Abstract
A silencing vector for cotton (Gossypium hirsutum) was developed from the geminivirus Cotton leaf crumple virus (CLCrV). The CLCrV coat protein gene was replaced by up to 500 bp of DNA homologous to one of two endogenous genes, the magnesium chelatase subunit I gene (ChlI) or the phytoene desaturase gene (PDS). Cotyledons of cotton cultivar ‘Deltapine 5415’ bombarded with the modified viral vectors manifested chlorosis due to silencing of either ChlI or PDS in approximately 70% of inoculated plants after 2 to 3 weeks. Use of the green fluorescence protein gene showed that replication of viral DNA was restricted to vascular tissue and that the viral vector could transmit to leaves, roots, and the ovule integument from which fibers originate. Temperature had profound effects on vector DNA accumulation and the spread of endogenous gene silencing. Consistent with reports that silencing against viruses increases at higher temperatures, plants grown at a 30°C/26°C day/night cycle had a greater than 10-fold reduction in viral DNA accumulation compared to plants grown at 22°C/18°C. However, endogenous gene silencing decreased at 30°C/26°C. There was an approximately 7 d delay in the onset of gene silencing at 22°C/18°C, but silencing was extensive and persisted throughout the life of the plant. The extent of silencing in new growth could be increased or decreased by changing temperature regimes at various times following the onset of silencing. Our experiments establish the use of the CLCrV silencing vector to study gene function in cotton and show that temperature can have a major impact on the extent of geminivirus-induced gene silencing.
Cotton (Gossypium spp.) is an economically important crop for both fiber and oil production in many warm areas of the world. Cotton's long-standing association with humans derives primarily from its long, strong, spinnable fibers, or seed hairs. Each fiber is a remarkable unicell that elongates to >2.25 cm and deposits a thick secondary wall (Kim and Triplett, 2001). Gossypium hirsutum is most commonly grown today, with smaller amounts of higher fiber quality Gossypium barbadense grown in compatible environments. Both are allotetraploids with large genomes that are recalcitrant to functional genomics approaches (Wendel and Cronn, 2003). To clothe an increasing world population and provide a sustainable raw material that is competitive with synthetic fiber, many agronomic properties of cotton, such as yield, drought tolerance, and pest resistance, need to be improved (Smith and Cothern, 1999). There is also a need to improve several properties of cotton fibers, including their length and strength, as well as the uniformity of all fiber properties. It is critical to identify genes that affect the molecular regulation of fiber development to begin to understand and modify these qualities (Arpat et al., 2004; Haigler et al., 2005).
Currently, complete sequencing of cotton genomes is just beginning (Chen et al., 2007). In the meantime, the basis for cotton functional genomics is provided by an ever-expanding set of Gossypium ESTs and derived unigene sets accessible on the Web (Udall et al., 2006), as well as by microarray analyses based on these sequences (Arpat et al., 2004; Gou et al., 2007; Udall et al., 2007; Wu et al., 2007). The availability of genomic sequences for Arabidopsis (Arabidopsis thaliana) and poplar (Populus spp.) has been extremely useful for identifying and annotating putative orthologs in the cotton EST databases. Even though analogies can be drawn between cotton fiber differentiation and the formation of leaf trichomes (Rong et al., 2005) and secondary-walled xylem cells (Haigler et al., 2005), ultimately the function of putative orthologous genes needs to be tested directly in cotton. The use of RNA interference in cotton has been hindered by inefficient production of stably transformed plants (Wilkins et al., 2004). In addition, cotton fibers express genes with no known homologs in other plants, and these may confer some of its unique properties. Because the most direct way of testing fiber gene functions is to positively or negatively manipulate their expression level, and transformation of cotton is difficult (Wilkins et al., 2004), a transient gene manipulation system is sorely needed.
Virus-induced gene silencing (VIGS) offers the opportunity to test gene function through silencing, using homologous cDNA sequences cloned from cotton. In geminivirus vectors, only partial gene sequences are needed to initiate gene silencing, ranging from about 90 to 150 bp if inserted as part of a viral gene (Peele et al., 2001; Jordan et al., 2007), or up to approximately 400 to 800 bp if inserted as a gene replacement (Kjemtrup et al., 1998). Silencing typically begins within a few weeks of inoculation and can extend throughout the flowering period. VIGS can also be used to test the function of essential genes because the targeted seedlings have already germinated and produced leaves before silencing begins (Peele et al., 2001; Jordan et al., 2007). Combinations of genes can be down-regulated simultaneously from the same vector (Peele et al., 2001; Turnage et al., 2002), allowing transcripts of multiple gene family members to be eliminated. This ability could be extremely useful in cotton because of its allotetraploid genome.
Cotton leaf crumple virus (CLCrV) is in the genus Begomovirus, family Geminiviridae (Brown and Nelson, 1984). It is endemic to the southwestern United States and northwestern Mexico (Brown and Nelson, 1987). CLCrV causes foliar crumpling and mosaic, floral hypertrophy, and stunted growth, resulting in reduced cotton fiber yield and quality (Butler et al., 1986; Brown and Nelson, 1987). CLCrV, like other begomoviruses, requires the coat protein gene for transmission by the whitefly Bemisia tabaci (Briddon et al., 1989; Azzam et al., 1994; Liu et al., 1999). The only other method of inoculating begomoviruses lacking a coat protein gene is under laboratory conditions, where the cloned viral genome components can be inoculated by gene gun bombardment (Kanevski et al., 1992), Agrobacterium (Elmer et al., 1988), or by rubbing DNA onto leaves dusted with carborundum (Ascencio-Ibanez and Settlage, 2007).
The organization of the CLCrV genome is typical of other bipartite begomoviruses and consists of two 2.5-kb circular DNA molecules referred to as the DNA-A and -B component, respectively (for review, see Gutierrez, 1999; Hanley-Bowdoin et al., 1999; Rojas et al., 2005). DNA-A and DNA-B share a region of homology of approximately 200 bp known as the common region. This common region contains the origin of viral DNA replication and is immediately upstream of the two, bidirectional promoters of each DNA component, which are involved in viral and complementary gene expression. The viral AL1 protein nicks double-stranded DNA to initiate replicational release of a single-stranded viral DNA molecule (Laufs et al., 1995). Host DNA replication machinery is required for both single- and double-stranded DNA synthesis and it is the single-stranded form that is encapsidated into viral particles that remain in the nucleus. Viral DNA behaves as mobile episomes, shuttling in and out of the cell nucleus and then between cells, tissues, and organs to establish systemic infection (Noueiry et al., 1994; Sanderfoot and Lazarowitz, 1996).
To date, only the viral-sense transcriptional unit of DNA-A or -B has been modified for gene silencing (Kjemtrup et al., 1998; Peele et al., 2001; Turnage et al., 2002; Fofana et al., 2004). Geminiviruses are themselves susceptible to gene silencing, but also encode suppressors of gene silencing and in some cases persist in the presence of siRNAs (Chellappan et al., 2004; Vanitharani et al., 2004; Bisaro, 2006; Zrachya et al., 2007). Several plant genes are known to be required for geminivirus-mediated VIGS in Arabidopsis (Muangsan et al., 2004) but the degree of functional conservation of these genes in other plants, and of silencing pathways in general, is not well understood.
