and g, inflorescences; d and h, siliques.
AtPRMT4a and AtPRMT4b Exhibit Nearly Identical Expression Patterns
The expression of AtPRMT4a and AtPRMT4b was evaluated by reverse transcription (RT)-PCR using total RNAs extracted from root, stem, leaf, inflorescence, silique, and callus. As shown in Figure 3B, AtPRMT4a and AtPRMT4b were constitutively expressed in all of the tissues tested. In order to determine the spatial and temporal expression patterns of AtPRMT4a and AtPRMT4b exactly, the fragments containing 1,066 and 1,082 bp before the start codon of AtPRMT4a and AtPRMT4b genomic sequences, respectively, were used to drive the GUS reporter gene, and the two constructs were transformed into the wild-type accession Columbia (Col-0) plants. Consistent with the RT-PCR data, GUS staining indicated that AtPRMT4a and AtPRMT4b showed a similar expression pattern and were ubiquitously expressed in all tissues throughout development. In addition, AtPRMT4a and AtPRMT4b were highly expressed in the shoot and root apical meristem as well as in the vasculature (Fig. 3C). Given that AtPRMT4a and AtPRMT4b exhibited nearly identical biochemical properties and subcellular distribution, and that these two genes showed indistinguishable expression patterns, we speculate that these two proteins might function redundantly.
AtPRMT4a and AtPRMT4b Form Homodimers and Heterodimers in Vitro and in Vivo
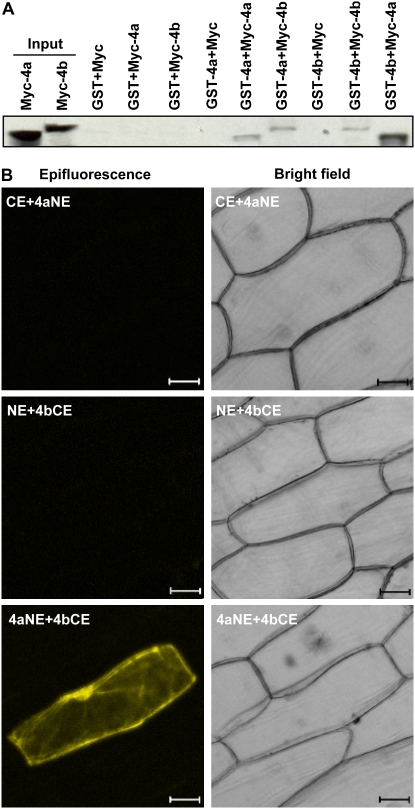
Previous studies have suggested that the domain of the PRMTs having the methyltransferase activity is also responsible for forming homodimers or large homooligomers (Zhang et al., 2000; Wang et al., 2001). A typical dimerization arm was also found in both AtPRMT4a and AtPRMT4b (Fig. 1). To investigate whether AtPRMT4a and AtPRMT4b are capable of forming homodimers or heterodimers, GST pull-down experiments were performed using in vitro translated and recombinant AtPRMT4a and AtPRMT4b proteins. GST was used as a negative control. [35S]Met-labeled AtPRMT4a interacted with GST-AtPRMT4a but not GST, indicating the capability of AtPRMT4a to form homodimers. Similarly, [35S]Met-labeled AtPRMT4b also interacted with GST-AtPRMT4b but not GST, suggesting that AtPRMT4b could form homodimers as well. Given their similarity, it is not surprising that an interaction between AtPRMT4a and AtPRMT4b was also detected by reciprocal assays, suggesting that both proteins formed heterodimers (Fig. 4A).
Figure 4.
AtPRMT4a and AtPRMT4b form homodimers and heterodimers. A, In vitro pull-down assays of the interactions between AtPRMT4s. GST-AtPRMT4a and GST-AtPRMT4b were expressed in E. coli and bound to glutathione-Sepharose 4B beads. The bound GST fusion proteins were incubated with in vitro translated [35S]Met-labeled Myc, Myc-AtPRMT4a, and Myc-AtPRMT4b in various combinations as indicated above. GST was used as a negative control. 4a, AtPRMT4a; 4b, AtPRMT4b. B, BiFC analysis demonstrated the in vivo interaction between AtPRMT4a and AtPRMT4b. Epifluorescence and bright-field images are shown for onion epidermal cells transiently cotransfected with constructs encoding the indicated fusion proteins. AtPRMT4a, AtPRMT4b, pSPYNE, and pSPYCE are indicated as 4a, 4b, NE, and CE, respectively. Bars = 50 μm. [See online article for color version of this figure.]
To further visualize the interaction between AtPRMT4a and AtPRMT4b in living plant cells, we used bimolecular fluorescence complementation (BiFC), which is based on the formation of a fluorescent complex when fragments of two interacting proteins come together (Walter et al., 2004). To this end, the coding sequences of AtPRMT4a and AtPRMT4b were cloned into the pSPYNE-35S (NE) and pSPYCE-35S (CE) vectors, which expressed AtPRMT4a-YFPN (4aNE) and PRMT4b-YFPC (4bCE) fusion proteins: AtPRMT4a fused to the N-terminal 155 amino acids of yellow fluorescent protein (YFP) and AtPRMT4b fused to the C-terminal 84 amino acids of YFP, respectively. Transient transfection results showed that onion epidermal cells that only cotransfected with AtPRMT4a-YFPN and AtPRMT4b-YFPC exhibited a strong YFP signal (Fig. 4B), whereas constructs that cotransfected with pSPYNE-35S and AtPRMT4b-YFPC or pSPYCE-35S and AtPRMT4a-YFPN did not exhibit a fluorescence signal. These results indicated an interaction between AtPRMT4a and AtPRMT4b and suggested that they might form heterodimers in vivo.
atprmt4a atprmt4b Double Mutants Are Late Flowering
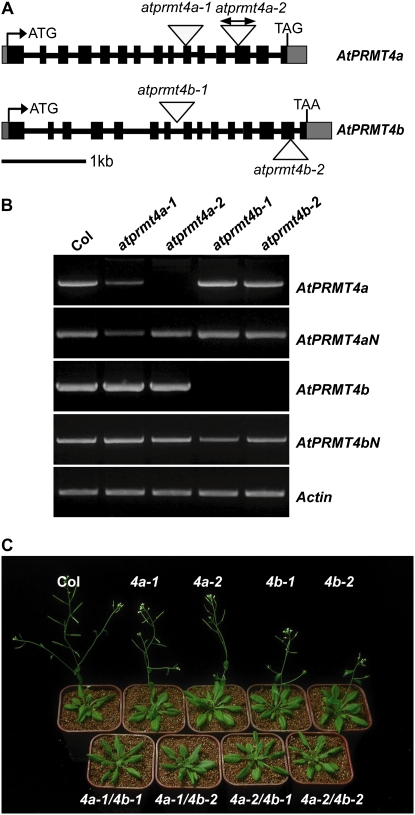
To investigate the functions of AtPRMT4a and AtPRMT4b in Arabidopsis development, we obtained several T-DNA insertion mutant alleles from the Signal Collection at the Salk Institute (Alonso et al., 2003). Both AtPRMT4a and AtPRMT4b genes contain 15 exons and display a similar genome structure, indicating a recent duplication of the two genes (Fig. 5A). Two atprmt4a homozygotes (atprmt4a-1 and atprmt4a-2) and two atprmt4b homozygotes (atprmt4b-1 and atprmt4b-2) were identified. The atprmt4a-1 allele contains a T-DNA insertion in the 10th exon, and atprmt4a-2 has two insertions in the 13th exon (Fig. 5A, top); atprmt4b-1 and atprmt4b-2 contain a T-DNA insertion in the seventh intron and the 14th exon, respectively (Fig. 5A, bottom). The location of each insertion was confirmed by PCR and sequencing of the junction regions. Full-length AtPRMT4a in atprmt4a-2 and AtPRMT4b in atprmt4b were not detected by RT-PCR analysis (Fig. 5B), but the N-terminal ends of AtPRMT4a and AtPRMT4b were still retained, and there was still a low level of full-length AtPRMT4a in atprmt4a-1.
