Abstract
Oilseeds are the main source of lipids used in both food and biofuels. The growing demand for vegetable oil has focused research toward increasing the amount of this valuable component in oilseed crops. Globally, soybean (Glycine max) is one of the most important oilseed crops grown, contributing about 30% of the vegetable oil used for food, feed, and industrial applications. Breeding efforts in soy have shown that multiple loci contribute to the final content of oil and protein stored in seeds. Genetically, the levels of these two storage products appear to be inversely correlated with an increase in oil coming at the expense of protein and vice versa. One way to overcome the linkage between oil and protein is to introduce a transgene that can specifically modulate one pathway without disrupting the other. We describe the first, to our knowledge, transgenic soy crop with increased oil that shows no major impact on protein content or yield. This was achieved by expressing a codon-optimized version of a diacylglycerol acyltransferase 2A from the soil fungus Umbelopsis (formerly Mortierella) ramanniana in soybean seed during development, resulting in an absolute increase in oil of 1.5% (by weight) in the mature seed.
Acyl-CoA:diacylglycerol acyltransferase (DGAT; EC2.3.1.20) is an integral membrane protein found in plants, fungi, and animals (Harwood, 1996; Sandager et al., 2002; Turkish and Sturley, 2007). The enzyme is responsible for transferring an acyl group from acyl-CoA to the sn-3 position of 1,2-diacylglycerol (DAG) to form triacylglycerol (TAG), the final product in the formation of storage lipid. Feeding studies and enzyme assays performed in multiple plant species indicate DGAT activity may be one of the limiting steps in TAG formation during seed development and that increasing this activity may result in an increase in stored lipid in seeds (Perry and Harwood, 1993; Cahoon et al., 2007).
Two distinct gene families encode proteins with DGAT activity. The first gene family, designated DGAT1, is related to the acyl-CoA:cholesterol acyltransferase family from which it diverged to prefer a DAG substrate (Cases et al., 1998). The second gene family, designated DGAT2, shares no homology with the DGAT1 family but shares homology with a broader family of genes that transfer acyl groups from CoA to neutral lipids, including monoacylglycerol, DAG, and fatty alcohol species (Turkish and Sturley, 2007). Within this broad family, the acyl-CoA:monoacylglycerol acyltransferase subgroup represents an alternative route for the production of DAG that does not involve phosphorylated intermediates, as in the Kennedy pathway, and plays a critical role in dietary fat absorption within the intestinal enterocyte in mammals (Yen et al., 2002; Turkish and Sturley, 2007). Another subgroup, the acyl-CoA:wax alcohol acyltransferase family, is involved in producing wax esters, which are major components in the surface lipids of plant cuticles, mammalian skin, and the exoskeleton coating of insects (Yen et al., 2005). The final subgroup, the DGAT2 family, is involved in TAG production in yeast, plant seeds, and animals (where it is believed to be responsible for the majority of TAG synthesis). Both DGAT families (1 and 2) are present in plants, animals, and fungi, and while they appear to represent redundant activities, their localization and temporal expression within the organisms determine the role they play (Shockey et al., 2006; Turkish and Sturley, 2007).
In plants, Shockey et al. (2006) showed that DGAT2 is more highly expressed in seed and is likely the main contributor to TAG production in Vernicia fordii. Similar conclusions were reached by Kroon et al. (2006) in their study of developing castor (Ricinus communis) seed. A similar study of castor development by He et al. (2005) reported DGAT1 may play a dominant role in ricinoleic acid production. Determining the contributions of DGAT1 and DGAT2 in oil production within the seed is a challenging task, because the elimination of one gene by mutagenesis or gene silencing tends to result in at least partial compensation by the other (Routaboul et al., 1999; Zou et al., 1999). The functional convergence of these two protein classes ensures TAG production is enabled within the seed to sustain the storage of this energetically important molecule.
Two studies conducted under greenhouse conditions, in Arabidopsis (Arabidopsis thaliana) and maize (Zea mays), have demonstrated that increasing DGAT activity during seed development can impact the final oil content in seed (Jako et al., 2001; Zheng et al., 2008). In these studies, endogenous DGAT1 genes were overexpressed in their native hosts and achieved absolute increases in oil of 9% to 12% for Arabidopsis and approximately 1% for maize. In this report, we describe the transgenic expression of a fungal DGAT2A gene from Umbelopsis ramanniana that was optimized for expression in soybean (Glycine max), analysis of the developing and mature seed, and description of the phenotype observed in multi-year, multi-location field trials for a commercial quality event. The data presented support the commercial viability of increasing oil in soybean by increasing DGAT activity in the seed.