Because CLCrV is indigenous to the United States, we modified it as a VIGS vector for cotton. We show here that deleting the coat protein gene, which is absolutely required for insect transmission, does not impair systemic movement in cotton, thereby obviating any chance for escape or transmission of vector sequences. We also report silencing of two different endogenous genes when 250- to 500-bp sequences, homologous to only one of the G. hirsutum homeologs, are used as coat protein gene replacements. We document widespread and long-lasting transmission of silencing signals in cotton vegetative and maternal reproductive tissues, and show that endogenous gene silencing is more efficient at a relatively cool temperature (e.g. 18°C–22°C) for cotton growth.
RESULTS
The Coat Protein Is Not Needed for Systemic CLCrV Infection in Cotton
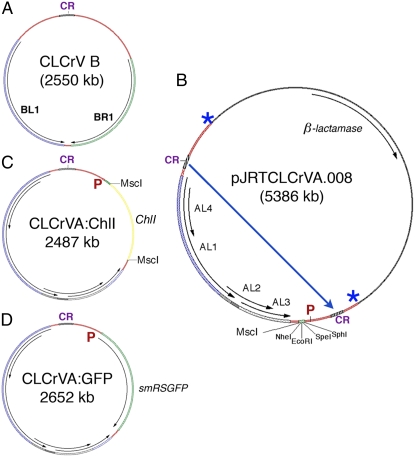
CLCrV had previously been modified to enable inoculation of plants with the wild-type virus using biolistic bombardment of cotton (Idris and Brown, 2004) with an Escherichia coli plasmid vector containing a tandemly repeated viral DNA-A or -B component. Figure 1A shows the B component of CLCrV represented in episomal form, which is the active form in plants. Episomes are released from plasmid DNA by rolling circle replication (Stenger et al., 1991) following bombardment. Figure 1B shows the parent E. coli plasmid used to manipulate viral DNA. In this case the viral DNA-A component has been engineered to contain a multiple cloning site (MCS) in place of the coat protein open reading frame. This plasmid was used as the empty vector control. Figure 1C shows the episomal form of the same viral DNA shown in Figure 1B, but also containing a silencing fragment, and in Figure 1D the entire GFP gene was cloned into the MCS.
Figure 1.
Plasmids used for bombardment. A, Wild-type CLCrV B component showing divergent transcription of the BR1 and BL1 genes from the common region (CR). The CLCrV DNA-A component (not shown) has a similar coding region and five divergently transcribed genes. Both components are needed for systemic infection. B, E. coli plasmid containing the A component of the empty VIGS vector. The coat protein (AR1) open reading frame has been replaced with an MCS with MscI, NheI, EcoR1, SpeI, and SphI recognition sites for cloning gene silencing fragments behind the AR1 promoter and before the putative polyA site. The duplicated common regions in this plasmid allow replicational release of unit length episomes, such as in A, C, and D, following introduction into plant cells. The long blue arrow shows sites in the coding region that are nicked by the AL1 Rep protein for initiation of rolling circle replication and then AL1-mediated ligation to circularize the viral DNA. Overlapping arrows in the viral DNA show AL1, AL4, AL2, and AL3 genes. E. coli sequence has the β-lactamase gene conferring ampicillin resistance. P, AR1 promoter. Asterisks show the junctions between viral and plasmid DNA. C, CLCrV A component of the VIGS vector for silencing ChlI. It is the same as the viral DNA part of the plasmid shown in B except that a 500-bp gene fragment from the cotton ChlI gene has been cloned into the MscI site for transcription by the AR1 promoter (P). This episome contains a single common region and is small enough to move between cells. D, CLCrV A component used as an expression vector for rsSMGFP. It is the same as C except that a full-length smRSGFP gene has been cloned into the MCS in sense orientation with respect to the AR1 promoter (P). [See online article for color version of this figure.]
One tool for bombardment used in these experiments was a custom-made particle inflow gun (PIG). This was used to inoculate cotyledon-stage seedlings with wild-type CLCrV using 1-μm-diameter DNA-coated gold microprojectiles. Previous experiments using the PIG showed that both Nicotiana benthamiana and Arabidopsis had high frequencies of infection and minimal damage at 30 psi of helium. Conditions for cotton bombardment required increasing the pressure used to propel the microprojectiles to 60 psi and placing the seedlings 4 cm beneath the microprojectile source. The total amount of DNA coated onto microprojectiles was varied from 0.5 to 10 μg each of the DNA-A and -B components (1–20 μg DNA, total). Optimal silencing efficiencies (100% of bombarded plants) using the PIG were found using 7 to 10 μg of each component. The infection rate declined with lower amounts of DNA-A and was about 50% for 5 μg and 30% to 50% for 0.5 μg each component. A commercially available particle delivery system (Bio-Rad PDS1000-He) was also used to inoculate seedlings and, with 0.5 μg of each component, also resulted in an average infection rate of 70%. Most data reported here used the PIG to inoculate plants with 5 μg of each DNA component (approximately 1 μg/shot).
ChlI and PDS Provide Two Visible Markers for Endogenous Gene Silencing
The magnesium chelatase subunit I gene (ChlI) was used to visualize the timing and extent of endogenous gene silencing. Because nearly exact homology is needed for effective silencing, reverse transcription (RT)-PCR was used to amplify a 500-bp ChlI fragment from G. hirsutum. This gene fragment, which was only 78.2% homologous to the Arabidopsis ChlI gene, was cloned in the antisense direction into pJRTCLCrVA.008 (Fig. 1B) to produce a DNA-A component transcribing the ChlI gene fragment using the coat protein gene promoter (Fig. 1C; CLCrV:ChlI). Because functional virus requires both the DNA-A and -B components, the silencing vector that includes both components is referred to here as CLCrV:ChlI. Bombardment of CLCrV:ChlI into seedlings resulted in silencing of ChlI, evidenced by loss of chlorophyll as early as 13 d postinoculation (dpi) at 25°C/23°C. Both the VIGS DNA-A component, carrying ChlI, and DNA-B component replicated out of the respective plasmid as viral episomes (Fig. 1, A and C) and moved together to establish infection.
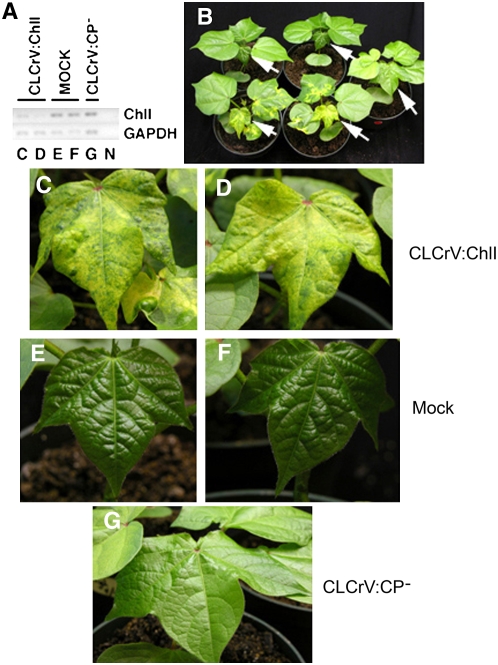
To verify reduction in ChlI mRNA in silenced leaves, a different region of the same cotton ChlI gene used in the vector was amplified from silenced tissues by RT-PCR. Figure 2A shows the molecular evidence for silencing. Almost undetectable ChlI transcript was in CLCrV:ChlI leaves (Fig. 2A, lanes C and D) that showed extensive chlorosis (Fig. 2, C and D). In contrast, leaves after mock inoculation (Fig. 2A, lanes E and F) or inoculation with empty vector (Fig. 2A, lane G) had higher ChlI transcript levels and minimal symptoms (Fig. 2, E–G).