Figure 5.
Late-flowering phenotype of atprmt4a atprmt4b double mutants. A, AtPRMT4a and AtPRMT4b gene structures. Black boxes and gray boxes represent exons and the 5′ or 3′ untranslated region, respectively. Introns are indicated as lines. The positions of T-DNA insertions are indicated with triangles in each allele. B, RT-PCR analysis of full length and N terminus of AtPRMT4a and AtPRMT4b transcripts in wild-type Col-0 and each mutant. Actin2/7 was used as a control for constitutive expression. C, atprmt4a atprmt4b double mutants are late flowering. Flowering phenotypes are shown for atprmt4a, atprmt4b, and atprmt4a atprmt4b in LD. atprmt4a, atprmt4b, and atprmt4a atprmt4b are indicated as 4a, 4b, and 4a/4b.
Compared with wild-type Col-0 plants, atprmt4a-1, atprmt4a-2, atprmt4b-1, and atprmt4b-2 had no obvious phenotype. Given the largely similar biochemical characteristics and overlapping patterns of distribution and expression between AtPRMT4a and AtPRMT4b, it is possible that both enzymes could be functionally redundant. Therefore, four double mutants, atprmt4a-1 atprmt4b-1, atprmt4a-1 atprmt4b-2, atprmt4a-2 atprmt4b-1, and atprmt4a-2 atprmt4b-2, were constructed by crossing between individual atprmt4a and atprmt4b alleles. In contrast to the wild-type plants and the atprmt4a and atprmt4b single mutants, these double mutant plants were all late flowering under long days (LD; Fig. 5C). Except for delayed flowering, there were no other obvious growth and development defects in the mutant plants under these normal growth conditions. Due to the same phenotype of the four atprmt4a atprmt4b double mutants, atprmt4a-2 atprmt4b-1 was used in subsequent analyses and abbreviated as atprmt4a atprmt4b.
atprmt4a atprmt4b Double Mutants Respond Normally to Photoperiod, GA, and Vernalization Treatments Similar to Autonomous Pathway Mutants
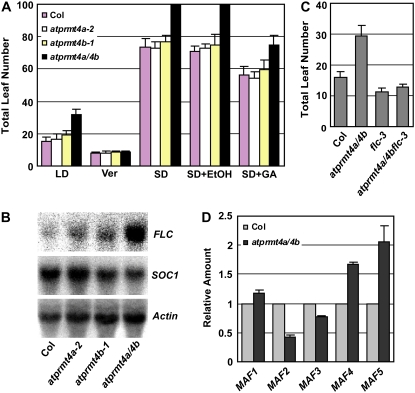
To dissect the role of AtPRMT4a and AtPRMT4b in the control of flowering time, we investigated which pathways were affected by the loss of AtPRMT4a and AtPRMT4b. Arabidopsis is a facultative LD plant that flowers earlier under LD than short days (SD). By comparing the flowering time of the wild-type Col-0 with the atprmt4a, atprmt4b single and atprmt4a atprmt4b double mutants grown under both LD and SD, a similar response to the photoperiod was observed in both the wild-type plants and the atprmt4a atprmt4b double mutants, indicating that AtPRMT4a and AtPRMT4b are not involved in the photoperiod pathway (Fig. 6A). Vernalization is the process by which a prolonged exposure to low temperature promotes flowering in Arabidopsis. After 10 weeks of vernalization (indicated as Ver), the atprmt4a and atprmt4b single mutants and the atprmt4a atprmt4b double mutant plants flowered as early as the wild-type Col-0. This indicates that lesions in AtPRMT4a and AtPRMT4b do not cause a defective response to vernalization (Fig. 6A). The plant growth regulator GA is essential for the promotion of flowering of Arabidopsis under SD. To determine the effect of GA on flowering time in atprmt4a atprmt4b plants, 100 μm GA was applied to growing plants in SD until they flowered (indicated as SD+GA). Similar to the wild-type controls, an exogenous supplement of GA could partially rescue the late-flowering phenotype of atprmt4a atprmt4b double mutants (Fig. 6A).
Figure 6.
The late-flowering phenotype of atprmt4a atprmt4b is FLC dependent. A, Flowering time of wild-type Col-0 and atprmt4 mutants was assessed by total leaf number after plants stopped producing new leaves under different conditions or treatments. Vernalization (Ver) treatment indicates plants grown at 4°C under SD (8 h of light/16 h of dark) conditions for 10 weeks before transfer to 23°C under LD. SD+GA indicates that 100 μm GA was sprayed once per week on the plants until flowering under SD conditions. Under SD, atprmt4a atprmt4b plants did not flower even after producing 100 leaves, and the assessment was terminated at this point. Error bars represent sd. B, Flowering time of atprmt4a atprmt4b flc-3 triple mutants was measured by total leaf numbers when the plants stopped producing new leaves. The plants were grown at 23°C under LD. Error bars represent sd. C, Transcription levels of FLC and SOC1 in Col-0 and atprmt4 mutants. Total RNAs extracted from young seedlings of Col-0 and mutants with four to five rosette leaves were analyzed by RNA-blot analysis. Actin2/7 was used as a control for constitutive expression. D, Transcription levels of MAF genes in atprmt4a atprmt4b. Real-time PCR analysis of the expression of MAF genes in seedlings with four to five rosette leaves of wild-type Col-0 and mutant plants. Error bars represent sd. Actin2/7 was used as a control for constitutive expression. atprmt4a/4b indicates atprmt4a atprmt4b. [See online article for color version of this figure.]