RESULTS
Expression of UrDGAT2A in Seed Increases Oil in Soybean
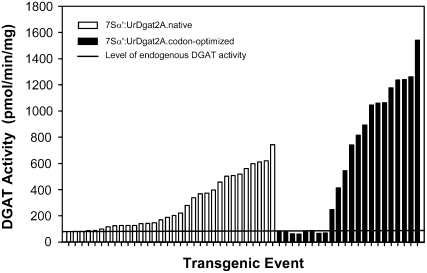
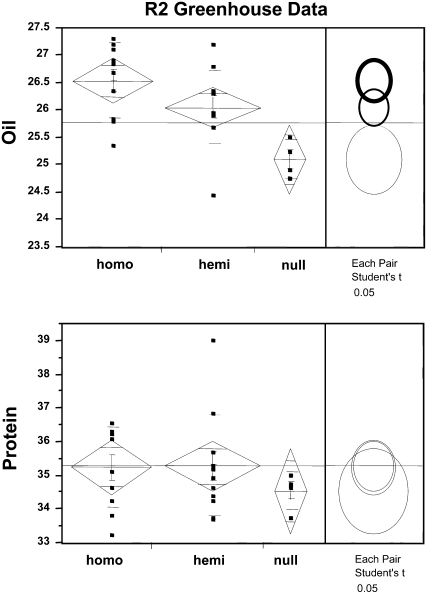
We previously reported the identification of the DGAT2 gene family by purification of two proteins (DGAT2A and DGAT2B) from U. ramanniana and confirmed their activity by enzyme assay following expression of the genes in insect cells (Lardizabal et al., 2001). The fungal codons of UrDGAT2A were modified to maximize expression of the gene in plants, and the optimized gene was expressed in soy under control of the seed-specific promoter 7S-α prime (Chen et al., 1986; Lardizabal et al., 2006). An increase in DGAT activity up to 20-fold over nontransgenic seed (1,540 versus 79 pmol min−1 mg−1 total protein) was detected in developing R1 embryos from different transgenic events. By comparison, expression of the native UrDGAT2A gene using the same promoter gave only up to a 10-fold increase in DGAT activity (Fig. 1). Homozygous R2 seed from multiple events (produced in the greenhouse) were analyzed for oil and protein content by near infrared transmittance. Statistical analysis of the data showed a 1% to 2% oil increase in homozygous seed with no significant change in protein content. A difference in the level of oil between homozygous, hemizygous, and null seed was also observed (Fig. 2), indicating a positive correlation between gene dosage and phenotype. Further testing was required to assess the impact on crop yield.
Figure 1.
DGAT activity assays of developing R1 expressing the native UrDGAT2A gene (white) compared with the optimized UrDGAT2A gene (black). Values are reported as pmol min−1 mg−1 total protein. The solid line represents the endogenous DGAT activity found in soybean.
Figure 2.
Statistical analysis of oil and protein in mature R2 seed showing the relationship between gene dosage and the effect on oil and protein. Analysis was performed using JMP software.
Developmental Analysis
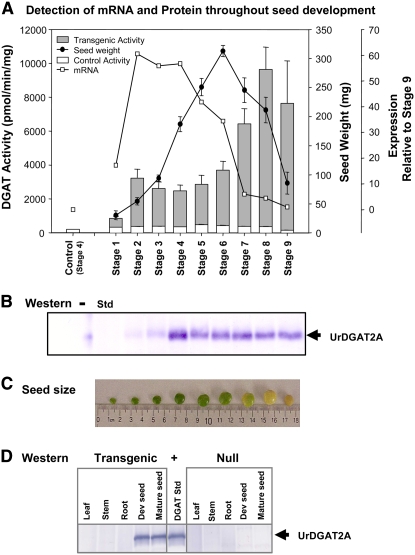
Homozygous seed was further characterized at nine different stages during seed development (Fig. 3, A–C). The stages were selected based on seed weight and color (1–6, early development; 7, mid-development; 8–9, late development). DGAT activity, seed weight, UrDGAT2A mRNA level (by reverse transcription-PCR), and UrDGAT2A protein presence (by western) were also determined (Fig. 3, A and B). Data showed that UrDGAT2A mRNA increased as the immature green seed developed (up to stage 4) but declined before seed expansion was complete (stage 7) and was very low after the seed turned yellow and began to desiccate (stages 8–9). In contrast to mRNA, DGAT activity and UrDGAT2A protein levels increased early and reached a peak mid-development that remained throughout seed desiccation and in mature seed (Fig. 3, A and B). The presence of DGAT activity in transgenic seed (Fig. 3A, gray bars) during desiccation (stage 8) and in mature seed (stage 9) differ from native DGAT activity levels in soybean (Fig. 3A, white bars). Endogenous DGAT activity levels in soybean were similar to those reported by Weselake et al. (1993) for oilseed rape (Brassica napus) and safflower (Carthamus tinctorius) seeds, where they exhibited a peak in activity early in development that decreased as the seed matured.
Figure 3.
A, Total DGAT activity reported as pmol min−1 mg−1 total protein (white, control; gray, transgenic) and abundance of UrDGAT2A mRNA at nine different stages of soybean development (relative to the control at stage 9) as measured by weight (sd of five seeds). B, Western analysis of developing seed shows that UrDGAT2A protein accumulates early in development and persists through later stages of development after the mRNA has diminished. Std, Purified UrDGAT2A. C, Size and color of soybean seed at different stages of development. D, Western analysis of different soybean tissues shows UrDGAT2A is found only in developing and mature transgenic seed.
