Abstract
Manganese (Mn) deficiency is an important plant nutritional disorder in many parts of the world. Barley (Hordeum vulgare) genotypes differ considerably in their ability to grow in soils with low Mn2+ availability. Differential genotypic Mn efficiency can be attributed to differences in Mn2+ uptake kinetics in the low nanomolar concentration range. However, the molecular basis for these differences has not yet been clarified. We present here the identification and characterization of the first barley gene encoding a plasma membrane-localized metal transport protein able to transport Mn2+. The gene is designated HvIRT1 (for IRON-REGULATED TRANSPORTER1) because it belongs to the ZIP gene family and has a high similarity to rice (Oryza sativa) OsIRT1. A novel yeast uptake assay based on inductively coupled plasma-mass spectrometry analysis of 31 different metal and metalloid ions showed that the HvIRT1 protein, in addition to Mn2+, also transported Fe2+/Fe3+, Zn2+, and Cd2+. Both Mn and iron deficiency induced an up-regulation of HvIRT1 in two barley genotypes differing in Mn efficiency, but the expression levels in all cases were highest (up to 40%) in the Mn-efficient genotype. The higher expression of HvIRT1 correlated with an increased Mn2+ uptake rate. We conclude that HvIRT1 is an important component controlling Mn2+ uptake in barley roots and contributes to genotypic differences in Mn2+ uptake kinetics.
Manganese (Mn) is an essential trace element for plants, and Mn is known to have a specific and nonredundant role in three key enzymes. These include (1) oxalate oxidase, which catalyzes the conversion of oxalate and oxygen into hydrogen peroxide and carbon dioxide (Requena and Bornemann, 1999); (2) the Mn-containing superoxide dismutase, located in the mitochondria, which protects the tissue from oxidative stress (Bowler et al., 1991; Alscher et al., 2002); and (3) the oxygen-evolving complex of PSII (Britt, 1996; Clemens et al., 2002; Rutherford and Boussac, 2004). In addition, Mn is an unspecific activator of a number of different enzymes, such as decarboxylases and dehydrogenases in the tricarboxylic acid cycle, Phe ammonia-lyase in the shikimic acid pathway, and in several glycosyltransferases in the Golgi apparatus (Marschner, 1995; Nunan and Scheller, 2003). Mn deficiency causes interveinal chlorosis and reduction in the content of fructans and structural carbohydrates, resulting in slack leaves (Pearson and Rengel, 1997). Consequently, Mn-deficient plants are more susceptible to low-temperature stress and pathogen infections (Marschner, 1995), and crop yields decline significantly under Mn deficiency (Hebbern et al., 2005).
The ability to grow in soils containing low levels of plant-available Mn2+ varies greatly among different plant species and among genotypes within the same species, a phenomenon commonly referred to as differential Mn efficiency (Ascher-Ellis et al., 2001). Large differences have been observed among barley (Hordeum vulgare) genotypes (Graham et al., 1983; Hebbern et al., 2005), but even though Mn efficiency has been investigated for decades, the exact physiological and molecular bases for the tolerance to low Mn2+ availability remain unknown (Graham, 1988; Ascher-Ellis et al., 2001). However, Pedas and co-workers (2005) identified a high-affinity transport system mediating Mn2+ influx at concentrations up to 130 nm and showed that the Mn-efficient genotype Vanessa had a transport capacity (Vmax) almost four times higher than the Mn-inefficient genotype Antonia. The physiological relevance of the observed difference in uptake rates for Mn2+ was demonstrated by online inductively coupled plasma-mass spectrometry (ICP-MS) measurements of Mn2+ depletion showing a severalfold higher net uptake of Mn2+ in Vanessa compared with Antonia. The molecular basis for Mn2+ transport into root cells from the soil solution is still poorly understood (Pittman, 2005).
The first identified plant member of the ZIP family, AtIRT1 (for IRON-REGULATED TRANSPORTER1) from Arabidopsis (Arabidopsis thaliana), transports a wide range of trace elements, including Fe2+/Fe3+, Cd2+, Zn2+, Co2+, and Mn2+ (Korshunova et al., 1999). Using a knockout mutant of AtIRT1, it was shown that this transporter is a key component in regulating iron (Fe) acquisition at low soil Fe availability (Vert et al., 2002). The chlorotic phenotype of the Atirt1 mutants could not be rescued by applying exogenous Mn2+, but the mutants showed a significant reduction in root Mn2+ concentrations relative to the wild type when grown under Fe limitation. This indicates that AtIRT1 functions as a Mn2+ transporter in planta and that it is the primary pathway for Mn2+ transport during Fe deficiency. Several members of the ZIP transporter family from other plant species than Arabidopsis also appear to have the ability to transport Mn2+, including MtZIP4 and MtZIP7 from Medicago truncatula (López-Millán et al., 2004), LeIRT1 and LeIRT2 from tomato (Solanum lycopersicum; Eckhardt et al., 2001), and PsRIT1 from pea (Pisum sativum; Cohen et al., 2004). Some of these transporters are most likely localized in the plasma membrane, but their metal ion specificity remains to be clarified. In rice (Oryza sativa), OsIRT1 transports Fe2+/Fe3+ (Bughio et al., 2002) but has not yet been shown to transport Mn2+, and no other Mn2+ transporter has been reported in monocotyledonous species.
In this study, yeast was used as a tool to isolate and characterize the first Mn2+ transporter from barley. The gene had a high similarity with OsIRT1 and was accordingly named HvIRT1. Heterologous expression indicated that HvIRT1 also had specificity for Fe2+/Fe3+, Zn2+, and Cd2+. Transient expression localized HvIRT1 to the plasma membrane, suggesting a role in metal ion uptake from the soil solution. Expression patterns of HvIRT1 and corresponding Mn2+ uptake rates in intact barley roots were positively correlated. Furthermore, HvIRT1 expression levels in all cases were considerably higher (up to 40%) in the Mn-efficient genotype Vanessa compared with the Mn-inefficient genotype Antonia. This suggests an important role of HvIRT1 in Mn2+ acquisition and in controlling differential Mn efficiency among barley genotypes.
RESULTS
Isolation and Sequence Analysis of a Putative Root Mn2+ Transporter
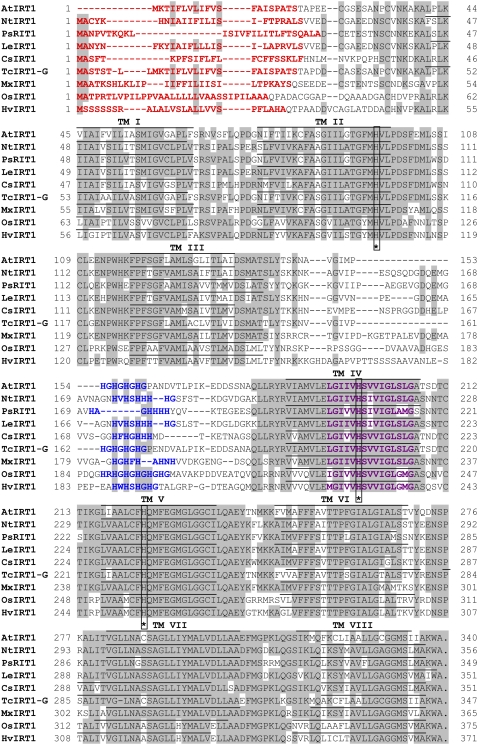
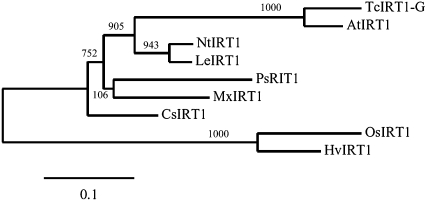
To identify transport proteins with specificity for Mn2+, a yeast screen using the Δsmf1 mutant was employed, containing a null mutation in the gene for high-affinity Mn2+ uptake (Supek et al., 1996). We produced a cDNA library from roots of the barley genotypes Vanessa and Antonia grown at insufficient or ample Mn2+ supply. The library was used for transformation of the Δsmf1 mutant, and primary transformants were plated on medium containing Mn2+, resulting in more than 107 colonies. Approximately 5% of the primary transformants were restreaked on Mn2+-limited medium (controlled by 10 mm EGTA). On the order of 500 colonies were able to complement the Δsmf1 mutant. Among these positive transformants, about 200 were analyzed, and 40% of the clones were found to carry the same barley cDNA clone, whereas the remaining 60% were false positives identified by the absence of growth on Mn2+-limited medium after retransformation, giving a fairly safe screen. The barley cDNA clone had high similarity to the rice IRT1 gene and was consequently named HvIRT1, in accordance with the founding member of the IRT family isolated from Arabidopsis (Eide et al., 1996). After identification from the yeast screening, the HvIRT1 gene was cloned from both the Mn-efficient genotype Vanessa and the Mn-inefficient genotype Antonia, and no differences in the coding gene sequences were seen. Thus, the differential Mn2+ uptake kinetics for the two genotypes observed previously (Pedas et al., 2005) were not caused by different isoforms. The open reading frame of the HvIRT1 gene was 1,113 bp in length, corresponding to a polypeptide of 371 amino acids. The predicted HvIRT1 protein exhibited 69% and 55% amino acid sequence identity to OsIRT1 and AtIRT1, respectively. In agreement with the known structure of other IRT1 family members, the HvIRT1 protein was predicted to contain eight transmembrane domains, a very short C-terminal tail, and a hydrophilic region between transmembrane domains III and IV directed toward the cytosolic side of the membrane (Fig. 1). This region contains the conserved His residues that are anticipated to be involved in the formation of a cytoplasmic metal ion-binding site (Guerinot, 2000). However, the proposed ZIP signature sequence located in transmembrane domain IV (Eng et al., 1998) does not fully match with the three IRT1 protein sequences reported for rice, pea, and barley. Therefore, an extended version of the ZIP signature sequence is presented here and the additions are indicated in boldface: [LIVFAM] [GAS] [LIVMD] [LIVSCG] [LIVFAS] H [SAN] [LIVFA] [LIVFMAT] [LIVDE] G [LIVF] [SANG] [LIFVFM] [GS], where [] refer to one of the amino acids between brackets being possible (Fig. 1). Thus, the sequence analysis of the barley HvIRT1 gene suggests that it encodes a metal ion transport protein belonging to the ZIP family. A phylogenetic tree obtained after comparing HvIRT1 with the sequences of eight other known plant IRT1 members (Fig. 2) revealed that HvIRT1 is most closely related to OsIRT1. Furthermore, the two IRT1 proteins from rice and barley fall into a subgroup separated from the dicotyledonous IRT1 proteins, indicating their evolutionary divergence.