The physiological characteristics of atprmt4a atprmt4b are similar to those of late-flowering mutants in the autonomous pathway in which FLC expression is up-regulated. Accordingly, expression of FLC was examined in the wild-type plants, atprmt4a-2, atprmt4b-1, and atprmt4a atprmt4b. Consistent with our genetic observations, the mRNA level of FLC was increased in atprmt4a atprmt4b, and the expression of SOC1, which was down-regulated by FLC, was lowered by the loss of AtPRMT4a and AtPRMT4b (Fig. 6B). Furthermore, the null FLC mutant flc-3 could largely suppress the late-flowering phenotype of atprmt4a atprmt4b, although atprmt4a atprmt4b flc-3 triple mutants did not flower as early as flc-3 mutants, indicating that other flowering repressors in addition to FLC may be involved (see below; Fig. 6C). Therefore, we concluded that AtPRMT4a and AtPRMT4b redundantly control the floral transition predominantly through negatively regulating FLC transcription.
Besides FLC, there are five FLC homologs in Arabidopsis: MADS-AFFECTING FLOWERING1 (MAF1)/FLOWERING LOCUS M (FLM) to MAF5 (Scortecci et al., 2001; Ratcliffe et al., 2003). MAF2 functions as a floral repressor that prevents vernalization in response to short cold periods (Ratcliffe et al., 2003). Previously, we and others showed that MAF4 and MAF5 were elevated in both mutants of histone acetyltransferase AtHAC1 and SET-DOMAIN GROUP26 plants (Deng et al., 2007; Xu et al., 2008). Therefore, we also tested the mRNA levels of MAF1 to MAF5 by real-time PCR. The expression levels of MAF4 and MAF5 were up-regulated in atprmt4a atprmt4b, whereas MAF1 and MAF3 were unaffected. Unexpectedly, the mRNA of MAF2 was decreased by AtPRMT4a and AtPRMT4b mutations (Fig. 6D).
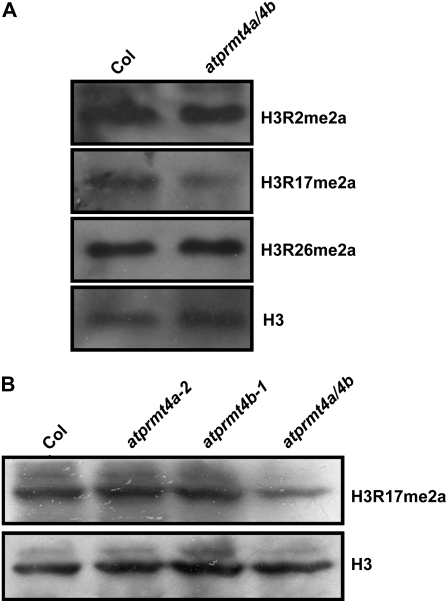
The Global Levels of Asymmetrically Dimethylated H3R17 Were Decreased in atprmt4a atprmt4b Double Mutants
As AtPRMT4a and AtPRMT4b could dimethylate histone H3 asymmetrically at Arg-2, Arg-17, and Arg-26 in vitro, we were interested in whether the AtPRMT4a and AtPRMT4b mutations affect the global levels of H3 asymmetrical dimethylation at Arg-2, Arg-17, and Arg-26. Accordingly, total histones from 12-d-old seedlings were probed with antibodies against H3R2me2a, H3R17me2a, and H3R26me2a. The global levels of H3R17me2a in atprmt4a atprmt4b were decreased compared with those in wild-type plants (Fig. 7A), whereas H3R2me2a and H3R26me2a were roughly the same as in the wild type. In addition, in the atprmt4a and atprmt4b single mutants, as shown in Figure 7B, the levels of H3R17me2a were not affected. These results, consistent with data from the genetic analysis, further demonstrated the redundancy of AtPRMT4a and AtPRMT4b, and both methyltransferases may be the major methyltransferases responsible for H3R17me2a in Arabidopsis. The global levels of H3R17me2a determined by the immunological methods employed, however, were not completely abolished in the atprmt4a atprmt4b double mutants. Other AtPRMTs may also be responsible for the methylation at this site, since we have observed increased H3R17me2a levels when histone H3 was incubated with AtPRMT1b and AtPRMT10 in histone methylation assays (data not shown).
Figure 7.
Global asymmetric dimethylation of H3R17 was decreased in atprmt4a atprmt4b double mutants. A, Equal amounts of total histones extracted from young seedlings of Col-0 and atprmt4a atprmt4b with four to five rosette leaves under LD were separated by 15% SDS-PAGE, transferred onto polyvinylidene difluoride membranes, and immunoblotted with the indicated antibodies. One was immunoblotted with H3R2me2a and H3R26me2a antibodies subsequently; the other was immunoblotted with H3R17me2a and H3 subsequently. B, Total histones extracted from young seedlings of Col-0 and atprmt4a atprmt4b were immunoblotted with anti-H3R17me2a antibody, and the membrane was subsequently stripped and immunoblotted with anti-H3 antibody to demonstrate equal loading. atprmt4a/4b indicates atprmt4a atprmt4b.
The Expression Levels of the Known FLC Regulators Were Not Changed in atprmt4a atprmt4b Double Mutants
In mammals, methylation of histone H3R17 by CARM1/PRMT4 was shown to play an important role in transcriptional activation (Bauer et al., 2002). Therefore, H3R17 asymmetric dimethylation in plants may also promote transcription. As a result, the elevated FLC expression in atprmt4a atprmt4b double mutants could be due to down-regulation of the autonomous pathway components. To test this possibility, the expression levels of eight FLC repressors of the autonomous pathway were examined in atprmt4a atprmt4b: FCA, FY, FPA, FLK, FVE, FLD, LUMINIDEPENDENS (LD), and REF6. Real-time PCR results showed that lesions in both AtPTMT4a and AtPRMT4b did not change the expression of any of these eight genes (data not shown), indicating that decreased levels of H3R17me2a in atprmt4a atprmt4b did not affect the transcription of these FLC repressors.
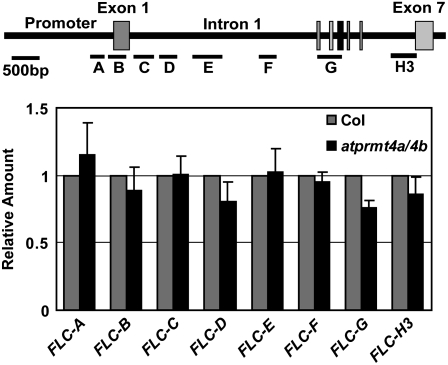
Many autonomous pathway components regulate FLC expression by chromatin modifications (He and Amasino, 2005). Therefore, we performed chromatin immunoprecipitation (ChIP) assays to compare the chromatin modification status of H3R17me2a between wild-type Col-0 and atprmt4a atprmt4b double mutants. Our results showed that histone H3R17me2a of FLC chromatin was not decreased in atprmt4a atprmt4b double mutants, indicating that AtPRMT4a and AtPRMT4b did not directly target FLC (Fig. 8). Moreover, no significant changes of H3K4me2, H3K4me3, and H3ac modifications at FLC chromatin were detected between wild-type Col-0 and atprmt4a atprmt4b double mutants (data not shown). Taken together, AtPRMT4a and AtPRMT4b affect flowering time mainly through FLC, but they might not directly act through chromatin modification of the FLC locus or known autonomous pathway genes at the transcriptional level.