The presence of active UrDGAT2A in mature soybean seed enabled purification of the transgenic protein for characterization. Using mature seed as starting material, UrDGAT2A was initially purified from a combination of the 20,000g floating fat pad and the 200,000g membrane fraction obtained from high speed centrifugation of the 20,000g supernatant. Using these two fractions, 1 to 3 mg of UrDGAT2A (at 75% purity) was obtained from 50 g of seed. To obtain protein with even higher purity, only the fat pad was used as starting material. This protocol eliminated several protein bands in the final preparation and improved protein purity to >95%. The specific activity of UrDGAT2A protein purified from transgenic soybean was approximately the same as the fungal purified protein (Lardizabal et al., 2001). We also studied the kinetics of purified UrDGAT2A with respect to a labeled 1-[14C]18:1-CoA donor and the unsaturated 18-carbon DAG substrates di18:1, di18:2, and di18:3. At saturating levels of 18:1-CoA, no significant differences were observed in the kinetic parameters of the three DAG acceptors (Table I). Similarly, at saturating levels of DAG species, no significant differences were observed with respect to 1-[14C]18:1-CoA (Table I). UrDGAT2A does not appear to demonstrate any preference with regard to these substrates, which comprise the majority of fatty acids found in soybean oil.
Table I.
Kinetic analysis of UrDGAT2A
UrDGAT2A (>95% pure) from mature transgenic soybean was assayed at a fixed concentration of 1-[14C]18:1-CoA (50 μm) and a range of DAG concentrations (0–1,000 μm) and at a fixed concentration of DAG (1,000 μm) and a range of 1-[14C]18:1-CoA concentrations (0.24–74 μm). Assay results (performed in triplicate) were plotted then analyzed using the Km equation (v = Vmax × S/(Km + S). Kinetic values and their ses were obtained for each data set using the Regression Wizard in SigmaPlot.
| Substrates
|
Kinetic Values
|
||
|---|---|---|---|
| Varied | Fixed | Vmax | Km |
| Fixed 1-[14C]18:1-CoA, varied DAG | |||
| 18:1 DAG | 18:1-CoA | 1,049 ± 30 | 36.4 ± 6.8 |
| 18:2 DAG | 18:1-CoA | 998 ± 26 | 19.5 ± 4.8 |
| 18:3 DAG | 18:1-CoA | 1,316 ± 48 | 33.0 ± 7.7 |
| Fixed DAG, varied 1-[14C]18:1-CoA | |||
| 18:1-CoA | 18:1 DAG | 1,135 ± 47 | 19.4 ± 2.0 |
| 18:1-CoA | 18:2 DAG | 895 ± 42 | 13.0 ± 1.7 |
| 18:1-CoA | 18:3 DAG | 1,165 ± 64 | 17.9 ± 2.5 |
Protein Localization
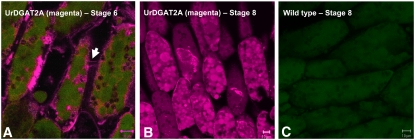
Protein localization was assessed using both confocal microscopy and transmission electron microscopy (TEM). Confocal results showed UrDGAT2A was present in the endoplasmic reticulum (ER), the membranes of oil bodies, and the plasmodesmata connecting cells (Fig. 4). At stage 6, UrDGAT2A was mainly localized in the ER, in some small oil bodies, and in the plasmodesmata (Fig. 4A). As seed development progressed through stages 7 and 8, the signal for UrDGAT2A was stronger and more frequently observed in the membranes of oil bodies (Fig. 4B). Also, oil body size and subcellular localization were different in cotyledons expressing UrDGAT2A compared to wild type. In transgenics, although the majority of oil bodies were still localized at the periphery of the cell, their size was 1.2- to 2.3-fold larger, with a variable number of significantly larger oil bodies randomly dispersed throughout the cell. Larger oil bodies were prominent in stages 7 and 8 for which oil bodies with a 13- to 14-μm radius were commonly observed (Table II). Protein bodies appeared to be smaller than those observed in wild-type cells but were more numerous. They were also frequently observed to be 1.4- to 2.5-fold smaller than the oil bodies. In addition, the size of storage vacuoles was smaller in cotyledon cells expressing UrDGAT2A. The distribution of lipid and protein from the abaxial to the adaxial region of the cotyledon appeared identical for wild-type and UrDGAT2A seed, with protein maximal in the abaxial region and lipid maximal in the adaxial region of the cotyledon.
Figure 4.
A, Immunolocalization of transgenic seed at stage 6 shows the presence of UrDGAT2A (red fluorescence) in the ER, plasmodesmata connecting cells (arrow), and small oil bodies of cotyledon cells. B, Immunolocalization of UrDGAT2A at stage 8 shows the presence of large oil bodies in transgenic seed. C, Immunolocalization of control seed shows the absence of UrDGAT2A (red fluorescence). Confocal three-dimensional projection micrographs of single (B) or merged (A and C) channels showing UrDGAT2A immunolabeling in red and autofluorescence in green.
Table II.
Determination of oil body size
A sampling of 20 cells from both wild-type and UrDGAT2A samples at stages 6 to 8 of seed development were randomly selected from the adaxial region of the cotyledon that had been stained for lipids with Nile Red. The following ranges in oil body diameter, measured using the Zeiss LSM510 software, were observed.
| Stage | Wild Type | Transgenic |
|---|---|---|
| mm | ||
| 6 | 0.72–1.95 | 1.10–6.80 |
| 7 | 0.75–2.47 | 1.22–13.99 |
| 8 | 0.90–2.53 | 2.52–14.1 |
TEM results corroborated the confocal results and in addition revealed that the dotted-like pattern observed in confocal microscopy resulted from alternating regions of ER with and without UrDGAT2A. This data is consistent with the findings of Shockey et al. (2006) and Cahoon et al. (2007) who showed that native DGAT1 and DGAT2 were localized to distinct subdomains of the ER. In addition, results showed UrDGAT2A was uniformly distributed in the oil body membrane. Some micrographs suggested oil body coalescence, which would explain larger oil body formation. A significant improvement in membrane perseveration was achieved by preparing the samples with high pressure freezing. Using this technique, we were also able to observe the presence of UrDGAT2A in the plasmodesmata (Fig. 4A, arrow).