Figure 1.
Comparison of the putative amino acid sequences of HvIRT1 and other plant IRT1 proteins. ClustalW alignment of the IRT1 proteins from various plant species was carried out using T-COFFEE (http://www.ch.embnet.org/software/Tcoffee.html). Conserved residues are shaded in gray. Dashes indicate gaps. Transmembrane domains (TM) were identified using TMHMM (version 2.0; http://www.cbs.dtu.dk/services/TMHMM/) and are shown as lines above the sequences. The potential signal peptides were identified using TargetP (version 1.1; http://www.cbs.dtu.dk/services/TargetP/) and are highlighted in red. The highly conserved motif for ZIP proteins in the fourth transmembrane domain is indicated in violet. The His-rich sequence located in the variable region between transmembrane domains III and IV is highlighted in blue, and fully conserved His residues within the sequences are indicated by asterisks. Species designations are as follows: At, A. thaliana (AtIRT1); Nt, N. tabacum (NtIRT1); Ps, P. sativum (PsRIT1); Le, S. lycopersicum (formerly Lycopersicon esculentum; LeIRT1); Cs, Cucumis sativus (CsIRT1); Tc, T. caerulescens (TcIRT1-G); Mx, Malus xiaojinensis (MxIRT1); Os, O. sativa (OsIRT1); Hv, H. vulgare (HvIRT1).
Figure 2.
Phylogenetic analysis of nine IRT1 protein sequences from various plant species. Alignment of full-length sequences is as described for Figure 1. The phylogenetic tree was drawn with MEGA version 4.0 (http://www.megasoftware.net; Tamura et al., 2007). Numbers represent bootstrap values.
Yeast Functional Complementation
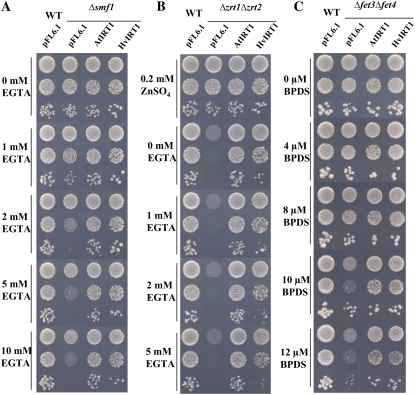
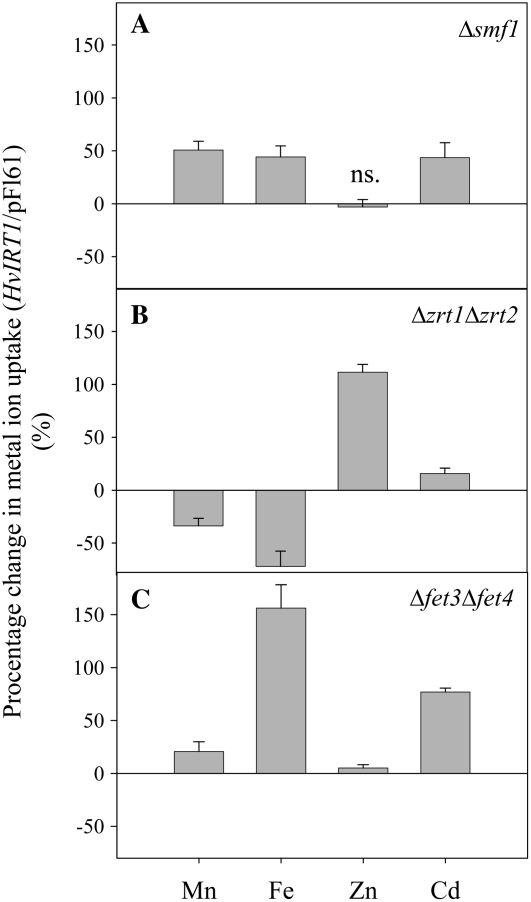
AtIRT1 has been shown previously to be involved in the transport of Fe2+/Fe3+, Zn2+, Mn2+, and Cd2+ (Korshunova et al., 1999), why AtIRT1 was used as a positive control in this study. Transformation of the yeast Δsmf1 mutant with HvIRT1 or AtIRT1 showed that both genes had the ability to restore growth on Mn2+-limited medium controlled by the chelator EGTA (Fig. 3A). Over the range of EGTA concentrations tested, the transformants containing the empty vector grew poorly in medium containing 1 to 2 mm EGTA and did not grow at all in medium containing 5 mm and higher. The transformants containing HvIRT1 and AtIRT1 grew almost as well as wild-type yeast at all EGTA concentrations tested. Since AtIRT1 is known to transport Zn2+ and Fe2+/Fe3+, we also examined HvIRT1 for those transport properties. HvIRT1 and AtIRT1 were both able to restore the severe growth defect of the Zn2+ transport-defective Δzrt1Δzrt2 double mutant (Fig. 3B). However, the complementation test showed that the cells expressing AtIRT1 grew faster than those expressing HvIRT1, which might suggest that HvIRT1 encodes a transporter with a lower uptake rate or affinity for Zn2+ than AtIRT1. Likewise, HvIRT1 and AtIRT1 were able to restore the growth defect of the Fe2+/Fe3+ transport-defective Δfet3Δfet4 double mutant with similar efficiency, demonstrating the importance of HvIRT1 for Fe2+/Fe3+ transport (Fig. 3C). Thus, the complementation study suggests that HvIRT1 is involved in the transport of Mn2+, Zn2+, and Fe2+/Fe3+.
Figure 3.
Complementation by AtIRT1 and HvIRT1 of different yeast mutant strains defective in metal uptake. Serial dilutions (1.0, 0.1, and 0.01 OD600) of Δsmf1 (defective in Mn2+ uptake; A), Δzrt1Δzrt2 (defective in Zn2+ uptake; B), and Δfet3Δfet4 (defective in Fe2+ uptake; C) yeast cells transformed with the empty vector pFL61, AtIRT1 cDNA clone, and HvIRT1 cDNA clone inserted in the pFL61 vector were spotted on selective medium with reduced content of free metals controlled by bathophenanthroline disulfonic acid and EGTA (see “Materials and Methods”). Plates were incubated for 3 to 5 d at 30°C.
Yeast Trace Element Uptake Studies
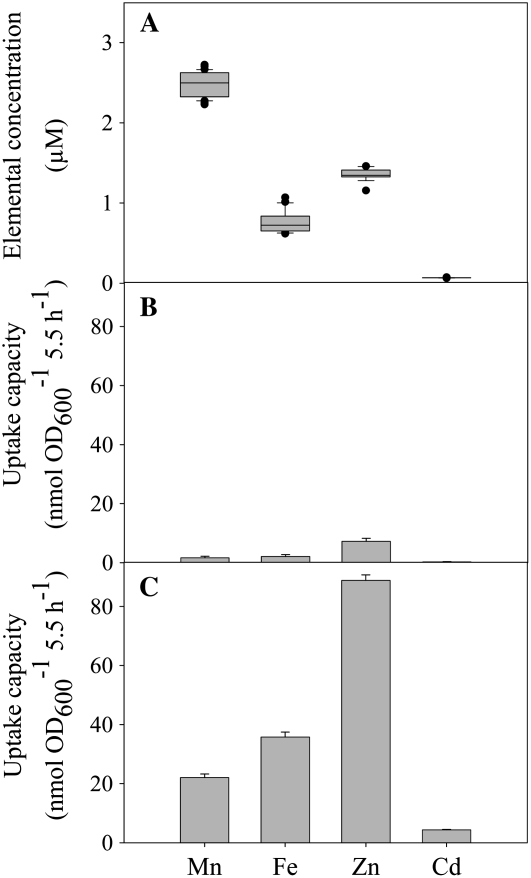
ICP-MS was used to study the HvIRT1 specificity for 31 different trace elements and metalloids across the periodic table. Yeast cells transformed with either empty vector or HvIRT1 were harvested from log-phase cultures. The cells were washed and used for a 5.5-h uptake study. The following elements showed no differences between HvIRT1- and empty vector-transformed yeast cells in any of the three mutant strains (data not shown): aluminum, arsenic, silver, barium, beryllium, boron, calcium, cobalt, chromium, copper (Cu), europium, holmium, lanthanum, magnesium, molybdenum, sodium, nickel, lead, antimony, scandium, selenium, strontium, thorium, thallium, uranium, vanadium, and ytterbium. On the contrary, significant differences were observed for the uptake of Mn2+, Zn2+, Fe2+/Fe3+, and Cd2+ (Figs. 4 and 5). Background values resulting from unspecific adsorption were determined by performing the uptake experiment below 5°C and subtracting these values from those measured at 30°C (Fig. 4, B and C). In general, the values obtained below 5°C were less than 10% of those measured at 30°C. In order to ensure optimal cell viability and still avoid metal toxicity, the initial trace element concentrations were not equimolar. Widely different uptake rates were observed for the individual elements, with no clear relation to the differences in their initial concentration (Fig. 4). Taking Zn2+ as an example, the uptake rate was significantly higher than that of Mn2+, even though the initial concentration at time zero was more than 50% lower (Fig. 4A).