Figure 8.
Asymmetric dimethylation of H3 at Arg-17 of FLC chromatin was similar in wild-type and atprmt4a atprmt4b double mutant plants. ChIP was performed with 5-d-old seedlings using antibody against H3R17me2a. Representative real-time PCR results are shown. FLC-A to FLC-H3 represent different regions of FLC chromatin as described previously (Bastow et al., 2004; Liu et al., 2007). Actin was used as an internal control. atprmt4a/4b indicates atprmt4a atprmt4b. Error bars represent sd.
DISCUSSION
In this study, we identified and characterized two homologs of the human protein Arg methyltransferase CARM1 from Arabidopsis, AtPRMT4a and AtPRMT4b. Both methyltransferases shared not only structural homology but also functional similarities with respect to enzyme activities, subcellular localization, and expression patterns. The presence of a dimerization arm suggested that AtPRMT4a and AtPRMT4b might form homodimers or heterodimers, and this was experimentally verified by GST pull-down and BiFC assays. Consistent with these results, genetic analysis showed that a mutation in AtPRMT4a or AtPRMT4b alone did not cause severe developmental defects, whereas atprmt4a atprmt4b double mutants, in which AtPRMT4 genes were mutated simultaneously, showed a delayed-flowering phenotype. Furthermore, global levels of H3R17me2a were greatly reduced in the double mutants but unchanged in the atprmt4a and atprmt4b single mutants compared with the wild type, demonstrating the requirement as well as the redundancy of AtPRMT4a and AtPRMT4b for methyltransferase activity.
Stringent control of the floral transition is important for maximum reproductive success in plants. Various external and internal cues are involved in this process and are mainly categorized into photoperiod, vernalization, GA, and autonomous pathways. We showed that atprmt4a atprmt4b double mutants flower late under both LD and SD conditions; vernalization and GA treatments rescue the late-flowering phenotype of the double mutants, suggesting that delayed flowering was independent of the photoperiod, vernalization, and GA pathways. Furthermore, the FLC mRNA levels were up-regulated in the atprmt4a atprmt4b double mutants, and atprmt4a atprmt4b flc-3 triple mutants flowered earlier than atprmt4a atprmt4b double mutants but similarly to flc-3, a null allele mutation of FLC. Therefore, in addition to AtPRMT5 and AtPRMT10, which promote flowering by repressing FLC in a nonredundant manner (Niu et al., 2007; Pei et al., 2007; Wang et al., 2007; Schmitz et al., 2008), we identified AtPRMT4a and AtPRMT4b as new components in the autonomous pathway, which cooperatively are required for proper flowering time control in Arabidopsis.
Histone modification is believed to be a conserved mechanism in eukaryotes (Fuchs et al., 2006). In mammalian cells, H3R17me2a is generally a mark of transcriptional activity (Bauer et al., 2002). Therefore, asymmetric H3R17 dimethylation in plants may also be such a mark. In this regard, the increased FLC expression in atprmt4a atprmt4b double mutants might have been due to down-regulation of the autonomous pathway repressors. However, in the atprmt4a atprmt4b double mutants, in which global levels of H3R17me2a were greatly reduced, the expression of these repressors did not appear to be affected, as shown by the real-time PCR results, suggesting that other unknown repressors might exist as the direct targets of AtPRMT4a and AtPRMT4b. Previous studies have demonstrated that chromatin modifications are associated with flowering time control (He and Amasino, 2005). If AtPRMT4a and AtPRMT4b modify FLC chromatin directly, it would be expected to activate FLC expression in the wild-type plants. However, loss of function of AtPRMT4a and AtPRMT4b caused elevated levels of FLC mRNA, indicating that AtPRMT4a and AtPRMT4b affect flowering time by repressing FLC. Moreover, our ChIP results of the FLC locus indicated that there were no significant differences in the levels of H3R17me2a, H3K4me2, H3K4me3, and H3ac between the atprmt4a atprmt4b double mutants and the wild-type plants. These results indicate that FLC chromatin was not the direct target of AtPRMT4a and AtPRMT4b and that they might function beyond the transcriptional level.
In support of this possibility, convincing evidence has emerged from studies with mammalian CARM1. In addition to histone H3, CARM1 could methylate nonhistone proteins, such as transcription coactivators, RNA-binding proteins, and splicing factors. For instance, methylation of the KIX domain in CBP/p300 by CARM1 interferes with the binding of CBP/p300 to CREB (for cAMP-responsive element binding protein), which ultimately blocks CREB activation (Xu et al., 2001). In this case, CARM1 functions as a repressor affecting protein-protein interactions in the cAMP signaling pathway. In the case of PC12 cells, CARM1-directed methylation of the RNA-binding protein HuD regulates the proliferation of PC12 cells by affecting the degradation of p21cip1/waf1 mRNA (Fujiwara et al., 2006). Recently, the splicing factor CA150, which couples transcription to splicing, was identified as the substrate for CARM1, and methylation of CA150 abrogates its interaction with the spinal muscular atrophy protein SMN and its function in alternative splicing (Cheng et al., 2007). Besides Arg methylation, in high-throughput peptide arrays, Rathert et al. (2008) has identified several nonhistone substrates of G9a, which was previously regarded as a histone H3K9-specific methyltransferase. All of these findings now reshape our thinking that Arg methylation and Lys methylation extend to nonhistone proteins involved in various biological processes. In Arabidopsis, AtPRMT4a and AtPRMT4b share conserved methyltransferase activities with their counterpart CARM1 (Fig. 2, A and B). Therefore, it is possible that AtPRMT4a and AtPRMT4b might down-regulate FLC expression by methylating nonhistone targets. In the autonomous pathway, the RNA-binding proteins FCA, FPA, and FLK are good candidates for AtPRMT4a and AtPRMT4b because they all contain the RNA recognition motifs. These RNA-binding proteins could be potential substrates for AtPRMT4a and AtPRMT4b, and their abilities in mRNA binding, protein-protein interaction, and their stability may be affected by Arg methylation, which will ultimately affect the FLC expression.
MATERIALS AND METHODS
Plant Materials
Experiments were performed using Arabidopsis (Arabidopsis thaliana) accession Col-0. Mutant lines from the SALK collection were supplied by the Arabidopsis Biological Resource Center (Ohio State University). atprmt4a-1 (SALK_030782), atprmt4a-2 (SALK_033423), atprmt4b-1 (SALK_097442), and atprmt4b-2 (SAIL_62_B03) mutants were screened according to http://signal.salk.edu/tdnaprimers.html with the following primer sets: CX119 (5′-GAGTTGCCTGAGAAGGCCGAT-3′), CX120 (5′-TAAGGAAACACCGTGGAACCG-3′), and CX101 (5′-GCGTGGACCGCTTGCTGCAACT-3′) were for atprmt4a-1; CX121 (5′-CGTCCCTGTTTAGTGAATGGCA-3′), CX122 (5′-GCTGAGCGTTTGCGTTTTGTT-3′), and CX101 were for atprmt4a-2; CX123 (5′-ACCTTGGTGCGCTGATCCATA-3′), CX124 (5′-TGCCAAACATGTGTATGCGGT-3′), and CX101 were for atprmt4b-1; and CX584 (5′-CGGGATCCATTCTTATTTATTGAGATGGCA-3′), CX359 (5′-CGCGGATCCCAATTATCATCTCTCACCTTCA-3′), and CX1778 (5′-TAGCATCTGAATTTCATAACCAATCTCGATACAC-3′) were for atprmt4b-2. atprmt4a atprmt4b double mutants were generated and screened based on molecular markers.