Figure 2.
Visual and molecular evidence of CLCrV-induced gene silencing. A, Semiquantitative RT-PCR showing down-regulation of the ChlI gene in silenced plants kept at 25°C/23°C for 29 d. The top and bottom sections show ChlI accumulation after 27 PCR cycles or expression of GAPDH (internal control) after 23 PCR cycles, respectively. Letter beneath each lane corresponds to the section showing the leaf used for RNA isolation, except N is the no-RT-PCR control. B, Plants oriented with the leaves used for RT-PCR facing front. Arrows show the leaf used for each RNA extraction. Top row: mock inoculated; bottom row: ChlI-silenced; and middle row: right, empty vector. C and D, CLCrV:ChlI-inoculated leaves. E and F, Mock-inoculated plants bombarded with the CLCrV B component, which does not replicate in the absence of the A component. G, Empty vector-inoculated plants bombarded with pJRTCLCrV.008 and the B component. Chlorophyll appears to be lighter than in E and F because the leaf is horizontal and reflects more light (see B for comparison).
The phytoene desaturase gene (PDS) is another commonly used marker gene for VIGS and also causes loss of chlorophyll and carotenoids (Rotenberg et al., 2006; Senthil-Kumar et al., 2007). A 326-bp PDS gene fragment isolated from cotton was cloned into the CLCrV vector, and plants were bombarded with CLCrV:PDS, CLCrV:ChlI, or the empty vector. Figure 3 shows that the general pattern of cells and tissues with PDS silencing was similar to that of ChlI, but the extent of silencing was not as great. To determine if the size of the fragment affected the extent of silencing, the ChlI insert was reduced to 254 bp. The extent of silencing achieved using this fragment was reduced compared to the 500-bp ChlI fragment (data not shown).
Figure 3.
Silencing of PDS and ChlI in leaves along the plant vertical axis, 47 dpi at 25°C/23°C. Top row shows CLCrV-mediated silencing with a 326-bp PDS gene fragment from cotton. In the bottom row, equivalent leaves from a plant bombarded with CLCrV containing a 500-bp cotton ChlI gene fragment are shown. This picture was taken with all leaves positioned together on a light table and black was substituted for the white background using Adobe Photoshop.
The CLCrV Vector Was Found in Young and Mature Leaves, Roots, and Ovules
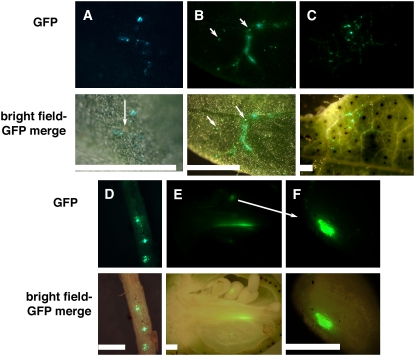
Although geminivirus-mediated VIGS facilitates extensive gene silencing, in situ hybridization using viral DNA-A as the probe has shown that very few of the silenced cells actually accumulate detectable viral DNA (Peele et al., 2001). To track exactly where and when the CLCrV silencing vector produced endogenous gene silencing signals, a small, 729-bp red-shifted GFP gene (Davis and Vierstra, 1998) with cell-autonomous expression was inserted into the CLCrV vector MCS in place of a silencing fragment. For plants grown at 25°C/23°C, CLCrV:GFP showed expression in small groups of cotyledon cells 4 dpi. Figure 4A shows one such group adjacent to a clump of gold particles, suggesting that these were the first cells to support viral DNA replication from the bombarded plasmid DNA. Since these cells were not part of veins, at least initially viral DNA replication and gene expression occurred in nonvascular tissues.
Figure 4.
CLCrV:GFP expression is limited to vascular tissue and extends to the ovule integument. Top section shows fluorescence from the smRSGFP-expressing CLCrV vector and the bottom section shows the merged brightfield-GFP photo. Plants were grown at 25°C/23°C and similar, mock-inoculated tissues were negative for GFP-like fluorescence (data not shown) unless mentioned. A, Cotyledon 9 dpi. Arrow indicates gold particles. B, Cotyledon tissue showing viral DNA accumulation along some of the veins, indicating the beginning of systemic movement. Arrows show glandular trichomes that were also autofluorescent in control leaves of the same age. Otherwise, controls showed no fluorescence. C, Older leaf showing GFP-expressing cells associated with vascular tissue. Only some of the veins showed GFP fluorescence, demonstrating that veins are not inherently autofluorescent. D, Root tissue with three regions of CLCrV:GFP DNA accumulation. E and F, Longitudinal section of a 3-DPA ovary showing green fluorescence along part of the central column and in one of the ovules, which is magnified in F. The ovule showed one area with strong GFP fluorescence. Bars = 2 mm.
Research in bean (Phaseolus vulgaris) showed that a GFP-expressing geminivirus (Bean dwarf mosaic virus) could also be localized in leaf cells following microinjection, but that systemic movement was restricted to the vascular tissue (Sudarshana et al., 1998). We observed a similar pattern with CLCrV:GFP and found that after 9 dpi, GFP expression was observed almost exclusively in or near the cotton plant vasculature, including the midvein and secondary and tertiary veins (Fig. 4, B and C). Early in the infection cycle, bright fluorescence was detected in both mature and young leaves. However, at approximately 45 to 50 dpi, GFP fluorescence became predominant in the rapidly expanding areas of the youngest leaves and more transient, diminishing as a leaf region matured. In addition, GFP fluorescence was sometimes observed in the outer boll wall (data not shown), the central column of the ovary (Fig. 4E), and the ovule integuments (Fig. 4F). For unknown reasons, plant-to-plant variation in GFP fluorescence occurred, and GFP fluorescence in some plants became undetectable in the aboveground tissues prior to or during boll formation. Surprisingly, some of these plants still maintained GFP expression in the roots (Fig. 4D).