Figure 4.
Trace element uptake by Δsmf1 yeast cells. A, Box plot of the initial concentration of the trace elements analyzed before the start of the experiment (12 replicates). B and C, Uptake rates were measured at 30°C. Background values resulting from unspecific adsorption were determined in the cold below 5°C. Data represent triplicate means ± se of yeast mutant Δsmf1 transformed with HvIRT1 measured at 5°C (B) or 30°C (C).
Figure 5.
The uptake rates of Mn2+, Fe2+/Fe3+, Zn2+, and Cd2+ were measured for 5.5 h in yeast mutants transformed with HvIRT1 and normalized on the basis of the uptake rates of similar yeast mutants transformed with empty vector (pFL61). Three yeast mutants were used: Δsmf1, defective in Mn2+ uptake (A), Δzrt1Δzrt2, defective in Zn2+ uptake (B), and Δfet3Δfet4, defective in Fe2+ uptake (C). Unspecific adsorption of metal ions was adjusted by subtracting the uptake values from similar experiments performed at 5°C. The experiment was replicated three times with different transformation lines with similar results, and representative data are shown. Data are means ± se of three independent yeast samples. ns. refers to no significant difference in uptake rate between HvIRT1 and empty vector transformants (P < 0.05).
Δsmf1 cells transformed with HvIRT1 showed approximately 50% higher uptake rates of Mn2+, Fe2+/Fe3+, and Cd2+ when compared with yeast cells transformed with empty vector, whereas no difference was seen in Zn2+ uptake (Fig. 5A). The same experiment carried out with the Zn2+-deficient Δzrt1Δzrt2 yeast strain resulted in an approximately 100% higher uptake rate of Zn2+ and a 15% higher Cd2+ uptake rate in HvIRT1-transformed cells compared with empty vector transformants, while Mn2+ and Fe2+/Fe3+ uptake rates were higher for yeast cells transformed with empty vector (Fig. 5B). The Fe2+/Fe3+ uptake rate was 150% higher in the Fe2+/Fe3+ uptake-defective Δfet3Δfet4 mutant when transformed with HvIRT1 compared with empty vector (Fig. 5C). In comparison, the Cd2+ and Mn2+ uptake rates were increased by 70% and 20%, respectively. Thus, the yeast uptake assays using ICP-MS support the results obtained from the complementation assays, confirming that HvIRT1 transports Mn2+, Zn2+, Fe2+/Fe3+, and Cd2+ when expressed in yeast.
Cellular Localization of HvIRT1-GFP Fusion Protein
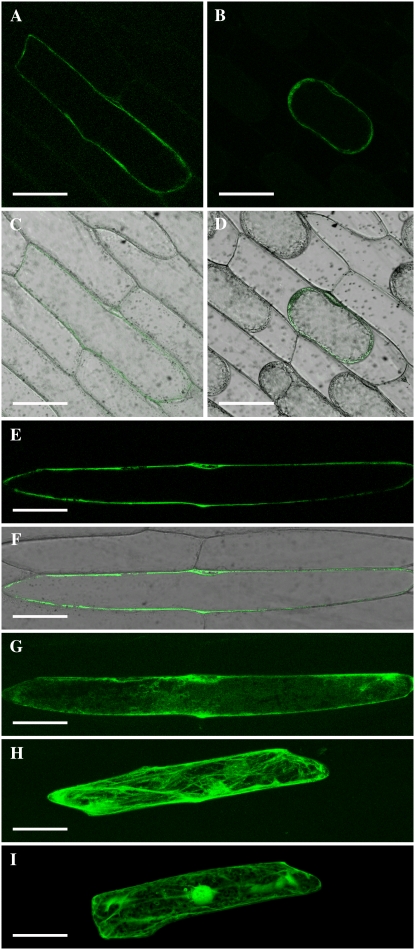
A HvIRT1:GFP fusion construct was transiently expressed in epidermal cells of onion (Allium cepa) bulb scales under the control of two times the cauliflower mosaic virus (2×CaMV) 35S promoter. This resulted in a high expression level with substantial accumulation of the fusion protein in the endoplasmic reticulum (ER), as evidenced by a well-defined ER network (Fig. 6H), and by staining of the nuclear envelope when looking at an optical section (data not shown). In addition, GFP labeled the border of the cell, indicating plasma membrane localization of HvIRT1:GFP (Fig. 6H). In order to test if there indeed is HvIRT1:GFP protein at the plasma membrane, we incubated the cells with cycloheximide to block the protein synthesis and thereby reduce the amount of protein accumulating within the ER. Figure 6 (A–G) shows cells expressing HvIRT1:GFP after incubation with cycloheximide for 5 h. In these cells, there was a clear signal at the plasma membrane, and by comparing Figure 6, G and H, it becomes evident that the ER network had disappeared. In order to confirm the localization at the plasma membrane, we performed plasmolysis of the cells (Fig. 6, B and D); here, the GFP signal followed the plasma membrane when the protoplasts retracted from the cell wall. Cells expressing free GFP were used as controls (Fig. 6I); these cells showed a clear accumulation of GFP in the nucleus and the cytoplasm. Based on these results, we conclude that HvIRT1 localizes to the ER and the plasma membrane.
Figure 6.
HvIRT1 localizes to the plasma membrane. HvIRT1:GFP fusion protein was transiently expressed in onion epidermal cells (A–H). Eighteen hours of expression of the fusion protein resulted in massive protein buildup in the ER (H). This effect was reduced by shortening the expression time to 5 h followed by 5 h of treatment with cycloheximide to block protein synthesis (G). Optical sections of cells show localization of HvIRT1 to the plasma membrane (A and E). This localization was confirmed by plasmolyzing the cell from A in a mannitol solution for 30 min (B). Expression of free GFP resulted in an entirely different localization pattern, with staining of the cytosol and the entire nucleus (I). A to F, Optical sections through cells. A, B, and E, GFP staining. C, D, and F, Transmission and GFP channel overlay. G to I, Whole cell projections of the GFP staining show the differences in localization patterns between constructs and treatments. Bars = 100 μm.