Plant Growth Conditions, Flowering Time, and Plant Transformation
Plant growth, flowering time analysis, and plant transformation were performed as reported previously (Niu et al., 2007; Pei et al., 2007). Total numbers of rosette and cauline leaves were counted as measurement of flowering time. At least 15 plants were measured and averaged for each measurement.
DNA Constructs
Construction of AtPRMT4a and AtPRMT4b Prokaryotic Expression Vectors
GST-AtPRMT4a and GST-AtPRMT4b fusion constructs were made by conventional molecular cloning methods. Briefly, total RNAs were isolated from Arabidopsis inflorescence using Trizol reagent (Invitrogen) and RNAs were reverse transcribed using SuperScript II reverse transcriptase (Invitrogen) or Moloney murine leukemia virus reverse transcriptase (Invitrogen). RT products were used to perform PCR using the following primer pairs: CX137 (5′-catgccATGGAGATTCCTTCTCTGAATA-3′; NcoI site underlined) and CX138 (5′-catgccatggTAGTTCTAGTTCTAGAGCTGAG-3′; NcoI site underlined) were used for AtPRMT4a cDNA corresponding to 91 to 1,688 bp of NM_124279.4 (amino acids 1–528); PCR products were digested with NcoI and subsequently cloned into modified pGEX-4T-2-TEV vector (XF153) at the NcoI site, resulting in pGEX-AtPRMT4a (XF146). Similarly, CX358 (5′-cgcggatccATGGAGGTATCTTCTGTGAAAA-3′; BamHI site underlined) and CX359 (5′-cgcggatccCAATTATCATCTCTCACCTTCA-3′; BamHI site underlined) were used for AtPRMT4b cDNA corresponding to 92 to 1,734 bp of NM_111573.4 (amino acids 1–534). PCR products were digested with BamHI and subsequently cloned into modified pGEX-4T-2 vector at the BamHI site, resulting in pGEX-AtPRMT4b (XF277). All inserts were confirmed by DNA sequencing.
Construction of AtPRMT4 Promoter∷GUS
Genomic DNA fragments containing 1,066 and 1,082 bp before the start codon of AtPRMT4a and AtPRMT4b genomic sequences were cloned into the PstI-NcoI-digested pCAMBIA1391Z-GUS vector. Primers used for the AtPRMT4a promoter were CX426 (5′-aaaactgcagCTCAAAATGTAATTAGAAATGTTGT-3′; PstI site underlined) and CX427 (5′-catgccatggTGTTTTGAGAATACAGTATGTGT-3′; NcoI site underlined). Primers used for the AtPRMT4b promoter were CX422 (5′-aaaactgcAGAAAGTGAACCACACCATTGGG-3′; PstI site underlined) and CX423 (5′-catgccatggTTTTACAAAACGATGCGAATGTA-3′; NcoI site underlined).
Construction of GFP-AtPRMT4a and GFP-AtPRMT4b Vectors
AtPRMT4a full-length cDNA was digested from pGEX-AtPRMT4a (XF146) with NcoI and was subcloned into pAVA321 at BglII and XbaI sites using blunt ligation, thereby creating an intermediate construct (XF198) with an N-terminal translational fusion of GFP to AtPRMT4a driven by the cauliflower mosaic virus 35S promoter. A plant transformation construct was produced by ligating the HindIII-SacI fragment of the intermediate vector (XF198) into the HindIII and SacI sites of pCAMBIA1300 (XF199).
AtPRMT4b full-length cDNA was digested from pGEX-AtPRMT4b (XF277) with BamHI and subcloned into the BglII site after the GFP sequence of pAVA321 (XF275), then the fragment was digested with PstI and inserted into the PstI-digested site of pCAMBIA1300 to generate the plant transformation vector (XF276).
Histochemical GUS Staining
For histochemical GUS staining, homozygous transgenic plants containing AtPRMT4a Promoter∷GUS and AtPRMT4b Promoter∷GUS constructs were grown under LD. Histochemical GUS staining was performed by incubating whole plants or different tissues in GUS staining buffer [100 mm NaPO4 with 0.1% Triton X-100, 0.1 m EDTA, 1 mg mL−1 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, 0.05 mm K3(Fe[CN]6), and 0.05 mm K4(Fe[CN]6)] for 12 to 16 h followed by clearing with 70% ethanol and 30% acetic acid.
Protein Arg Methyltransferase Assay
Recombinant proteins were expressed in Escherichia coli strain BL21 (RIL or Rosseta), and methyltransferase activity assays were performed as described (Niu et al., 2007; Pei et al., 2007).
Western Blotting
Histones were extracted from 12-d-old seedlings as described previously (Yan et al., 2007). Histones were separated by 15% SDS-PAGE and then transferred onto an Immobilon P SQ polyvinylidene difluoride membrane (Millipore). The membrane was sequentially probed with anti-asymmetrically dimethylated H3R2 (αH3R2me2a; Abcam; ab8046), anti-asymmetrically dimethylated H3R17 (αH3R17me2a; Abcam; ab8284), anti-asymmetrically dimethylated H3R26 (αH3R26me2a; Upstate; 07-215), and anti-histone H3 (Upstate; 06–755) antibodies. Supersignal West Dura substrate (Pierce) was used to detect the horseradish peroxidase-conjugated secondary antibodies.
GST Pull-Down Assays of AtPRMT4a and AtPTRMT4b Interaction
In vitro translation was performed as described in Promega Technical Manual Number 045. AtPRMT4a and AtPRMT4b proteins were synthesized from plasmid templates and radiolabeled with [35S]Met using the T7 TNT coupled transcription and translation system (Promega). GST fusion proteins of AtPRMT4a and AtPRMT4b were immobilized onto glutathione-Sepharose 4B beads (Amersham Biosciences) and incubated with the translated products at 4°C for 2 h with constant gentle mixing. The mixture was centrifuged, and the pellet was extensively washed with phosphate-buffered saline containing 140 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, 0.5 mm dithiothreitol, 0.1 mm leupeptinin, 1 μg mL−1 aprotinin, and 0.1% Nonidet P-40. After the final wash, the pellet was resuspended in 1× SDS loading buffer and separated by 10% SDS-PAGE. The gel was treated with Amplifier (Amersham Biosciences) and dried. The radioactivity of the labeled proteins was visualized by fluorography.