Lower Temperatures Enhanced Geminivirus-Mediated VIGS
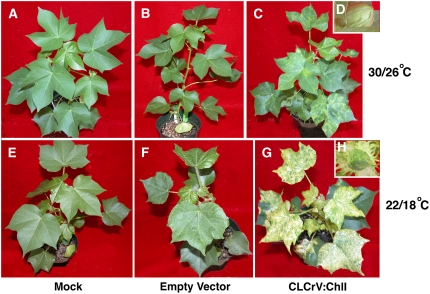
To try to increase the efficiency of silencing, bombarded plants were grown in greenhouses with tight temperature control in three trials to compare ChlI silencing at relatively high (30°C/26°C) and low (22°C/18°C) temperature regimes. Although silencing was delayed at the cooler temperatures, the extent of silencing was dramatically increased and was significantly more stable throughout the life of the plant (Fig. 5, G and H compared to C and D). Importantly, CLCrV:ChlI-mediated gene silencing occurred in the ovary wall as variegated tissue at lower temperature (Fig. 5H), but this was variable between plants. In contrast, bolls of plants grown at 30°C/26°C did not show conclusive evidence of ChlI silencing (Fig. 5D), probably because plants inoculated at the seedling stage could not routinely sustain silencing until the bolls formed. Plants inoculated with the empty vector were only slightly more symptomatic at lower temperatures (compare Fig. 5, F and B). However, growth of the cotton plants was slower at 22°C/18°C (compare Fig. 5, E and A) and the number of days to flowering increased. To decrease the time to flowering and boll formation, bombarded cotton seedlings were kept in a growth chamber at a constant temperature of 26°C, 16/8 h light/dark photoperiod until 26 dpi, at which time the plants had begun to silence. They were then transferred to the greenhouses with either low or high temperature regimes. The 26°C preincubation did not significantly affect the extent of silencing at either temperature regime compared to continuous incubation at high or low temperature. Silencing in newly formed tissues was still enhanced at low temperatures (data not shown).
Figure 5.
The efficiency of gene silencing is affected by temperature. Plants mock-bombarded (A and E), bombarded with an empty vector (B and F), or bombarded with CLCrV:ChlI (C, D, G, and H) were transferred at 2 dpi to a high or low temperature day/night cycle (30°C/26°C or 22°C/18°C, respectively). Mock-inoculated plants (A and E) showed slower growth at 22°C/18°C, but leaf morphology was normal. Empty vector-inoculated plants at high temperature (B) were similar to mock, but at low temperature, downward leaf curling and lighter veins were seen (F). CLCrV:ChlI-inoculated plants at high temperature showed a mixture of punctate and diffuse silencing (C). The diffuse silencing ranged from yellow to light green and disappeared with time. Plants grown at lower temperature (G) also produced punctate silencing, but diffuse silencing was more extensive, ranging from yellow to white, and persisting over time. There was some downward leaf curling at the edges, but symptoms were otherwise not noticeable. The outer walls of developing bolls did not show silencing at the high temperature regime (D), whereas variegation of bolls was observed with the lower temperature regime (H).
To further investigate the relationship between temperature change and silencing, two sets of plants were kept at a constant intermediate (25°C) or low temperature regime (22°C/18°C) until 59 dpi and then switched to the reciprocal temperature regimes for 7 d. Figure 6 shows that new growth formed during the 7-d period showed silencing characteristic for the new temperature, and that the change in silencing effectiveness was immediate for both treatments. These results suggest that CLCrV-induced endogenous gene silencing is quite sensitive to temperature and verify that temperature was the main variable in the experiment.
Figure 6.
Temperature rapidly changes the extent of gene silencing in new tissue. The same plant is shown in the top two or bottom two photos. Plants on the left were 59 dpi at 20°C/18°C (top left) or 25°C/23°C (bottom left). At 59 dpi, the plants were switched to new temperature regimes for 7 d (right). Leaves within the red circle show new growth at higher temperatures and have reduced chlorosis, the phenotype for ChlI silencing, while the new leaf at low temperatures has areas of increased, confluent silencing. The experiment used identical Percival chambers, except for temperature. Arrows on the bottom photos point to the same leaf for reference.
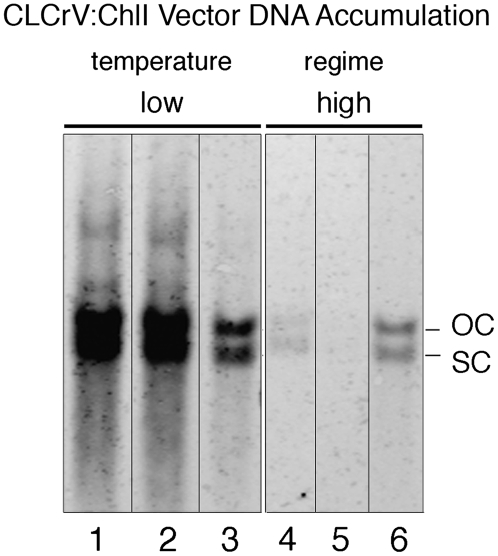
To determine if the lack of ChlI silencing at high temperatures was due to a breakdown in the host gene silencing response or to a lack of CLCrV:ChlI vector, leaf tissue that showed at least some silencing was used for DNA isolation and probed with viral vector DNA. Figure 7 shows that much less CLCrV:ChlI vector DNA was present in plants grown at 30°C/26°C compared to 22°C/18°C. Analysis of the blot using Image J showed that there was a 12-fold difference between DNA accumulation in the two treatments.
Figure 7.
Vector DNA accumulation is reduced at higher temperature. Leaves showing visible evidence of silencing from the CLCrV:ChlI vector were collected 47 dpi and used for DNA isolation. Lanes 1 to 3 show DNA from three different leaves from different plants grown at lower temperature (22°C/18°C, as in Fig. 5G) while lanes 4 to 6 show DNA isolated from plants grown at higher temperature (30°C/26°C; see Fig. 5C). A digoxigenin-labeled DNA probe corresponding to the coding region and part of AL1 was used. Ethidium bromide staining of the gel showed that equal amounts of genomic DNA were loaded in each lane (data not shown). OC, Open circular form of CLCrVA:ChlI; SC, supercoiled.
DISCUSSION
This report describes a geminivirus-induced gene silencing vector that functions in cotton, an important, sustainable source of fiber and oil. Because cotton is difficult to transform, development of a transient gene silencing method greatly extends the power of molecular analysis for testing gene function. Genes with homology to previously characterized genes can now be tested for predicted loss-of-function phenotypes to verify or refute the possible conservation of function. Using data from several cotton EST projects (Udall et al., 2006), gene sequences now labeled as “unknown or hypothetical protein” can be tested to determine phenotypes associated with their loss of function. As previously described (Robertson, 2004), such analyses should always include an empty virus VIGS vector to control for any possible effects of the modified viral DNA infection itself.
Geminivirus-mediated VIGS in Arabidopsis (Turnage et al., 2002) and the first version of the geminivirus VIGS vector for N. benthamiana (Kjemtrup et al., 1998) are more symptomatic than the widely used tobacco rattle virus (TRV) VIGS vector (Ratcliff et al., 2001; Dong et al., 2007), and the viruses they are derived from produce severe symptoms. However, symptoms of wild-type CLCrV and its vector are not severe in cotton (Figs. 2, 5, and 6). Previous studies demonstrated that genetic changes, such as up-regulation of DNA replication machinery, are cell autonomous in geminivirus infections (Nagar et al., 2002), but more recent studies using microarrays (Ascencio-Ibañez et al., 2008) show that genes in pathogen response pathways that are regulated by diffusible compounds are also up-regulated. The GFP experiments demonstrated that only cells in the vascular tissue become infected, suggesting that genes expressed only in the mesophyll, epidermal, and other tissues may be more straightforward to analyze by VIGS. We also found that GFP-containing vectors showed fewer symptoms than no-insert VIGS controls, likely because the addition of nonhomologous foreign sequence to the viral genome further attenuated symptoms. Even though symptoms appear to be attenuated, it is essential to control for background changes in gene expression due to the VIGS vector. When analyzing RBR function in N. benthamiana, we used a no-insert (empty) VIGS vector as a negative control and a VIGS vector containing ChlI as the positive control to gauge the timing and extent of silencing (Jordan et al., 2007).