Seed Analysis
To examine the transgenic trait in even greater depth, a single-copy, commercial-quality event with a 1.5% oil increase was selected for additional studies. To determine the degree of processing of the UrDGAT2A protein in soybean, traditional Edman sequencing was attempted on the purified protein. Sequencing of the N terminus failed, as it had with the native, fungal-purified UrDGAT2A, indicating it was blocked. Instead, sequence of the purified transgenic protein was analyzed using enzyme digestion, isotopic labeling, and matrix-assisted laser desorption ionization time-of-flight mass spectrometry and the overall sequence was confirmed by peptide mass fingerprinting (Courchesne and Patterson, 1999). The N-terminal Met was found to be truncated, and the new N-terminal amino acid, Ala, was blocked by acetylation. The C terminus was confirmed to be intact.
No significant changes in the content of protein or carbohydrates were detected in mature seed of the transgenic event relative to controls. We did observe a 15% decrease (on a percentage of dry weight basis) in neutral detergent fiber (NDF; significant at P < 0.05) in the transgenic positive relative to the control but saw no difference in acid detergent fiber. A small increase was observed in the amount of 18:1 fatty acid (+1.13%) with a corresponding decrease in the amount of 18:2 and 18:3 fatty acids (−0.72%, −0.40% = −1.12%) in the final TAG pool that were statistically significant (P < 0.05). However, this small compositional difference is well within the range of normal soybean fatty acid compositions observed in different varieties (http://www.codexalimentarius.net).
Field Trials
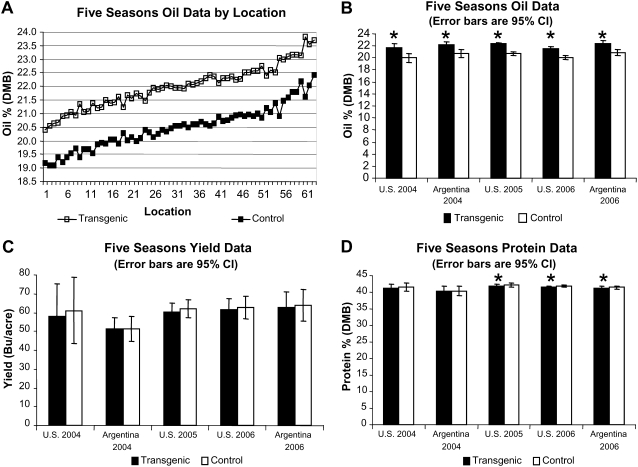
To determine whether oil increases observed in greenhouse-grown R2 seed translated to increases in the field, the homozygous commercial-quality event containing a single insertion of the UrDGAT2A gene (same event used for seed analysis) was grown at 63 locations over 3 years along with the corresponding control lines and nontransgenic checks. The transgenic DGAT event was evaluated for oil, protein, and yield in five seasons of field trials in the United States and Argentina from 2004 to 2006. All field trials were conducted as split-plot designs with the transgenic event as the whole plot and the negative and positive isolines as the split-plot. We did not observe any agronomic differences between the transgenic soybean plants expressing UrDGAT2A and the control plants during growth. They were identical with respect to germination, seedling vigor, plant height, time to flowering, and time to harvest.
At all locations and generations, oil was increased significantly over the negative isoline by an average of 1.5% (P < 0.05), indicating generational stability of the oil phenotype (Fig. 5, A and B). Though there appeared to be a slight decrease in the average yield in 4/5 field trials, grain yield observed over five seasons in both the United States and Argentina showed no statistical difference between the transgenic event and its negative isoline control (Fig. 5C). No major impact on protein level was observed in transgenic seed, indicating the oil increase associated with this transgene does not carry a (1:1) corresponding decrease in protein in contrast to results obtained from traditional breeding (Fig. 5D; Burton, 1984; Wilcox, 1998). In addition, when crossed into high oil soy germplasm, transgenic events increase oil in that germplasm, indicating the endogenous and transgenic mechanisms for achieving a high oil phenotype are complementary. Progeny of events crossed into a high protein germplasm also maintain the high protein phenotype while exhibiting an increase in oil level corresponding to the transgene donor parent (data not shown).
Figure 5.
A, Higher oil levels were observed for transgenic seed relative to the negative isoline over five seasons of oil data consisting of 63 multi-replication locations indicating environmental stability for the transgene trait. B, The across-locations analysis for oil shows that in five multi-location field trials in both the United States and Argentina, transgenic seed was significantly higher in oil than the negative isoline control. Data represents results for consecutive, sequential generations. This seed was not planted in Argentina during the 2005 field season. C, The across-locations analysis for yield shows no significant differences between the transgenic line and the negative isoline. D, Seed protein was significantly impacted in three of five field trails, but the largest significant difference was 0.44%. *, Values are significantly different (P < 0.05) from control. DMB, Dry matter basis.