BiFC Analysis
The full-length coding regions of AtPRMT4a and AtPRMT4b were PCR amplified from plasmid templates containing the corresponding cDNAs using specific primers as follows: CX1339 (5′-ggactagtATGGAGATTCCTTCTCTGAATAAGCAGCAAG-3′; SpeI site underlined) and CX1340 (5′-acgcgtcgacGAGCTGAGCGTTTGCGTTTTG-3′; SalI site underlined) for AtPRMT4a; CX0358 (5′-cgcggatccATGGAGGTATCTTCTGTGAAAA-3′; BamHI site underlined) and CX1341 (5′-ccgctcgagGAGCTGGGCACTTGGGTTC-3′; XhoI site underlined) for AtPRMT4b. The PCR product of AtPRMT4a was then inserted into SpeI/SalI restriction sites of pSPYNE-35S, and AtPRMT4b was inserted into BamHI/XhoI restriction sites of the pSPYCE-35S vector to generate the expression vectors of pSPYNE-35S-AtPRMT4a (AtPRMT4a-YFPN, 4aNE, XF0613) and pSPYCE-35S-AtPRMT4b (AtPRMT4b-YFPC, 4bCE, XF0618). These constructions were subsequently verified by DNA sequencing. Onion (Allium cepa) epidermal layers were placed on petri dishes containing half-strength MS medium (2.15 g L−1 Murashige and Skoog salt and vitamins [PhytoTechnology Laboratories], pH 5.8). The pSPYNE-35S (YFPN, NE) or pSPYCE-35S (YFPC, CE) empty vectors and the AtPRMT4a-YFPN and AtPRMT4b-YFPC expression vectors were precipitated onto gold particles (Bio-Rad). After being washed and resuspended in 100% ethanol, the gold-coated particles were transfected in different combinations into onion epidermal layers using the model PDS-1000/He Biolistic Particle Delivery System (Bio-Rad). The transfected onion epidermal layers were incubated at 23°C for 24 h in the dark before being applied to confocal fluorescence microscopy (Zeiss).
ChIP
Five-day-old seedlings were harvested from Murashige and Skoog plates containing 3% Suc grown under LD at 23°C. ChIP assay was performed as described previously (Johnson et al., 2002; Gendrel et al., 2005; Pei et al., 2007). Immunoprecipitations were performed with anti-H3R17me2a (Abcam; ab8284), anti-H3ac (Upstate; 06-599), anti-H3K4me2 (Upstate; 07-030), and anti-H3K4me3 (Upstate; 07-473) antibodies. PCR analysis was performed using specific primers corresponding to different regions of FLC chromatin as described previously (Bastow et al., 2004; Liu et al., 2007). Actin was used as an internal control.
Analysis of Transcription Levels
Total RNAs were isolated from seedlings with four to five visible rosette leaves (12 d) grown on Murashige and Skoog salt and vitamin (PhytoTechnology Laboratories) plates with 3% Suc using the Trizol reagent (Invitrogen) according to the manufacturer's instructions. The first-strand DNA was reverse transcribed using SuperScript II (Invitrogen) or Moloney murine leukemia virus (Invitrogen) reverse transcriptase followed by semiquantitative or quantitative PCR using hot-start ExTaq polymerase (TaKaRa). Real-time PCR was performed using the Chromo4 real-time PCR instrument (MJ) and SYBR Green I (Invitrogen; S-7567). Primers were as follows: CX137 and CX138 for AtPRMT4a; CX137 and CX1190 (5′-TCCTCAATTTTTCCCTTGATGACTGTG-3′) for AtPRMT4aN; CX358 and CX359 for AtPRMT4b; CX358 and CX1220 (5′-ATGTTTGGCACCAGCCTGGGCCG-3′) for AtPRMT4bN; and CX415 (5′-CTCAGCACCTTCCAACAGATGTGGA-3′) and CX416 (5′-CCAAAAAAATGAACCAAGGACCAAA-3′) for Actin2/7 (NM_121018), the constitutive expression control. Primers used for FLC repressors were described previously (Niu et al., 2007; Pei et al., 2007). Primers used for AtPRMT4a and AtPRMT4b expression patterns were as follows: CX119 and CX122 for AtPRMT4a; CX584 (5′-cgggatccATTCTTATTTATTGAGATGGCA-3′) and CX359 for AtPRMT4b. For total RNA-blot analyses, 20 μg of total RNAs of each sample was separated on a 1% denaturing agarose/formaldehyde gel and transferred onto N+ nylon membrane (Hybond-N+; Amersham-Pharmacia). The primers for the FLC and SOC1 probes were described previously (Niu et al., 2007; Pei et al., 2007), with CX415 and CX416 used for Actin2/7.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NP_199713 for AtPRMT4a and NP_187349 for AtPRMT4b.
Acknowledgments
We thank Aiying Zhang and Qingbao Zhu for technical support and Falong Lu, Ayaz Ahmad, Xian Deng, and Xia Cui in X.C.'s laboratory for comments on the manuscript. We also thank Dr. Dongqiao Shi from Dr. Weicai Yang's laboratory and Dr. Xia Wang at the Institute of Genetics and Developmental Biology for help with confocal microscopy. We thank Dr. R. Amasino for providing flc-3 and fld-4 seeds. We also thank the Arabidopsis Biological Resource Center at The Ohio State University for providing SALK T-DNA insertion lines.
This work was supported by the National Basic Research Program of China (grant nos. 2007CB948202 and 2005CB522400), the National Natural Science Foundation of China (grant nos. 30430410 and 30621001 to X.C.), and the Chinese Academy of Sciences (grant nos. CXTD–S2005–2 and KSCX2–YW–N–047 to X.C.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Xiaofeng Cao (xfcao@genetics.ac.cn).
Some figures in this article are displayed in color online but in black and white in the print edition.