With the CLCrV silencing system, plants grown in a temperature-controlled greenhouse at low temperatures (22°C/18°C) showed substantially better silencing than those grown at higher (30°C/26°C) temperatures (Fig. 5). For naturally occurring plant virus infections, symptom severity and the molecular processes involved in infection are known to be affected by temperature, with lower temperatures inhibiting silencing and higher temperatures favoring silencing and even viral elimination in some cases (Szittya et al., 2003). For both RNA and DNA viruses, siRNA accumulation has been shown to increase at higher temperatures (Szittya et al., 2003; Chellappan et al., 2005). This observation along with decreased viral DNA accumulation led to the proposal that posttranscriptional gene silencing (PTGS) caused reduction or elimination of viral DNA at high temperatures (Chellappan et al., 2005). RDR6 is a known virulence factor for Cabbage leaf curl virus in Arabidopsis (Muangsan et al., 2004) and is involved in both PTGS and RNA-directed DNA methylation (Herr et al., 2005), which could also adversely impact DNA viruses (Wang et al., 2005; Bisaro, 2006). There is also recent evidence for a role for RDR6 in long-distance siRNA movement (Brosnan et al., 2007). The antiviral activity of RDR6 has been shown to increase with temperature in N. benthamiana (Qu et al., 2005), providing a mechanism for the lesser accumulation of viral DNA at higher temperature.
Although the higher temperatures employed here appear to have increased PTGS of the viral DNA vector, the efficiency of endogenous ChlI silencing from VIGS was reduced at higher versus lower temperatures. Fauquet's group showed that siRNAs increase with temperature while viral DNA accumulation dropped at 30°C compared to 25°C (Chellappan et al., 2005). Reduced or arrested viral replication is therefore likely to decrease ChlI silencing by preventing transcription of the ChlI silencing fragment, which is needed to provide siRNAs for endogenous gene silencing. Although this might seem obvious, it is not clear why the endogenous ChlI silencing is so extensive at low temperatures. In another report of temperature effects on VIGS, TRV-VIGS of endogenous tomato (Solanum lycopersicum) genes, PDS and EIN, was increased and more persistent (until fruiting) at low temperature, although onset of silencing was delayed up to 6 weeks at 15°C compared to 21°C (Fu et al., 2006). A comparison of viral RNA levels was not reported, so it is unknown whether the VIGS vector was eliminated at 21°C. Similarly to tomato, ChlI silencing in cotton was more transient at high temperature. A second report also showed that TRV-mediated VIGS in tomato decreased when temperature increased but that VIGS from a DNA satellite was relatively stable at temperatures from 20°C to 32°C (Cai et al., 2007). It remains to be determined whether temperature will have dramatic effects on the efficiency of VIGS in other virus-host combinations. The finding that TRV-mediated VIGS in N. benthamiana was more tolerant to temperature variation than tomato (Nethra et al., 2006) suggests that temperature should be optimized for each virus-host combination. The rapid increase in silencing of developing cotton leaves following a shift from 25°C/23°C to 22°C/18°C (Fig. 6) indicates that the temperature-affected endogenous gene silencing pathway in cotton is dynamic and malleable, a finding that offers new opportunities for silencing optimization and research.
An important goal for cotton research is to be able to modify properties of the cotton fiber, which arise from ovular epidermal cells near the time of flower opening. Because the ChlI VIGS could not be used to track silencing in tissues lacking chlorophyll, we used GFP expression to determine if the CLCrV vector was capable of infecting the ovule integument, which is derived from maternal tissue. GFP fluorescence from geminivirus vectors invading the maternal tissue of developing ovules has now been observed in two plants, cotton (this report, Fig. 4) and bean (Sudarshana et al., 1998). In both cases, the viruses are known to be vascular tissue specific. Localization of virus in the cotton ovule integument is consistent with previous reports on the continuity between strands of phloem in the central column and each individual ovule (Gore, 1935; Van Lersel et al., 1995). The results shown in Figure 4 suggest that cotton fibers might be subject, in future experiments, to gene silencing at an early stage in their development. Although CLCrV:GFP showed GFP expression in the integument, the frequency of infection was very low (e.g. four ovules within bolls of 10 plants grown at 25°C/23°C postinoculation). Perhaps the efficiency could be increased if plants were subjected to lower temperature shortly before or just after flowering. In addition, Agrobacterium-based delivery methods currently under development could add versatility by allowing inoculation into specific leaves, including subtending leaves that are a major carbon source for developing bolls.
MATERIALS AND METHODS
Cotton Plant Growth
Cotton (Gossypium hirsutum ‘Deltapine 5415’ for experiments) seeds were germinated in MetroMix potting soil in 4-inch square pots with four seeds per pot. Seedlings were grown until they had initiated the first true leaves (5–10 d), then biolistically inoculated and transplanted to individual pots (2–4 dpi). Plants were kept in a custom-built, walk-in growth chamber with a 16/8 h photoperiod at 900 μmol m−2 s−1 and a 25°C/23°C temperature regime for all experiments except the low and high temperature comparisons. Plants were fertilized twice weekly with Miracle Gro (Miracle Gro Products, Inc.). The temperature comparison experiments were carried out in two North Carolina State University Phytotron temperature-controlled (±1°C) greenhouses with either 30°C/26°C or 22°C/18°C day/night temperature cycles, relative humidity 40% to 50%, or in Percival growth chambers as stated. The Phytotron plants received ambient day length with a 3-h interruption during the dark period with 11 to 12 μmol m−2 s−1 of incandescent light to trigger long day responses throughout the year. Three experiments were performed consecutively from April to December. Plants were started individually in 225-mL styrofoam cups with two-thirds pea gravel, one-third peat-lite (WR Grace Co.) potting mixture, and transplanted to 1,650-mL pots 2 dpi. Plants were watered daily and fertilized three times a week with weak Hoagland solution (Saravitz et al., 2008).
Particle Bombardment
Seedlings were bombarded with 1-μm-diameter gold microprojectiles (InBio) coated with a mixture of 5 μg each of the A and B components of CLCrV as described (Kjemtrup et al., 1998). Experiments at University of Arizona used a Bio-Rad PDS1000 while those at North Carolina State University used a homemade PIG (details available upon request) attached to a high-pressure helium tank. DNA-coated microprojectiles were loaded onto the filter end of a Millipore Swinnex filter and each seedling was placed directly under the outlet, about 4 cm beneath it, and bombarded once. For all experiments except the temperature comparisons, four seedlings/4-inch square pot were shot one plant at a time with an outlet pressure of 30 to 60 psi, with 60 psi becoming the standard for routine work.