DISCUSSION
We report the first, to our knowledge, transgenic expression of DGAT2 in any plant for the purpose of increasing oil content in seed and the first transgenic high oil soybean. This was achieved using a fungal DGAT2 from U. ramanniana that was optimized for expression in soy. Introduction of the transgene led to a 1.5% increase (by weight) in seed oil with no significant impact on yield or protein content. To determine whether the additional carbon found in oil came at the expense of another seed component, we also measured carbohydrates (Suc, raffinose, and stachyose) and acid and NDF. The only difference observed was a decrease in NDF (15% on a dry weight basis; P < 0.05), which could have contributed to the oil increase in transgenic seed. We detected a similar reduction (13%) in NDF in the nontransgenic high oil soybean line DKB31-51, which was grown as a positive control for oil levels; however, the difference in this case did not meet the significance test. A trend toward lower fiber was observed in 5/6 events; therefore, these results could indicate that a reduction in fiber may support increased carbon flow to oil (data not shown). Another possibility is the loss in carbon came from several sources whose differences were not detectable. In addition to changes in the NDF fraction, the only other measurable difference we observed in transgenic soybean was a significant increase in oil body size. Tzen et al. (1993) reported that oil bodies from different species had average diameters that were different, but the differences were within a narrow range (0.6–2.0 μm). While we observed a similar range of oil body sizes in control soybean (0.72–2.5 μm), the oil bodies in our transgenic soy were significantly larger (1–14 μm). Relationships between oil body size and oil content have been observed in several species (Lambert, 2000; Mantese et al., 2006) where there is a tendency for higher oil genotypes to contain slightly larger oil bodies than lower oil cultivars.
Other transgenic strategies designed to increase oil in plant seeds have also been reported. In Arabidopsis, Zou et al. (1997) observed that overexpression of yeast SLC1 (lysophosphatidic acid acyltransferase) resulted in an increase in seed oil, while Jako et al. (2001) found that overexpression of AtDGAT1 increased seed oil content and weight. Until recently, however, this had not been demonstrated in a major crop. Zheng et al. (2008) reported that expression in maize of an ancestral maize DGAT1 allele, PH09B, under control of the oleosin promoter resulted in an increase in kernel oil of about 1% (by weight). We have also observed that increasing DGAT activity in maize, by expression of UrDGAT2A in the germ, elevated kernel oil content (D. Hawkins, D.L. Val, J. Oakes, and J. Deikman, unpublished data). It will be important to determine whether the increase in kernel oil in these transgenic maize plants has impacted starch, protein, or grain yield.
The significance of producing soybean with higher levels of oil is due to its position as the major source of plant seed oil in the United States. Using the March 10, 2008 price for soybean oil published by the Chicago Board of Trade ($0.61/lb; http://www.cbot.com), this incremental increase in oil translates to an additional $17.9 U.S. dollars per metric ton of soybeans. In the United States alone, 70.4 million metric tons of soybeans were harvested in 2007 to 2008 (http://www.fas.usda.gov/oilseeds_arc.asp), translating to an increased crop value of more than $1.26 billion U.S. dollars.
In summary, increasing the amount of oil in soybean adds significant value to the crop if it can be accomplished without a yield penalty or a significant decrease in protein content. Transgenic expression of a codon-optimized UrDGAT2A has produced the first high-oil soybean crop that accomplishes these objectives. The mechanism for achieving this goal was to significantly increase the amount of DGAT activity specifically in developing seed.
MATERIALS AND METHODS
Enzyme Assays
UrDGAT2A activity is measured as described by Lardizabal et al. (2006) with the following changes. When assaying developing seed of transgenic plants, CHAPS (0.06%) replaced Triton X-100 as the detergent to maintain the endogenous DGAT activity present in the seed. This allows for a more realistic comparison between total DGAT activity levels in different events and was used to select the most active events for advancement at R1. When assaying fractions during the purification of DGAT from transgenic seed, phosphatidic acid is replaced with phosphatidyl choline to reconstitute DGAT activity.
Purification from Transgenic Plants to >95%
Starting with 50 g of ground mature seed suspended in buffer, centrifugation (20,000g, 30 min) yielded a floating fat pad, liquid fraction containing soluble and membrane-associated proteins, and a pellet. The purest form of UrDGAT2A was obtained using only the fat pad for purification. DGAT activity was solubilized from the fat pad with 5% Triton X-100 (5.5:1 detergent: protein, w/w) and ultracentrifugation (200,000g, 1 h). Two different resins were used in the purification: Reactive Yellow 86 (Sigma R-8504) and ceramic hydroxyapatite (Bio-Rad 157-0020). The high speed supernatant from detergent solubilization was loaded onto 500 mL of Reactive Yellow 86 in buffer A (100 mm Tricine, pH 7.5, 0.1% Triton X-100, 10% glycerol) containing 75 mm NaCl, washed with loading buffer, and eluted with buffer A containing 500 mm NaCl. The eluate was concentrated approximately 12-fold in an Amicon stirred cell (YM30 membrane) and applied to a 20-mL ceramic hydroxyapatite column. The majority of UrDGAT2A activity was collected in the flow-through, while a significant amount of protein bound the column and was removed. The sample was dialyzed to lower the salt concentration and applied to a 20-mL Reactive Yellow 86 column in buffer A with 75 mm NaCl and eluted in buffer A with a gradient of 75 to 500 mm NaCl. The yield varied for each preparation, but 1 to 3 mg UrDGAT2A protein with >95% purity was typically recovered.