HvIRT1 Expression versus Mn2+ Uptake during Mn, Fe, Zn, and Cu Deficiency
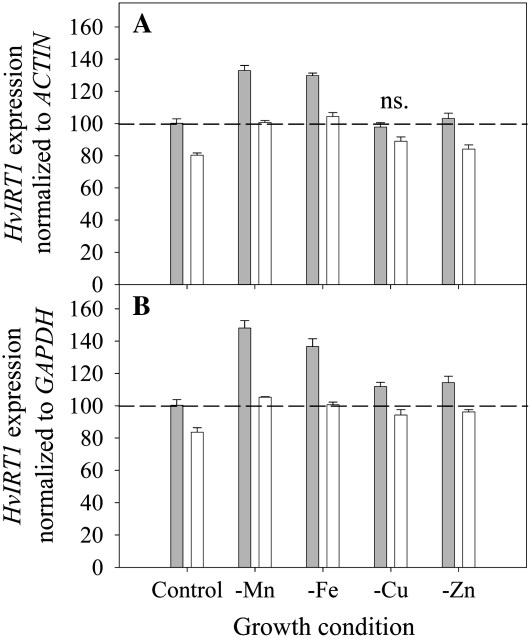
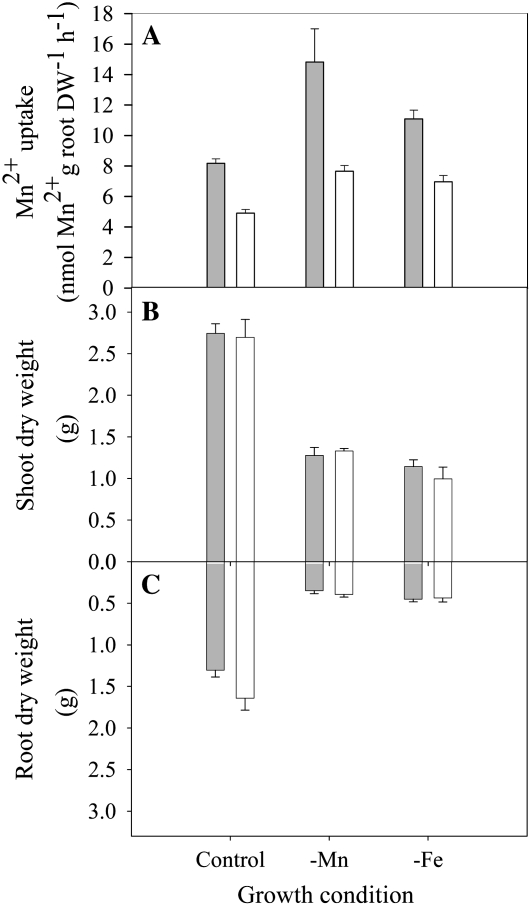
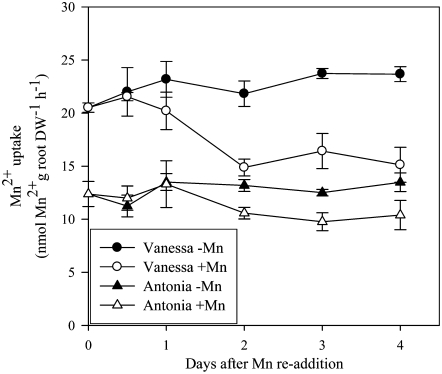
Expression of HvIRT1 in roots was induced by Mn or Fe deficiency and responded to a much lesser extent to deprivation of Zn and Cu (Fig. 7). Under Mn or Fe deficiency, the expression level of HvIRT1 increased 40% to 50% in the Mn-efficient genotype Vanessa compared with 25% in the Mn-inefficient genotype Antonia. Significantly higher expression of HvIRT1 in Vanessa compared with Antonia was also evident under control conditions as well as under Zn deficiency (Fig. 7). The observed differences in the expression response of HvIRT1 were reflected by an up to 65% higher Mn2+ uptake rate in roots of Vanessa compared with Antonia under control conditions (Fig. 8A). Under Mn deficiency, the Mn2+ uptake rate in Vanessa was 95% higher than that in Antonia, whereas under Fe-deficient conditions, the difference was 60% in favor of Vanessa. Relative to the control conditions, the increase in Mn2+ uptake rate was highest for Vanessa, as the Mn2+ uptake rate increased 80% and 40% in Mn- and Fe-deficient plants, respectively. For Antonia, the corresponding increases were 55% and 40%. The differences in uptake rate were not related to differences in biomass production, as no significant differences between the two genotypes were observed (Fig. 8, B and C). Resupply of Mn2+ to Mn-deficient plants caused Mn2+ uptake rates to drop in both genotypes, most distinctly in Vanessa (Fig. 9). Although ample Mn2+ supply resulted in a decreased difference in Mn2+ uptake rates between the two genotypes, Vanessa maintained a significantly higher Mn2+ uptake rate than Antonia, consistent with the results obtained for plants with steady-state Mn status (Fig. 8). Elemental tissue analysis showed a clear effect of the nutrient deficiency treatments in both genotypes (Table I). Noticeably, a specific micronutrient deficiency induced a higher uptake of other micronutrients, leading to twice as high Fe, Zn, and Cu concentrations in Mn-deficient plants compared with control plants. Similarly, in Fe- and Cu-deficient plants, the Mn and Zn concentrations were more than doubled. The stress level, measured as the quantum yield efficiency of PSII caused by individual nutrient deficiencies, was determined by measuring the chlorophyll a fluorescence induction kinetics. The resulting Fv/Fm values (for maximum photochemical efficiency of PSII in the dark-adapted state) within each treatment were similar between the genotypes, being 0.8, 0.5, 0.5, 0.7 and 0.8 for control and Mn-, Fe-, Cu-, and Zn-deficient conditions, respectively, indicating similar stress levels between the genotypes (data not shown).
Figure 7.
Reverse transcription-PCR analysis of HvIRT1 expression in intact roots of the Mn-efficient genotype Vanessa (gray bars) and the Mn-inefficient genotype Antonia (white bars) grown under control conditions or exposed to Mn, Fe, Cu, or Zn deficiency. Data were normalized with respect to the expression of the two reference genes ACTIN (A) or GAPDH (B). Furthermore, data are indexed relative to Vanessa in the control treatment. Error bars represent se (n = 3). ns. refers to no significant difference in expression level of HvIRT1 between genotypes (P < 0.05) in the same nutrient treatment.
Figure 8.
The effects of nutrient deficiencies on biomass production and Mn2+ uptake rates were determined in the Mn-efficient genotype Vanessa (gray bars) and the Mn-inefficient genotype Antonia (white bars). A, Influence of Mn and Fe deficiency on 54Mn2+ uptake in Vanessa and Antonia. Data are means ± se (n = 4). B and C, Influence of Mn and Fe deficiency on shoot (B) and root (C) dry weights of Vanessa and Antonia. Data are means ± se (n = 6).
Figure 9.
Mn2+ uptake rates of Mn-deficient plants after Mn2+ resupply. 54Mn2+ uptake rates (nmol Mn2+ g−1 root dry weight [DW] h−1) in 33-d-old plants of the Mn-efficient genotype Vanessa (circles) and the Mn-inefficient genotype Antonia (triangles). Black symbols denote plants exposed to continuous Mn deficiency (0 μg of Mn2+ per bucket per day), while white symbols show data for plants resupplied with Mn2+ (4.4 μg of Mn2+ per bucket per day on the first day, 22 μg of Mn2+ per bucket per day on the following days, resembling Mn2+ concentrations of 20 and 100 nm, respectively). Plants were Mn deficient (shoot Mn of 3–4 μg g−1 dry weight) before resupply of Mn2+ at time zero. Each data point represents a mean ± se (n = 4).
Table I.
Element concentration (μg g−1 dry weight) in the youngest leaf of the Mn-efficient genotype Vanessa and the Mn-inefficient genotype Antonia plants grown under Mn-, Fe-, Cu-, and Zn-deficient and control conditions
Numbers in boldface refer to the element concentration corresponding to nutrient deficiency. The results are presented as means ± se (n = 3).
| Genotype and Element | Growth Condition
|
|||||
|---|---|---|---|---|---|---|
| Control | −Mn | −Fe | −Cu | −Zn | ||
| Vanessa | Mn | 13.24 ± 0.5 | 3.45 ± 0.5 | 24.02 ± 4.9 | 29.89 ± 8.0 | 9.75 ± 0.2 |
| Fe | 58.54 ± 1.5 | 76.79 ± 3.7 | 12.40 ± 0.9 | 91.20 ± 11.1 | 52.98 ± 9.3 | |
| Cu | 13.24 ± 0.6 | 30.11 ± 2.6 | 9.00 ± 0.5 | 0.40 ± 0.1 | 9.98 ± 0.5 | |
| Zn | 37.76 ± 1.3 | 86.69 ± 2.8 | 73.64 ± 8.6 | 73.43 ± 15.4 | 11.89 ± 0.7 | |
| Antonia | Mn | 13.64 ± 0.9 | 3.78 ± 1.1 | 34.36 ± 7.2 | 36.07 ± 3.2 | 10.73 ± 0.8 |
| Fe | 37.04 ± 4.2 | 69.23 ± 8.4 | 14.09 ± 0.8 | 87.89 ± 11.3 | 51.33 ± 6.5 | |
| Cu | 8.68 ± 0.6 | 22.24 ± 3.0 | 9.14 ± 0.2 | 0.63 ± 0.1 | 9.28 ± 0.6 | |
| Zn | 26.24 ± 3.1 | 72.51 ± 9.3 | 72.08 ± 10.2 | 81.86 ± 7.6 | 13.96 ± 0.8 | |
DISCUSSION
Differences in Mn efficiency among barley genotypes are related to the high-affinity uptake system for Mn2+ operating in the low nanomolar concentration range (Pedas et al., 2005). However, the molecular basis for this phenomenon is not known. In this study, we report on the cloning and characterization of a barley Mn2+ transporter, HvIRT1, belonging to the ZIP family. ZIP members have been cloned from other plant species (Eide et al., 1996; Eckhardt et al., 2001; Bughio et al., 2002; Lombi et al., 2002; Cohen et al., 2004; Li et al., 2006; Enomoto et al., 2007; Waters et al., 2007), but so far only AtIRT1 from Arabidopsis has been documented to be involved in Mn2+ uptake in planta (Vert et al., 2002). Sequence analysis of the known IRT1 proteins revealed that the predicted HvIRT1 protein contains all of the characteristic features of the ZIP family (Fig. 1). HvIRT1 is predicted to have eight transmembrane domains, with the highly conserved ZIP signature in the fourth transmembrane domain (Eng et al., 1998) and a putative metal-binding His-rich region between domains III and IV (Guerinot, 2000). However, a few additional modifications to the ZIP signature are proposed (Fig. 1), including those in HvIRT1.