Construction of a CLCrV A Component Vector
The DNA-A component dimer, pCLCrV-H250 (Idris and Brown, 2004), was recloned to contain a single AR1 gene by PCR amplification of an 1,185-bp fragment from pCLCrV-H250 using the primer sequences 5′-AGTTCTAGAATCACCTTCCACTATGAGAC-3′ and 5′-TCAGAATTCCCTTAACGTGCGATAGATTCTGGGC-3′. This fragment, containing the common region and 669 bp of AL1, was digested with EcoRI and XbaI and ligated into pBluescript II SK+ (Stratagene) to make pJRTCLCrVA.002. pCLCrV-H250 was cut with XbaI and a 2,628-bp fragment containing AL2, AL3, AR1, and the remaining portion of AL1 was ligated into pJRTCLCrVA.002 to make pJRTCLCrVA.003. To produce a frameshift mutation in the coat protein gene, pJRTCLCrVA.003 was digested with BsmI, trimmed with T4 DNA polymerase, and then religated to make pJRTCLCrVA.007.
To insert the MCS, pJRTCLCrVA.007 was digested with HindIII and StuI, releasing a 1,772-bp fragment that was ligated into a HincII- and HindIII-cut pBluescript to make pJRTCLCrVA.004. Two PCR reactions were used to amplify regions of pJRTCLCrVA.007 that flanked AR1 and add an MCS. The first PCR reaction used primer sequences 5′-GCAAGCTTACCTGAACTTCCAAGTCTG-3′ and 5′-GTGAATTCGCTAGCGTTAACTGGCCATAATCCTGTGTATGCAACGTTGAA-3′ to amplify a 297-bp fragment that spanned AL3, introduced half of the MCS, and retained the AR1 stop codon and putative polyadenylation site. This product was digested with HindIII and EcoRI and ligated into pJRTCLCrVA.004 to produce pJRTCLCrVA.005. The second PCR reaction used the primers 5′-GCGAATTCACTAGTCTGCAGGCATGCCATTTTGCTCTATACCCAT-3′ and 5′-GCGGAGCTCCACTTGGGATAGGTTAAGAA-3′ to produce a 454-bp fragment that contained the common region, the start codon of AR1, and the remaining MCS. This product was digested with EcoRI and SacI and ligated into pJRTCLCrVA.005 to complete the MCS and produce pJRTCLCrVA.008 (GenBank EU541443). This plasmid contained the entire viral genome and two common regions, with AR1 replaced by an MCS, inserted between the AR1 start and stop codons.
Isolation of ChlI and PDS Gene Fragments from Cotton
From young leaves, 100 mg total RNA was isolated using the Spectrum Total Plant RNA kit (Sigma). A 500-bp fragment of the cotton ChlI gene (GenBank accession EU541445) was obtained by RT-PCR using primers 5′-GCATGGCCATTCGGTGACCCTTATAAC-3′ and 5′-GCTTGGCCAATCAAACCGTGCTCTTTC-3′. The PCR product was digested with MscI and ligated into pJRTCLCrVA.008 to produce pJRTCLCrVA.009 (GenBank accession EU541444), an A component vector for silencing the ChlI gene. The PDS gene fragment was similarly obtained by RT-PCR using the primers 5′-GCCGCATGCGCCTGAAGACTGGAGAGAGATT-3′ and 5′-GCTACTAGTGCTTTACTCTGATCCGCAGATA-3′ (GenBank accession no. EU541446) and was cloned into pJRTCLCrVA.008 to construct pJRTCLCrVA.027.
Insertion of a Full-Length GFP Gene into CLCrV
PCR was used to amplify a 729-bp fragment from psmRSGFP (GenBank accession U70496; Davis and Vierstra, 1998). This was digested with SphI and EcoRV and ligated to SphI- and MscI-cut pJRTCLCRVA.008 to produce pJRT010, containing full-length smRSGFP placed in frame with the start and stop codons of AR1. The presence of the SphI site upstream of smRSGFP resulted in the addition of Ala and Cys codons between Met-1 and Ser-4 at the 5′ end of the open reading frame. The EcoRV/MscI ligation resulted in the addition of Asp and Pro codons immediately before the stop codon of the coding sequence.
Semiquantitative RT-PCR to Determine Transcript Levels of ChlI
Total RNA was extracted as above and 250 ng was used as a template for RT using ImpromII (Promega). One microliter of each cDNA reaction was used for semiquantitative PCR with primers 5′-GGCTCAGAAGCTTGCTGCTAAAGA-3′ and 5′-AACAGTTGTGGACTTCCCAGTTCC-3′, which amplified a 123-bp fragment of ChlI with no homology to the silencing vector fragment. A 165-bp fragment of cotton GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE A SUBUNIT (GAPDH; accession TC66315) was used as a reference gene and was amplified by PCR using 5′-ATCAAGGGCACCATGACTACCACT-3′ and 5′-ACCAGTTGAAGTCGGGACGATGTT-3′. Threshold cycles, or the cycles for which a 5-μL reaction yielded a DNA product that could be detected by ethidium bromide staining for ChlI and GAPDH, were empirically determined at 27 and 23, respectively.
Virus Detection
Cotton DNA was extracted by a modified protocol for Plant DNEASY (Qiagen) as described (Horne et al., 2004). Genomic DNA samples were digested with DpnI to eliminate input DNA, quantified, and 50 ng used for PCR. Using the primers 5′-CATGATCGAATCGTAAAAATAGATCCG-3′and 5′-GCCTAATGGGTATAGAGCAAAATG-3′, insert fragment sizes of 120, 627, and 800 bp were generated from pJRTCLCrVA.008, pJRTCLCrVA.009, and pJRTCLCrVA.010, respectively. DNA gel-blot hybridization was done using standard procedures and a 948-bp viral DNA probe corresponding to the common region and AL1 gene was labeled with digoxigenin-dUTP using the manufacturer's instructions (Roche Biomedical).
Photography
Photos were taken with a Nikon Coolpix digital camera and adjusted in Adobe Photoshop 7.1 (Adobe). Fluorescence was detected using a Leica MZFLIII dissecting microscope connected to a Qimaging Micropublisher 3.3 CCD camera. The red-shifted smRSGFP protein (excitation 495 nm and emission 510 nm) was imaged using a filter set with 450 to 490 nm excitation and 500 to 550 emission. All overlays were produced by setting the opacity of the GFP image to 50%, flattening with the second image, and using gamma correction to readjust the pixel histogram. Quantification of DNA in Figure 7 used the Gel Analysis tool of Image J version 1.40G.
Acknowledgments
We thank Dr. Alan Wenck for construction of the PIG, and Drs. Steve Spiker and Ron Sederoff for donating it to our lab. We are grateful to Taliesin Cochran for help with the plants and to Drs. Linda Hanley-Bowdoin and George Allen for critical evaluation of the manuscript. The smRSGFP plasmid, originally constructed by the R. Vierstra lab, was obtained from the Arabidopsis Biological Resource Center.
This work was supported by grants from Cotton Incorporated (to D.R., C.H.H., and J.K.B.) and a Fellowship from Cotton Incorporated (to J.R.T.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Dominique Robertson (niki_robertson@ncsu.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
Open Access articles can be viewed online without a subscription.