Antibody Development and Western Analysis
Rabbit antibodies used to detect UrDGAT2A in the developing seed profile were generated to a small KLH-linked peptide (Zymed), KYGQTKDEIIRELHDS, near the C-terminal end of UrDGAT2A (Fig. 3C). Goat antibodies generated to the full-length UrDGAT2A (expressed and purified from E. coli as an N-HIS fusion) were used to detect protein in different soy (Glycine max) tissues (Fig. 3D). Western analysis was performed as described (Burnette, 1981), using PVDF as the blotting membrane, and a 1:5,000 dilution of the primary and secondary antibodies. Blots were developed with 1-Step NBT/BCIP reagent for alkaline phosphatase (Pierce no. 34042).
Developmental Analysis
Single-copy homozygous events expressing UrDGAT2A and a null segregant were used in the experiment. Plants were grown in the greenhouse, and seed was collected throughout development and stored at −80C. Nine stages were chosen for analysis based on a combination of weight, color, and stage of development. Five seeds from each stage were ground together in liquid nitrogen to a fine powder, and three aliquots were resuspended in buffer for analysis. Enzyme assays and westerns were performed as described above. For transcript-level measurements, total RNA was isolated from the same original powdered seed material, using the ABI Prism 6100 PrepStation instrument and associated buffers (Applied Biosystems). Approximately 10 mg of each frozen ground sample was mixed with 200 μL of 2× ABI Lysis buffer followed by an equal volume phosphate-buffered saline (PBS) on ice, then the samples were prefiltered according to the manufacturer's instructions using the preconfigured “Pre-Filter” method on the instrument. Filtered samples were mixed with 0.5 volume 100% ethanol, and 100 μL of the mixture was loaded onto the purification plate. The manufacturer's preconfigured method “ab.rnawash” was used to complete the RNA isolation. Real-time PCR was performed in a 96-well format using the ABI 7700 SDS instrument (Applied Biosystems) and analyzed with SDS software. Each reaction consisted of 20 ng of total RNA, 800 nm each primer, 150 nm probe, 1× Universal Master Mix (Applied Biosystems), and 1× Multiscribe Reverse Transcriptase (Applied Biosystems) in a 25-μL volume. The reverse transcription reaction was performed at 48°C for 30 min, using the gene-specific PCR primers, followed immediately by PCR at 95°C for 10 min and 40 cycles of 95°C for 15 s and 60°C for 1 min. The UrDGAT2A probe/primer set was (5′-CTGAGAACGGAGGCAGACCT-3′, Probe FAM-AGCTGCGTTCCTGAGCCACCTGA-TAMRA, 5′-TCCAGCAAAGAGCTTCCACC-3′). Relative expression was normalized to 18S rRNA expression (5′-CGTCCCTGCCCTTTGTACAC-3′, probe VIC-CCGCCCGTCGCTCCTACCGAT-TAMRA, 5′-CGAACACTTCACCGGATCATT) and the relative values calibrated to the Stage 9 sample, using the comparative threshold method (Applied Biosystems, User Bulletin No. 2, December 11, 1997).
Microscopy
Cotyledons from wild-type (negative controls) and transgenic seed at stages 6, 7, and 8 were analyzed by confocal laser scanning microscopy and TEM. For confocal microscopy, DGAT2A was labeled using AlexaFluor 532 with and without additional counterstaining. Immunolabeling and spectra collection from independent staining of cotyledon sections was performed with Nile Red (for lipid bodies) and ER Tracker Blue-White DPX (for ER labeling), and double and triple labeling of immunolabeled sections followed by linear unmixing. For immunofluorescence labeling, cryo-sections (60 μm) fixed in 3.7% formaldehyde in PBS buffer, pH 7.2, were permeabilized in 0.05% Triton X-100, washed in PBS with 10 mm Gly (three times), briefly incubated in 4% pectinase, 2% cellulase in PBS, pH 7.8 (to thin cell walls), blocked in undiluted goat serum for 15 min, and incubated overnight in 1:100 rabbit anti-DGAT2A diluted in 10% goat serum, 90% PBS with 0.05% Triton X-100; the sections were then washed in PBS with 10 mm Gly (three times), incubated in undiluted goat serum, and incubated with 1:500 AlexaFluor 532 goat anti-rabbit-conjugate secondary antibody. Sections were then washed in PBS 10 mm Gly (three times), counterstained with Hoechst 33342 or 4′,6-diamino-phenylindole, washed, and mounted for immediate observation with a two-photon confocal Zeiss LSM510 META (488-nm laser, 17%, BP 500–530 IR or BP 500–550 IR; 543-nm laser, 25%, LP 560; Chameleon laser at 760 nm, 11%, BP 435–485 IR). Controls were used to determine sample autofluorescence, absence of crossover, or bleed-through at the settings used for data collection, potential unspecific binding, and other artifacts. Linear unmixing confirmed the signal origin from double- and triple-labeled samples. Oil bodies were selectively stained by Nile Red (Greenspan et al., 1985; Chameleon laser at 790 nm, 10%, BP 565–615 IR). ER and phospholipid bilayer membranes in general were stained with ER Tracker Blue-White DPX (1:1,000, 30 min, in the dark; Chameleon laser at 760 nm, 12%, BP 480–520 IR filter, selected to exclude crossover from autofluorescence signal above 560 nm). For immunogold labeling, cotyledons were ultrarapidly frozen by high pressure freezing (Liberton et al., 2006), ultra-sectioned, immunogold labeled with goat anti-rabbit-gold conjugate secondary antibody, silver enhanced, contrasted with uranyl acetate, and viewed with a Leo 912AB TEM. Wild-type sections and sections incubated in the absence of primary or secondary antibodies were used as negative controls.