Open Access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Amasino RM (2005) Vernalization and flowering time. Curr Opin Biotechnol 16 154–158 [DOI] [PubMed] [Google Scholar]
- An W, Kim J, Roeder RG (2004) Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117 735–748 [DOI] [PubMed] [Google Scholar]
- Ananthanarayanan M, Li S, Balasubramaniyan N, Suchy FJ, Walsh MJ (2004) Ligand-dependent activation of the farnesoid X-receptor directs arginine methylation of histone H3 by CARM1. J Biol Chem 279 54348–54357 [DOI] [PubMed] [Google Scholar]
- Ausin I, Alonso-Blanco C, Jarillo JA, Ruiz-Garcia L, Martinez-Zapater JM (2004) Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat Genet 36 162–166 [DOI] [PubMed] [Google Scholar]
- Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C (2004) Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427 164–167 [DOI] [PubMed] [Google Scholar]
- Bauer UM, Daujat S, Nielsen SJ, Nightingale K, Kouzarides T (2002) Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Rep 3 39–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, Richard S (2005) Arginine methylation an emerging regulator of protein function. Mol Cell 18 263–272 [DOI] [PubMed] [Google Scholar]
- Cakouros D, Daish TJ, Mills K, Kumar S (2004) An arginine-histone methyltransferase, CARMER, coordinates ecdysone-mediated apoptosis in Drosophila cells. J Biol Chem 279 18467–18471 [DOI] [PubMed] [Google Scholar]
- Chen D, Huang SM, Stallcup MR (2000) Synergistic, p160 coactivator-dependent enhancement of estrogen receptor function by CARM1 and p300. J Biol Chem 275 40810–40816 [DOI] [PubMed] [Google Scholar]
- Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR (1999) Regulation of transcription by a protein methyltransferase. Science 284 2174–2177 [DOI] [PubMed] [Google Scholar]
- Chen SL, Loffler KA, Chen D, Stallcup MR, Muscat GE (2002) The coactivator-associated arginine methyltransferase is necessary for muscle differentiation: CARM1 coactivates myocyte enhancer factor-2. J Biol Chem 277 4324–4333 [DOI] [PubMed] [Google Scholar]
- Cheng D, Cote J, Shaaban S, Bedford MT (2007) The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol Cell 25 71–83 [DOI] [PubMed] [Google Scholar]
- Covic M, Hassa PO, Saccani S, Buerki C, Meier NI, Lombardi C, Imhof R, Bedford MT, Natoli G, Hottiger MO (2005) Arginine methyltransferase CARM1 is a promoter-specific regulator of NF-kappaB-dependent gene expression. EMBO J 24 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daujat S, Bauer UM, Shah V, Turner B, Berger S, Kouzarides T (2002) Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Curr Biol 12 2090–2097 [DOI] [PubMed] [Google Scholar]
- Deng W, Liu C, Pei Y, Deng X, Niu L, Cao X (2007) Involvement of the histone acetyltransferase AtHAC1 in the regulation of flowering time via repression of FLOWERING LOCUS C in Arabidopsis. Plant Physiol 143 1660–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Messaoudi S, Fabbrizio E, Rodriguez C, Chuchana P, Fauquier L, Cheng D, Theillet C, Vandel L, Bedford MT, Sardet C (2006) Coactivator-associated arginine methyltransferase 1 (CARM1) is a positive regulator of the cyclin E1 gene. Proc Natl Acad Sci USA 103 13351–13356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Yi P, Wong J, O'Malley BW (2006) Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol Cell Biol 26 7846–7857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel A, Yadav N, Lee J, Branscombe TL, Clarke S, Bedford MT (2002) The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J Biol Chem 277 3537–3543 [DOI] [PubMed] [Google Scholar]
- Fuchs J, Demidov D, Houben A, Schubert I (2006) Chromosomal histone modification patterns: from conservation to diversity. Trends Plant Sci 11 199–208 [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Mori Y, Chu DL, Koyama Y, Miyata S, Tanaka H, Yachi K, Kubo T, Yoshikawa H, Tohyama M (2006) CARM1 regulates proliferation of PC12 cells by methylating HuD. Mol Cell Biol 26 2273–2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel AV, Lippman Z, Martienssen R, Colot V (2005) Profiling histone modification patterns in plants using genomic tiling microarrays. Nat Methods 2 213–218 [DOI] [PubMed] [Google Scholar]
- Han SK, Song JD, Noh YS, Noh B (2007) Role of plant CBP/p300-like genes in the regulation of flowering time. Plant J 49 103–114 [DOI] [PubMed] [Google Scholar]
- He Y, Amasino RM (2005) Role of chromatin modification in flowering-time control. Trends Plant Sci 10 30–35 [DOI] [PubMed] [Google Scholar]
- He Y, Michaels SD, Amasino RM (2003) Regulation of flowering time by histone acetylation in Arabidopsis. Science 302 1751–1754 [DOI] [PubMed] [Google Scholar]
- Jiang D, Yang W, He Y, Amasino RM (2007) Arabidopsis relatives of the human lysine-specific Demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell 19 2975–2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L, Cao X, Jacobsen S (2002) Interplay between two epigenetic marks: DNA methylation and histone H3 lysine 9 methylation. Curr Biol 12 1360–1367 [DOI] [PubMed] [Google Scholar]
- Kenzior AL, Folk WR (1998) AtMSI4 and RbAp48 WD-40 repeat proteins bind metal ions. FEBS Lett 440 425–429 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Hyun Y, Park JY, Park MJ, Park MK, Kim MD, Lee MH, Moon J, Lee I, Kim J (2004) A genetic link between cold responses and flowering time through FVE in Arabidopsis thaliana. Nat Genet 36 167–171 [DOI] [PubMed] [Google Scholar]
- Kim J, Lee J, Yadav N, Wu Q, Carter C, Richard S, Richie E, Bedford MT (2004) Loss of CARM1 results in hypomethylation of thymocyte cyclic AMP-regulated phosphoprotein and deregulated early T cell development. J Biol Chem 279 25339–25344 [DOI] [PubMed] [Google Scholar]
- Koh SS, Chen D, Lee YH, Stallcup MR (2001) Synergistic enhancement of nuclear receptor function by p160 coactivators and two coactivators with protein methyltransferase activities. J Biol Chem 276 1089–1098 [DOI] [PubMed] [Google Scholar]
- Koh SS, Li H, Lee YH, Widelitz RB, Chuong CM, Stallcup MR (2002) Synergistic coactivator function by coactivator-associated arginine methyltransferase (CARM) 1 and beta-catenin with two different classes of DNA-binding transcriptional activators. J Biol Chem 277 26031–26035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeda Y (2004) Genetic regulation of time to flower in Arabidopsis thaliana. Annu Rev Plant Biol 55 521–535 [DOI] [PubMed] [Google Scholar]
- Krones-Herzig A, Mesaros A, Metzger D, Ziegler A, Lemke U, Bruning JC, Herzig S (2006) Signal-dependent control of gluconeogenic key enzyme genes through coactivator-associated arginine methyltransferase 1. J Biol Chem 281 3025–3029 [DOI] [PubMed] [Google Scholar]
- Lee DY, Teyssier C, Strahl BD, Stallcup MR (2005) Role of protein methylation in regulation of transcription. Endocr Rev 26 147–170 [DOI] [PubMed] [Google Scholar]
- Lee J, Bedford MT (2002) PABP1 identified as an arginine methyltransferase substrate using high-density protein arrays. EMBO Rep 3 268–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Sayegh J, Daniel J, Clarke S, Bedford MT (2005) PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. J Biol Chem 280 32890–32896 [DOI] [PubMed] [Google Scholar]
- Lee YH, Coonrod SA, Kraus WL, Jelinek MA, Stallcup MR (2005) Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc Natl Acad Sci USA 102 3611–3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim MH, Kim J, Kim YS, Chung KS, Seo YH, Lee I, Hong CB, Kim HJ, Park CM (2004) A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS C. Plant Cell 16 731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Quesada V, Crevillen P, Baurle I, Swiezewski S, Dean C (2007) The Arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Mol Cell 28 398–407 [DOI] [PubMed] [Google Scholar]