IRT1 homologs have been identified for other plants species (Eckhardt et al., 2001; Vert et al., 2002; Ishimaru et al., 2006), and searching public databases has identified several barley EST sequences with high homology to IRT1 (TC145252, TC151006, TC138364, and TC153445), but to our knowledge, no full-length IRT1 homolog has been identified from barley.
HvIRT1 restored the growth of the Δsmf1 yeast mutant deficient in Mn2+ uptake (Fig. 3), suggesting that HvIRT1 is an integral plasma membrane protein, at least when expressed in yeast. This was further substantiated by transient expression of a HvIRT1:GFP fusion protein in onion epidermal cells, showing that the protein was localized to the ER and the plasma membrane (Fig. 6). HvIRT1 was predicted to contain a signal peptide (Fig. 1) targeting the protein to the secretory pathway, and the relatively high abundance in the ER suggests accumulation in the secretory system before reaching the final destination in the plasma membrane. OsIRT1 and AtIRT1 are also localized in the plasma membrane (Vert et al., 2002; Ishimaru et al., 2006), whereas the positions of the other IRT1 proteins remain to be identified.
All IRT1 proteins appear to have specificity for transporting Fe2+/Fe3+ except in Thlaspi caerulescens and tobacco (Nicotiana tabacum), where the elemental preferences remain to be determined. In addition, IRT1s from Arabidopsis, tomato, and pea have the ability to transport Mn2+ (Korshunova et al., 1999; Eckhardt et al., 2001; Lombi et al., 2002; Vert et al., 2002: Cohen et al., 2004; Enomoto et al., 2007), but HvIRT1 is the first transport protein with a proven ability to transport Mn2+ that has been isolated from a graminaceous plant species. Functional expression of HvIRT1 cDNA in the yeast double mutants Δfet3Δfet4 and Δzrt1Δzrt2, defective in Fe2+/Fe3+ and Zn2+ uptake, respectively, show that the HvIRT1 protein has the ability to transport, besides Mn2+, Fe2+/Fe3+ and Zn2+ (Fig. 3).
The transport specificity of HvIRT1 was further examined in a yeast uptake assay using ICP-MS (see “Materials and Methods”) for analysis of 31 different metals and metalloids. This constituted a sensitive screen for all tested elements, even those in the subnanomolar range, such as cadmium, strontium, antimony, lanthanum, thorium, and uranium. However, only Mn2+, Fe2+/Fe3+, Zn2+, and Cd2+ transport were affected by the expression of HvIRT1 in yeast (Figs. 4 and 5). AtIRT1 has previously been shown to transport Fe2+/Fe3+, Mn2+, Zn2+, Co2+, and Cd2+ (Korshunova et al., 1999; Vert et al., 2002), while IRT1 from rice only is able transport Fe2+/Fe3+ and Cd2+ (Bughio et al., 2002; Nakanishi et al., 2006). An interesting observation was made with the yeast uptake assay, as the empty vector transformants showed a higher uptake of Mn and Fe than HvIRT1-transformed cells when expressed in the Δzrt1Δzrt2 yeast mutant (Fig. 5B). However, studies have shown that Zn deficiency in yeast induces an up-regulation of several genes involved in various cell processes (Lyons et al., 2000; De Nicola et al., 2007). One gene highly up-regulated is Fet4, and Fet4p being localized to the plasma membrane is suggested to be involved in the transport of Fe2+, Mn2+, Zn2+, Co2+, and Cu2+ (Li and Kaplan, 1998; Hassett et al., 2000; Waters and Eide, 2002). This can potentially explain the difference in Fe2+/Fe3+ and Mn2+ uptake rates between HvIRT1- and empty vector-transformed Δzrc1Δzrc2 yeast cells, as the HvIRT1-transformed yeast cells have a higher Zn supply and thereby a lower expression level of Fet4 and consequently a lower capacity for Fe2+ and Mn2+ uptake.
The transcript level of HvIRT1 was mainly influenced by Mn and Fe deficiency (Fig. 7). Similar responses have been observed for OsIRT1 (Bughio et al., 2002; Ishimaru et al., 2006), while AtIRT1 was unaffected by micronutrient deficiencies other than Fe (Eide et al., 1996; Korshunova et al., 1999). The HvIRT1 expression level was significantly (P < 0.01) higher in the Mn-efficient genotype Vanessa compared with the Mn-inefficient genotype Antonia in all treatments except Cu deficiency (Fig. 7). The genotypic difference in HvIRT1 transcript abundance corresponded to the observed higher Mn2+ uptake rates in Vanessa relative to Antonia both under steady-state Mn provision (Fig. 8) and during resupply of Mn2+ to Mn-deficient plants (Fig. 9). The genotypic differences in HvIRT1 transcript levels also fit with the fact that Vanessa has a considerably higher Vmax value for Mn2+ uptake than Antonia (Pedas et al., 2005). Also, transcript levels of IRT1-G in T. caerulescens correlated markedly with the Vmax for root Cd2+ influx and shoot cadmium accumulation and were related to genotypic differences (Lombi et al., 2002). The substrate specificity of TcIRT1-G has not yet been determined, but the putative ability to transport Cd2+ is in accordance with the reported specificities of other IRT1 homologs.
Resupply of Mn2+ to Mn-deficient plants caused lower Mn2+ uptake rates (Fig. 9), indicating a down-regulation of Mn2+ transport proteins at ample Mn2+ supply. Also, the root-to-shoot translocation of Mn2+ seems to be reduced under Mn sufficiency (Tsukamoto et al., 2006). OsIRT1 has not yet been documented to transport Mn2+, but an OsIRT1 promoter-GUS experiment showed GUS activity in root phloem cells, particularly in the companion cells, and in root epidermis and exodermis. This indicates an important role for IRT1 in primary Fe2+/Fe3+ uptake, phloem loading, and long-distance transport to sink organs (Ishimaru et al., 2006). The leaf Mn concentration in Fe-deficient barley plants was up to 150% higher than in control plants (Table I), suggesting a strong up-regulation of Mn2+ transport proteins involved in both primary Mn2+ uptake and root-to-shoot translocation during Fe deficiency. The high similarity with OsIRT1 suggests that HvIRT1 may also be involved in Mn2+-translocation during Fe deficiency. This is supported by the parallel increases in HvIRT1 expression, root Mn2+ uptake, and foliar Mn concentration during Fe deficiency. However, further analysis of the specific cell tissue localization of HvIRT1 is needed before this can be confirmed.
The genotypic differences in Mn2+ uptake kinetics shown previously (Pedas et al., 2005) might be due to the expression of different isoforms of Mn2+ transporters rather than differential expression levels of Mn2+ transport proteins. However, screening of the barley root cDNA library for genes encoding putative plasma membrane-localized Mn2+ transport proteins only resulted in one isolated gene, HvIRT1. The observed difference in induction level upon deficiency treatments between the expression level of HvIRT1 and the Mn2+ uptake capacity might be related to the transport capacity of the HvIRT1 protein, as a higher protein level can be followed by an even higher transport capacity. The IRT1 homolog from Arabidopsis is shown to be posttranscriptionally regulated, and in addition, a transcription factor is also involved in regulating the protein level of AtIRT1 (Connolly et al., 2002; Colangelo and Guerinot, 2004). Whether such regulation mechanisms exist in barley is currently unknown.
Therefore, HvIRT1 represents the only plasma membrane-localized Mn2+ transport protein that has been found in barley, and it is concluded that HvIRT1 is important for Mn2+ uptake in barley roots. Moreover, HvIRT1 seems to be a central component in differential Mn efficiency among barley genotypes.
MATERIALS AND METHODS
Yeast Strains
A wild-type Saccharomyces cerevisiae strain (BY4741) and five deletion mutants in the same genetic background were used in this study (Table II). Two double mutants were produced by crossing haploid single-deletion mutants: Y06461 (ΔYMR319c) crossed with Y16192 (ΔYMR058w), and Y04622 (ΔYLR130c) crossed with Y14087 (ΔYLR130c). Subsequent sporulation according to standard procedures generated the Δfet3Δfet4 and Δzrt1Δzrt2 double mutants.
Table II.
List of S. cerevisiae strains used in this study
Euroscarf is a collection of single-deletion mutants in S. cerevisiae (Frankfurt).
| Mutant | Background | Mating Type | Genotype | Reference |
|---|---|---|---|---|
| Wild type | BY4741 | MATa | his3Δ1; leu2Δ0; met15Δ0; ura3Δ0 | Euroscarf Y00000 |
| Δsmf1 | BY4741 | MATa | his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; YOL122c∷kanMX4 | Euroscarf Y06272 |
| Δfet4 | BY4741 | MATa | his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; YMR319c∷kanMX4 | Euroscarf Y06461 |
| Δfet3 | BY4742 | MATα | his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; YMR058w∷kanMX4 | Euroscarf Y16192 |
| Δzrt1 | BY4741 | MATa | his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; YGL255w∷kanMX4 | Euroscarf Y04622 |
| Δzrt2 | BY4742 | MATα | his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; YLR130c∷kanMX4 | Euroscarf Y14087 |
| Δfet3Δfet4 | BY4742/BY4741 | MATα | his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; YMR058w∷kanMX4; YMR319c∷kanMX4 | This study |
| Δzrt1Δzrt2 | BY4741/BY4742 | MATα | his3Δ1; leu2Δ0; met15Δ0; lys2Δ0; ura3Δ0; YGL255w∷kanMX4; YLR130c∷kanMX4 | This study |
Functional Complementation in Yeast
Yeast strains were transformed with HvIRT1, AtIRT1 vector construct, or the empty vector pFL61. Transformants were selected on uracil-deficient medium and grown in synthetic medium containing 2% Glc, 50 mm succinic acid/Tris base, pH 5.5, 0.7% yeast nitrogen base (YNB) without amino acids (Difco), and 0.3% appropriate amino acids. Agar was added to 2% for solid plate medium (Sherman, 1991). For medium deprived of Mn, Zn, or Fe, YNB without metals (BIO 101 Systems) supplemented with various metals was used, and various additions of the chelators bathophenanthroline disulfonic acid (Fluka) and EGTA (Sigma) were added, as specified in “Results.” Plates were incubated at 30°C for 3 to 5 d.