Construction of Plasmid
The plasmid pMON70900 contains two separate T-DNAs, each flanked by left and right border elements. One cassette carries the gene-of-interest (UrDGAT2A), and the other carries a marker (CP4) used to select events during the transformation process. The gene-of-interest cassette utilizes the seed-specific promoter 7Sa' from soybean and the E9 3′ untranslated region from Pisum sativum to drive expression of the codon-optimized UrDGAT2A gene. The CP4 selectable marker is contained in a second T-DNA cassette and is driven by the figwort mosaic virus 35S promoter and petunia (Petunia hybrida) heat shock protein 70 5′ untranslated region with transit peptide EPSPS from Arabidopsis (Arabidopsis thaliana). The first generation of transformed seed was screened for absence of the T-DNA containing the selectable marker so that only marker-free events were carried forward.
Plant Transformation, Zygosity Assays, and Linkage Southerns
Soybean was transformed as described by Martinell et al. (2002). Leaves of developing R1 plants were analyzed in a PCR-based assay (http://www.twt.com/invader_chemistry/invaderchem.htm) designed to quantitate the amount of a specific DNA target to determine if the plants were homozygous, heterozygous, or null for the transgene. Events where the transgene and the selectable marker were linked (closely colocalized on the same chromosome) were identified by Southern analysis and eliminated.
Field Trial Data
Field trials were conducted as two-row plots seeded at nine seeds per foot in 12-foot rows with a 3-foot border on 30-inch-row spacing. Seed was sampled from every plot and submitted for proximate analysis using near-infrared transmittance. Oil and protein data were reported on a dry matter basis. Outlier analysis based on deleted studentized residuals using SAS was performed on all data prior to statistical analysis. Statistical analyses were run using mixed model procedures in SAS. The analysis was performed using a split-plot model to compare positive to negative isolines within the transgenic event. The analysis was run across locations, with locations, replications within locations, and their interactions with the fixed effects considered random effects. Event, isoline, and their interactions were considered fixed effects.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers bankit1118767 and EU934741.
Acknowledgments
The authors thank Joel Ream, Elizabeth Hill, Greg Keithly, and the Agracetus soy transformation team (Middleton, WI) for their contributions to the development of this seed trait; Bosong Xiang for the N- and C-terminal analyses of the protein; Steve Modiano, Ron Colletti, and Wayne Brown for analysis of metabolites; Jihong Liang for the estimates of crop value; and Dr. Howard Berg from the D. Danforth Plant Science Center (St. Louis) for high pressure freezing of the developing seed samples prior to microscopy.
This work was supported by Renessen, a joint-venture between Monsanto Company Inc. and Cargill Inc.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Kathryn Lardizabal (kathy.lardizabal@monsanto.com).
Open Access articles can be viewed online without a subscription.
References
- Burnette WN (1981) ‘Western blotting’: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem 112 195–203 [DOI] [PubMed] [Google Scholar]
- Burton JW (1984) Breeding soybeans for improved protein quantity and quality. In R Shibles, ed, Proceedings of the World Soybean Research Conference III. Westview Press, Boulder, CO, pp 361–367
- Cahoon EB, Shockey JM, Dietrich CR, Gidda SK, Mullen RT, Dyer JM (2007) Engineering oilseeds for sustainable production of industrial and nutritional feedstocks: solving bottlenecks in fatty acid flux. Curr Opin Plant Biol 10 236–244 [DOI] [PubMed] [Google Scholar]
- Cases S, Smith SJ, Zheng YW, Myers HM, Lear SR, Sande E, Novak S, Collins C, Welch CB, Lusis AJ, et al (1998) Identification of a gene encoding an acyl-CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc Natl Acad Sci USA 95 13018–13023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZL, Schuler MA, Beachy RN (1986) Functional analysis of regulatory elements in a plant embryo-specific gene. Proc Natl Acad Sci USA 83 8560–8564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne PL, Patterson SD (1999) Identification of proteins by matrix-assisted laser desorption/ionization mass spectrometry using peptide and fragment ion masses. In AJ Link, ed, Methods in Molecular Biology, Vol 112. Humana Press, Totowa, NJ, 487–511 [DOI] [PubMed]
- Greenspan P, Mayer EP, Fowler SD (1985) Nile Red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol 100 965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood JL (1996) Recent advances in the biosynthesis of plant fatty acids. Biochim Biophys Acta 1301 7–56 [DOI] [PubMed] [Google Scholar]