References
- Arpat AB, Waugh M, Sullivan JP, Gonzales M, Frisch D, Main D, Wood T, Leslie A, Wing RA, Wilkins TA (2004) Functional genomics of cell elongation in developing cotton fibers. Plant Mol Biol 54 911–929 [DOI] [PubMed] [Google Scholar]
- Ascencio-Ibanez JT, Settlage SB (2007) DNA abrasion onto plants is an effective method for geminivirus infection and virus-induced gene silencing. J Virol Methods 142 198–203 [DOI] [PubMed] [Google Scholar]
- Ascencio-Ibáñez JT, Sozzani R, Lee T-J, Chu T-M, Wolfinger RD, Cella R, Hanley-Bowdoin L (2008) Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol 148 436–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzam O, Frazer J, de la Rosa D, Beaver JS, Ahlquist P, Maxwell DP (1994) Whitefly transmission and efficient ssDNA accumulation of bean golden mosaic geminivirus require functional coat protein. Virology 204 289–296 [DOI] [PubMed] [Google Scholar]
- Bisaro DM (2006) Silencing suppression by geminivirus proteins. Virology 344 158–168 [DOI] [PubMed] [Google Scholar]
- Briddon RW, Watts J, Markham PG, Stanley J (1989) The coat protein of beet curly top virus is essential for infectivity. Virology 172 628–633 [DOI] [PubMed] [Google Scholar]
- Brosnan CA, Mitter N, Christie M, Smith NA, Waterhouse PM, Carroll BJ (2007) Nuclear gene silencing directs reception of long-distance mRNA silencing in Arabidopsis. Proc Natl Acad Sci USA 104 14741–14746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JK, Nelson MR (1984) Geminate particles associated with cotton leaf crumple disease in Arizona. Phytopath 74 987–990 [Google Scholar]
- Brown JK, Nelson MR (1987) Host range and vector relationships of cotton leaf crumple virus. Plant Dis 71 522–524 [Google Scholar]
- Butler GD, Brown JK, Henneberry TJ (1986) Effect of cotton seedling infection by cotton-leaf crumple-virus on subsequent growth and yield. J Econ Entomol 79 208–211 [Google Scholar]
- Cai X, Wang C, Xu Y, Xu Q, Zheng Z, Zhou X (2007) Efficient gene silencing induction in tomato by a viral satellite DNA vector. Virus Res 125 169–175 [DOI] [PubMed] [Google Scholar]
- Chellappan P, Vanitharani R, Fauquet C (2004) Short interfering RNA accumulation correlates with host recovery in DNA virus-infected hosts, and gene silencing targets specific viral sequences. J Virol 78 7465–7477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellappan P, Vanitharani R, Ogbe F, Fauquet CM (2005) Effect of temperature on geminivirus-induced gene silencing. Plant Physiol 138 1830–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Scheffler BE, Dennis E, Triplett BA, Zhang T, Guo W, Chen X, Stelly DM, Rabinowicz PD, Town CD, et al (2007) Toward sequencing cotton (Gossypium) genomes. Plant Physiol 145 1303–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis SJ, Vierstra RD (1998) Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol Biol 36 521–528 [DOI] [PubMed] [Google Scholar]
- Dong Y, Burch-Smith TM, Liu Y, Mamillapalli P, Dinesh-Kumar SP (2007) A ligation-independent cloning tobacco rattle virus vector for high-throughput virus-induced gene silencing identifies roles for NbMADS4-1 and -2 in floral development. Plant Physiol 145 1161–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmer JS, Sunter G, Gardiner WE, Brand L, Browning CK, Bisaro DM, Rogers SG (1988) Agrobacterium-mediated inoculation of plants with tomato golden mosaic virus DNAs. Plant Mol Biol 10 225–234 [DOI] [PubMed] [Google Scholar]
- Fofana IB, Sangare A, Collier R, Taylor C, Fauquet CM (2004) A geminivirus-induced gene silencing system for gene function validation in cassava. Plant Mol Biol 56 613–624 [DOI] [PubMed] [Google Scholar]
- Fu DQ, Zhu BZ, Zhu HL, Zhang HX, Xie YH, Jiang WB, Zhao XD, Luo KB (2006) Enhancement of virus-induced gene silencing in tomato by low temperature and low humidity. Mol Cells 21 153–160 [PubMed] [Google Scholar]
- Gore UR (1935) Morphogenetic studies on the inflorescence of cotton. Bot Gaz 97 118–138 [Google Scholar]
- Gou JY, Wang LJ, Chen SP, Hu WL, Chen XY (2007) Gene expression and metabolite profiles of cotton fiber during cell elongation and secondary cell wall synthesis. Cell Res 17 422–434 [DOI] [PubMed] [Google Scholar]
- Gutierrez C (1999) Geminivirus DNA replication. Cell Mol Life Sci 56 313–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigler CH, Zhang DH, Wilkerson CG (2005) Biotechnological improvement of cotton fibre maturity. Physiol Plant 124 285–294 [Google Scholar]
- Hanley-Bowdoin L, Settlage SB, Orozco BM, Nagar S, Robertson D (1999) Geminiviruses: models for plant DNA replication, transcription and cell cycle regulation. Crit Rev Plant Sci 18 71–106 [PubMed] [Google Scholar]
- Herr AJ, Jensen MB, Dalmay T, Baulcombe DC (2005) RNA polymerase IV directs silencing of endogenous DNA. Science 308 118–120 [DOI] [PubMed] [Google Scholar]
- Horne EC, Kumpatla SP, Patterson KA, Gupta M, Thompson SA (2004) Improved high-throughput sunflower and cotton genomic DNA extraction and PCR fidelity. Plant Mol Biol Rep 22 83a–83i [Google Scholar]
- Idris AM, Brown JK (2004) Cotton leaf crumple virus is a distinct western hemisphere begomovirus species with complex evolutionary relationships indicative of recombination and reassortment. Phytopathology 94 1068–1074 [DOI] [PubMed] [Google Scholar]
- Jordan CV, Shen W, Hanley-Bowdoin L, Robertson D (2007) Geminivirus-induced gene silencing of the tobacco retinoblastoma-related gene results in cell death and altered development. Plant Mol Biol 65 163–175 [DOI] [PubMed] [Google Scholar]
- Kanevski IF, Thakur S, Cosowsky L, Sunter G, Brough C, Bisaro D, Maliga P (1992) Tobacco lines with high copy numbers of replicating recombinant geminivirus vectors after biolistic DNA delivery. Plant J 2 457–463 [Google Scholar]
- Kim HJ, Triplett BA (2001) Cotton fiber growth in planta and in vitro: models for plant cell elongation and cell wall biogenesis. Plant Physiol 127 1361–1366 [PMC free article] [PubMed] [Google Scholar]
- Kjemtrup S, Sampson KS, Peele CG, Nguyen LV, Conkling MA, Thompson WF, Robertson D (1998) Gene silencing from plant DNA carried by a Geminivirus. Plant J 14 91–100 [DOI] [PubMed] [Google Scholar]
- Laufs J, Schumacher S, Geisler N, Jupin I, Gronenborn B (1995) Identification of the nicking tyrosine of geminivirus Rep protein. FEBS Lett 377 258–262 [DOI] [PubMed] [Google Scholar]