Identification of the Mn2+ Transporter Protein
A cDNA library inserted into the vector pFL61 (Minet et al., 1992) was produced from mRNA extracted from the barley (Hordeum vulgare) genotypes Vanessa and Antonia supplied with varying levels of Mn2+, ranging from sufficient to insufficient levels, achieving all scenarios potentially important for Mn efficiency. The yeast strain S. cerevisiae Δsmf1 (Table II) was transformed by electroporation as described (http://www.biw.kuleuven.be/dp/logt/protocol/yeastelectroporation.htm) with the barley root cDNA library. The pFL61 vector was modified with the Gateway system (Invitrogen), and it contains the replication origin of the yeast 2 μ plasmid, the Ura3 gene, and the promoter and terminator of the yeast phosphoglycerate kinase gene for the expression of the foreign cDNA in S. cerevisiae. Transformants were selected on uracil-deficient synthetic medium with 2% Glc, 50 mm succinic acid/Tris base, pH 5.5, 0.7% YNB without amino acids (Difco), and 0.3% amino acids His, Leu, and Met. Transformants were harvested from the plates, pooled, and resuspended in glycerol. From this pool, aliquots were replated on medium without Mn2+ to select for yeast cells complementing the Mn2+-defective mutant. Mn2+-deficient medium was composed as above except that the YNB (Difco) were replaced with YNB without metals and without amino acids (BIO 101 Systems) supplemented with 0.9 mm CaCl2, 2 mm MgSO4, 160 nm CuSO4, 740 nm FeCl3, and 1.4 μm ZnSO4. The agar used contained a significant amount of Mn2+; therefore, the ion activity of Mn2+ was decreased using 10 to 15 mm EGTA. Plasmid cDNA from yeast transformants was extracted (Hoffman and Winston, 1987) and amplified in Escherichia coli by standard procedures. Plasmids, which after retransformation still were able to complement Δsmf1, were sequenced, and the gene sequences were BLASTed against various databases to identify similarities to known gene families.
Cloning of AtIRT1 into Yeast Expression Vector
AtIRT1 (Eide et al., 1996) was subcloned into the pFL61 vector to conduct yeast complementation as a positive control. A PCR-based cloning strategy was used to clone AtIRT1 and insert the cDNA into the Gateway-adapted pFL61 vector. Primers with attB1 and attB2 sequences including gene-specific sequences were used to amplify the open reading frame: AtIRT1 forward, 5′-ACAAGTTTGTACAAAAAAGCAGGCTTCATGAAAACAATCTTCCTCGTA-3′; AtIRT1 reverse, 5′-GACCACTTTGTACAAGAAAGCTGGGTCTTAAGCCCATTTAGCGATAAT-3′. The PCR product was amplified using LA Taq (Takara), and the in vitro BP clonase recombinant reaction into the pDONR221 vector (Invitrogen) was carried out according to the manufacturer's directions followed by sequencing. The coding sequence of AtIRT1 (At4g19690) was cloned from cDNA from Arabidopsis (Arabidopsis thaliana) roots. After sequencing, AtIRT1 cDNA was transferred by LR recombinant reaction according to the manufacturer's instructions into the pFL61 vector, identical to the vector used for the cDNA barley library.
Yeast Metal Uptake Studies
Yeast transformants were precultured overnight in 2 mL of the medium prepared as above, and a liquid culture was inoculated overnight, achieving an optical density value measured at 600 nm (OD600) in the range of approximately 1.0 to 1.2. A growth assay was made for observing growth rates between yeast strains transformed with different constructs, so the harvesting was done at the optimal time point for the individual transformants. Cells were then pelleted by centrifugation and washed three times in ice-cold Milli-Q water, and the resulting pellets were resuspended in Milli-Q water. Ten milliliters of the cell suspensions with similar content of cells (measured and adjusted with OD600) was added to a 250-mL Erlenmeyer flask (polypropylene; Nalgene) containing 100 mL of growth medium made as above. The growth medium was further spiked with 0.01 μg mL−1 different trace elements: aluminum, arsenic, silver, barium, beryllium, boron, calcium, cobalt, cadmium, chromium, Cu, europium, Fe, holmium, lanthanum, magnesium, Mn, molybdenum, sodium, nickel, lead, antimony, scandium, selenium, strontium, thorium, thallium, uranium, vanadium, ytterbium, and Zn (P/N 4400-ICP-MSCS calibration standard; CPI International). The OD600 was 1.6 (cells up-concentrated from OD600 of 1.0–1.2 by reducing the amount of liquid) at the start of the experiment, and the growth rate was recorded by measuring the OD600 at the end of the experiment. The uptake study was done at 30°C and 5°C (mimicking yeast absorption and zero uptake, respectively), with shaking horizontally at 150 rpm for 5.5 h. At time zero and at the end of the experiment, sample aliquots of 1 mL were taken and immediately put on ice. The samples were pelleted by centrifugation at 10,000g for 5 min, and 500 μL of the supernatant was transferred to a new tube. A total of 4.5 mL of 1.75% HNO3 was added, and the samples were stored until measurement by ICP-MS (Agilent 7500ce; Agilent Technologies). For every run of ICP-MS, 10 blanks and 10 samples of two different certified reference materials (apple [Malus domestica] leaf, standard reference material 1515; durum wheat [Triticum durum] flour, reference material 8436; National Institute of Standards and Technology) were included to estimate the accuracy and precision of the analysis. The ICP-MS apparatus was configured with the octopole reaction system to reduce polyatomic interference and increase accuracy. Data were accepted when the accuracy of the certified reference material value for each element was higher than 90%.
Plant Material
Seeds of two barley (Hordeum vulgare) genotypes differing in Mn efficiency were germinated at 21°C in vermiculite. After 5 d, uniform seedlings were selected and transferred to light-impermeable 4-L black buckets with four plants per bucket. The buckets were filled with a chelate-buffered solution prepared in double ionized water as specified by Pedas et al. (2005), with the exception of Mn2+. pH was kept at 6.0 using 0.5 mm MES-Tris, pH 6.0. To induce deficiencies of Zn, Mn, or Cu, plants were grown for 4 weeks in solutions without these nutrients. For the Fe-deficient treatment, 2-week-old control plants were transferred to nutrient solution without Fe and grown for 2 weeks more. With the exception of plants exposed to Mn deficiency, 22 and 44 μg of Mn2+ (resembling Mn2+ concentrations of 100 and 200 nm, respectively) was added every second day in the form of MnCl2 for the first and last 2 weeks of growth, respectively. The plants used for 54Mn2+ uptake experiments did not receive any Mn2+ addition on the same day that the measurements were performed. A number of plants were induced with Mn deficiency, and they were used to examine how the Mn2+ uptake rates were influenced by changing the Mn2+ available concentration compared with plants with continuous Mn deficiency. These plants were grown as follows. For 2 weeks, plants were grown at control conditions. Then, the plants were without Mn2+ additions for 19 d, inducing Mn deficiency, controlled by measuring chlorophyll a fluorescence. At this point, the experiment started by taking plants to analyze the Mn2+ uptake rate as described below. A Mn2+ addition of 4.4 μg per bucket (achieving 20 nm Mn2+) was then added to half of the plants, and every morning for the following 3 d, an additional Mn2+ addition of 22 μg was added to same plants, leaving half of the plants with continuous Mn deficiency. Sample times were at day zero (before adding Mn2+) and at 12 h and 1, 2, 3, and 4 d after the first Mn addition. Plants were grown in a controlled growth chamber with a 250 to 280 μmol m−2 s−1 photon flux density, 75% to 80% humidity, and a 20°C/15°C (16 h/8 h) day/night temperature regime.
Chlorophyll a Fluorescence
Induction of the individual micronutrient deficiency and its effect on PSII were recorded with chlorophyll a fluorescence measurements (Kriedmann et al., 1985) using a Handy PEA (Plant Efficiency Analyzer; Hansatech Instruments) on four leaves per bucket. A flash of saturating light (3,000 μmol photons m−2 s−1) lasting 2 s was applied to leaves that had been dark adapted for 25 min. The fluorescence data were analyzed, and the Fv/Fm ratio was calculated using the Handy PEA software (version 1.30).