- He X, Chen GQ, Lin JT, McKeon TA (2005) Regulation of diacylglycerol acyltransferase in developing seeds of castor. Lipids 39 865–871 [DOI] [PubMed] [Google Scholar]
- Jako C, Kumar A, Wei Y, Zou J, Barton DL, Giblin EM, Covello PS, Taylor DC (2001) Seed-specific over-expression of an Arabidopsis cDNA encoding a diacylglycerol acyltransferase enhances seed oil content and seed weight. Plant Physiol 126 861–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon JTM, Wei W, Simon WJ, Slabas AR (2006) Identification and functional expression of a type 2 acyl-CoA:diacylglycerol acyltransferase (DGAT2) in developing castor bean seeds which has high homology to the major triglyceride biosynthetic enzyme of fungi and animals. Phytochemistry 67 2541–2549 [DOI] [PubMed] [Google Scholar]
- Lambert RJ (2000) High-oil corn hybrids. In AR Hallauer, ed, Specialty Corns, Ed 2. CRC Press, Baton Rouge, LA, pp 131–154
- Lardizabal KD, Hawkins D, Thompson G, inventors. November 14, 2006. Diacylglycerol acyltransferase proteins. US Patent No. US7135617
- Lardizabal KD, Mai JT, Wagner NW, Wyrick A, Voelker T, Hawkins D (2001) DGAT2 is a new diacylglycerol acyltransferase gene family. J Biol Chem 276 38862–38869 [DOI] [PubMed] [Google Scholar]
- Liberton M, Berg RH, Heuser J, Roth R, Pakrasi HB (2006) Ultrastructure of the membrane systems in the unicellular cyanobacterium Synechocystis sp strain PCC 6803. Protoplasma 227 129–138 [DOI] [PubMed] [Google Scholar]
- Mantese AJ, Medan D, Hall AJ (2006) Achene structure, development and lipid accumulation in sunflower cultivars differing in oil content at maturity. Ann Bot (Lond) 97 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinell B, Julson LS, Emler CA, Huang Y, McCabe DE, Williams EJ, inventors. May 7, 2002. Soybean Agrobacterium transformation method. US Patent No. US6384301
- Perry HJ, Harwood JL (1993) Use of [2-3H]glycerol precursor in radiolabelling studies of acyl lipids in developing seeds of Brassica napus. Phytochemistry 34 69–73 [Google Scholar]
- Routaboul JM, Benning C, Bechtold N, Caboche M, Lepiniec L (1999) The TAG1 locus of Arabidopsis encodes for a diacylglycerol acyltransferase. Plant Physiol Biochem 37 831–840 [DOI] [PubMed] [Google Scholar]
- Sandager L, Gustavsson MH, Ståhl U, Dahlqvist A, Wiberg E, Banas A, Lenman M, Ronne H, Stymne S (2002) Storage lipid synthesis is non-essential in yeast. J Biol Chem 277 6478–6482 [DOI] [PubMed] [Google Scholar]
- Shockey JM, Gidda SK, Chapital DC, Kuan JC, Dhanoa PK, Bland JM, Rothstein SJ, Mullen RT, Dyer JM (2006) Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 18 2294–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkish A, Sturley S (2007) Regulation of triglyceride metabolism I. Eukaryotic neutral lipid synthesis: “Many ways to skin ACAT or a DGAT. Am J Physiol Gastrointest Liver Physiol 292 G953–G957 [DOI] [PubMed] [Google Scholar]
- Tzen J, Cao YZ, Laurent P, Ratnayake C, Huang AH (1993) Lipids, proteins, and structure of seed oil bodies from diverse species. Plant Physiol 101 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weselake RJ, Pomeroy MK, Furukawa TL, Goldern JL, Little DB, Laroche A (1993) Developmental profile of diacylglycerol acyltransferase in maturing seeds of oilseed rape and safflower and microspore-derived cultures of oilseed rape. Plant Physiol 102 565–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox JR (1998) Increasing seed protein with eight cycles of recurrent selection. Crop Sci 38 1536–1540 [Google Scholar]
- Yen CLE, Brown CH, Monetti M, Farese RV (2005) A human skin multifunctional O-acyltransferase that catalyzes the synthesis of acylglycerols, waxes and retinyl esters. J Lipid Res 46 2388–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen CLE, Stone SJ, Cases S, Zhou P, Farese RV (2002) Identification of a gene encoding MGAT1, a monoacylglycerol acyltransferase. Proc Natl Acad Sci USA 99 8512–8517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P, Allen WB, Roesler K, Williams ME, Zhang S, Li J, Glassman K, Ranch J, Nubel D, Solawetz W, et al (2008) A phenylalanine in DGAT is a key determinant of oil content and composition in maize. Nat Genet 40 367–372 [DOI] [PubMed] [Google Scholar]
- Zou JT, Katavic V, Giblin EM, Barton DL, MacKenzie SL, Keller WA, Hu X, Taylor DC (1997) Modification of seed oil content and acyl composition in the brassicaceae by expression of a yeast sn-2 acyltransferase. Plant Cell 9 909–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Wei Y, Jako C, Kumar A, Selvaraj G, Taylor DC (1999) The Arabidopsis thaliana TAG1 mutant has a mutation in a diacylglycerol acyltransferase gene. Plant J 19 645–653 [DOI] [PubMed] [Google Scholar]