- Lu F, Li G, Cui X, Liu C, Wang XJ, Cao X (2008) Comparative analysis of JmjC domain-containing proteins in higher plants and human reveals potential histone demethylases in Arabidopsis and rice. J Integr Plant Biol 50 886–896 [DOI] [PubMed] [Google Scholar]
- Ma H, Baumann CT, Li H, Strahl BD, Rice R, Jelinek MA, Aswad DW, Allis CD, Hager GL, Stallcup MR (2001) Hormone-dependent, CARM1-directed, arginine-specific methylation of histone H3 on a steroid-regulated promoter. Curr Biol 11 1981–1985 [DOI] [PubMed] [Google Scholar]
- Macknight R, Bancroft I, Page T, Lister C, Schmidt R, Love K, Westphal L, Murphy G, Sherson S, Cobbett C, et al (1997) FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell 89 737–745 [DOI] [PubMed] [Google Scholar]
- Majumder S, Liu Y, Ford OH III, Mohler JL, Whang YE (2006) Involvement of arginine methyltransferase CARM1 in androgen receptor function and prostate cancer cell viability. Prostate 66 1292–1301 [DOI] [PubMed] [Google Scholar]
- Matsuda H, Paul BD, Choi CY, Shi YB (2007) Contrasting effects of two alternative splicing forms of coactivator-associated arginine methyltransferase 1 on thyroid hormone receptor-mediated transcription in Xenopus laevis. Mol Endocrinol 21 1082–1094 [DOI] [PubMed] [Google Scholar]
- Miao F, Li S, Chavez V, Lanting L, Natarajan R (2006) Coactivator-associated arginine methyltransferase-1 enhances nuclear factor-kappaB-mediated gene transcription through methylation of histone H3 at arginine 17. Mol Endocrinol 20 1562–1573 [DOI] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (2001) Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13 935–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler TC, Yu X, Shalitin D, Parikh D, Michael TP, Liou J, Huang J, Smith Z, Alonso JM, Ecker JR, et al (2004) Regulation of flowering time in Arabidopsis by K homology domain proteins. Proc Natl Acad Sci USA 101 12759–12764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouradov A, Cremer F, Coupland G (2002) Control of flowering time: interacting pathways as a basis for diversity. Plant Cell (Suppl) 14 S111–S130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeem H, Cheng D, Zhao Q, Underhill C, Tini M, Bedford MT, Torchia J (2007) The activity and stability of the transcriptional coactivator p/CIP/SRC-3 are regulated by CARM1-dependent methylation. Mol Cell Biol 27 120–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu L, Lu F, Pei Y, Liu C, Cao X (2007) Regulation of flowering time by the protein arginine methyltransferase AtPRMT10. EMBO Rep 8 1190–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh B, Lee SH, Kim HJ, Yi G, Shin EA, Lee M, Jung KJ, Doyle MR, Amasino RM, Noh YS (2004) Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 16 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Y, Niu L, Lu F, Liu C, Zhai J, Kong X, Cao X (2007) Mutations in the type II protein arginine methyltransferase AtPRMT5 result in pleiotropic developmental defects in Arabidopsis. Plant Physiol 144 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe OJ, Kumimoto RW, Wong BJ, Riechmann JL (2003) Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15 1159–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathert P, Dhayalan A, Murakami M, Zhang X, Tamas R, Jurkowska R, Komatsu Y, Shinkai Y, Cheng X, Jeltsch A (2008) Protein lysine methyltransferase G9a acts on non-histone targets. Nat Chem Biol 4 344–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves PH, Coupland G (2000) Response of plant development to environment: control of flowering by daylength and temperature. Curr Opin Plant Biol 3 37–42 [DOI] [PubMed] [Google Scholar]
- Schmitz RJ, Sung S, Amasino RM (2008) Histone arginine methylation is required for vernalization-induced epigenetic silencing of FLC in winter-annual Arabidopsis thaliana. Proc Natl Acad Sci USA 105 411–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schomburg FM, Patton DA, Meinke DW, Amasino RM (2001) FPA, a gene involved in floral induction in Arabidopsis, encodes a protein containing RNA-recognition motifs. Plant Cell 13 1427–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurter BT, Koh SS, Chen D, Bunick GJ, Harp JM, Hanson BL, Henschen-Edman A, Mackay DR, Stallcup MR, Aswad DW (2001) Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry 40 5747–5756 [DOI] [PubMed] [Google Scholar]
- Scortecci KC, Michaels SD, Amasino RM (2001) Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J 26 229–236 [DOI] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C (2003) FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113 777–787 [DOI] [PubMed] [Google Scholar]
- Simpson GG, Gendall AR, Dean C (1999) When to switch to flowering. Annu Rev Cell Dev Biol 15 519–550 [DOI] [PubMed] [Google Scholar]
- Stallcup MR, Kim JH, Teyssier C, Lee YH, Ma H, Chen D (2003) The roles of protein-protein interactions and protein methylation in transcriptional activation by nuclear receptors and their coactivators. J Steroid Biochem Mol Biol 85 139–145 [DOI] [PubMed] [Google Scholar]
- Teyssier C, Ou CY, Khetchoumian K, Losson R, Stallcup MR (2006) Transcriptional intermediary factor 1alpha mediates physical interaction and functional synergy between the coactivator-associated arginine methyltransferase 1 and glucocorticoid receptor-interacting protein 1 nuclear receptor coactivators. Mol Endocrinol 20 1276–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Padilla ME, Parfitt DE, Kouzarides T, Zernicka-Goetz M (2007) Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature 445 214–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, Nake C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40 428–438 [DOI] [PubMed] [Google Scholar]
- Wang H, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst P, et al (2001) Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 293 853–857 [DOI] [PubMed] [Google Scholar]
- Wang X, Zhang Y, Ma Q, Zhang Z, Xue Y, Bao S, Chong K (2007) SKB1-mediated symmetric dimethylation of histone H4R3 controls flowering time in Arabidopsis. EMBO J 26 1934–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Zhao Z, Dong A, Soubigou-Taconnat L, Renou JP, Steinmetz A, Shen WH (2008) Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol Cell Biol 28 1348–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Chen H, Du K, Asahara H, Tini M, Emerson BM, Montminy M, Evans RM (2001) A transcriptional switch mediated by cofactor methylation. Science 294 2507–2511 [DOI] [PubMed] [Google Scholar]
- Yadav N, Lee J, Kim J, Shen J, Hu MC, Aldaz CM, Bedford MT (2003) Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc Natl Acad Sci USA 100 6464–6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Zhang Y, Niu L, Yuan Y, Cao X (2007) Identification and characterization of two closely related histone H4 arginine 3 methyltransferases in Arabidopsis thaliana. Biochem J 408 113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhou L, Cheng X (2000) Crystal structure of the conserved core of protein arginine methyltransferase PRMT3. EMBO J 19 3509–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zika E, Fauquier L, Vandel L, Ting JP (2005) Interplay among coactivator-associated arginine methyltransferase 1, CBP, and CIITA in IFN-gamma-inducible MHC-II gene expression. Proc Natl Acad Sci USA 102 16321–16326 [DOI] [PMC free article] [PubMed] [Google Scholar]