54Mn2+ Uptake Measurements
Experiments were started at times corresponding to the photoperiod of the plants, except for the sample time 12 h after Mn2+ resupply. The barley plants were gently removed from the nutrient solution. The roots were rinsed in 18.2 MΩ Milli-Q water for 10 min and placed in 700-mL polypropylene beakers containing a pretreatment solution (2 mm MES-Tris, pH 6.0, 0.2 mm CaSO4, 12.5 μm H3BO4, and 0.5 nm MnCl2) for 30 min. A new set of beakers was prepared with 700 mL of uptake solution consisting of 5 mm MES-Tris, pH 6.0, 0.2 mm CaSO4, 12.5 μm H3BO4, radiolabeled 54Mn2+ (1,409.44 MBq mg−1; Perkin-Elmer Life Science Products), and nonlabeled MnCl2 to yield the desired total Mn2+ concentration of 50 nm and 100 nm for the steady-state uptake measurements (control and Mn- and Fe-deficient conditions) and for the response experiments (resupply of Mn2+ to Mn-deficient plants), respectively. Before initiating the uptake measurements, the Mn2+ concentration in the uptake solutions was checked by ICP-MS. At the end of the 2-h uptake period, plants were gently moved and rinsed for 10 min in Milli-Q water to remove the nutrient solution-water film around the roots. All of the solutions were aerated, and the experiments were carried out at 20°C under artificial light with a photon flux density of 250 to 280 μmol m−2 s−1. In parallel, similar experiments were carried out in a cooling room (4°C–5°C) with no light in ice-cold (2°C) solutions mimicking the Mn2+ absorption in the root apoplasm. This fraction was subtracted from the data obtained under 20°C and light, estimating the actual uptake during the 2-h uptake period. After the final rinse in Milli-Q water, the roots were blotted dry with paper towels, freeze dried (Christ Alpha 2-4; Martin Christ), and weighed. The roots were dissolved in 10 mL of concentrated HNO3. The radioactivity of 54Mn2+ taken up by the roots was measured by γ spectrometry using a Ge(Li) detector (Princeton Gamma-Tech; 28% relative efficiency, 1.8-keV energy resolution) placed in a 10-cm lead shield and connected to a Canberra multiport multichannel analyzer. The γ spectra were evaluated with Canberra Genie 2000 software.
Determination of Trace Element Concentrations in Plant Material
To ensure that the micronutrient deficiency had been induced in the plants, multielemental analyses of leaf tissue were performed and compared with the threshold limits found by Reuter et al. (1997). The plant samples were freeze dried (Christ Alpha 2-4; Martin Christ), digested using ultrapure acids, and analyzed by ICP-MS as described previously (Pedas et al., 2005).
Analysis of HvIRT1 Expression in Planta
Total RNA was extracted from approximately 250 mg of fresh plant tissues using the FastRNA Pro Green Kit (Q BIOgene) and a Fast Prep (FP120). Total RNA was treated with RQ1 RNase-free DNase (Promega) to remove contaminating genomic DNA. The RNA was checked for purity, integrity, and quantity using RNA gel electrophoresis and spectrophotometry. One microgram of total RNA was used as a template for cDNA synthesis using Moloney murine leukemia virus reverse transcriptase with dT18 oligonucleotide primers according to the manufacturer's directions (New England Biolabs). Prior to PCR, cDNA was diluted 1:5 in sterile water (BIBCO; Invitrogen). The PCR program used consisted of 27 cycles (45 s at 94°C, 1 min at 55°C, and 1.5 min at 72°C), and the control reactions specific for the HvACTIN and HvGAPDH transcripts were allowed to proceed for only 25 cycles. The number of cycles was optimized so that the PCRs were not saturated. The primers used were HvIRT1 forward (5′-CCAGATGTTTGAGGGGATGG-3′) and reverse (5′-GATAGACACAAGACACACCC-3′; fragment size, 409 bp); HvACTIN forward (5′-GGCCGTGCTTTCCCTCTA-3′) and reverse (5′-TCTCTGCGCCAATCGTGA-3′; fragment size, 350 bp); and HvGAPDH forward (5′-CAAGGACTGGAGRGGTGG-3′) and reverse (5′-CCCACTCGTTGTCRTACC-3′; fragment size, 376 bp). Amplified DNA was separated on 1.5% agarose gels and visualized using ethidium bromide. For each sample, the amount of the HvIRT1 transcript, quantified by ImageQuant (version 5.0), was expressed relative to the amount of the ACTIN and GAPDH transcripts, respectively.
Subcellular Localization of a HvIRT1:GFP Construct in Onion Epidermal Cells
A PCR-based cloning strategy was used to generate HvIRT1 DNA with a mutated stop codon for C-terminal fusion to the GFP6 gene. The primers used were HvIRT1 forward (5′-ACAAGTTTGTACAAAAAAGCAGGCTTCATGTCGTCGTCGTCGTCG-3′) and reverse (5′-GACCACTTTGTACAAGAAAGCTGGGTCTGCCGCCCATTTGGCCATGAC-3′). The PCR product was amplified using LA Taq (Takara), and the in vitro BP clonase recombinant reaction into the pDONR221 vector (Invitrogen) was carried out according to the manufacturer's directions followed by sequencing. After sequencing, HvIRT1 DNA was transferred by LR recombinant reaction according to the manufacturer's instructions into the pMDC83 vector, consisting of 2×CaMV 35S promoter, a NOS terminator, and the GFP6 gene (Curtis and Grossniklaus, 2003). The pMDC83-HvIRT1 vector was then digested with EcoRI and HindIII, resulting in the 2×CaMV 35S promoter-HvIRT1-GFP6-NOS terminator fragment. A construct of the pIPKTA9-GFP vector harboring the CaMV 35S promoter, the GFP gene, and the 35S terminator was kindly provided by Dr. Michael Krogh Jensen (University of Copenhagen). The construct contains an EcoRI site 5′ of the promoter and a HindIII site 3′ of the terminator. A sequential digestion of the pIPKTA9-GFP vector was made by EcoRI and HindIII, and the 2×CaMV 35S promoter-HvIRT1-GFP6-NOS terminator fragment from pMDC83-HvIRT1 was then ligated into the pIPKTA9 vector. pIPKTA9-GFP and pIPKTA9-HvIRT1-GFP were used to transiently express GFP and HvIRT-GFP, respectively, in onion (Allium cepa) epidermal cells. The transient expression and localization were done as described by Jensen et al. (2007) with a few modifications. Briefly, 1 × 1 cm2 of onion bulb scales was placed on agar (1× Murashige and Skoog medium, 0.7% agar, and 3% Suc, pH 5.8) with the inner epidermis facing up. They were bombarded using the PDS-1000/He biolistic particle delivery system (Bio-Rad). Approximately 6 μg of expression vector was coated onto 1.5 mg of 1.6-μm gold particles and transferred into the cells. After bombardment, petri dishes containing the onion bulb squares were placed in the dark at room temperature for 18 to 24 h. Where indicated, the cells were treated with 10 μg mL−1 cycloheximide (Sigma) for 5 h after only 5 h in the dark. After treatment, the cells were washed in water before the epidermis was peeled, and transformed cells were visualized using a Leica TCS SP2/MP confocal laser scanning microscope (Leica Microsystems). Excitation for GFP was 488 nm, and emission was detected between 500 and 575 nm. For plasmolysis, the cells were incubated in a 1 m mannitol solution for 30 min.
Accession Numbers
The GenBank accession numbers for the sequences described in this article are as follows. Barley: HvIRT1, EU545802; HvACTIN, TC131547; and HvGAPDH, X60343; Arabidopsis: AtIRT1, NM_118089; tobacco: NtIRT1, AB263746; pea: PsRIT1, AFO65444; tomato: LeIRT1, AF246266; rice: OsIRT1, AB070226; Malus xiaojinensis: MsIRT1, AY193886; Cucumis sativus: CsIRT1, AAT01414; T. caerulescens: TcIRT1-G, AJ320253.
Acknowledgments
The technical assistance of Bente Broeng and Mette Sylvan is gratefully acknowledged. We thank Dr. Michael Krogh Jensen for generously providing the pIPKTA9-GFP vector.
This work was supported by grants from the Ministry of Food and Fisheries (grant no. RES03–11), the Ministry of Science, Technology, and Innovation (grant nos. 274–06–0325 and 23–04–0241), EU-FP6 PHIME (grant no. FOOD.CT–2006–016253), and the Danish Cereal Breeding Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Pai Pedas (pp@life.ku.dk).
Open Access articles can be viewed online without a subscription.