- Liu S, Briddon RW, Bedford ID, Pinner MS, Markham PG (1999) Identification of genes directly and indirectly involved in the insect transmission of African cassava mosaic geminivirus by Bemisia tabaci. Virus Genes 18 5–11 [DOI] [PubMed] [Google Scholar]
- Muangsan N, Beclin C, Vaucheret H, Robertson D (2004) Geminivirus VIGS of endogenous genes requires SGS2/SDE1 and SGS3 and defines a new branch in the genetic pathway for silencing in plants. Plant J 38 1004–1014 [DOI] [PubMed] [Google Scholar]
- Nagar S, Hanley-Bowdoin L, Robertson D (2002) Host DNA replication is induced by geminivirus infection of differentiated plant cells. Plant Cell 14 2995–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nethra P, Nataraja KN, Rama N, Udayakumar M (2006) Standardization of environmental conditions for induction and retention of post-transcriptional gene silencing using tobacco rattle virus vector. Curr Sci 90 431–435 [Google Scholar]
- Noueiry AO, Lucas WJ, Gilbertson RL (1994) Two proteins of a plant DNA virus coordinate nuclear and plasmodesmal transport. Cell 76 925–932 [DOI] [PubMed] [Google Scholar]
- Peele C, Jordan CV, Muangsan N, Turnage M, Egelkrout E, Eagle P, Hanley-Bowdoin L, Robertson D (2001) Silencing of a meristematic gene using geminivirus-derived vectors. Plant J 27 357–366 [DOI] [PubMed] [Google Scholar]
- Qu F, Ye X, Hou G, Sato S, Clemente TE, Morris TJ (2005) RDR6 has a broad-spectrum but temperature-dependent antiviral defense role in Nicotiana benthamiana. J Virol 79 15209–15217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff F, Martin-Hernandez AM, Baulcombe DC (2001) Technical advance: tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J 25 237–245 [DOI] [PubMed] [Google Scholar]
- Robertson D (2004) VIGS vectors for gene silencing: many targets, many tools. Annu Rev Plant Physiol Plant Mol Biol 55 495–519 [DOI] [PubMed] [Google Scholar]
- Rojas MR, Hagen C, Lucas WJ, Gilbertson RL (2005) Exploiting chinks in the plant's armor: evolution and emergence of geminiviruses. Annu Rev Phytopathol 43 361–394 [DOI] [PubMed] [Google Scholar]
- Rong J, Bowers JE, Schulze SR, Waghmare VN, Rogers CJ, Pierce GJ, Zhang H, Estill JC, Paterson AH (2005) Comparative genomics of Gossypium and Arabidopsis: unraveling the consequences of both ancient and recent polyploidy. Genome Res 15 1198–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotenberg D, Thompson TS, German TL, Willis DK (2006) Methods for effective real-time RT-PCR analysis of virus-induced gene silencing. J Virol Methods 138 49–59 [DOI] [PubMed] [Google Scholar]
- Sanderfoot AA, Lazarowitz SG (1996) Getting it together in plant virus movement: cooperative interactions between bipartite geminivirus movement proteins. Trends Cell Biol 6 353–358 [DOI] [PubMed] [Google Scholar]
- Saravitz CH, Downs RJ, Thomas JF (2008) Phytotron Procedural Manual. Technical Bulletin 244 (Revised). http://www.ncsu.edu/phytotron/manual.pdf
- Senthil-Kumar M, Hema R, Anand A, Kang L, Udayakumar M, Mysore KS (2007) A systematic study to determine the extent of gene silencing in Nicotiana benthamiana and other Solanaceae species when heterologous gene sequences are used for virus-induced gene silencing. New Phytol 176 782–791 [DOI] [PubMed] [Google Scholar]
- Smith CW, Cothern JT (1999) Cotton. John Wiley and Sons, New York
- Stenger DC, Revington GN, Stevenson MC, Bisaro DM (1991) Replicational release of geminivirus genomes from tandemly repeated copies: evidence for rolling-circle replication of a plant viral DNA. Proc Natl Acad Sci USA 88 8029–8033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarshana MR, Wang HL, Lucas WJ, Gilbertson RL (1998) Dynamics of bean dwarf mosaic geminivirus cell-to-cell and long-distance movement in Phaseolus vulgaris revealed, using the green fluorescent protein. Mol Plant Microbe Interact 11 277–291 [Google Scholar]
- Szittya G, Silhavy D, Molnar A, Havelda Z, Lovas A, Lakatos L, Banfalvi Z, Burgyan J (2003) Low temperature inhibits RNA silencing-mediated defence by the control of siRNA generation. EMBO J 22 633–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnage MA, Muangsan N, Peele CG, Robertson D (2002) Geminivirus-based vectors for gene silencing in Arabidopsis. Plant J 30 107–114 [DOI] [PubMed] [Google Scholar]
- Udall JA, Flagel LE, Cheung F, Woodward AW, Hovav R, Rapp RA, Swanson JM, Lee JJ, Gingle AR, Nettleton D, et al (2007) Spotted cotton oligonucleotide microarrays for gene expression analysis. BMC Genomics 8 81–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udall JA, Swanson JM, Haller K, Rapp RA, Sparks ME, Hatfield J, Yu Y, Wu Y, Dowd C, Arpat AB, et al (2006) A global assembly of cotton ESTs. Genome Res 16 441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lersel MW, Harris WM, Oosterhuis DM (1995) Phloem in developing cotton fruits: 6(5)carboxyfluorescein as a tracer for functional phloem. J Exp Bot 46 321–328 [Google Scholar]
- Vanitharani R, Chellappan P, Pita JS, Fauquet CM (2004) Differential roles of AC2 and AC4 of cassava geminiviruses in mediating synergism and suppression of posttranscriptional gene silencing. J Virol 78 9487–9498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Buckley KJ, Yang X, Buchmann RC, Bisaro DM (2005) Adenosine kinase inhibition and suppression of RNA silencing by geminivirus AL2 and L2 proteins. J Virol 79 7410–7418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel JF, Cronn RC (2003) Polyploidy and the evolutionary history of cotton. Adv Agron 87 137–186 [Google Scholar]
- Wilkins TA, Mishra R, Trolinder NL (2004) Agrobacterium-mediated transformation and regeneration of cotton. Journal of Food, Environment and Agriculture 2 179–197 [Google Scholar]
- Wu Y, Llewellyn DJ, White R, Ruggiero K, Al-Ghazi Y, Dennis ES (2007) Laser capture microdissection and cDNA microarrays used to generate gene expression profiles of the rapidly expanding fibre initial cells on the surface of cotton ovules. Planta 226 1475–1490 [DOI] [PubMed] [Google Scholar]
- Zrachya A, Glick E, Levy Y, Arazi T, Citovsky V, Gafni Y (2007) Suppressor of RNA silencing encoded by Tomato yellow leaf curl virus-Israel. Virology 358 159–165 [DOI] [PubMed] [Google Scholar]