References
- Alscher RG, Erturk N, Heath LSF (2002) Role of superoxide dismutase (SODx) in controlling oxidative stress in plants. J Exp Bot 53 1331–1341 [PubMed] [Google Scholar]
- Ascher-Ellis JS, Graham RD, Hollamby GJ, Paull J, Davies P, Huang C, Pallotta MA, Howes N, Khabez-Saberi H, Jefferies SP, et al (2001) Micronutrients. In MP Reynolds, JI Ortiz-Monasterio, A McNab, eds, Application of Physiology in Wheat Breeding. CIMMYT, Mexico, pp 219–240
- Bowler C, Slooten L, Vanderbranden S, Rycke RD, Botterman J, Sybesma C, Montagu MV, Inzé D (1991) Manganese superoxide dismutase can reduce cellular damage mediated by oxygen radicals in transgenic plants. EMBO J 10 1723–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt RD (1996) Oxygen evolution. In C Yocum, ed, Oxygenic Photosynthesis: The Light Reactions. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 137–159
- Bughio N, Yamaguchi H, Nishizawa NK, Nakanishi H, Mori S (2002) Cloning an iron-regulated metal transporter from rice. J Exp Bot 53 1677–1682 [DOI] [PubMed] [Google Scholar]
- Clemens KL, Force DA, Britt RD (2002) Acetate binding at the photosystem II oxygen evolving complex: an S(2)-state multiline signal ESEEM study. J Am Chem Soc 124 10921–10933 [DOI] [PubMed] [Google Scholar]
- Cohen CK, Garvin DF, Kochian LV (2004) Kinetic properties of a micronutrient transporter from Pisum sativum indicate a primary function in Fe uptake from the soil. Planta 218 784–792 [DOI] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML (2004) The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly EL, Fett JP, Guerinot ML (2002) Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell 14 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M, Grossniklaus U (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in plants. Plant Physiol 133 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nicola R, Hazelwood LA, De Hulster EAF, Walsh MC, Knijnenburg TA, Reinders MJT, Walker GM, Pronk JT, Daran JM, Daran-Lapujade P (2007) Physiological and transcriptional responses of Saccharomyces cerevisiae to zinc limitation in chemostat cultures. Appl Environ Microbiol 73 7680–7692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt U, Marques AM, Buckhout TJ (2001) Two iron-regulated cation transporters from tomato complement metal uptake-deficient yeast mutants. Plant Mol Biol 45 437–448 [DOI] [PubMed] [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng BH, Guerinot ML, Eide D, Saier MH Jr (1998) Sequence analyses and phylogenetic characterization of the ZIP family of metal ion transport proteins. J Membr Biol 166 1–7 [DOI] [PubMed] [Google Scholar]
- Enomoto Y, Hodoshima H, Shimada H, Shoji K, Yoshihara T, Goto F (2007) Long-distance signals positively regulate the expression of iron uptake genes in tobacco roots. Planta 227 81–89 [DOI] [PubMed] [Google Scholar]
- Graham RD (1988) Genotypic differences in tolerance to manganese deficiency. In RD Graham, RJ Hannam, NC Uren, eds, Manganese in Soils and Plants. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 261–276
- Graham RD, Davies WJ, Sparrow DHB, Ascher JS (1983) Tolerance of barley and other cereals to manganese-deficient calcareous soil of South Australia. In BC Loughman, ed, Genetic Aspects of Plant Nutrition. Martinus Nijhoff, The Hague, The Netherlands, pp 339–344
- Guerinot ML (2000) The ZIP family of metal transporters. Biochim Biophys Acta 1465 190–198 [DOI] [PubMed] [Google Scholar]
- Hassett R, Dix DR, Eide DJ, Kosman DJ (2000) The Fe(II) permease Fet4p functions as a low affinity copper transporter and supports normal copper trafficking in Saccharomyces cerevisiae. Biochem J 351 477–484 [PMC free article] [PubMed] [Google Scholar]
- Hebbern CA, Pedas P, Schjoerring JK, Knudsen L, Husted S (2005) Genotypic differences in manganese efficiency: a field trial with winter barley (Hordeum vulgare L.). Plant Soil 272 233–244 [Google Scholar]
- Hoffman CS, Winston F (1987) A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57 267–272 [DOI] [PubMed] [Google Scholar]
- Ishimaru Y, Suzuki M, Tsukamoto T, Suzuki K, Nakazono M, Kobayashi T, Wada Y, Watanabe S, Matsuhashi S, Nakanishi H, et al (2006) Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J 45 335–346 [DOI] [PubMed] [Google Scholar]
- Jensen MK, Rung JH, Gregersen PL, Gjetting T, Fuglsang AT, Jøhnk N, Lyngkjær MF, Collinge DB (2007) The HvNAC6 transcription factor: a positive regulator of penetration resistance in barley and Arabidopsis. Plant Mol Biol 65 137–150 [DOI] [PubMed] [Google Scholar]
- Korshunova YO, Eide D, Clark WG, Guerinot ML, Pakrasi HB (1999) The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol Biol 40 37–44 [DOI] [PubMed] [Google Scholar]
- Kriedmann PE, Graham RD, Wiskich JT (1985) Photosynthetic dysfunction and in vivo changes in chlorophyll a fluorescence from manganese deficient wheat leaves. Aust J Agric Res 36 157–169 [Google Scholar]
- Li L, Kaplan J (1998) Defects in the yeast high affinity iron transport system result in increased metal sensitivity because of the increased expression of transporters with a broad transition metal specificity. J Biol Chem 273 22181–22187 [DOI] [PubMed] [Google Scholar]
- Li P, Qi JL, Wang L, Huang QN, Han ZH, Yin LP (2006) Functional expression of MxIRT1, from Malus xiaojinensis, complements an iron uptake deficient yeast mutant for plasma membrane targeting via membrane vesicles trafficking process. Plant Sci 171 52–59 [Google Scholar]
- Lombi E, Tearall KL, Howarth JR, Zhao FJ, Hawkesford MJ, McGrath SP (2002) Influence of iron status on cadmium and zinc uptake by different ecotypes of the hyperaccumulator Thlaspi caerulescens. Plant Physiol 128 1359–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Millán AF, Ellis DR, Grusak MA (2004) Identification and characterization of several new members of the ZIP family of metal ion transporters in Medicago truncatula. Plant Mol Biol 54 583–596 [DOI] [PubMed] [Google Scholar]
- Lyons TJ, Gasch AP, Gaither LA, Botstein D, Brown PO, Eide DJ (2000) Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc Natl Acad Sci USA 97 7957–7962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner H (1995) Mineral Nutrition of Higher Plants. Academic Press, San Diego, pp 325–329
- Minet M, Dufour ME, Lacroute F (1992) Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J 2 417–422 [DOI] [PubMed] [Google Scholar]
- Nakanishi H, Ogawa I, Ishimaru Y, Mori S, Nishizawa NK (2006) Iron deficiency enhances cadmium uptake and translocation mediated by the Fe2+ transporters OsIRT1 and OsIRT2 in rice. Soil Sci Plant Nutr 52 464–469 [Google Scholar]
- Nunan KJ, Scheller HV (2003) Solubilization of an arabinan arabinosyltransferase activity from mung bean hypocotyls. Plant Physiol 132 331–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JN, Rengel Z (1997) Genotypic differences in the production and partitioning of carbohydrates between roots and shoots of wheat grown under zinc or manganese deficiency. Ann Bot (Lond) 80 803–808 [Google Scholar]
- Pedas P, Hebbern CA, Schjoerring JK, Holm PE, Husted S (2005) Differential capacity for high-affinity manganese uptake contributes to differences between barley genotypes in tolerance to low manganese availability. Plant Physiol 139 1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman JK (2005) Managing the manganese: molecular mechanisms of manganese transport and homeostasis. New Phytol 167 733–742 [DOI] [PubMed] [Google Scholar]
- Requena L, Bornemann S (1999) Barley (Hordeum vulgare) oxalate oxidase is a manganese-containing enzyme. Biochem J 343 185–190 [PMC free article] [PubMed] [Google Scholar]
- Reuter DJ, Edwards DG, Wilhelm NS (1997) Temperate and tropical crops. In DJ Reuter, JB Robinson, eds, Plant Analysis: An Interpretation Manual, Ed 2. CSIRO Publishing, Victoria, Australia, pp 81–279
- Rutherford AW, Boussac A (2004) Water photolysis in biology. Science 303 1782–1784 [DOI] [PubMed] [Google Scholar]
- Sherman F (1991) Getting started with yeast. Methods Enzymol 194 3–21 [DOI] [PubMed] [Google Scholar]
- Supek F, Supekova L, Nelson H, Nelson N (1996) A yeast manganese transporter related to the macrophage protein involved in conferring resistance to mycobacteria. Proc Natl Acad Sci USA 93 5105–5110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tsukamoto T, Nakanishi H, Kiyomiya S, Watanabe S, Matsuhashi S, Nishizawa NK, Mori S (2006) 52Mn translocation in barley monitored using a positron-emitting tracer imaging system. Soil Sci Plant Nutr 52 717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dédaldedéchamp F, Gaymard F, Guerinot ML, Briat JF, Curie C (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for the plant growth. Plant Cell 14 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters BM, Eide DJ (2002) Combinatorial control of yeast FET4 gene expression by iron, zinc and oxygen. J Biol Chem 277 33749–33757 [DOI] [PubMed] [Google Scholar]
- Waters BM, Lucena C, Romera FJ, Jester GG, Wynn AN, Rojas CL, Alcántara E, Pérez-Vicente R (2007) Ethylene involvement in the regulation of the H+-ATPase CsHA1 gene and of the new isolated ferric reductase CsFRO1 and iron transporter CsIRT1 genes in cucumber plants. Plant Physiol Biochem 45 293–301 [DOI] [PubMed] [Google Scholar]